Background: Cytochromes P450 2A13 and 2A6 are involved in nicotine metabolism and tobacco-related lung cancer initiation.
Results: CYP2A13 and CYP2A6 structures were solved with nicotine and CYP2A13 with NNK.
Conclusion: Structure series reveals basis for nicotine and NNK selectivity and access to buried active site.
Significance: Understanding CYP2A binding and oxidation of pharmacological substrates can aid in drug development efforts.
Keywords: Carcinogenesis, Cytochrome P450, Drug Metabolism, Membrane Enzymes, Structural Biology, NNK, Nicotine, Substrate Access
Abstract
Cytochromes P450 (CYP) from the 2A subfamily are known for their roles in the metabolism of nicotine, the addictive agent in tobacco, and activation of the tobacco procarcinogen 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK). Although both the hepatic CYP2A6 and respiratory CYP2A13 enzymes metabolize these compounds, CYP2A13 does so with much higher catalytic efficiency, but the structural basis for this has been unclear. X-ray structures of nicotine complexes with CYP2A13 (2.5 Å) and CYP2A6 (2.3 Å) yield a structural rationale for the preferential binding of nicotine to CYP2A13. Additional structures of CYP2A13 with NNK reveal either a single NNK molecule in the active site with orientations corresponding to metabolites known to form DNA adducts and initiate lung cancer (2.35 Å) or with two molecules of NNK bound (2.1 Å): one in the active site and one in a more distal staging site. Finally, in contrast to prior CYP2A structures with enclosed active sites, CYP2A13 conformations were solved that adopt both open and intermediate conformations resulting from an ∼2.5 Å movement of the F to G helices. This channel occurs in the same region where the second, distal NNK molecule is bound, suggesting that the channel may be used for ligand entry and/or exit from the active site. Altogether these structures provide multiple new snapshots of CYP2A13 conformations that assist in understanding the binding and activation of an important human carcinogen, as well as critical comparisons in the binding of nicotine, one of the most widely used and highly addictive drugs in human use.
Introduction
Lung cancer is responsible for more deaths than any other cancer (1). Although the highest risk factor for lung cancer is cigarette smoking, 19.3% of adults in the United States smoke cigarettes (2). Much of the difficulty in smoking cessation is the addictive properties of nicotine (3). Upon inhalation of tobacco smoke, nicotine rapidly circulates systemically via the bloodstream, with the majority of nicotine metabolism occurring in the liver via cytochrome P450 (CYP)2 2A6 (3). In addition to nicotine, tobacco smoke also contains several procarcinognic nitrosamines with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) being one of the most prevalent and procarcinogenic (4). NNK is converted to its carcinogenic form by hydroxylation of either carbon α to the nitrosamino group (5–7) with the resulting metabolites forming DNA adducts, often in tumor suppressor or proto-oncogenes like Ras and p53 (8). In humans, both the hepatic CYP2A6 and CYP2A13 in the respiratory tract can activate NNK to some extent, but CYP2A13 does so with 61- to 214-fold greater catalytic efficiency than CYP2A6 (7, 9). Activation of NNK in the lung results in DNA adducts and lung tumors in mice (10) and is thought to be a potent determinant for lung adenocarcinoma in smokers. The catalytic efficiency of CYP2A13 with nicotine is also 22-fold higher than CYP2A6 (11), although rapid nicotine distribution suggests that hepatic CYP2A6 is likely primarily responsible for most phase I metabolism of nicotine in vivo (3). Although the CYP2A6 and CYP2A13 enzymes are 94% identical, the bases for differential binding and metabolism of nicotine and NNK are unknown.
Structures are available for CYP2A6 with the largely in vitro substrate coumarin and inhibitors (12–14) and of CYP2A13 bound to the in vitro substrate indole or inhibitor pilocarpine (14, 15). However, no structures are available for substrates with significant pharmacological relevance, much less the key addictive and procarcinogenic agents in tobacco. Several studies have used existing CYP2A13 structures as a basis for docking studies to predict NNK binding in CYP2A13 (15–18). In part to test these predictions, we have determined structures of human CYP2A13 bound to NNK by x-ray crystallography. In addition, we sought to investigate the structural differences in nicotine binding between CYP2A13 and CYP2A6 by determining the structures of both complexes. In total, four new crystal structures with these ligands not only identify amino acid residues important for the functional differences between these two enzymes in the binding and metabolism of nicotine and biotransformation of NNK into a carcinogen, but they also provide unexpected evidence for multiple ligand binding modes and the location of a channel in CYP2A13 that may allow ligand access/egress. The compilation of these structural vignettes is a stop motion perspective on CYP2A enzyme conformational changes controlling the binding and metabolism of nicotine and NNK, two substrates responsible for human tobacco addiction and lung cancer.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
(S)-Nicotine (Sigma-Aldrich), NNK (Toronto Research Chemicals, Toronto, Canada), and Anatrace detergents (Affymetrix, Maumee, OH) were obtained as indicated.
Protein Expression, Purification, and Crystallization
Truncated, His-tagged human CYP2A6 and CYP2A13 were expressed recombinantly in Escherichia coli, purified as described (14, 15, 19), and crystallized using hanging drop vapor diffusion.
CYP2A6/Nicotine
The CYP2A6/nicotine crystal was grown from a drop formed by mixing 200 μm CYP2A6 with 50 mm nicotine in potassium phosphate buffer (50 mm potassium phosphate, pH 7.4, 20% glycerol, 1 mm EDTA, 0.5 m NaCl) 1:1 with precipitant solution A (30% PEG 3350, 0.175 m Tris, pH 8.5, and 0.2 m ammonium sulfate) and then equilibrated against the same precipitant solution. The crystal was immersed in a 7:3 mixture of synthetic mother liquor and ethylene glycol and flash-cooled in liquid nitrogen.
CYP2A13/Nicotine
CYP2A13/nicotine crystals were grown from a drop consisting of 200 μm CYP2A13 with 50 mm nicotine in potassium phosphate buffer and 2% Anapoe-35, mixed 2:1 with precipitant solution A and cryoprotected as above.
CYP2A13/NNK in P1 Space Group
Crystals were grown from a drop consisting of 200 μm CYP2A13 with 50 mm NNK in potassium phosphate buffer and 0.2% Triton X-405, mixed 2:1 with precipitant solution A and cryoprotected as above.
CYP2A13/NNK in I422 Space Group
CYP2A13 (33.5 nmol) with 50 mm NNK was mixed 1:1 with cytochrome b5 in a phosphate/sucrose buffer (0.25 m potassium phosphate, 0.5 m sucrose), concentrated to 1 ml, and run on a size exclusion column (Superdex 200; GE Healthcare) in the same buffer. The complex eluted in the void volume and was concentrated to ∼500 μm. The crystals grew from a 2-μl drop containing this protein solution mixed 1:1 with precipitant solution B (4.0 m sodium formate, pH 7.8). The cryoprotectant consisted of 6 m sodium formate.
Data Collection, Molecular Replacement, Refinement, and Validation
Native data sets were collected at the Stanford Synchrotron Radiation Lightsource (Stanford, CA) on Beamlines 9-2 or 11-1 using a 0.98 Å wavelength at a temperature of 100 K. The data were processed using Mosflm and Scala (20, 21) with the statistics shown (Table 1). All four structures were solved by molecular replacement using Phaser (21).
TABLE 1.
CYP2A13 and CYP2A6 data collection and refinement statistics
| CYP2A6/nicotine | CYP2A13/nicotine | CYP2A13/NNK (P1) | 2A13/NNK (I422) | |
|---|---|---|---|---|
| Data collection | ||||
| Space group | P1211 | P1 | P1 | I422 |
| Cell dimensions | ||||
| a, b, c (Å) | 71.2, 194.0, 90.3 | 71.7, 120.3, 153.7 | 71.7, 119.4, 153.8 | 122.8, 122.8, 192.2 |
| α, β, γ (°) | 90.0, 102.7, 90.0 | 100.6, 100.8, 93.7 | 100.7, 100.9, 93.6 | 90.0, 90.0, 90.0 |
| Resolution (Å)a | 97.0-2.3 (2.36-2.30) | 117.6-2.5 (2.63-2.50) | 83.6-2.35 (2.41-2.35) | 97.1-2.1 (2.15-2.10) |
| Redundancya | 3.9 (3.9) | 2.0 (2.0) | 4.0 (4.0) | 14.7 (14.8) |
| Rpima | 0.045 (0.420) | 0.066 (0.452) | 0.048 (0.342) | 0.027 (0.372) |
| Mn(I/sd)a | 11.9 (1.9) | 7.1 (1.8) | 9.3 (2.4) | 18.3 (2.1) |
| Completenessa (%) | 99.9 (99.9) | 96.7 (88.1) | 97.5 (96.7) | 100.0 (100.0) |
| Total/unique reflectionsa | 409,663/105,683 (30,105/7804) | 329,774/164,366 (43,876/21,915) | 787,673/198,013 (57,791/14,523) | 639,843/43,580 (46,911/3,175) |
| Refinement | ||||
| Resolution (Å) | 97.02-2.30 | 102.58-2.50 | 69.77-2.35 | 51.90-2.10 |
| No. reflections | 100,359 | 156,132 | 187,903 | 41,386 |
| Matthews coefficient/% solvent | 2.79/55.8 | 2.91/57.7 | 2.89/57.4 | 3.31/62.8 |
| Molecules per asymmetric unit, details of liganded state and conformation | Four (four closed with nicotine) | Eight (four closed with nicotine, two closed without nicotine, and two different open conformations without nicotine; mol. G, H) | Eight (six closed with one NNK each and two different open conformations similar to CYP2A13/nicotine; mol. G, H) | One (two NNK molecules and closed conformation) |
| Rwork/Rfree | 20.3/26.4 | 22.4/28.5 | 21.4/27.3 | 20.1/23.6 |
| Number of atoms/B factor | 15,432/41.2 | 30,625/48.2 | 30,811/47.9 | 3936/40.9 |
| Protein | 15,074/41.3 | 30,178/48.3 | 30,104/48.1 | 3794/40.8 |
| Heme | 172/32.5 | 344/34.7 | 344/34.9 | 43/32.3 |
| Ligand | 48/56.5 | 48/60.4 | 90/65.0 | 30/47.1 |
| Water | 138/36.3 | 49/31.9 | 261/38.3 | 69/45.1 |
| Glycerol | 6/33.3 | 12/47.9 | ||
| rmsd | ||||
| Bond lengths (Å) | 0.023 | 0.018 | 0.019 | 0.024 |
| Bond angles (°) | 1.915 | 1.692 | 1.694 | 1.918 |
| Error in coordinates by Luzzati plot (Å) | 0.292 | 0.339 | 0.314 | 0.288 |
| Residues in Ramachandran plot regions: favorable/additional allowed/generously allowed/disallowed (%) | 90.2/9.4/0.2/0.2 | 87.8/11.5/0.4/0.3 | 88.3/11.0/0.4/0.3 | 91.8/7.8/0.2/0.2 |
a The values in parentheses are for the highest resolution shell.
The search model for the CYP2A6 nicotine complex was a CYP2A6 inhibitor structure (Protein Data Bank code 2FDV) (12). The search model for the CYP2A13 nicotine complex was CYP2A6 with coumarin (Protein Data Bank code 1Z10) (13). Six CYP2A13 molecules (molecules A–F) were initially located by molecular replacement (Matthews coefficient 3.88 and 68% solvent). However, it was readily apparent upon examination of the crystal packing and initial electron density maps that two additional molecules of CYP2A13 were present, and they were manually added to the overall model as molecules G and H (Matthews coefficient 2.91 and 57% solvent). These two additional molecules clearly had alterations in the placement of the F, F′, G′, and G helices, resulting in a more open conformations of CYP2A13, which is likely why they were not identified by molecular replacement originally. The original six CYP2A13 molecules (A–F) were closed, with four (A–D) containing electron density in the active site that could clearly be modeled as nicotine (see Fig. 1B). Two of the closed molecules (E and F) did not have sufficient electron density in the 2|Fo| − |Fc| map at 1 σ to model nicotine.
FIGURE 1.
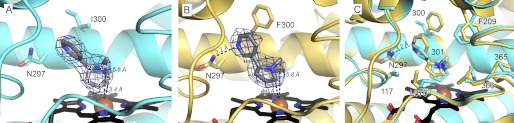
Nicotine binding to CYP2A6 (blue) and CYP2A13 (yellow). A and B, CYP2A6 (A, blue ribbons and sticks) and CYP2A13 (B, yellow ribbons and sticks) with composite omit σA-weighted 2|Fo| − |Fc| electron density (blue mesh) contoured at 1.0 σ around nicotine (gray sticks). C, overlay of CYP2A6 and CYP2A13 active sites, colored as in A and B, with important active site residues labeled. All panels show heme as black sticks with a red sphere (iron). The dashed lines represent hydrogen bonding to Asn-297 or distances from ligand sites of metabolism to the heme iron.
For the CYP2A13/NNK P1 structure, the search models were CYP2A6 bound to coumarin (Protein Data Bank code 1Z10) (13) and molecule G from the CYP2A13/nicotine structure above. As in the case with the CYP2A13/nicotine structure, six molecules (A–F) were in the closed conformation, and two molecules (G and H) were in more open conformations. Molecules A–F all had clear electron density for NNK in the active sites.
The search model for the CYP2A13/NNK I422 structure was CYP2A13 (Protein Data Bank code 2P85) (15). Although a solution initially containing a 1:1 ratio of CYP2A13 to cytochrome b5 was used to grow the crystals, and density weakly suggestive of cytochrome b5 was present in the channels of the P450 crystalline lattice, it was not strong enough to model cytochrome b5.
Model building and refinement were accomplished iteratively using COOT (22) and Refmac5 (23) in the CCP4i suite (21). Validation was accomplished using WHAT_CHECK (24) and PROCHECK (25). Refinement and Ramachandran plot statistics are in Table 1. Coordinates have been submitted to the Protein Data Bank (CYP2A6/nicotine, Protein Data Bank code 4EJJ; CYP2A13/nicotine, Protein Data Bank code 4EJG; CYP2A13/NNK (P1), Protein Data Bank code 4EJH; and CYP2A13/NNK (I422), Protein Data Bank code 4EJI). Probe-occupied voids were defined using VOIDOO (26) with a 1.4 Å probe and a grid mesh of 0.4 Å as described (19, 27).
Spectral Ligand Binding Assay
The spectral ligand binding assay was performed as described previously (28, 29). The results are the averages of two different titrations.
Site-directed Mutagenesis
Internal single and multiple mutations were made in the modified CYP2A6 gene in the expression vector pKK2A6dH using synthetic oligonucleotides that incorporated the desired nucleotide substitutions using the QuikChange site-directed mutagenesis method as previously reported (19). The proteins were expressed and purified also as described (19). The specific contents for the CYP2A6 I300F and I300F/G301A mutants were 9.1 and 8.6 nmol/mg. All of the other proteins had specific contents >13.9 nmol/mg. The proteins were active in phenacetin O-dealkylation as reported (19).
RESULTS
Dissociation Constants for Nicotine and NNK
Nicotine and NNK binding to both CYP2A13 and CYP2A6 resulted in classic type I spectral shifts indicative of displacement of the iron-coordinated water molecule. NNK bound with similar affinities to CYP2A13 (Kd = 4.4 μm) and CYP2A6 (Kd = 1.4 μm). In contrast, the KD for nicotine binding to CYP2A6 was 8.7-fold higher than to CYP2A13 (Kd values 470 and 54 μm, respectively) (30). Because a subset of active site residue substitutions between these two CYP2A enzymes have proved key in controlling the differential binding and metabolism of other CYP2A ligands (19, 29), we subsequently explored the effects of selected mutations on nicotine affinity. For example, substitution of the CYP2A6 Ile-300 residue with the Phe found at this position in CYP2A13 decreased the nicotine Kd from 470 to 234 μm. However, the CYP2A6 I300F/G301A double mutant, which simultaneously replaces two CYP2A6 residues with the corresponding CYP2A13 residues, reverts the Kd to 419 μm, suggesting that the contributions of individual mutations are not simply additive. Simultaneous substitution of four of the ten active site residues differing between the two wild type enzymes (CYP2A6 I208S/I300F/G301A/S369G) resulted in a Kd of 196 μm, or ∼3.6-fold higher than that observed for wild type CYP2A13. The addition of the V117A substitution further increases the affinity to 144 μm, or ∼2.6-fold higher Kd than for the CYP2A13 wild type enzyme.
Structure Determination and Overall Structures
Data collection and refinement statistics for structures determined of CYP2A13 and CYP2A6 with nicotine and two different structures of CYP2A13 with NNK are provided in Table 1. The four molecules present in the asymmetric unit of the CYP2A6/nicotine structure were very similar and contained strong density corresponding to nicotine binding in the active site (Fig. 1A). The CYP2A13/nicotine structure contained eight molecules (A–H) with four molecules containing nicotine (Fig. 1B). The CYP2A13/NNK structure from the P1 space group also contained a total of eight molecules, with six containing NNK. In both the CYP2A13/nicotine and the CYP2A13/NNK P1 structures, molecules G and H had significant relocation of the F, F′, G′, and G helices compared with the closed conformation, yielding intermediate and open conformations, respectively. The CYP2A13/NNK structure derived from crystallization in the space group I422 consisted of only one CYP2A13 molecule in the asymmetric unit. In this case the protein was in the closed conformation, but with two molecules of NNK present in/near the active site.
Comparisons among the protein conformations observed in the different complexes reveal that the backbone structures of the closed molecules of both CYP2A13 and CYP2A6 are very similar, regardless of ligand, with an rmsd for core Cα atoms of 0.25–0.65 Å. The intermediate conformation called molecule G in the CYP2A13/nicotine structure is very similar to the intermediate conformation modeled as molecule G in the CYP2A13/NNK (P1) structure (Cα rmsd 0.411 Å). Similarly the open conformation modeled as molecule H in the CYP2A13/nicotine structure is very similar to the open conformation modeled as molecule H in the CYP2A13/NNK (P1) structure (Cα rmsd 0.488 Å). In contrast, both intermediate and open conformations are distinct from the closed conformation, with molecule G having a Cα rmsd of 0.96 Å compared with the closed structures and the open conformation molecule H demonstrating an even higher Cα rmsd of 1.3 Å compared with the closed structures.
Comparison of CYP2A13 and CYP2A6 with Nicotine
Both CYP2A13 and CYP2A6 oxidize nicotine at various locations on the methylpyrrolidine ring. Consistent with this, nicotine binds with the methylpyrrolidine ring oriented toward the heme in both enzymes (Fig. 1, A and B). The dominant oxidation reaction for both enzymes is 5′-hydroxylation to form 5′-hydroxynicotine (31), which is in equilibrium with the nicotine Δ1′(5′)-iminium ion and then further oxidized to cotinine and eliminated in urine (32). However, both enzymes can also oxidize nicotine on the methyl to ultimately generate nornicotine (31, 33), and at least CYP2A6 is also able to oxidize the 2′ position to generate the aminoketone precursor of NNK (34). In the CYP2A13 complex with nicotine (Fig. 1B), the pyrrolidine is oriented so that the 5′ and methyl carbons are closest to the heme iron at 4.0 and 5.8 Å, respectively. The pyridine ring of nicotine forms a 3.2 Å hydrogen bond with Asn-297 in CYP2A13. In the CYP2A6 nicotine complex (Fig. 1A), the pyrrolidine 5′ and methyl carbons are at similar distances from the heme iron (3.4 and 5.8 Å, respectively), but the 4′-carbon is even closer at only 2.5 Å. Another difference is that in CYP2A6 the pyridine nitrogen is too far (3.8 Å) from Asn-297 for even a weak hydrogen bonding interaction.
Although the overall orientations of nicotine in the two active sites are similar, comparison by overlapping them (Fig. 1C) emphasizes the differences in the relative orientations. In CYP2A13 the methylpyrrolidine ring is oriented more parallel to the heme plane, whereas in CYP2A6 the orientation is closer to perpendicular. The sp3 hybridization of C2′ and the (S) stereochemistry of the substrate then dictate different planes for the pyridine rings and differential interactions with Asn-297. Because Asn-297 is a key orienting feature for the CYP2A active site with several other ligands (6, 12–15), it is notable that in CYP2A6 the Asn-297 side chain is rotated slightly farther away from nicotine compared with the CYP2A13 structure. Differences in the details of nicotine orientation are likely also mediated by several other substantial differences between the two active sites involving both conserved and nonconserved residues. First, in CYP2A13 the larger Phe at position 300 is nonetheless torsioned away from the active site, providing more space in this region compared with the Ile at this position in CYP2A6. This change appears in concert with differential positioning of the conserved Phe-209, which in CYP2A6 occludes slightly more active site volume than in CYP2A13. Thus residues at both positions 300 and 209 may sequester nicotine slightly closer to the heme in CYP2A6. Another important difference is observed in the position of the conserved Leu-370 residue. The orientation directly toward nicotine in CYP2A6 would seem to be correlated with the more vertical orientation of the pyrrolidine ring, whereas rotation of this ring to be more parallel to the heme in CYP2A13 would be facilitated by the redirection of Leu-370 away from the active site center as observed in CYP2A13. Both a preceding Gly only present in CYP2A13 and/or reduction in the adjacent side chain size at position 117 from a Val in CYP2A6 to the Ala in CYP2A13 may facilitate this rotation of Leu-370. In aggregate, these residues and slight changes caused by residues 365 and 366 result in a CYP2A13 active site that is larger (330 Å3) compared with that of CYP2A6 (247 Å3) with the same ligand.
Structures of CYP2A13 with NNK Bound
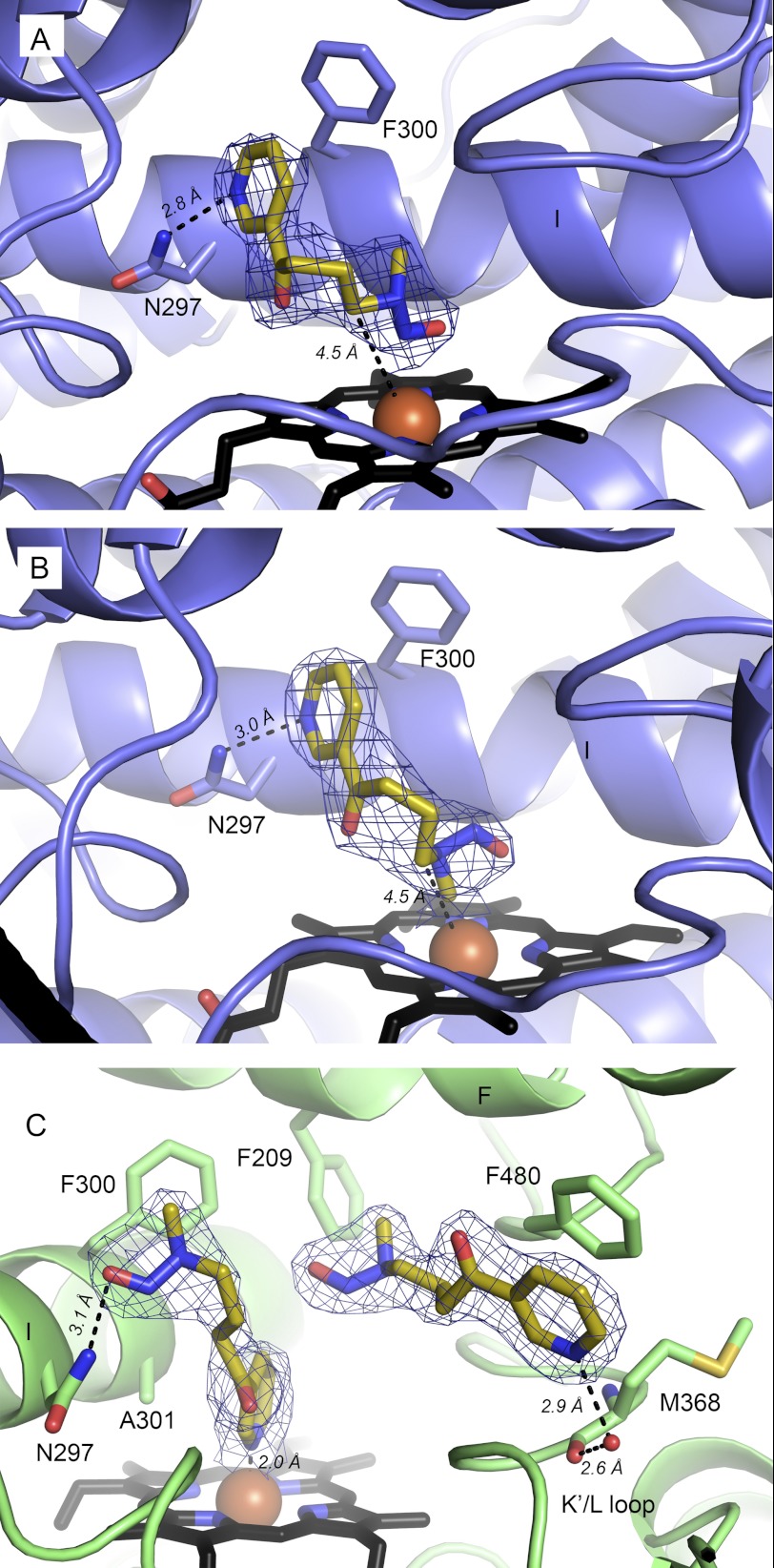
NNK can be oxidized by CYP2A13 at either carbon α to the nitrosamine. The dominant metabolite (5, 7, 9) results from hydroxylation of the α-methylene, but the α-methyl can also be hydroxylated with about a 3-fold lower Kcat/Km (5, 7, 9). NNK bound to CYP2A13 in two different orientations in different molecules of the CYP2A13/NNK P1 structure and in yet another orientation in the CYP2A13/NNK I422 structure. In the P1 structure, the two NNK orientations were very similar with respect to the placement of the pyridine ring, with the pyridine nitrogen serving as a hydrogen bond acceptor for Asn-297 (Fig. 2, A and B). As in the CYP2A13/nicotine structure above, Phe-300 appears to pack well with the pyridine ring. In contrast, this NNK structure revealed two different orientations of the nitrosamine end of the substrate. The orientation in Fig. 2A clearly shows the terminal nitrosamine nitrogen located most directly above the heme iron with the α-methylene and α-methyl carbons 4.5 and 5.3 Å from the heme iron, respectively. The methylene distance and blocking of the methyl by the nitrosamine bulk in this orientation are consistent with the preference of CYP2A13 for α-methylene hydroxylation. In this orientation, the methyl would have to rotate toward the iron for hydroxylation. The second NNK orientation in the P1 structure (Fig. 2B) differs from the first in that the nitrosamine end is torsioned so that the methyl carbon is very close to the heme iron (2.2 Å) and the α-methylene is more distal with a distance of 4.5 Å. A slight shift of NNK away from the heme would appear to be required prior to metabolism because the 2.2 Å distance is too close to allow the simultaneous presence of the catalytic iron(IV) oxo intermediate. Overall, the active site volumes containing both of these NNK orientations are similar, ranging from 280 to 306 Å3.
FIGURE 2.
Orientations of NNK (yellow sticks) found in molecules of CYP2A13. A and B, CYP2A13/NNK orientations most consistent with methylene (A) and methyl (B) oxidation, respectively. C, CYP2A13/NNK I422 structure showing with two NNK molecules bound. For all panels, NNK is shown as sticks with yellow carbon atoms, and mesh is the composite omit σA-weighted 2|Fo| − |Fc| electron density contoured at 1.0 σ around NNK.
The I422 crystals yielded a very different scenario for NNK binding to CYP2A13. NNK in the active site adopts a nonproductive orientation with the pyridine ring coordinated to the heme iron (Fig. 2C). This NNK molecule maintains a hydrogen bond to Asn-297, albeit via the nitrosamine oxygen. In addition to the molecule of NNK interacting directly with the heme iron, a second molecule of NNK was also present. This second NNK molecule is more distant from the heme and located above the K′-L loop. The volume occupied by this second NNK was not available in the P1 CYP2A13/NNK structures. The excess volume becomes available through movement of the Phe-480 side chain, which rotates to π stack with the pyridine ring of the second NNK (Fig. 2C). The pyridine nitrogen of this second NNK is part of a water-bridged hydrogen bonding network to the backbone carbonyl of Met-368. Reorganization of Phe-480 and a smaller shift of the Met-368 side chain result in an active site volume of 598 Å3, almost double that observed in the CYP2A13/NNK P1 crystals, but without significant differences in the backbone.
Structures of CYP2A13 with an Access/Egress Channel
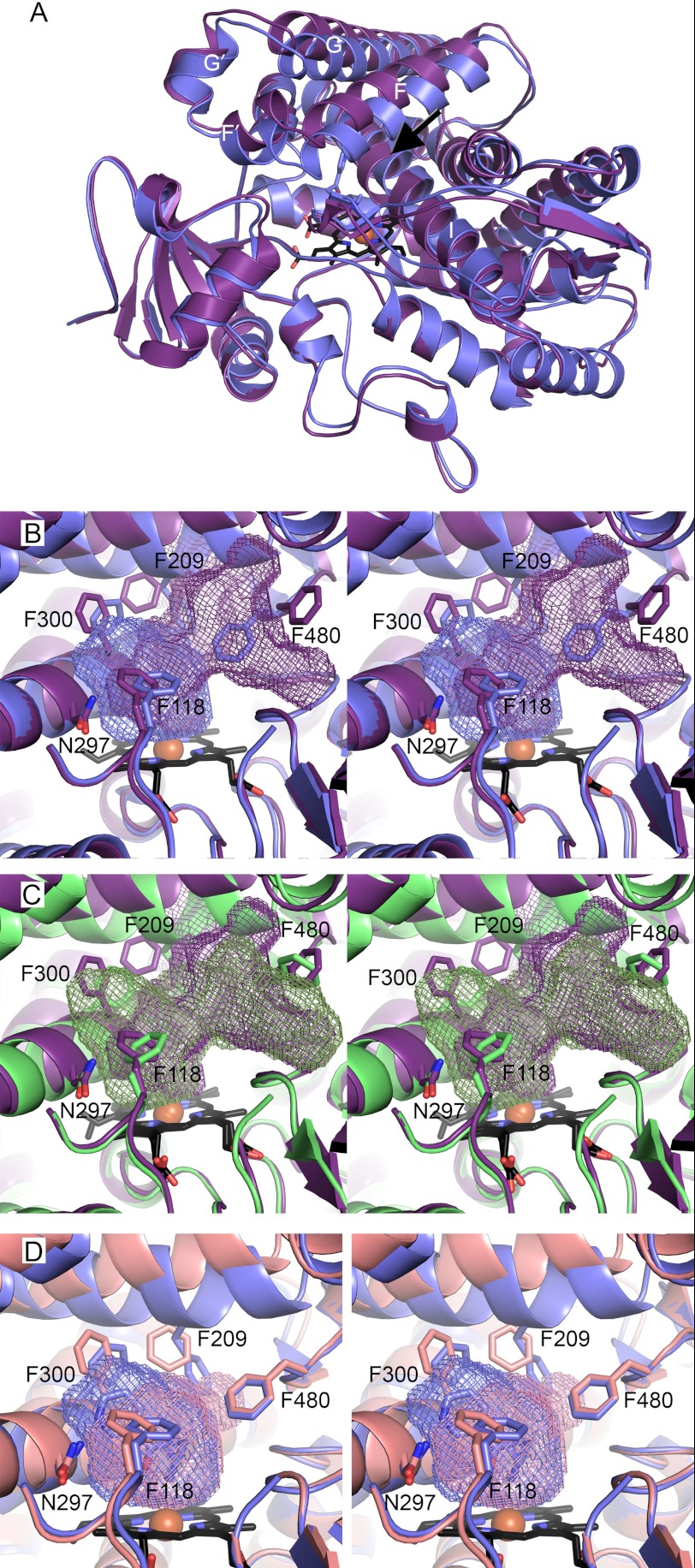
Comparison of the overall structures of different CYP2A molecules in the various structures indicates some significant differences. A canonical “closed” conformation is present for CYP2A6 with nicotine (all four molecules in the asymmetric unit), for the single molecule in the CYP2A13/2×NNK I422 structure and for six of the eight molecules in the CYP2A13/nicotine and CYP2A13/NNK P1 structures. However, the last two molecules in the CYP2A13/nicotine and the CYP2A13/NNK P1 structures were substantially different and appear to form a series of conformations from open to closed. This potentially indicates a route by which substrates and metabolites might pass in and out of the otherwise enclosed active site, although of course these conformational changes could also occur in the absence of ligand transit.
Molecule H is similar in both the CYP2A13/nicotine and CYP2A13/NNK P1 structures with an rmsd of 0.488 Å and has the most pronounced alterations compared with the canonical closed conformation. The previously enclosed ∼300 Å3 active site opens up enough to have a channel all the way to the surface passing between helix F and the β4 sheet (Fig. 3A). At the backbone level, the most significant differences are observed as shifts in the F, F′, G′, and G helices away from the heme and a bulge in helix I over the heme (tip of arrow in Fig. 3A). The entrance/exit of the channel on the protein exterior is lined with hydrophilic residues Gln-210, Glu-304, Thr-308, Thr-212, Asp-218, and His-477. Raising the active site “roof” composed of the F-G helices, in concert with substantial repositioning of key side chains, have significant effects on the active site topology as well. First, there are substantial changes on either side of the newly formed exit from the active site proper (Fig. 3B). Significantly, the F-helix residue Phe-209 is not only shifted upward by ∼3 Å along with the whole F helix but also torsioned away from the new channel. On the opposite side of the new channel, Phe-480 is torsioned in the opposite direction. The movement of Phe-480 opens the same large volume observed over the K′-L loop as in the CYP2A13/2×NNK I422 structure, space that was occupied in that case by the second molecule of NNK (Fig. 3C). Although the cavity is opened up on one face allowing access to the protein surface, on the opposite face of the active site adjustments are observed that would serve to discourage ligand binding. The I helix kink and corresponding adjustments in the Phe-300 side chain and shifts in Phe-118 toward the active site reduce ligand access to Asn-297 and occlude volume otherwise occupied by the pyridine rings of NNK and nicotine. This results in an active site with insufficient capacity to bind either nicotine or NNK in the orientations consistent with metabolism of these substrates as observed herein. Thus this 883 Å3 cavity is the net result of both opening a channel to the protein surface and simultaneously obscuring the most distal part of the active site proper, a concerted process for ligand egress akin to getting toothpaste out of the tube by opening the cap and squeezing from the bottom.
FIGURE 3.
Comparison of CYP2A13 conformations (ribbons) and cavities (mesh). A, comparison of CYP2A13/NNK in the canonical closed orientation (blue) and the most open conformation (molecule H, purple). The black arrow indicates channel with the bulge in the I helix just past the arrow tip. B, stereo image of active site of A with key side chains (sticks) and active site cavities. C, stereo image comparison of CYP2A13/NNK open conformation (molecule H, purple) with CYP2A13/2×NNK I422 closed structure (green) reveals the second NNK molecule is located in space that becomes the base of the channel to the surface in molecule H. D, stereo image comparison of CYP2A13/NNK P1 closed conformation containing NNK (blue) and the intermediate conformation (molecule G, pink) reveals in the intermediate conformation that the primary void is smaller on one side with the I helix shift, Phe-300, Phe-209, and Phe-118 occluding part of the active site but a small new bulge at the base of the channel to the surface in the open conformation. B and C, ligands omitted for clarity.
An intermediate protein conformation is observed in the form of molecule G from both the CYP2A13/nicotine and CYP2A13/NNK P1 structures. In this case, the overall protein backbone of G is most similar to the “open” molecule H conformation, but key side chains once again occupy different positions to yield yet another new active site topology. In this case the resulting enclosed active site cavity is smaller than ever reported for CYP2A13 (247 Å3; Fig. 3D). The overall decrease in active site volume is primarily due to a ∼2 Å bulge of one turn of the I helix backbone, which repositions Phe-300 and Ala-301 to reduce the volume available near the I helix immediately over the active site into the cavity (Fig. 3A). Additionally Phe-209 and Phe-118 adopt positions that occlude part of the cavity normally occupied by substrates in a way very similar to the “open” conformation in molecule H. Although repositioning of Phe-209 opens a small volume corresponding to the base of the channel in molecule H, on the opposite side of where the channel base would be, Phe-480 is still oriented toward the active site instead of torsioned away as seen when the channel is open. Thus only the beginning of the channel to the surface is observed in molecule G.
DISCUSSION
Important Residues for Differential Nicotine and NNK Binding and Metabolism
Differences in the levels and function of human CYP2A6 have been associated with smoking behavior and drug clearance (3, 35), and CYP2A13 with lung cancer risk (36). The human CYP2A enzymes 2A13 and 2A6 are 94% identical at the amino acid sequence level, but there are substantial differences in their metabolism of the key pharmacologically important agents in tobacco, primarily the addictive agent nicotine and a key procarcinogen, NNK. Although the primary nicotine metabolite for both enzymes results from 5′-oxidation, CYP2A13 demonstrates 5–23-fold higher in vitro enzymatic efficiency (11, 31). Nicotine also binds to CYP2A13 with a binding affinity 8.7-fold greater than CYP2A6. Similarly, although CYP2A13 readily generates carcinogenic metabolites from hydroxylation at both carbons α to the nitrosamine, CYP2A6 only does so poorly and at only one of these positions (9, 37).
The structures of both human CYP2A enzymes presented herein suggest that these functional differences are related to important changes in the active sites that affect substrate affinity and height and orientation over the heme. A key interaction between CYP2A13 and the pyridine rings of both nicotine and NNK is a hydrogen bond that is not present for CYP2A6 with nicotine and likely has a substantial contribution to both the observed enzyme affinities and ligand orientations. Mutation of Asn-297 in CYP2A6 was noted to increase the Km for coumarin (38). Structures of CYP2A6 with coumarin and methoxsalen show that both ligands hydrogen bond to Asn-297 but do so via a ketone oxygen substituent on a ring system rather than to the ring system itself, as occurs for nicotine and NNK in CYP2A13. This is consistent with a steric rationale for nicotine and NNK positioning (see below).
The overall orientations of nicotine in both active sites are consistent with metabolism on the pyrrolidine ring. Although both enzymes can form a minor nornicotine metabolite resulting from methyl hydroxylation, both complexes suggest that a reorientation might be required to generate this metabolite. Instead, the primary 5′ site of metabolism is positioned more directly over the iron. Assuming the observed binding orientations are retained during the catalytic cycle, the stabilizing hydrogen bond between the pyridine nitrogen and Asn-297 and the orientation of the plane of the pyrrolidine ring in CYP2A13 would appear to present the 5′-carbon to a catalytic Fe(IV)=O species with a more favorable geometry for hydrogen abstraction and rebound. The identity and placement of residues at positions 300, 208, and 209 above the pyridine ring facilitate a nicotine position slightly higher above the heme in CYP2A13 than is observed in CYP2A6, seemingly enabling the hydrogen bonding interaction with the heteroatom within the ligand ring for CYP2A13, but not CYP2A6. The importance of the identity of the residue at position 300 is also supported by the substantially increased nicotine binding affinity in the CYP2A6 I300F mutant enzyme. Similarly, changes in the identity and/or position of residues at positions 117, 369, 370, and perhaps 366 nearer the pyrrolidine ring appear to modulate both the vertical placement of the overall nicotine molecule and the plane of the pyrrolidine ring and thus the entire molecule. This is also supported by binding studies where incorporation of five CYP2A13 residues into the CYP2A6 active site (CYP2A6 V117A/I208S/I300F/G301A/S369G) increased the affinity for nicotine from 470 μm for the parent CYP2A6 to only ∼2.6-fold greater (144 μm) than wild type CYP2A13 (54 μm).
Comparison between structures of CYP2A6 with various ligands and of CYP2A13 with NNK in productive orientations, along with site-directed mutagenesis information, suggests possible reasons why hepatic CYP2A6 does not readily activate the NNK procarcinogen to initiate liver cancer in smokers. Although NNK binds to both enzymes with similar affinities, the orientation of NNK in CYP2A6 may not be consistent with metabolism. In particular, the projection of Ile-300 above the pyridine ring of nicotine in CYP2A6 may also limit the ability of the NNK pyridine to adopt a position that would facilitate hydrogen bonding with Asn-297. Other residues identified by site-directed mutagenesis to alter NNK metabolism include 117 (7), 208 (7), 297 (6), 372 (7), and 465 (7). Most of these residues appear to play steric roles in the differential binding of nicotine and likely do so for NNK as well.
The overall orientations of NNK in the CYP2A13/NNK P1 structure are consistent with NNK metabolism, with the nitrosamine end oriented over the heme. One orientation of the nitrosamine chain is most consistent with α-methylene hydroxylation (4.5 Å from iron) and has the α-methyl group further away. The second orientation of NNK observed in this structure is more consistent with α-methyl hydroxylation product with the methyl group only 2.2 Å from the heme. However, in this case, the methyl group located very close to the heme would have to move slightly distally for oxygen to bind and generate the catalytic species.
These observed orientations of NNK are generally well predicted by docking studies, all of which suggested hydrogen bonding between the NNK pyridine ring and Asn-297 (6, 7, 15, 18). However, the positions of atoms in the nitrosamine end of the molecule appear to vary slightly between studies, most of which showed binding modes for both metabolites (6, 7, 15). Flexibility in this region is also evident in the structures herein. Notably, an NNK study by Xu et al. (18) also included molecular dynamics simulations and specifically stated that poses with the pyridine ring oriented toward the heme were disregarded because this pose was not consistent with metabolism. This is an assumption that may have been made by other studies as well, yet the CYP2A13/NNK I422 structure described below indicates that this orientation does exist, despite the fact that metabolism does not appear to occur from NNK in this orientation.
Simultaneous Binding of Two NNK Molecules
Surprisingly, a separate I422 structure of the CYP2A13/NNK complex contained two molecules of NNK: one in a nonproductive orientation in the active site and one more distal from the heme. The space to bind the distal NNK molecule is made available through torsion of Phe-480 without significant backbone alterations. The reorientation of this single side chain nearly doubles the active site volume (598 Å3) from that observed in previous structures containing a single ligand (∼300 Å3). This type of Phe repositioning is very similar to two different structures of CYP2E1 where repositioning of either Phe-298 or Phe-478 on opposite sites of the active site can substantially increase the active site size to accommodate larger ligands (14, 39). Simultaneous binding of more than one ligand has previously been observed for CYP3A4, CYP2C8, CYP2B4, and CYP21A2 (40–43). Unlike the structures of CYP3A4 with ketoconazole (43) or CYP2C8 with 9-cis-retinoic acid (40), the second NNK ligand molecule does not pack or interact with the NNK in the active site. Instead the second molecule of NNK occupies a separate portion of the active site with the pyridine ring π-stacking with Phe-480. This location is reminiscent of the second molecule of 17α-hydroxyprogesterone in CYP21A2, which is located in a channel to the protein surface (41).
Monitoring CYP2A13 binding of NNK binding by spectral shifts that occur upon ligand titration reveal no experimental evidence for cooperative binding. However binding of the distal NNK would not be likely to alter the iron-coordinated water molecule like the more proximal molecule would. Thus spectral binding shifts may only be responsive to the NNK molecule in the active site, as has been observed for fluorol-7GA binding to CYP3A4 (44). Additionally, the proximal NNK molecule in the nonproductive orientation would be expected to yield a type II spectral shift because the pyridine nitrogen appears to coordinate to iron, but in solution only a type I spectral shift is observed, even at high concentrations, when CYP2A13 is titrated with NNK. Thus it appears that simultaneous binding of two NNK molecules in this orientation is not the predominant situation in the purified two-component system in solution. However, the space occupied by the distal NNK molecule over the K-L loop is also observed as part of the void available in the “open” unliganded molecule H of both the CYP2A13 structures with nicotine and NNK (P1).
Access/Egress Channel
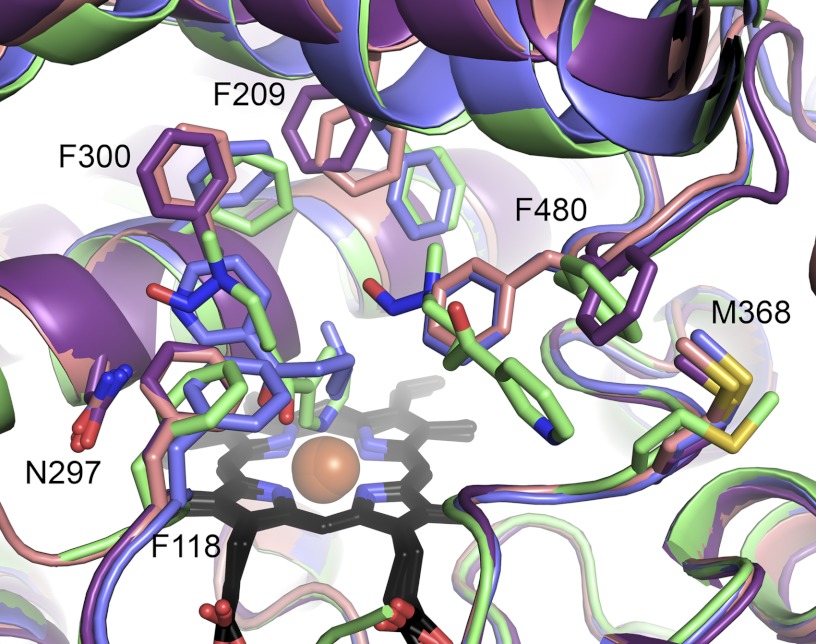
Molecules G and H of both CYP2A13 P1 structures present two previously undescribed conformations. Molecule G appears to be an intermediate conformation between the closed CYP2A13 conformation and the open conformation observed for molecule H. The channel in molecule H is very similar to the channel observed in CYP2D6 (Protein Data Bank code 2F9Q) where the active site also opens between the F helix and β4 sheet (supplemental Fig. S1) (17). At the backbone level, molecules G and H are very similar to each other, both demonstrating a striking shift of the F-G helices, with helix F shifting 3 Å away from the active site compared with the closed conformation (Fig. 4). These three conformations have a progressive shift in the location of the side chain of Phe-209 and a bimodal orientation of Phe-480, which open up opposite sides of the channel. The flexibility of the Phe-480 side chain is notable in that a similar torsion of Phe-480 is also observed in the CYP2A13/2×NNK I422 structure (Fig. 4, green). In the intermediate and open structures, reorientation of Phe-300, Phe-118, and a bulge in helix I directly over the heme are repositioned to reduce the active site volume near, and ligand access to, Asn-297.
FIGURE 4.
Comparison of CYP2A13 conformations. Shown are CYP2A13/NNK P1 closed (blue), intermediate (pink, molecule G), and open (purple, molecule H) conformations versus CYP2A13/2×NNK I422 closed structure (green). Several phenylalanine residues have substantially different conformations between molecules. The main difference between the intermediate (pink) and open (purple) conformation is Phe-480 reorientation.
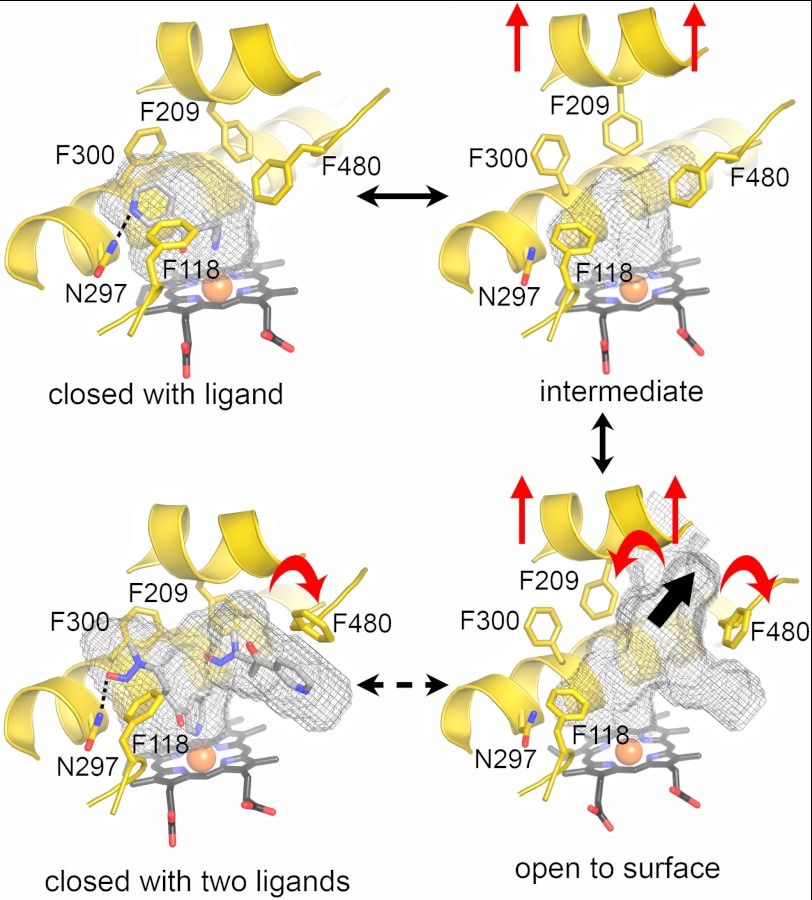
The conformations observed herein for CYP2A13 can be used to suggest a trajectory for ligand entry/exit and the attendant structural changes as illustrated (Fig. 5). In the closed, ligand-bound state, the F-G helices are in their lowered position, Phe-480 and Phe-209 close off exit from the active site, and the positions of Phe-300, Phe-118, and helix I immediately over the active site are compatible with ligand (nicotine or NNK) hydrogen bonding to Asn-297 in orientations consistent with the observed metabolites. In the intermediate conformation, the F-G helices are raised, and the position of 209 begins to open one side of the channel at the same time Phe-300, Phe-290, and helix adopt orientations that occlude ligand interaction with Asn-297, but Phe-480 still largely closes off exit from the active site. Finally, in the fully open conformation, the F-G helices are still raised, and Phe-300, Phe-118, and part of the I helix still occlude ligand access to Asn-297, but Phe-209 rotates further and Phe-480 rotates dramatically to open up the channel from the active site to the protein surface. It is unclear where the structure containing two NNK molecules fits within this proposed continuum. The backbone of helices F-G and the position of Phe-209 are clearly most similar to the closed conformation, but the positions of Phe-300 and Phe-118 are intermediate, and the position of Phe-480 is clearly in the open orientation. Thus although the structure with two molecules is overall most similar to the closed liganded conformation, the flexibility of the β4 sheet system encompassing Phe-480 is apparent.
FIGURE 5.
Conformations of CYP2A13 and possible entry/exit scheme. CYP2A13 was observed in four different conformations: closed with ligand (CYP2A13 NNK P1), intermediate (CYP2A13 NNK P1, molecule G), open to the surface (CYP2A13 NNK P1, molecule H), and closed with two NNK ligands (CYP2A13 with NNK I4222). Solid black arrows between structures suggest a continuum that would facilitate ligand entry/exit from the CYP2A13 active site. The dashed arrow represents one possible route to the CYP2A13 structure with two ligands. Red arrows highlight differences between structures. The single thick black arrow highlights the lobe of the channel connecting to the protein surface.
In conclusion, the four structures herein of two different human membrane cytochrome P450 enzymes provide valuable information into the differential binding and metabolism of the highly addictive human drug nicotine, as well as the selective activation of a key tobacco procarcinogen responsible for lung tumor initiation in smokers. For a variety of CYP2A ligands, binding appears to be largely sterically controlled with Asn-297 as a key orienting feature. In addition, one structure of CYP2A13 revealed its potential to simultaneously bind two molecules of NNK. Finally, two previously undescribed conformations of CYP2A13 were elucidated in both nicotine and NNK crystals, one with an open channel leading from the active site to the protein surface between the F helix and β4 region and one intermediate between this open conformation and the previously observed closed conformation.
Acknowledgments
Thanks are due to Linda Blake who made the CYP2A6 V117A/I208S/I300F/G301A/S369G mutant and protein. Crystals were grown using the facilities of the Protein Structure Laboratory core facility at The University of Kansas (National Institutes of Health RR017708 and GM103420). Portions of this research were carried out at the Stanford Synchrotron Radiation Lightsource, a Directorate of SLAC National Accelerator Laboratory and an Office of Science User Facility operated for the U.S. Department of Energy Office of Science by Stanford University. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393) and the National Center for Research Resources (P41RR001209).
This work was supported, in whole or in part, by National Institutes of Health Grant GM076343.
The atomic coordinates and structure factors (codes 4EJG, 4EJH, 4EJI, and 4EJJ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).

This article contains supplemental Fig. S1.
- CYP
- cytochrome P450
- NNK
- 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- rmsd
- root mean square deviation.
REFERENCES
- 1. American Cancer Society (2011) Cancer Facts and Figures, American Cancer Society, Atlanta, GA [Google Scholar]
- 2. Centers for Disease Control and Prevention (2011) Vital signs. Current cigarette smoking among adults aged ≥18 years: United States, 2005–2010, in Morbidity and Mortality Weekly Report 61, 38–45 [Google Scholar]
- 3. Malaiyandi V., Sellers E. M., Tyndale R. F. (2005) Implications of CYP2A6 genetic variation for smoking behaviors and nicotine dependence. Clin. Pharmacol. Ther. 77, 145–158 [DOI] [PubMed] [Google Scholar]
- 4. Hecht S. S. (1999) DNA adduct formation from tobacco-specific N-nitrosamines. Mutat. Res. 424, 127–142 [DOI] [PubMed] [Google Scholar]
- 5. Jalas J. R., Ding X., Murphy S. E. (2003) Comparative metabolism of the tobacco-specific nitrosamines 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol by rat cytochrome P450 2A3 and human cytochrome P450 2A13. Drug Metab. Dispos. 31, 1199–1202 [DOI] [PubMed] [Google Scholar]
- 6. Schlicht K. E., Berg J. Z., Murphy S. E. (2009) Effect of CYP2A13 active site mutation N297A on metabolism of coumarin and tobacco-specific nitrosamines. Drug Metab. Dispos. 37, 665–671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. He X. Y., Shen J., Ding X., Lu A. Y., Hong J. Y. (2004) Identification of critical amino acid residues of human metabolic activation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-tobacco-specific carcinogen. Drug Metab. Dispos. 32, 1516–1521 [DOI] [PubMed] [Google Scholar]
- 8. Cloutier J. F., Drouin R., Weinfeld M., O'Connor T. R., Castonguay A. (2001) Characterization and mapping of DNA damage induced by reactive metabolites of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK) at nucleotide resolution in human genomic DNA. J. Mol. Biol. 313, 539–557 [DOI] [PubMed] [Google Scholar]
- 9. Su T., Bao Z., Zhang Q. Y., Smith T. J., Hong J. Y., Ding X. (2000) Human cytochrome p450 CYP2A13: Predominant expression in the respiratory tract and its high efficiency metabolic activation of a tobacco-specific carcinogen, 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone. Cancer Res. 60, 5074–5079 [PubMed] [Google Scholar]
- 10. Weng Y., Fang C., Turesky R. J., Behr M., Kaminsky L. S., Ding X. (2007) Determination of the role of target tissue metabolism in lung carcinogenesis using conditional cytochrome P450 reductase-null mice. Cancer Res. 67, 7825–7832 [DOI] [PubMed] [Google Scholar]
- 11. Bao Z., He X. Y., Ding X., Prabhu S., Hong J. Y. (2005) Metabolism of nicotine and cotinine by human cytochrome P450 2A13. Drug Metab. Dispos. 33, 258–261 [DOI] [PubMed] [Google Scholar]
- 12. Yano J. K., Denton T. T., Cerny M. A., Zhang X., Johnson E. F., Cashman J. R. (2006) Synthetic inhibitors of cytochrome P-450 2A6. Inhibitory activity, difference spectra, mechanism of inhibition, and protein cocrystallization. J. Med. Chem. 49, 6987–7001 [DOI] [PubMed] [Google Scholar]
- 13. Yano J. K., Hsu M. H., Griffin K. J., Stout C. D., Johnson E. F. (2005) Structures of human microsomal cytochrome P450 2A6 complexed with coumarin and methoxsalen. Nat. Struct. Mol. Biol. 12, 822–823 [DOI] [PubMed] [Google Scholar]
- 14. DeVore N. M., Meneely K. M., Bart A. G., Stephens E. S., Battaile K. P., Scott E. E. (2012) Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine. FEBS J 279, 1621–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Smith B. D., Sanders J. L., Porubsky P. R., Lushington G. H., Stout C. D., Scott E. E. (2007) Structure of the human lung cytochrome P450 2A13. J. Biol. Chem. 282, 17306–17313 [DOI] [PubMed] [Google Scholar]
- 16. He X. Y., Shen J., Hu W. Y., Ding X., Lu A. Y., Hong J. Y. (2004) Identification of Val117 and Arg372 as critical amino acid residues for the activity difference between human CYP2A6 and CYP2A13 in coumarin 7-hydroxylation. Arch. Biochem. Biophys. 427, 143–153 [DOI] [PubMed] [Google Scholar]
- 17. Rowland P., Blaney F. E., Smyth M. G., Jones J. J., Leydon V. R., Oxbrow A. K., Lewis C. J., Tennant M. G., Modi S., Eggleston D. S., Chenery R. J., Bridges A. M. (2006) Crystal structure of human cytochrome P450 2D6. J. Biol. Chem. 281, 7614–7622 [DOI] [PubMed] [Google Scholar]
- 18. Xu Y., Shen Z., Shen J., Liu G., Li W., Tang Y. (2011) Computational insights into the different catalytic activities of CYP2A13 and CYP2A6 on NNK. J. Mol. Graph. Model. 30, 1–9 [DOI] [PubMed] [Google Scholar]
- 19. DeVore N. M., Smith B. D., Urban M. J., Scott E. E. (2008) Key residues controlling phenacetin metabolism by human cytochrome P450 2A enzymes. Drug Metab. Dispos. 36, 2582–2590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Leslie A. G. (1998) MOSFLM 6.0, Cambridge, UK [Google Scholar]
- 21. Evans P. (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 22. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 24. Hooft R. W., Vriend G., Sander C., Abola E. E. (1996) Errors in protein structures. Nature 381, 272–272 [DOI] [PubMed] [Google Scholar]
- 25. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) PROCHECK. A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 [Google Scholar]
- 26. Kleywegt G. J., Zou J. Y., Kjeldgaard M., Jones T. A. (2001) International Tables for Crystallography, Volume F, pp. 353–356 and 366–367, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 27. Porubsky P. R., Meneely K. M., Scott E. E. (2008) Structures of human cytochrome P-450 2E1. Insights into the binding of inhibitors and both small molecular weight and fatty acid substrates. J. Biol. Chem. 283, 33698–33707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schlicht K. E., Michno N., Smith B. D., Scott E. E., Murphy S. E. (2007) Functional characterization of CYP2A13 polymorphisms. Xenobiotica 37, 1439–1449 [DOI] [PubMed] [Google Scholar]
- 29. DeVore N. M., Smith B. D., Wang J. L., Lushington G. H., Scott E. E. (2009) Key residues controlling binding of diverse ligands to human cytochrome P450 2A enzymes. Drug Metab. Dispos. 37, 1319–1327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stephens E. S., Walsh A. A., Scott E. E. (2012) Evaluation of inhibition selectivity for human cytochrome P450 2A enzymes. Drug Metab. Dispos., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Murphy S. E., Raulinaitis V., Brown K. M. (2005) Nicotine 5′-oxidation and methyl oxidation by P450 2A enzymes. Drug Metab. Dispos. 33, 1166–1173 [DOI] [PubMed] [Google Scholar]
- 32. Hukkanen J., Jacob P., 3rd, Benowitz N. L. (2005) Metabolism and disposition kinetics of nicotine. Pharmacol. Rev. 57, 79–115 [DOI] [PubMed] [Google Scholar]
- 33. von Weymarn L. B., Brown K. M., Murphy S. E. (2006) Inactivation of CYP2A6 and CYP2A13 during nicotine metabolism. J. Pharmacol. Exp. Ther. 316, 295–303 [DOI] [PubMed] [Google Scholar]
- 34. Hecht S. S., Hochalter J. B., Villalta P. W., Murphy S. E. (2000) 2′-Hydroxylation of nicotine by cytochrome P450 2A6 and human liver microsomes. Formation of a lung carcinogen precursor. Proc. Natl. Acad. Sci. U.S.A. 97, 12493–12497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Tan W., Chen G. F., Xing D. Y., Song C. Y., Kadlubar F. F., Lin D. X. (2001) Frequency of CYP2A6 gene deletion and its relation to risk of lung and esophageal cancer in the Chinese population. Int. J. Cancer 95, 96–101 [DOI] [PubMed] [Google Scholar]
- 36. Wang H., Tan W., Hao B., Miao X., Zhou G., He F., Lin D. (2003) Substantial reduction in risk of lung adenocarcinoma associated with genetic polymorphism in CYP2A13, the most active cytochrome P450 for the metabolic activation of tobacco-specific carcinogen NNK. Cancer Res. 63, 8057–8061 [PubMed] [Google Scholar]
- 37. He X. Y., Tang L., Wang S. L., Cai Q. S., Wang J. S., Hong J. Y. (2006) Efficient activation of aflatoxin B1 by cytochrome P450 2A13, an enzyme predominantly expressed in human respiratory tract. Int. J. Cancer 118, 2665–2671 [DOI] [PubMed] [Google Scholar]
- 38. Wu Z. L., Podust L. M., Guengerich F. P. (2005) Expansion of substrate specificity of cytochrome P450 2A6 by random and site-directed mutagenesis. J. Biol. Chem. 280, 41090–41100 [DOI] [PubMed] [Google Scholar]
- 39. Porubsky P. R., Battaile K. P., Scott E. E. (2010) Human cytochrome P450 2E1 structures with fatty acid analogs reveal a previously unobserved binding mode. J. Biol. Chem. 285, 22282–22290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Schoch G. A., Yano J. K., Sansen S., Dansette P. M., Stout C. D., Johnson E. F. (2008) Determinants of cytochrome P450 2C8 substrate binding. Structures of complexes with motelukast, troglitazone, felodipine, and 9-cis-retinoic acid. J. Biol. Chem. 283, 17227–17237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao B., Lei L., Kagawa N., Sundaramoorthy M., Banerjee S., Nagy L. D., Guengerich F. P., Waterman M. R. (2012) A three-dimensional structure of steroid 21-hydroxylase (cytochrome P450 21A2) with two substrates reveals locations of disease-associated variants. J. Biol. Chem. 287, 10613–10622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Gay S. C., Sun L., Maekawa K., Halpert J. R., Stout C. D. (2009) Crystal structures of cytochrome P450 2B4 in complex with the inhibitor 1-biphenyl-4-methyl-1H-imidazole. Ligand-induced structural response through alpha-helical repositioning. Biochemistry 48, 4762–4771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ekroos M., Sjögren T. (2006) Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. U.S.A. 103, 13682–13687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Davydov D. R., Rumfeldt J. A., Sineva E. V., Fernando H., Davydova N. Y., Halpert J. R. (2012) Peripheral ligand-binding site in cytochrome P450 3A4 located with fluorescence resonance energy transfer (FRET). J. Biol. Chem. 287, 6797–6809 [DOI] [PMC free article] [PubMed] [Google Scholar]