Background: CP32M is a newly designed HIV-1 fusion inhibitor.
Results: The crystal structure of CP32M and its potent activity against diverse HIV-1 variants were determined.
Conclusion: The crystal structure reveals the mechanistic insights of CP32M.
Significance: Our data provide important information for designing potent HIV-1 fusion inhibitors.
Keywords: Crystal Structure, Fusion Protein, HIV-1, Membrane Fusion, Virus Entry, CP32M, Fusion Inhibitor, Six-helix Bundle
Abstract
CP32M is a newly designed peptide fusion inhibitor possessing potent anti-HIV activity, especially against T20-resistant HIV-1 strains. In this study, we show that CP32M can efficiently inhibit a large panel of diverse HIV-1 variants, including subtype B′, CRF07_BC, and CRF01_AE recombinants and naturally occurring or induced T20-resistant viruses. To elucidate its mechanism of action, we determined the crystal structure of CP32M complexed with its target sequence. Differing from its parental peptide, CP621-652, the 621VEWNEMT627 motif of CP32M folds into two α-helix turns at the N terminus of the pocket-binding domain, forming a novel layer in the six-helix bundle structure. Prominently, the residue Asn-624 of the 621VEWNEMT627 motif is engaged in the polar interaction with a hydrophilic ridge that borders the hydrophobic pocket on the N-terminal coiled coil. The original inhibitor design of CP32M provides several intra- and salt bridge/hydrogen bond interactions favoring the stability of the helical conformation of CP32M and its interactions with N-terminal heptad repeat (NHR) targets. We identified a novel salt bridge between Arg-557 on the NHR and Glu-648 of CP32M that is critical for the binding of CP32M and resistance against the inhibitor. Therefore, our data present important information for developing novel HIV-1 fusion inhibitors for clinical use.
Introduction
Infection of human immunodeficiency virus type 1 (HIV-1) requires a fusion reaction between the viral and cellular membranes. This process is mediated by the viral envelope glycoprotein (Env), a trimeric complex consisting of three transmembrane gp41 subunits and three non-covalently attached gp120 surface subunits (1–4). The gp120 subunit is responsible for attachment to the target cells, which initiates the entry process by binding to the CD4 receptor and a co-receptor (CCR5 or CXCR4), whereas the gp41 subunit mediates the membrane fusion. In the current fusion model, the gp41 undergoes a dramatic transition from a native state into a pre-hairpin intermediate that releases its hydrophobic ectodomain in an extended conformation, thus allowing insertion of the fusion peptide into the targeting cell membrane (4, 5). This state may persist over a span of 15 min and then collapses into a low energy trimeric hairpin structure (six-helix bundle (6-HB))5 that juxtaposes the viral and host membranes. The crystal structure of 6-HB revealed that three N-terminal heptad repeat (NHR) regions of gp41 form a central trimeric coiled coil, whereas three C-terminal heptad repeat (CHR) segments pack as antiparallel helices into the hydrophobic grooves on the NHR trimer (6–8). Prominently, a deep hydrophobic pocket is present in the C-terminal portion of NHR helices, whereas three hydrophobic residues (Trp-628, Trp-631, and Ile-635) from the pocket-binding domain (PBD) of the CHR helix penetrate into the pocket causing an extensive interaction. It is believed that the deep pocket can serve as an ideal target site for anti-HIV agents that block the 6-HB formation (5, 9). It is becoming clear that the 6-HB structure is involved in the fusion mechanism of many enveloped viruses (1, 3, 10–12). During the extended pre-hairpin conformation, peptide or small molecule-based fusion inhibitors act by competitive binding to the exposed NHR or CHR, thus blocking the 6-HB formation dominant negatively (5, 13–15).
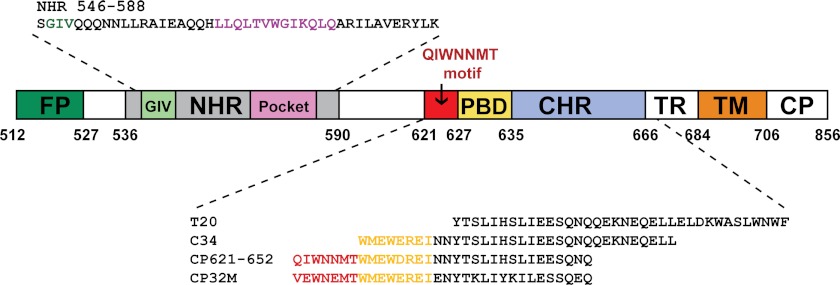
T20 (enfuvirtide (Fuzeon)) is the first and only clinically approved HIV-1 fusion inhibitor being used for the treatment of HIV/AIDS patients who fail to respond to current antiretroviral drugs (16–18). However, T20 requires frequent administration, and resistance to it develops rapidly. To overcome these challenges, a series of more potent and stable peptide fusion inhibitors has been developed (13, 15, 19); and some of these, such as sifuvirtide and T1249, have been evaluated by clinical trials (20–23). Notably, these peptide inhibitors were derived primarily from the gp41 CHR region, not including the upstream sequence of the PBD. In particular, peptide C34 (amino acids 628–661) was widely used as a design template (Fig. 1). This limitation might arise from the lack of structural and functional information available on this region. Indeed, all three pioneering structures of HIV-1 gp41 core have been determined by using biosynthetic or synthesized peptide fragments that lack the loop region between the putative NHR and CHR (6–8). However, our recent studies may challenge this belief (24–26). We found that the 621QIWNNMT627 motif, located immediately adjacent to the PBD, plays a critical role in 6-HB formation; and the CP621–652 peptide that contains this motif is a potent fusion inhibitor against the wild-type and T20-resistant HIV-1 strains (24). Based on this finding, we engineered CP621–652 into CP32M (Fig. 1), which shows very strong inhibition on HIV-1 with different genotypes and phenotypes, especially T20-resistant viruses (25).
FIGURE 1.
Schematic illustration of HIV-1 gp41 functional regions and NHR- or CHR-derived peptide sequences. Shown are the residue numbers of each region corresponding to their positions in gp160 of HIV-1HXB2. FP, fusion peptide; TM, transmembrane domain; TR, tryptophan-rich domain; CP, cytoplasmic domain. The residues corresponding to the NHR pocket region are marked in magenta; the residues for the PBD are marked in yellow; the critical motif adjacent to the N-terminal of PBD is marked in red; the hot spot GIV motif for drug resistance on NHR is marked in green.
In this study, we have further shown that CP32M possesses potent and broad inhibitory activity against a large panel of diverse HIV-1 variants. We determined the crystal structures of CP32M in complex with the NHR target. In stark contrast to the largely disordered N-terminal motif of the parental peptide, CP621–652, the engineered 621VEWNEMT627 motif of CP32M adopts two α-helix turns at the N terminus of the PBD, thereby forming a novel layer in the 6-HB structure that stabilize the pocket region. Several intra- and interhelical salt bridges and hydrogen bonds synthetically favor the helical conformation of CP32M and its interactions with NHR targets. Therefore, our data present mechanistic insights into CP32M and provide important information for designing HIV-1 fusion inhibitors.
EXPERIMENTAL PROCEDURES
Peptide Synthesis
Peptides CP32M and T20 were synthesized by a standard solid-phase Fmoc (N-(9-fluorenyl)methoxycarbonyl) method as described previously (25). All peptides were acetylated at the N terminus and amidated at the C terminus. They were purified by reversed-phase high-performance liquid chromatography (HPLC) and verified for >95% purity and correct amino acid composition by mass spectrometry. Concentrations of the peptides were determined by UV absorbance and a theoretically calculated molar extinction coefficient (280 nm) of 5500 m−1·cm−1 and 1490 m−1·cm−1 based on the number of tryptophan and tyrosine residues (all of the peptides tested contain Trp and/or Tyr), respectively.
Plasmids and Cells
Six CRF07_BC Env clones (CH064.20, CH070.1, CH091.9, CH110.2, CH119.10, and CH120.6) were generous gifts by Dr. Yiming Shao in the Chinese Center for Disease Control and Prevention, Beijing, China. A panel of Env clones derived from subtype B′ (B01, B02, B04, and 43-22), CRF01_AE (SHX335.24, YN192.31, AE01, AE03, GX2010.36, GX11.13, GX2010.36H, and BJ5.11), and CRF07_BC (BC02, BC03, BC05, BC07, BC14, SC19-15, BJ22-5, YN148R-9, XJ50-6, and HB5-3) were kindly provided by Dr. Youchun Wang from the National Institute for the Control of Pharmaceutical and Biological Products, Beijing, China. TZM-bl cells were contributed by J. C. Kappes and X. Wu through the AIDS Research and Reference Reagent Program (ARRRP), Division of AIDS, NIAID, National Institutes of Health. This reporter cell line stably expresses high levels of CD4, CCR5, and CXCR4 and contains Tat-responsive reporter genes for firefly luciferase and β-galactosidase under the control of an HIV-1 long terminal repeat promoter (27). The cells were maintained in Dulbecco's modified Eagle's medium (dMEM) growth medium (Invitrogen) containing 10% heat-inactivated fetal bovine serum (FBS). Cells were harvested using a trypsin/EDTA solution (Invitrogen). All cell lines were maintained at 37 °C in humidified air containing 5% CO2.
Inhibition of HIV-1 Single-cycle Infection
HIV-1 pseudoviruses were generated as described previously (28, 29). Briefly, 293T cells (5 × 106 cells in 15 ml of growth medium in a T-75 culture flask) were cotransfected with 10 μg of an Env-expressing plasmid and 20 μg of a backbone plasmid, pSG3Δenv, that encodes an Env-defective, luciferase-expressing HIV-1 genome using Lipofectamine 2000 reagent (Invitrogen). Pseudovirus-containing culture supernatants were harvested 48 h after transfection, filtered by 0.45-mm pore size, and stored at −80 °C in 1-ml aliquots until use. The 50% tissue culture infectious dose (TCID50) of a single thawed aliquot of each pseudovirus batch was determined in TZM-bl cells. The antiviral activity of CP32M or T20 was determined using TZM-b1 cells as described previously (21). Briefly, the peptides were prepared with 10 series dilutions in a 3-fold stepwise manner, mixed with 100 TCID50 viruses, and incubated for 1 h at room temperature. The mixture was added to TZM-bl cells (104/well) and incubated at 37 °C for 48 h. Luciferase activity was measured using luciferase assay regents (Promega, Madison, WI) and a luminescence counter (Promega) according to the manufacturer's instructions.
Inhibition of HIV-1 NL4-3 Variants
HIV-1 molecular clone NL4-3 wild type or variants carrying naturally occurring mutations (L33V, L34M, S35F, Q39R, L54M, and Q56K) and T20-resistant mutations (L33S, I37Q/V38Q, I37Q/V38M, and I37V/V38T) (30, 31) were kindly provided by Dr. Frank Kirchhoff from the Institute of Virology, University of Ulm, Ulm, Germany. The mutant viruses were generated by transient transfection of NL4-3 plasmids into 293T cells. The virus stocks were harvested 48 h post-transfection and quantified for TCID50. Inhibition of the peptides (CP32M and T20) on the NL4-3 mutants was performed as described for pseudoviruses. In brief, 100 TCID50 viruses were used to infect TZM-bl cells in the absence or presence of serially diluted peptides. Two days post-infection, the cells were harvested and lysed in reporter lysis buffer, and the luciferase activity was measured.
Protein Expression and Purification
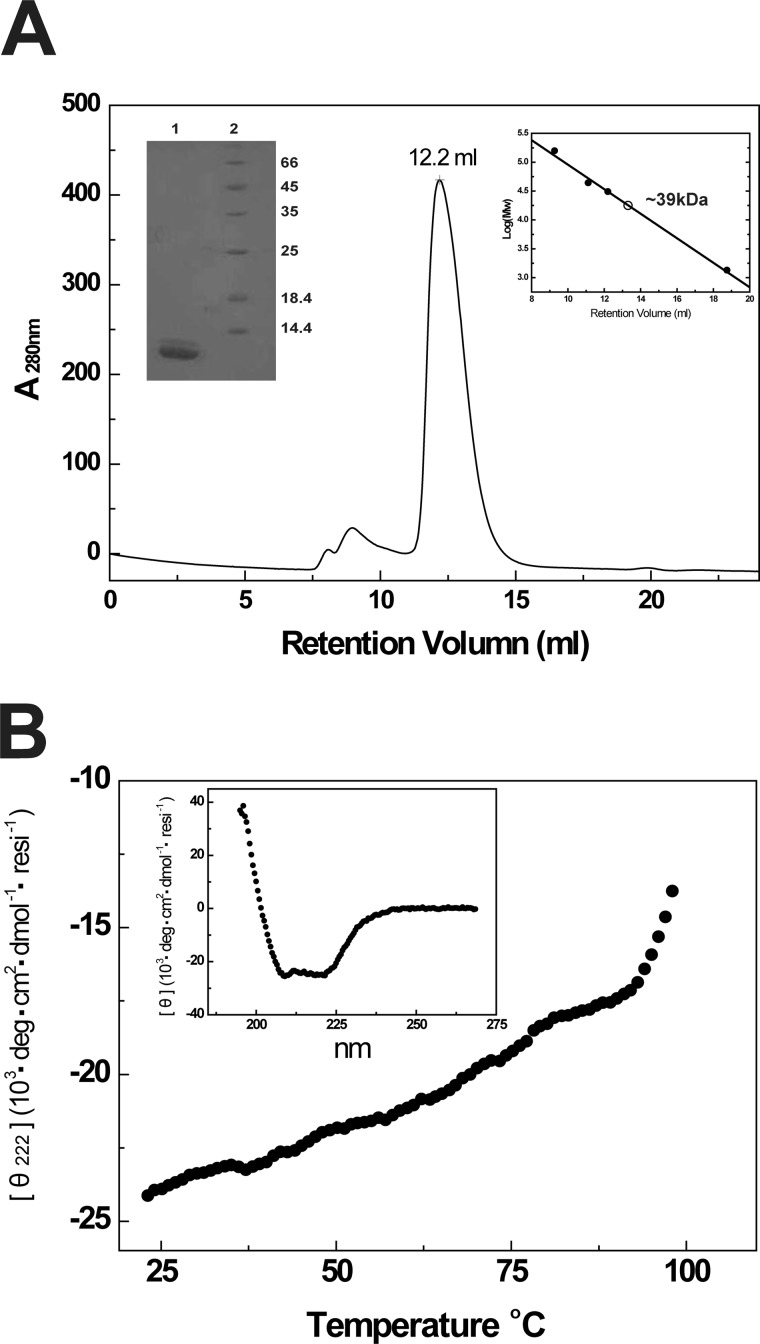
6-HB containing CP32M was prepared by expressing a NHR546–588/CP32M chimera. The cDNA encoding the chimera containing the N-terminal His6 tag, HIV-1 gp41 NHR546–588, linker “SGGRGG” (with a typsin cleavage site), and CP32M (MHHHHHHSSGLVPRGSAMADIGSEFSGIVQQQNNLLRAIEAQQHLLQLTVWGIKQLQARILAVERYLKSGGRGGVEWNEMTWMEWEREIENYTKLIYKILEESQEQ) was synthesized and inserted into expression vector pETH (derived from pET32a). The chimera was overexpressed in Escherichia coli strain B834(DE3). Bacteria were grown in LeMaster medium to an A600 nm of 1.0 and induced with IPTG (1 mm) at 18 °C overnight. The bacteria were harvested by centrifugation, suspended in lysis buffer (50 mm Tris-HCl, pH 7.5, 500 mm NaCl), and disrupted by ultrasonication. The inclusion body containing the fusion protein was collected by centrifugation and solubilized in denaturing buffer (100 mm NaH2PO4, 10 mm Tris-HCl, pH 8.0, 8 m urea) at 65 °C for 20 min. Protein refolding was conducted by dialysis against the buffer containing 50 mm Tris-HCl, pH 7.5, 100 mm NaCl at 4 °C overnight. The dialyzed sample was clarified by centrifugation and purified with nickel-nitrilotriacetic acid resin (Qiagen). The N-terminal His6 tag was eventually removed by thrombin (Sigma-Aldrich) digestion prior to ion exchange chromatography (HighTrap Q, GE Healthcare). 6-HB was finally purified by size-exclusion chromatography (Superdex 75 10/300 GL, GE Healthcare). The peak corresponding to the 6-HB size (∼39 kDa) was used for crystallization trials.
Circular Dichroism Spectroscopy
Circular dichroism (CD) spectroscopy was performed as described previously (24). The final concentration of NHR546–588/CP32M was 10 μm in PBS buffer, pH 7.2. The CD spectra were acquired on a Jasco spectropolarimeter (model J-815) using a 1 nm bandwidth with a 1 nm step resolution from 195 to 260 nm at room temperature. The spectra were corrected by subtraction of a blank corresponding to the solvent. Data were averaged over three accumulations. The α-helical content was calculated from the CD signal by dividing the mean residue ellipticity [θ] at 222 nm by the value expected for 100% helix formation (−33,000·degree·cm2·dmol−1). a thermal denaturation experiment was performed by monitoring the change in ellipticity [θ] at 222 nm at increasing temperature (20–98 °C) using a temperature controller. The temperature was increased at a rate of 1.2 °C/min; data were acquired using a 1 nm bandwidth at 222 nm at a frequency of 0.25 Hz. The melting curve was smoothened, and the midpoint of the thermal unfolding transition (Tm) values were taken as the maximum of the derivative d[θ]222/dT.
Crystallization and Structure Determination
The 6-HB assembled by the NHR546–588/CP32M chimera was crystallized by mixing equal volumes (0.8 μl) of purified protein (∼10 mg/ml) and reservoir solution in a hanging drop vapor diffusion system at 22 °C. Two crystal forms were found. Crystal form 1 crystallized using reservoir buffer containing 0.2 m MgSO4 and 20% PEG 3350; crystal form 2 crystallized using the reservoir buffer containing 0.1 m MgCl2, 0.1 m sodium acetate, pH 4.6, and 25% PEG 400). Trypsin was added to the sample (protease:sample = 1:100 in molar ratio) prior to the crystallization trails. The in situ trypsinization was essential for obtaining the measurable crystals. Cryocooling treatments for crystal forms 1 and 2 were achieved by soaking the crystal for 0.5–1 min in the reservoir solution containing 30% ethylene glycol and 15% glycerol. Subsequently, the crystals were flash-frozen in liquid nitrogen. Complete data sets were collected for crystal forms 1 and 2 at beamline PX III SLS (Villigen, Switzerland) using an x-ray of wavelength 0.9787 and 1.000 Å. Crystal form 1 belonged to space group P321, contained one-third of a 6-HB (one NHR546–588/CP32M chimera)/asymmetry unit, and diffracted the x-ray to the resolution limit of 2.0 Å. Crystal form 2 belonged to space group P21, contained one complete six-helix bundle (three NHR546–588/CP32M chimera)/asymmetry unit, and diffracted the x-ray to the resolution limit of 2.0 Å. The structures of both crystal forms were solved by molecular replacement (Phaser CCP4 suite) using HIV-1 gp41 core structure (Protein Data Bank ID 3F4Y) as the searching model. The initial electron density map was improved by manual model building (Coot). The structures were refined using PHENIX (32). The final atomic models have excellent refinement statistics and stereochemistry qualities (Table 3). Identifying a hydrogen bond, the distance from the donor hydrogen to the electronegative acceptor is ≤3.0Å, and the “donor-H-acceptor” angle is ≥120°. Identifying a salt bridge, the distance between the positively charged atom of the basic residue (Lys or Arg) and the negatively charged atom of the acidic residues (Glu or Asp) is ≤4 Å.
TABLE 3.
Data collection and refinement statistics for NHR546–588/CP32M complex
| Crystal form 1 | Crystal form 2 | |
|---|---|---|
| Data collection | ||
| Space group | P321 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 45.09, 45.09, 73.03 | 50.51, 45.50, 55.51 |
| α, β, γ (°) | 90.0, 90.0, 120.0 | 90.0, 107.65, 90.0 |
| X-ray source | PSI-SLS Beamline PX III | PSI-SLS Beamline PX III |
| Wavelength (Å) | 0.9787 | 1.0000 |
| Data range (Å) | 39.05-2.03 | 42.61-2.02 |
| Rsyma (last shell) | 0.079 (0.63) | 0.084 (0.75) |
| I/σI | 16.42 (2.89) | 7.8 (1.2) |
| Completeness (%) (last shell) | 99.8 (98.8) | 90.8 (63.3) |
| Redundancy (last shell) | 5.77 (5.61) | 1.97 (1.24) |
| Refinement | ||
| Resolution range (Å) | 39.05-2.03 | 42.61-2.02 |
| Reflections, cutoff, cross-validation | 5846, F > 1.99, 529 | 14944, F > 1.45, 748 |
| Rworkb/Rfreec (last shell) | 0.1969/0.2590 | 0.2216/0.2741 |
| Non-hydrogen protein atoms | 708 | 2181 |
| Protein | 686 | 2113 |
| Water | 22 | 68 |
| B-factors average | 42.17 | 41.19 |
| Protein (Å2) | 42.25 | 41.16 |
| Water (Å2) | 37.04 | 38.77 |
| r.m.s.d.d | ||
| Bond lengths (Å) | 0.014 | 0.002 |
| Bond angles (°) | 1.359 | 0.545 |
| Validation | ||
| MolProbity score | 1.82, rating 86th percentile among structures of comparable resolution | 1.64, rating 93rd percentile among structures of comparable resolution. |
| % Residues in favored regions, allowed regions, outliers in Ramachandran plot | 100.0, 0, 0 | 100.0, 0, 0 |
a Rsym = ΣhklΣj|Ihkl,j − Ihkl|/ΣhklΣjIhkl,j, where Ihkl is the average of symmetry-related observations of a unique reflection.
b Rwork = Σhkl‖Fobs(hkl)|−|Fcalc(hkl)‖/Σhkl|Fobs(hkl)|.
c Rfree = the cross-validation R-factor for 5% of reflections against which the model was not refined.
d Root mean square deviation.
RESULTS
Potent Inhibition of CP32M on Diverse HIV-1 Variants
Our previous studies demonstrated that CP32M had potent inhibitory activity against several representative primary HIV-1 isolates from multiple genotypes (subtypes A–G and group O) and phenotypes (R5, X4, and R5X4) (25). To advance CP32M for clinical development, we were interested to know whether CP32M acts effectively against the HIV-1 variants that currently predominate in AIDS epidemics in China, including CRF07_BC (B/C) and CRF01_AE (A/E) recombinants and B′ (also known as Tai B) (33, 34). A panel of 27 HIV-1 pseudoviruses with their Env showing high polymorphisms was constructed and used in single-cycle infection assays. As shown in Table 1, CP32M can inhibit CRF07_BC (B/C), CRF01_AE, and B′ with mean IC50 values at 12.03, 16.94, and 10.64 nm, respectively. In comparison, T20 has much lower inhibitory activity against these HIV-1 variants.
TABLE 1.
Inhibition of CP32M and T20 on subtypes B/C, A/E, and B′ HIV-1 variants
| Pseudovirus | Subtype | Co-R | IC50 |
|
|---|---|---|---|---|
| CP32M | T20 | |||
| nm | ||||
| CH64.20 | B/C | R5 | 6.13 ± 0.40 | 56.89 ± 12.19 |
| CH070.1 | B/C | R5 | 22.99 ± 7.09 | 153.40 ± 39.71 |
| CH091.9 | B/C | R5 | 8.95 ± 2.73 | 63.89 ± 17.23 |
| Ch110.2 | B/C | R5 | 9.47 ± 1.27 | 21.10 ± 4.91 |
| Ch119.10 | B/C | R5 | 11.32 ± 1.44 | 20.08 ± 4.01 |
| Ch120.6 | B/C | R5 | 42.66 ± 1.26 | 49.36 ± 3.51 |
| SC19-15 | B/C | R5 | 3.25 ± 0.54 | 7.38 ± 0.79 |
| BJ22-5 | B/C | R5 | 8.11 ± 1.08 | 26.70 ± 3.35 |
| BC07 | B/C | R5 | 8.93 ± 0.83 | 11.25 ± 2.48 |
| BC14 | B/C | R5 | 8.45 ± 1.13 | 94.4 ± 44.55 |
| YN148r-9 | B/C | R5 | 34.89 ± 4.41 | 87.42 ± 14.74 |
| XJ50-6 | B/C | R5 | 5.13 ± 0.87 | 74.39 ± 17 |
| HB5-3 | B/C | R5 | 8.05 ± 3.12 | 22 ± 2.49 |
| BC02 | B/C | R5 | 2.5 ± 0.3 | 2.90 ± 0.63 |
| BC03 | B/C | R5 | 5.38 ± 0.83 | 3.56 ± 0.34 |
| BC05 | B/C | R5 | 6.22 ± 0.72 | 4.33 ± 0.21 |
| SHX335.24 | A/E | R5 | 3.08 ± 0.40 | 44.73 ± 12.04 |
| YN192.31 | A/E | R5 | 15.81 ± 2.62 | 65.52 ± 5.7 |
| AE03 | A/E | R5 | 8.08 ± 0.91 | 7.44 ± 1.33 |
| GX2010.36 | A/E | R5 | 10.93 ± 1.97 | 19.42 ± 3.47 |
| GX11.13 | A/E | R5 | 8.12 ± 1.66 | 6.60 ± 0.22 |
| GX2010.36H | A/E | R5 | 22.99 ± 3.18 | 25.94 ± 0.28 |
| AE01 | A/E | R5 | 11.67 ± 6.83 | 26.62 ± 2.25 |
| BJ5.11 | A/E | R5 | 54.85 ± 9.25 | 317.97 ± 102.26 |
| B01 | B′ | R5 | 3.31 ± 0.29 | 49.50 ± 2.41 |
| B02 | B′ | R5 | 12.81 ± 1.51 | 8.85 ± 0.72 |
| B04 | B′ | R5 | 12.51 ± 2.07 | 7.10 ± 1.25 |
| 43–22 | B′ | R5 | 13.91 ± 1.94 | 11.62 ± 2.42 |
Some naturally occurring variations in the NHR region of gp41 have been observed in treatment-naive patients infected with various HIV-1 subtypes, showing resistance to HIV-1 fusion inhibitors (30). Here, we characterized the susceptibility of several naturally occurring variants on CP32M. Table 2 shows that L33V, L34M, and L54M mutations result in significant resistance to T20 (-fold change > 15) but have no or only mild effects on CP32M. Compared with the wild type, the susceptibility of the S35M mutant decreased 5.86-fold for T20, whereas it decreased 2.51-fold for CP32M. Furthermore, we tested CP32M by several well characterized T20-resisant viruses that carry a L33S point mutation or I37Q/V38Q, I37Q/V38M, and I37V/V38T double mutations (30, 31). The results demonstrate that CP32M can efficiently inhibit infection by these HIV-1 variants; in sharp contrast, T20 had no inhibitory activity at a concentration as high as 750 nm. These results indicate that CP32M has potent and broad anti-HIV activity.
TABLE 2.
Sensitivity of naturally occurring and induced HIV-1 variants to fusion inhibitors
| HIV-1 variant | CP32M |
T20 |
||
|---|---|---|---|---|
| IC50 | -Fold change | IC50 | -Fold change | |
| nm | nm | |||
| NL4–3 wt | 0.88 ± 0.16 | 1.00 | 41.97 ± 15.95 | 1.00 |
| Natural mutation | ||||
| L33V | 1.31 ± 0.15 | 1.49 | 665.40 ± 303.49 | 15.85 |
| L34M | 2.23 ± 0.26 | 2.53 | >750 | >17.87 |
| S35F | 2.21 ± 0.47 | 2.51 | 245.90 ± 5.52 | 5.86 |
| Q39R | 1.11 ± 0.11 | 1.26 | 18.45 ± 5.93 | 0.44 |
| L54M | 2.50 ± 0.16 | 2.84 | >750 | >17.87 |
| Q56K | 3.17 ± 0.08 | 3.60 | 84.57 ± 13.53 | 2.02 |
| T20-resistant | ||||
| L33S | 1.92 ± 0.31 | 2.18 | >750 | >17.87 |
| I37Q/V38Q | 1.17 ± 0.10 | 1.33 | >750 | >17.87 |
| I37Q/V38M | 1.64 ± 0.16 | 1.86 | >750 | >17.87 |
| I37V/V38T | 1.48 ± 0.17 | 1.69 | >750 | >17.87 |
Biophysical Characterization of a 6-HB Formed by NHR-CP32M Chimera
Our previous studies demonstrated that CP23M possesses outstanding antiviral activity and forms the 6-HB structure with improved thermostability and helical content (25). To elucidate the molecular determinants, we utilized the crystallographic approach. We assembled the 6-HB by expressing a chimera comprising a gp41 NHR target sequence (NHR546–588), a linker, and CP32M. The molecular mass of the purified NHR546–588/CP32M chimera determined by SDS-PAGE was ∼12 kDa, whereas the molecular weight measured by size-exclusion chromatography was ∼39 kDa (Fig. 2A, insert) corresponding to the size of a trimeric chimera. The dynamic light scattering analysis measured the hydrodynamic radius of the NHR546–588/CP32M chimera as 2.99 ± 0.44 nm corresponding to an apparent molecular mass of 43.9 ± 19 kDa, which indicates not only the size for a trimeric chimera but also the size for other possible oligomers of the chimera. The resolution of this experimental method cannot provide the defined oligomerization state of the chimera. CD spectroscopy demonstrated that the chimera maintains highly helical conformation with ultra-high thermostability in solution (helical content ∼80%, Tm ∼100 °C) (Fig. 2B, insert). The above biophysical studies confirmed that the recombinant chimera assemble into a 6-HB structure in solution.
FIGURE 2.
Assembly and biophysical characterizations of the 6-HB formed by gp41 NHR546–588/CP32M chimera. A, size-exclusion chromatography and SDS-PAGE analyses of HIV gp41 NHR546–588/CP32M chimera. Chromatographic profile shows UV absorbance at 280 nm. Upper right insert, the log (molecular weight) values of the standard proteins for size-exclusion column calibration (Superdex 75 10/300 GL) are plotted as the function of Ve (elution volume) (●). The data are fitted linearly to derive the standard curve. The molecular mass of HIV gp41 NHR546–588/CP32M chimera is calculated as∼39 kDa (○). Upper left insert, SDS-PAGE analysis of purified HIV gp41 NHR546–588/CP32M chimera. Lane 1, HIV gp41 NHR546–588/CP32M chimera (∼12kDa); lane 2, molecular weight standards. B, the melting curve of NHR546–588/CP32M chimera showing the thermostability of NHR546–588/CP32M chimera. Insert, normalized circular dichroism spectrum showing the α-helical conformation of the 6-HB formed by the chimera. resi, the number of the residues.
Crystallization and Structure Determination
The purified 6-HB consisting of the NHR546–588/CP32M chimera was subjected to crystallization trials. We discovered two different crystal forms. Crystal form 1 belonged to space group P321 and contained one NHR546–588/CP32M chimera (one-third 6-HB)/asymmetrical unit. Crystal form 2 belonged to space group P21 and contained three NHR546–588/CP32M chimeras assembling into a complete 6-HB/asymmetrical unit. The redundancy of the diffraction data collected for crystal form 2 is significantly lower than for crystal form 1, which coincides with the lower symmetry in crystal form 2 as compared with crystal form 1. The 3-fold symmetry found in crystal form 1 is not present in crystal form 2. This observation suggests the possible conformational differences between the two crystal structures. We solved both structures by molecular replacement. The final refined model has excellent refinement statistics and stereochemistry qualities (Table 3). The structures of crystal forms 1 and 2 were validated by MolProbity analysis (35). The MolProbity score for crystal form 1 is 1.82, ranking in the 86th percentile among structures of comparable resolution. The Ramachandran plot found all residues in the favored area. The MolProbity score for crystal form 2 is 1.64, ranking in the 93rd percentile among structures of comparable resolution. The Ramachandran plot found all residues in the favored area.
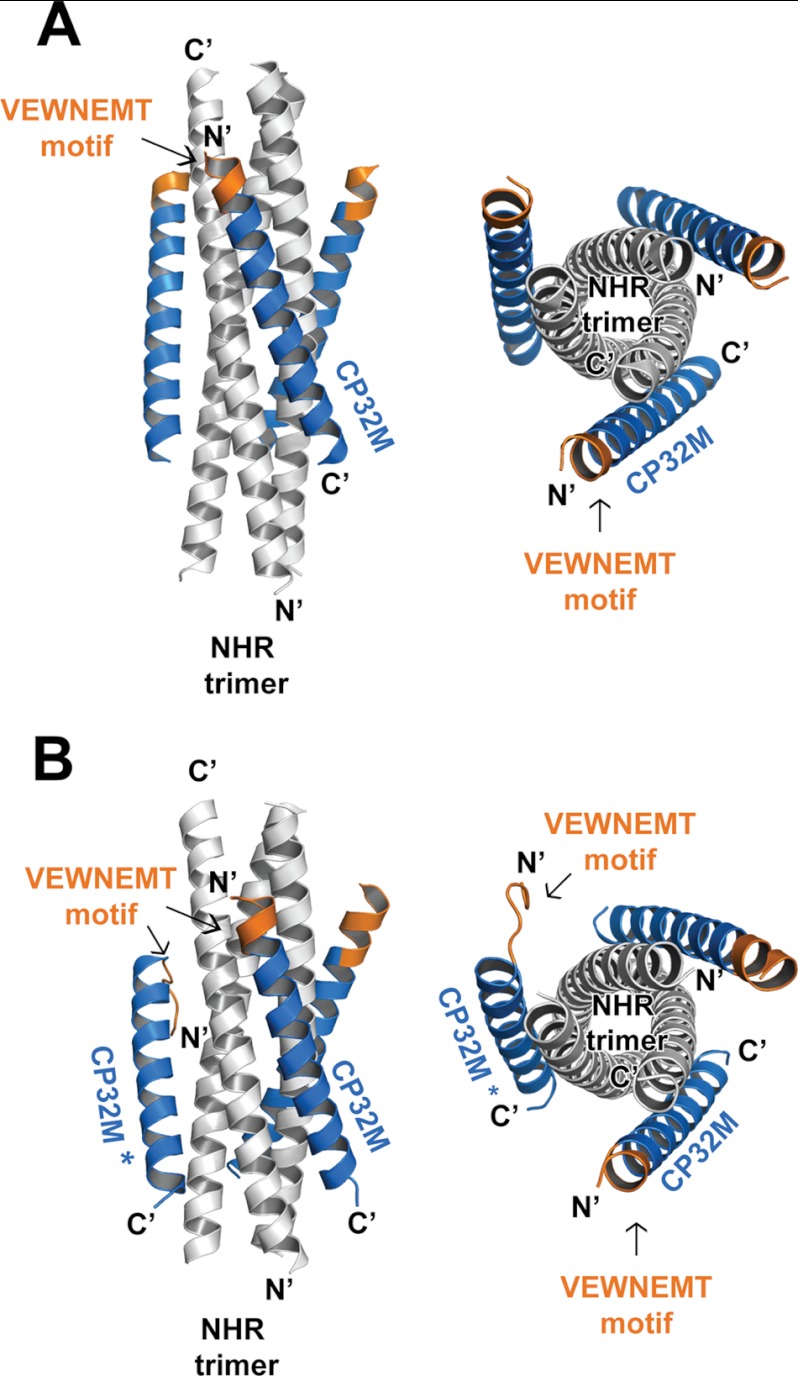
Overall Structure
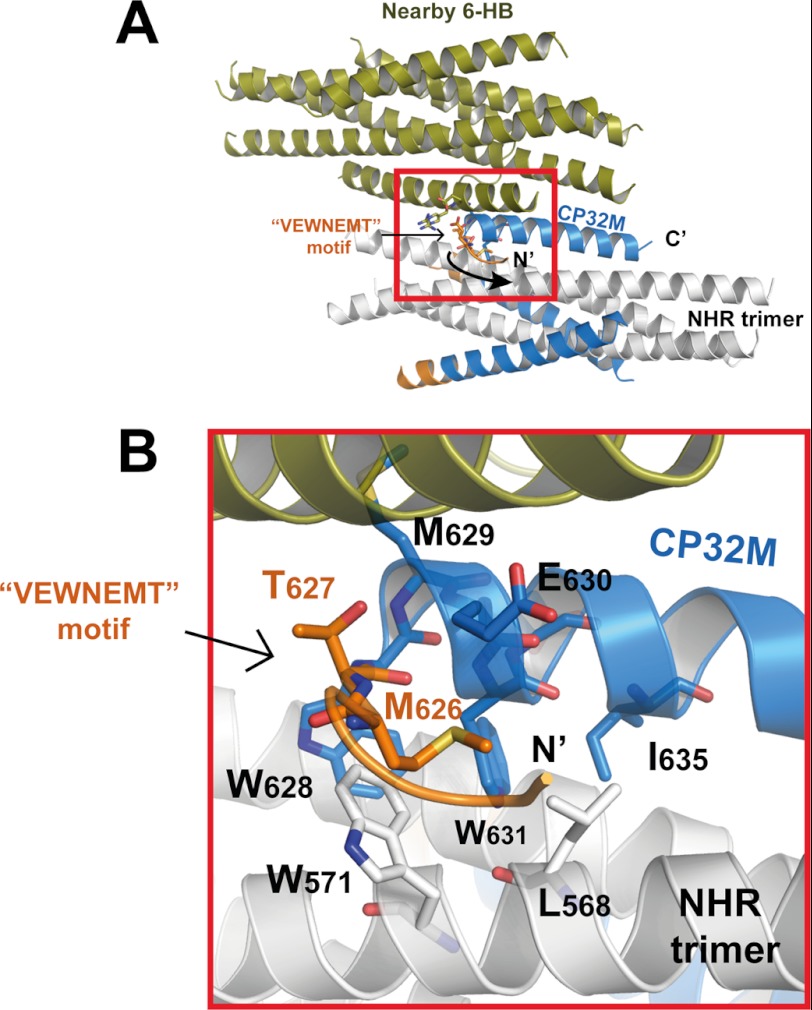
In both crystal form 1 and crystal form 2, CP32M and its NHR counterpart form a typical 6-HB structure as anticipated (Fig. 3, A and B). Three CP32M helices wrap around the outside of the NHR coiled coil trimer in an antiparallel orientation. The N-terminal QIWNNMT motif of the parental peptide CP621–652 was engineered to VEWNEMT in CP32M (25). Our previous crystallographic study had revealed that the QIWNNMT motif is largely disordered, but its residues, Met-626 and Thr-627, fold into a unique M-T hook structure that stabilizes the hydrophobic pocket on an NHR trimer (26). Surprisingly, the crystal structure of CP32M shows that the peptide is fully α-helical, and the N-terminal VEWNEMT motif folds into two α-helix turns targeting a novel region on the NHR trimer (see details of the results below). However, the conformation of the additional α-helix is not as stable as the rest of the 6-HB structure. The VEWNEMT motif is associated with a high temperature factor in both crystal forms. The average temperature factor of this motif in both crystal forms ranges from 60 to 61 Å2, whereas the average temperature factor of the 6-HBs ranges from 41 to 42 Å2. Interestingly, in crystal form 2, the VEWNEMT motif of one particular CP32M peptide is largely disordered. The N terminus of this peptide is directed away from the central NHR core (Fig. 3B), and thus the internal 3-fold symmetry of the 6-HB structure is lost (the space group for crystal form 1 is P321, whereas the space group for crystal form 2 is P21 ). As shown in Fig. 4A, the unusual conformation of the particular CP23M molecule is caused by the collision with a nearby 6-HB in crystal. Although most of the residues on the VEWNEMT motif are disordered in the electron density, the conserved residues Met-626 and Thr-627 fold into a M-T hook structure similar to that observed in CP621–652 (26) (Fig. 4B). This exceptional structure suggests that the M-T hook could be another possible conformation for the VEWNEMT motif of CP32M if the preferred α-helical conformation is disrupted. In such case, the conserved residues Met-626 and Thr-627 could still contribute to the stability of 6-HB structure.
FIGURE 3.
Overall structure of the 6-HB structures formed by HIV gp41 NHR546–588/CP32M chimera. Ribbon models of two 6-HB structures formed by NHR546–588/CP32M chimera. The NHR trimers are colored in gray, the CP32M peptides are colored in blue, and the VEWNEMT motifs are colored in orange and labeled. The same color scheme as for the 6-HB is used in all of the figures that follow (Figs. 4–7). The CP32M molecule with the disordered N-terminal motif is marked with an asterisk. A, crystal form 1 (space group P321). Left, side view; right, top view. B, crystal form 2 (space group P21). Left, side view; right, top view.
FIGURE 4.
Crystal packing interactions caused the conformational rearrangement of the N-terminal motif of one particular CP32M in crystal form 2. A, ribbon model of a 6-HB formed by the NHR546–588/CP32M chimera (positioned horizontally) collides with a nearby 6-HB (colored in olive), causing the conformational rearrangement of the N-terminal VEWNEMT motif of one particular CP32M. The collision site is highlighted with a red box. B, magnification of the red-boxed area in A. The VEWNEMT motif of the CP32M peptide is largely disordered, whereas the conserved residues Met-626 and Thr-627 adopt the M-T hook-like structure similar to that found in the crystal structure of CP621–625 (26). The M-T hook residues and the residues interacting with the M-T hook are shown in stick model with labels.
The Molecular Determinants Underlying the Stability of the 6-HB Structure
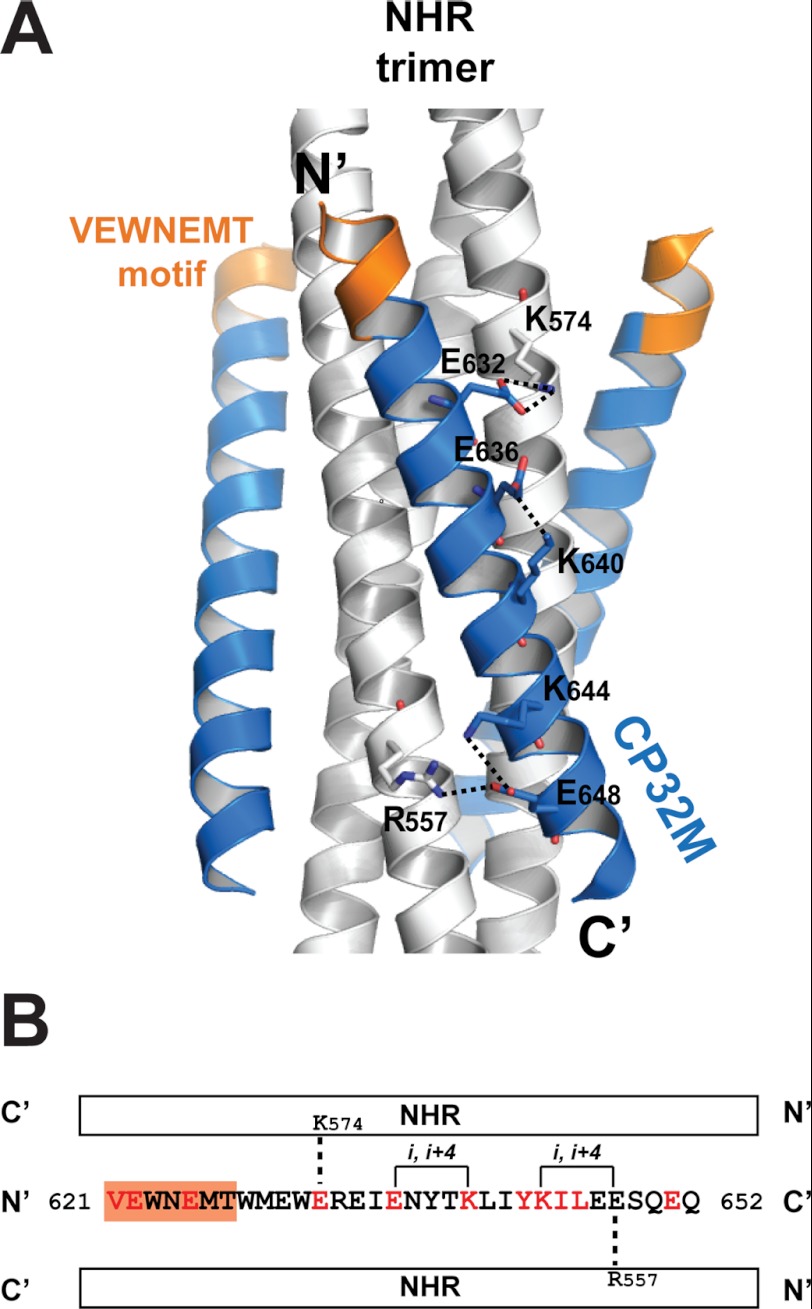
To stabilize the helical conformation of CP32M, charged residues were introduced to promote ion pairing between residues at the i and i + 4 positions during the inhibitor design (25). The crystal structure of CP32M confirms that the substitution of Asn-636 by a glutamic acid results in the pairing between Glu-636 and Lys-640. The distance from the Oϵ2 atom of Glu-636 to the hydrogen donated by the Nζ atom of Lys-640 is 2.14 Å (angle, 147.1°), indicating a hydrogen bond interaction (Fig. 5A). A lysine substitution of Ser-644 leads to the formation of a salt bridge between Lys-644 and Glu-648 (3.8 Å). Intriguingly, we observed a novel intermolecular salt bridge between Glu-648 and Arg-557 on the NHR helix (2.7Å), indicating a stronger salt bridging interaction. However, this salt bridge is not present in the crystal structure of CP621–652, regardless of the fact that the Glu-648 remained unchanged in the inhibitor design (see details in the results below). We previously identified a conserved salt bridge between Asp-632 on CHR and Lys-574 on NHR, which is critical for the HIV-1 entry and 6-HB stability (28, 29). In CP32M design, Asp-632 was changed to Glu-632 to improve this salt bridge interaction. The crystal structure reveals that the longer side chain of Glu-632 allows its carboxylate group to orient in a favored syn orientation to accept a hydrogen bond from the side chain of Lys-574 (2.04 Å, 147.5°), which contributes to the interhelical interaction between CP32M and its NHR target.
FIGURE 5.
The molecular determinants underlying the stability of the helical conformation of CP32M and the 6-HB structure. A, ribbon model of the 6-HB formed by NHR546–588/CP32M chimera with the charged residues forming the intra- and interhelical salt bridges/hydrogen bonds shown in stick model with labels. The salt bridges/hydrogen bonds are indicated by dashed lines. B, the sequence of CP32M with various features indicated. A single CP32M peptide interacts with two NHR helices (white bars). The residues changed during the inhibitor design are colored in red; the unchanged residues are colored in black. The VEWNEMT motif is highlighted by an orange background. The intrahelical salt bridges/hydrogen bonds observed in the crystal structures are indicated by solid lines; the interhelical salt bridges/hydrogen bonds observed in the crystal structures are indicated by dashed lines.
To summarize, our crystallographic study revealed the novel hydrogen bonds and salt bridges that contribute to the stability of the helical conformation of CP32M and its binding affinity to the NHR target. Especially, a single CP32M helix is able to salt bridge two NHR helices simultaneously, enhancing the overall stability of the 6-HB structure (Fig. 4B).
The VEWNEMT Motif Targets a Novel Region on NHR Trimer
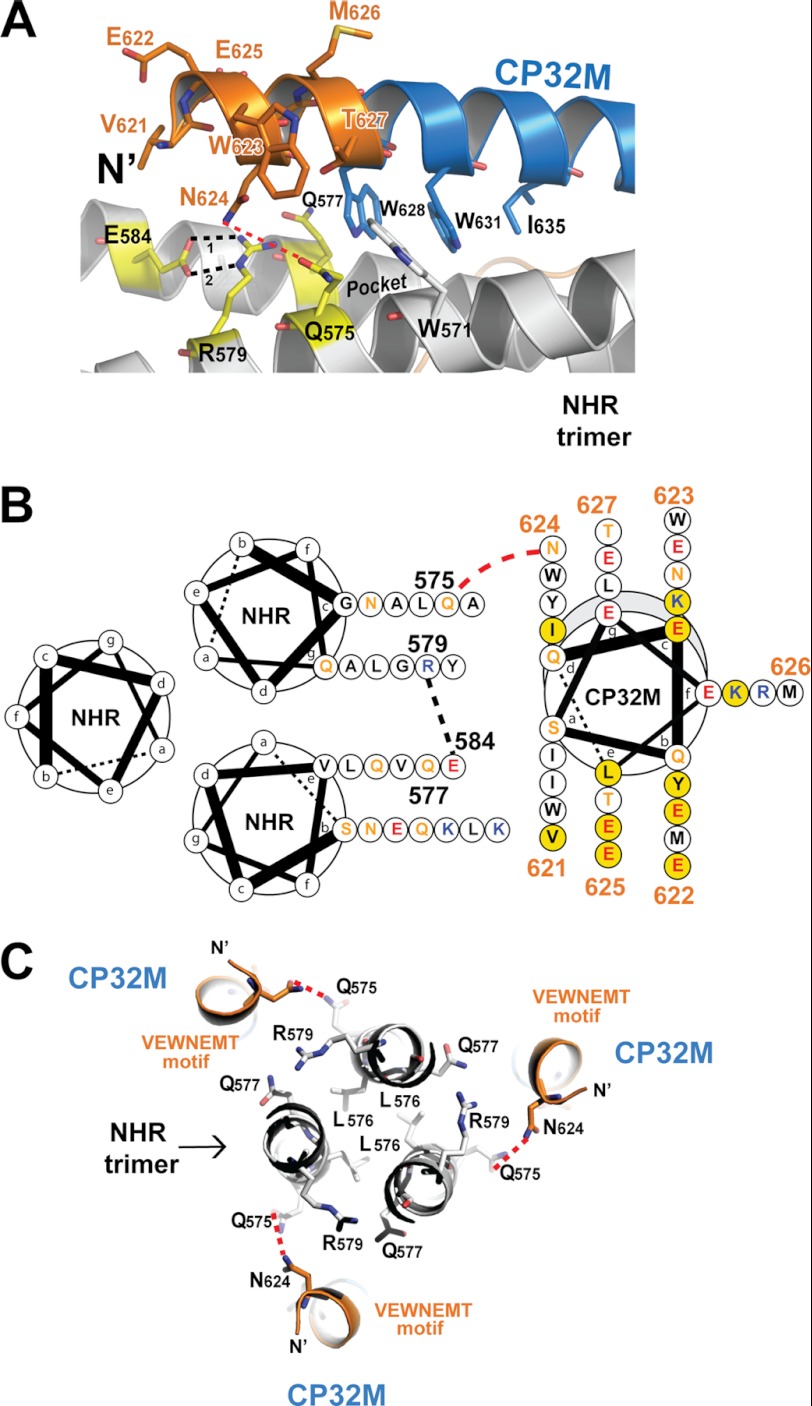
Except for one particular CP32M molecule in crystal form 2 clashing with a nearby molecule, resulting in a significant conformational rearrangement of the VEWNEMT motif, all other CP32M molecules in both crystal forms are fully α-helical. As shown in Fig. 6A, the N-terminal VEWNEMT motif extends the α-helical conformation by two turns, in which the side chains of Glu-622, Trp-623, Glu-625, and Met-626 face the exterior of the 6-HB; the side chains of Val-621, Asn-624, and Thr-627 face the NHR target. The targeting region on the NHR trimer involves a part of the hydrophobic pocket and a novel region adjacent to the pocket. The β-branched side chain of Thr-627 (at the “g” position of CP32M helix) is positioned on top of the bulky side chain of Trp-571, stabilizing the hydrophobic pocket below (Fig. 6, A and B). Residue Asn-624 (at the “d” position of CP32M helix) projects its side chain toward a remarkable hydrophilic region comprising Gln-575, Gln-577, Arg-579, and Glu-584 on the NHR trimer. This hydrophilic region is located next to the hydrophobic pocket, forming a high ridge that borders the deep pocket (Fig. 6A). The side chains of Arg-579 and Glu-584 (at the “g” and “e” positions on NHR helix) on the ridge are tightly connected to each other by two ideal hydrogen bonds (H-bond 1: 2.1Å, 144.3°; H-bond 2: 2.1 Å, 168.1°), stabilizing the interaction between the two NHR helices. Based on our current crystal structures, we could not identify the well defined intermolecular forces between Asn-624 and the residues on the hydrophilic ridge. However, the side chain of Asn-624 is close to the side chain of Gln-575 (at the “c” positions on NHR helix), but their distance (from the Nδ of Asn-642 to the Oϵ of Glu-575) ranges from 3.5 to 4.5 Å in both crystal forms, indicating a possible hydrogen bonding interaction. The molecular dynamic approach may be employed to further investigate the interaction between Asn-624 and the residues on the hydrophilic ridge (Fig. 6, A and B). Val-621 (at the “a” position of CP32M helix) orients the side chain toward the NHR trimer; however, the distance to its NHR counterpart (7–8 Å) is out of the range of any possible intermolecular forces.
FIGURE 6.
The VEWNEMT motif of CP32M targets a hydrophilic region on the NHR trimer and form a novel layer of the 6-HB structure. A, a portion of a ribbon model of the 6-HB structure formed by the NHR546–588/CP32M chimera (positioned horizontally). The residues on the N-terminal VEWNEMT motif of CP32M, the residues of the pocket-binding domain, and the residues of the NHR binding site of CP32M are shown in stick model with labels. The hydrophilic ridge comprising Gln-575, Gln-577, Glu-584, and Arg-579 is colored in yellow. The hydrophobic pocket on NHR is indicated. Hydrogen bonds 1 and 2 between Glu-584 and Arg-579 are indicated by black dashed lines. The possible hydrogen bond between Asn-624 and Gln-575 is indicated by a red dashed line. B, helical wheel presentation showing the interaction between CP32M and the NHR trimer. Three NHR helices and one CP32M helix are shown as helical wheel projections. The view is from the bottom of the 6-HB. Three NHR helices form the central coiled coil, and a CP32M helix is packed against the interhelical groove between two NHR helices. At the top of the complex, the N terminus of CP32M (gray shading) is slightly tilted toward the upper NHR helix, and at the bottom of the complex, the C-terminal of the CP32M is tilted toward the lower NHR helix. The color code for the residues is: black, hydrophobic; orange, uncharged; blue, positively charged; red, negatively charged. Residues on the VEWNEMT motif of CP32M are labeled with the residue numbers. Residues mutated from the parental sequence to generate CP32M are highlighted by a yellow background. Red dashed lines indicate the possible hydrogen bonds between Asn-624 on CP32M and Gln-575 on the NHR helices. Black dashed lines indicate the hydrogen bond between Glu-584 and Arg-579. C, the VEWNEMT motif of CP32M forms the novel layer of the 6-HB. Three Leu-576 residues form the hydrophobic core of the NHR trimer that is locked by three Glu-584–Arg-579 “hasps” (the side chain of Glu-584 comes from the front layer and thus is not visible here; see A). Gln-575, Gln-577, Glu-584, and Arg-579 form a hydrophilic region on the NHR trimer, which mediates the hydrophilic interaction with Asn-624 on the VEWNEMT motif of CP32M. The possible hydrogen bonds between Asn-624 and Gln-575 are indicated by red dashed lines.
Collectively, the VEWNEMT domain of CP32M and its NHR counterparts form an additional layer of the 6-HB structure (Fig. 5C). Three Leu-576 residues form the hydrophobic core of NHR trimer, which is locked tightly by three Arg-584–Glu-579 hasps (Glu-579 belongs to the adjacent front layer) (Fig. 6, A and C). The side chain of Thr-627 stabilizes the hydrophobic pocket on NHR, and the side chain of Asn-624 is in close contact with a hydrophilic ridge bordering the hydrophobic pocket on the NHR trimer, which contributes to the stabilization of the 6-HB structure.
The Structural Basis for the Inhibitor Efficacy and Drug Resistance
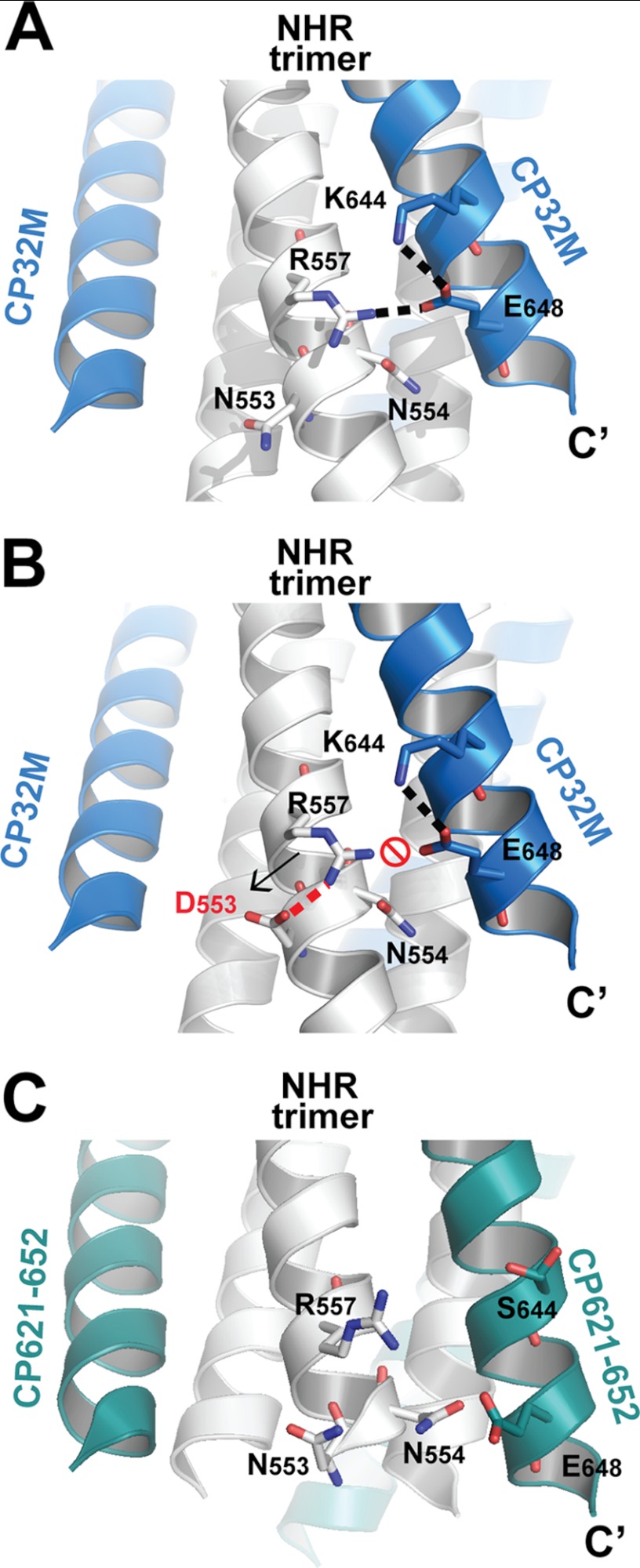
CP32M is highly effective against HIV-1 strains that are resistant to T20 (25). One major determinant for T20 resistance is the mutation at the 36GIV38 motif on the NHR of gp41 (reference the gp41 numbering of HIV-1HXB2 for comparison, corresponding to positions 547–549 in the gp160 numbering) (36). Our data demonstrate that HIV-1 bearing mutation V38T, V38Q, V38M, or V38A is highly resistant to T20, C34, and SFT but is barely resistant to CP32M (25, 37). The crystal structure of CP32M confirms that inhibitor does not bind the GIV motif, and thus the mutations occurring at this motif cannot influence the binding affinity of CP32M. Intriguingly, a combined mutation, V38A/N42D, induced ∼18-fold resistance against CP32M (25), whereas other substitutions of Asn-42 (equivalent to Asn-553), like V38A/N42T or N42S, did not contribute to CP32M resistance. These data suggest that Asn-42 is one of the key residues for CP32M resistance. The crystal structures of CP32M reveal the underlying mechanism. As shown in Fig. 7, Asn-553 on the NHR helix does not interact with CP32M and thus cannot contribute to the binding affinity of CP32M directly. An upstream residue, Arg-557, forms a salt bridge with Glu-648 on CP32M, favoring the electrostatic interaction between CP32M and the NHR target (Figs. 4, A and B, and 7A). When Asn-553 was replaced with the aspartic acid (modeled by Coot, Fig. 7B), the negatively charged side chain of Asp-553 could then form a salt bridge with the positively charge Arg-557, neutralizing the charge on Arg-557. Note that Arg-557 and N553D are situated at the i and i + 4 positions on the same NHR helix, the ideal positions for ion pairing; therefore this salt bridge may preform before the CP32M comes into play. In such case, Glu-648 of CP32M will then have little chance to pair with Arg-557 on NHR, thus disfavoring the binding of CP32M to the NHR target. Although Glu-648 remains unchanged in the original inhibitor design, the corresponding salt bridge between Arg-557 and Glu-648 (distance = 4.8 Å) in the crystal structure of CP621–652 cannot be identified (Fig. 7C). This is likely due to the presence of the uncharged Ser-644 in the parental sequence, which is unable to form a salt bridge with Glu-648. By contrast, in CP32M, mutation S644K not only leads to a salt bridge formation between Lys-644 and Glu-648 but also brings the side chain of Glu-648 into proximity with the side chain of Arg-557, leading to the formation of a salt bridge between the residues. Therefore, the substitution of Ser-644 by lysine offers two advantages for the efficacy of CP32M. First, it forms an intrahelical salt bridge to stabilize the helical conformation; second, it promotes the formation of an interhelical salt bridge to favor the binding of CP32M to the NHR target.
FIGURE 7.
Molecular basis of a key residue critical for CP32M resistance. A, a portion of a ribbon model of the 6-HB structure formed by the NHR546–588/CP32M chimera. Residues that are important to CP32M resistance and the interaction with NHR helix are shown in stick model. Glu-648 and Lys-644 form an intrahelical salt bridge on CP32M, and Glu-648 forms another interhelical salt bridge with Arg-557 on the NHR helix. The salt bridges are indicated by a dashed line. B, model (generated by Coot and PyMOL) of the 6-HB structure formed by the NHR546–588/CP32M chimera with mutation N553D (equivalent to N42D). The assumed salt bridge between Asp-553 and Arg-557 (indicated by a red dashed line) will neutralize the positive charge of Arg-557, thus disrupting the interhelical salt bridge between Arg-557 and Glu-648 on CP32M and disfavoring the electrostatic interaction between CP32M and its NHR target. C, a portion of a ribbon model of the 6-HB structure formed by T21/CP621–652 peptide (Protein Data Bank ID 3VGX). The NHR helices are colored in gray, and CP621–652 peptide is colored in green. The corresponding residues shown in A and B are shown here in stick model with labels. The corresponding salt bridge between Arg-557 and Glu-648 (distance 4.8 Å) cannot be identified in this structure.
Recent studies have identified the mutations of Gln-64, Glu-66, and Asn-113 (equivalent to Gln-575, Gln-577, and Asn-624, respectively) of gp41 as related to drug resistance against a number of HIV-1 fusion inhibitors such as T2635, C34, and CP32M (38, 39). Interestingly, these residues are involved in the interactions between the VEWNEMT motif of CP32M and its targeting sites on NHR. Eggink et al. (39) found that Asn-113 on the C-peptide of gp41 is in close contact with Gln-66 on the N-peptide based on a theoretical model of the gp41 ectodomain and the mutagenesis study. They concluded that the mutations at these positions to oppositely charged amino acids, such as Q66R/N113E allowing salt bridging, could speed up the formation of the fusogenic core of gp41 and thus decrease the time window for T2635 to act (39). By contrast, our crystal structures show that Asn-624 of CP32M is closer to Gln-575 (3.5–4.5Å) rather than Gln-577 (7–8 Å) on different NHR helices (Fig. 6, A and C), suggesting a possible hydrogen bond interaction between Asn-624 and Gln-575. This observation is supported by the mutagenesis study by Yu et al. (38) showing that the HIV-1-bearing mutation Q575A or Q575L that presumably disrupts the hydrogen bond interaction between Asn-624 and Gln-575 is highly resistant to CP32M. They suggested a model for the interaction between Asn-624 and Gln-575, which is similar to our crystal structures. Therefore, the interaction between the key residue, Asn-624, on the VEWNEMT motif of CP32M and the hydrophilic ridge on NHR is critical for the binding and antiviral activity of the inhibitor. Collectively, the structural data provide a molecular basis for the improved inhibitor efficacy of CP32M compared with its parental peptide as well as for the mechanisms underlying drug resistance.
DISCUSSION
In our previous studies, we designed CP32M as a peptide HIV-1 fusion inhibitor on the basis of the structural and functional information of gp41 and a recently identified anti-HIV peptide (CP621–652) containing a motif (621QIWNNMT627) that is critical for 6-HB formation and stability (24, 25). We showed that CP32M has great potential to serve as a unique anti-HIV drug for treatment of HIV variants that are resistant to the first generation fusion inhibitor T20. To develop CP32M for clinical use, we are currently performing preclinical studies of its safety and pharmacokinetic properties in animals. In this parallel leading study, we first characterized the anti-HIV spectrum of CP32M with a particular attention to HIV-1 strains that are currently circulating in China, including CRF07_BC (B/C) and CRF01_AE (A/E) recombinants and subtype B′ viruses that are responsible for a total nationwide infection of about 95% (33, 34). The results demonstrating that CP32M has potent inhibitory activity against diverse HIV-1 variants highlight its broad spectrum.
Resistance to T20 usually maps to the amino acid-(36–45) region of the peptide binding site in the NHR domain of the viral gp41, with the 36GIV38 motif being a hot spot for resistance (14, 36, 40–42). However, our previous studies demonstrated that CP32M could efficiently inhibit T20-resistant HIV-1 variants that carry single or double mutations in the amino acid-(36–45) region (e.g. V38A, V38A/N42T, or N42T/N43K) (20). In this study, we have also evaluated CP32M for its anti-HIV spectrum with a panel of HIV-1 variants that carry naturally occurring or induced T20-resistant mutations. From the results we conclude that CP32M possesses broad inhibitory activity on the diverse HIV-1 mutants. Among the tested variants, the L33S mutant was initially induced by peptide C34 and showed cross-resistance to T20, C34, and T1249 (31, 39, 43). Therefore, our results here indicate further that CP32M maintains a high genetic barrier for resistance. We previously explained that the CP32M-targeting site does not contain T20- and C34-resisant hot spots and that the N-terminal 621VEWNEMT627 motif plays a critical role in interactions between CP32M and its NHR target (25).
The discovery of virus-derived peptide fusion inhibitors has opened a bright avenue for exploring the mechanism of HIV-1 entry and developing antiviral therapeutics (14, 44–47). Several pioneering crystal structures deciphered the fusion-active 6-HB core of gp41, which delivered series stories about the membrane fusion of many viruses. On the basis of the gp41 core structure, a number of new generation peptide inhibitors with improved antiviral activity and pharmacokinetic profiles have been developed. However, there are limited structural data available for the engineered peptides. We recently reported the crystal structures of two fusion inhibitors (21, 26). The structure of sifuvirtide confirms its fully helical conformation stabilized by the multiply engineered salt bridges, providing direct evidence to support the rational inhibitor designing strategy (21). The structure of CP621–652, the parental peptide of CP32M, reveals the key motifs and residues responsible for its stability and anti-HIV activity (26). Importantly, our structures found that the N-terminal 621QIWNNMT627 motif of CP621–652 is highly flexible, but the residues Met-626 and Thr-627 form a unique hook-like structure (termed the M-T hook) that plays important roles in 6-HB stabilization and antiviral activity (26). In this present study, we continued our crystallographic studies for CP32M to visualize its molecular details at the atomic level.
In designing the rationale for CP32M (25), the N-terminal 621QIWNNMT627 motif of CP621–652 template was engineered by changing three residues. The first residue, Gln-621, at the “a” position in the heptad repeat was replaced by a hydrophobic valine to enhance its hydrophobic interaction with the NHR target. Ile-622 and Asn-625, which are located at the “b” and “e” positions in the helical wheel, were replaced by negatively charged glutamic acid to increase the hydrophilicity of the peptide. Surprisingly, our crystal structures show that the engineered VEWNEMT motif of CP32M becomes fully helical in one crystal form and predominantly helical in another crystal form. By adopting the helical conformation, the VEWNEMT motif folds into two α-helix turns, forming an extensive interaction layer with the pocket region of NHR helix. Previous structural studies have revealed that several distinctive layers of residues that are highly conserved in HIV-1 and SIV gp41, such as Thr/Trp, Gln-50, and the Gln-40/Asn-140 layers, which are critical to the stability of the 6-HB structure. The novel layer involving the residues in the VEWNEMT motif of CP32M presents another unique layer. Gln-575, Gln-577, Arg-579, and Glu-584 form the hydrophilic ridge, severing the C-terminal wall of the hydrophobic pocket. The hydrophilic ridge stabilizes the NHR helices by multiple interhelical hydrogen bonds and mediates the interaction with Asn-624 of the VEWNEMT motif of the CP32M helix via a possible hydrogen bond. Therefore, we believe that the novel layer can benefit the NHR binding stability of CP32M and thus confer antiviral activity. The VEWNEMT motif of one CP32M molecule in crystal form 2 is largely disordered, albeit the M-T hook structure is well conserved to be identical to that of CP621–652. It is obvious that this unusual conformation was caused by the crystallographic artifacts (Fig. 4), further suggesting the flexibility of the CHR region upstream of the pocket-binding domain. However, this feature may provide a valuable strategy for engineering peptide-based fusion inhibitors.
Indeed, three pioneering gp41 core structures were determined by taking advantage of the limited proteolysis treatments that eliminate flexible or unstructured peptide loop (6–8); thus the linking regions between the putative NHR and CHR of HIV gp41 are largely invisible. In other words, the upstream sequence of the PBD or the boundary of a C-helix could not be observed because of their intrinsic flexibility. Although the VEWNEMT motif in the CP32M was engineered to the α-helical conformation, it is apparently not as stable as the rest of the peptide. This is evidenced by the finding that the relatively weak crystal packing force could induce such a dramatic change that the α-helical conformation was completely disordered. The distance between the extreme N terminus of CP32M and its NHR counterpart is out of range of any type of molecular force (7–8 Å), suggesting that N-terminal residues Val-621 and Glu-622 may not contribute significantly to the binding affinity to the NHR target. This idea was tested by our mutagenesis experiments showing that the CP32M peptide, missing two N-terminal residues, does not suffer significant losses in antiviral activity (data not shown).
Interestingly, the analogous WNNET motif in SIV gp41 core appeared to be folded into a helical structure, whereas the amino acid sequence difference in this region is a glutamic acid instead of a Met-626. In the case of an M-T hook structure, position 626 demands a hydrophobic amino acid to be able to cap the hydrophobic pocket on HIV gp41 NHR trimer. Thus, adopting the M-T hook-like structure to stabilize the 6-HB is not an option for the WNNET motif of SIV gp41. In the structure of SIV gp41, the conformation of the PKWNNET motif and its interaction mode with the NHR are highly similar to that of the VEWNEMT motif of CP32M. The side chain of Thr-111 is positioned above the side chain of Trp-59 on the NHR pocket; the side chain of Asn-108 points down to a hydrophilic region comprising Arg-67, Glu-72, Asn-63, and Gln-65. These data suggest that the VEWNEMT motif of CP32M utilizes a similar mechanism for targeting NHR helices as that utilized by SIV gp41 CHR. As two residues in the N terminus of CP32M were substituted by glutamic acid, it will be interesting to investigate whether these negatively charged residues are the molecular determinants to the local conformation and thus affect the 6-HB structure and HIV-1 entry.
The first and second generation HIV-1 fusion inhibitors can easily induce drug resistance by single or multiple mutations at the inhibitor binding site, which obstructs the docking of the peptide (15, 19, 39, 48). By contrast, the third generation inhibitor T2635 has more difficulty inducing drug resistance. Combined mutations at various gp41 domains, especially the mutations outside the binding site, were found to be important for escaping T2635. The novel inhibitor CP32M belongs to the third generation inhibitor, which is highly effective against T20-resistant HIV-1 strains. The careful characterization of CP32M resistance is certainly essential for the further studies of the inhibitor. Nevertheless, the available data have revealed at least two important mutations that trigger CP32M resistance. The mutation positions 575 and 553 are located around the N- and C-terminal edges of the CP32M binding site on the NHR helices, suggesting that these residues may play key roles during the docking of CP32M. Asn-624 of the VEWNEMT motif is engaged in the hydrophilic interaction with the hydrophilic ridge on the NHR trimer and possibly forms a hydrogen bond with Gln-575 on the ridge. Therefore, substitution of Gln-575 by nonpolar residues could abolish the interaction, disfavoring the docking of the inhibitor. Our crystal structure demonstrates that Asn-553 is clearly out of the binding site for CP32M. Therefore, a number of Asn-553 substitutions (N42T and N42S) do not induce resistance against CP32M. Surprisingly, mutation N42D could cause dramatic resistance against CP32M. This is because the negatively charged aspartic acid side chain may form a salt bridge with Arg-557, thereby abolishing an important interhelical salt bridge between Arg-557 and Glu-648 on CP32M, which contributes significantly to the binding affinity of CP32M. This observation provides a novel mechanism for causing resistance against an HIV-1 fusion inhibitor. Mutation outside of the inhibitor binding site can indirectly influence the binding of the inhibitor to its target on the NHR trimer by rearranging the salt bridge interactions between the inhibitor and its target.
In conclusion, our studies have further demonstrated that CP32M is a potent fusion inhibitor against diverse HIV-1 variants. We determined the high resolution crystal structure of CP32M, which identifies the key motifs and residues underlying the improved antiviral activity and stability of the inhibitor and provides the structural basis for drug resistance. Therefore, the present work has multiple implications for understanding the mechanism of HIV-1 fusion and designing gp41 inhibitors.
This work was supported by Grants 81025009 from the National Outstanding Youth Award of the Natural Science Foundation of China (NSFC), 30870123 from the NSFC, 2010CB530100 from the National 973 Program of China, and 7122116 from the Beijing Natural Science Foundation.
The atomic coordinates and structure factors (codes 3VGY and 3VH7) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- 6-HB
- six-helix bundle
- NHR
- N-terminal heptad repeat
- CHR
- C-terminal heptad repeat
- PBD
- pocket-binding domain
- SIV
- simian immunodeficiency virus.
REFERENCES
- 1. Colman P. M., Lawrence M. C. (2003) The structural biology of type I viral membrane fusion. Nat. Rev. Mol. Cell Biol. 4, 309–319 [DOI] [PubMed] [Google Scholar]
- 2. Zhu P., Liu J., Bess J., Jr., Chertova E., Lifson J. D., Grisé H., Ofek G. A., Taylor K. A., Roux K. H. (2006) Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441, 847–852 [DOI] [PubMed] [Google Scholar]
- 3. Harrison S. C. (2008) Viral membrane fusion. Nat. Struct. Mol. Biol. 15, 690–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Eckert D. M., Kim P. S. (2001) Mechanisms of viral membrane fusion and its inhibition. Annu. Rev. Biochem. 70, 777–810 [DOI] [PubMed] [Google Scholar]
- 5. Chan D. C., Kim P. S. (1998) HIV entry and its inhibition. Cell 93, 681–684 [DOI] [PubMed] [Google Scholar]
- 6. Chan D. C., Fass D., Berger J. M., Kim P. S. (1997) Core structure of gp41 from the HIV envelope glycoprotein. Cell 89, 263–273 [DOI] [PubMed] [Google Scholar]
- 7. Tan K., Liu J., Wang J., Shen S., Lu M. (1997) Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc. Natl. Acad. Sci. U.S.A. 94, 12303–12308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Weissenhorn W., Dessen A., Harrison S. C., Skehel J. J., Wiley D. C. (1997) Atomic structure of the ectodomain from HIV-1 gp41. Nature 387, 426–430 [DOI] [PubMed] [Google Scholar]
- 9. Chan D. C., Chutkowski C. T., Kim P. S. (1998) Evidence that a prominent cavity in the coiled coil of HIV type 1 gp41 is an attractive drug target. Proc. Natl. Acad. Sci. U.S.A. 95, 15613–15617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bosch B. J., Martina B. E., Van Der Zee R., Lepault J., Haijema B. J., Versluis C., Heck A. J., De Groot R., Osterhaus A. D., Rottier P. J. (2004) Severe acute respiratory syndrome coronavirus (SARS-CoV) infection inhibition using spike protein heptad repeat-derived peptides. Proc. Natl. Acad. Sci. U.S.A. 101, 8455–8460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bullough P. A., Hughson F. M., Skehel J. J., Wiley D. C. (1994) Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 371, 37–43 [DOI] [PubMed] [Google Scholar]
- 12. Weissenhorn W., Carfí A., Lee K. H., Skehel J. J., Wiley D. C. (1998) Crystal structure of the Ebola virus membrane fusion subunit, GP2, from the envelope glycoprotein ectodomain. Mol. Cell 2, 605–616 [DOI] [PubMed] [Google Scholar]
- 13. Eggink D., Berkhout B., Sanders R. W. (2010) Inhibition of HIV-1 by fusion inhibitors. Curr. Pharm. Des. 16, 3716–3728 [DOI] [PubMed] [Google Scholar]
- 14. Ashkenazi A., Wexler-Cohen Y., Shai Y. (2011) Multifaceted action of Fuzeon as virus-cell membrane fusion inhibitor. Biochim. Biophys. Acta 1808, 2352–2358 [DOI] [PubMed] [Google Scholar]
- 15. Steffen I., Pöhlmann S. (2010) Peptide-based inhibitors of the HIV envelope protein and other class I viral fusion proteins. Curr. Pharm. Des. 16, 1143–1158 [DOI] [PubMed] [Google Scholar]
- 16. Kilby J. M., Hopkins S., Venetta T. M., DiMassimo B., Cloud G. A., Lee J. Y., Alldredge L., Hunter E., Lambert D., Bolognesi D., Matthews T., Johnson M. R., Nowak M. A., Shaw G. M., Saag M. S. (1998) Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat. Med. 4, 1302–1307 [DOI] [PubMed] [Google Scholar]
- 17. Lalezari J. P., Henry K., O'Hearn M., Montaner J. S., Piliero P. J., Trottier B., Walmsley S., Cohen C., Kuritzkes D. R., Eron J. J., Jr., Chung J., DeMasi R., Donatacci L., Drobnes C., Delehanty J., Salgo M. (2003) Enfuvirtide, an HIV-1 fusion inhibitor, for drug-resistant HIV infection in North and South America. N. Engl. J. Med. 348, 2175–2185 [DOI] [PubMed] [Google Scholar]
- 18. Lazzarin A., Clotet B., Cooper D., Reynes J., Arastéh K., Nelson M., Katlama C., Stellbrink H. J., Delfraissy J. F., Lange J., Huson L., DeMasi R., Wat C., Delehanty J., Drobnes C., Salgo M., and TORO 2 Study Group (2003) Efficacy of enfuvirtide in patients infected with drug-resistant HIV-1 in Europe and Australia. N. Engl. J. Med. 348, 2186–2195 [DOI] [PubMed] [Google Scholar]
- 19. Berkhout B., Eggink D., Sanders R. W. (2012) Is there a future for antiviral fusion inhibitors? Curr. Opin. Virol. 2, 50–59 [DOI] [PubMed] [Google Scholar]
- 20. He Y., Xiao Y., Song H., Liang Q., Ju D., Chen X., Lu H., Jing W., Jiang S., Zhang L. (2008) Design and evaluation of sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 283, 11126–11134 [DOI] [PubMed] [Google Scholar]
- 21. Yao X., Chong H., Zhang C., Waltersperger S., Wang M., Cui S., He Y. (2012) Broad antiviral activity and crystal structure of HIV-1 fusion inhibitor sifuvirtide. J. Biol. Chem. 287, 6788–6796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Eron J. J., Gulick R. M., Bartlett J. A., Merigan T., Arduino R., Kilby J. M., Yangco B., Diers A., Drobnes C., DeMasi R., Greenberg M., Melby T., Raskino C., Rusnak P., Zhang Y., Spence R., Miralles G. D. (2004) Short-term safety and antiretroviral activity of T-1249, a second-generation fusion inhibitor of HIV. J. Infect. Dis. 189, 1075–1083 [DOI] [PubMed] [Google Scholar]
- 23. Lalezari J. P., Bellos N. C., Sathasivam K., Richmond G. J., Cohen C. J., Myers R. A., Jr., Henry D. H., Raskino C., Melby T., Murchison H., Zhang Y., Spence R., Greenberg M. L., Demasi R. A., Miralles G. D. (2005) T-1249 retains potent antiretroviral activity in patients who had experienced virological failure while on an enfuvirtide-containing treatment regimen. J. Infect Dis. 191, 1155–1163 [DOI] [PubMed] [Google Scholar]
- 24. He Y., Cheng J., Li J., Qi Z., Lu H., Dong M., Jiang S., Dai Q. (2008) Identification of a critical motif for the human immunodeficiency virus type 1 (HIV-1) gp41 core structure: implications for designing novel anti-HIV fusion inhibitors. J. Virol. 82, 6349–6358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. He Y., Cheng J., Lu H., Li J., Hu J., Qi Z., Liu Z., Jiang S., Dai Q. (2008) Potent HIV fusion inhibitors against enfuvirtide-resistant HIV-1 strains. Proc. Natl. Acad. Sci. U.S.A. 105, 16332–16337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Chong H., Yao X., Qiu Z., Qin B., Han R., Waltersperger S., Wang M., Cui S., He Y. (2012) Discovery of critical residues for viral entry and inhibition through structural insight of HIV-1 fusion inhibitor CP621–652. J. Biol. Chem. 287, 20281–20289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Derdeyn C. A., Decker J. M., Sfakianos J. N., Wu X., O'Brien W. A., Ratner L., Kappes J. C., Shaw G. M., Hunter E. (2000) Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74, 8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. He Y., Liu S., Jing W., Lu H., Cai D., Chin D. J., Debnath A. K., Kirchhoff F., Jiang S. (2007) Conserved residue Lys574 in the cavity of HIV-1 Gp41 coiled-coil domain is critical for six-helix bundle stability and virus entry. J. Biol. Chem. 282, 25631–25639 [DOI] [PubMed] [Google Scholar]
- 29. He Y., Liu S., Li J., Lu H., Qi Z., Liu Z., Debnath A. K., Jiang S. (2008) Conserved salt bridge between the N- and C-terminal heptad repeat regions of the human immunodeficiency virus type 1 gp41 core structure is critical for virus entry and inhibition. J. Virol. 82, 11129–11139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chinnadurai R., Münch J., Kirchhoff F. (2005) Effect of naturally occurring gp41 HR1 variations on susceptibility of HIV-1 to fusion inhibitors. Aids 19, 1401–1405 [DOI] [PubMed] [Google Scholar]
- 31. Chinnadurai R., Rajan D., Münch J., Kirchhoff F. (2007) Human immunodeficiency virus type 1 variants resistant to first- and second-version fusion inhibitors and cytopathic in ex vivo human lymphoid tissue. J. Virol. 81, 6563–6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lu L., Jia M., Ma Y., Yang L., Chen Z., Ho D. D., Jiang Y., Zhang L. (2008) The changing face of HIV in China. Nature 455, 609–611 [DOI] [PubMed] [Google Scholar]
- 34. Liao L., Xing H., Shang H., Li J., Zhong P., Kang L., Cheng H., Si X., Jiang S., Li X., Shao Y. (2010) The prevalence of transmitted antiretroviral drug resistance in treatment-naive HIV-infected individuals in China. J. Acquir. Immune Defic. Syndr. 53, Suppl. 1, S10–S14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Rimsky L. T., Shugars D. C., Matthews T. J. (1998) Determinants of human immunodeficiency virus type 1 resistance to gp41-derived inhibitory peptides. J. Virol. 72, 986–993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Liu Z., Shan M., Li L., Lu L., Meng S., Chen C., He Y., Jiang S., Zhang L. (2011) In vitro selection and characterization of HIV-1 variants with increased resistance to sifuvirtide, a novel HIV-1 fusion inhibitor. J. Biol. Chem. 286, 3277–3287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yu X., Lu L., Cai L., Tong P., Tan S., Zou P., Meng F., Chen Y. H., Jiang S. (2012) Mutations of Gln-64 in the HIV-1 gp41 N-terminal heptad repeat render viruses resistant to peptide HIV fusion inhibitors targeting the gp41 pocket. J. Virol. 86, 589–593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Eggink D., Bontjer I., Langedijk J. P., Berkhout B., Sanders R. W. (2011) Resistance of human immunodeficiency virus type 1 to a third-generation fusion inhibitor requires multiple mutations in gp41 and is accompanied by a dramatic loss of gp41 function. J. Virol. 85, 10785–10797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Wei X., Decker J. M., Liu H., Zhang Z., Arani R. B., Kilby J. M., Saag M. S., Wu X., Shaw G. M., Kappes J. C. (2002) Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46, 1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Greenberg M. L., Cammack N. (2004) Resistance to enfuvirtide, the first HIV fusion inhibitor. J. Antimicrob. Chemother. 54, 333–340 [DOI] [PubMed] [Google Scholar]
- 42. Reeves J. D., Lee F. H., Miamidian J. L., Jabara C. B., Juntilla M. M., Doms R. W. (2005) Enfuvirtide resistance mutations: impact on human immunodeficiency virus envelope function, entry inhibitor sensitivity, and virus neutralization. J. Virol. 79, 4991–4999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Armand-Ugón M., Gutiérrez A., Clotet B., Esté J. A. (2003) HIV-1 resistance to the gp41-dependent fusion inhibitor C-34. Antiviral Res. 59, 137–142 [DOI] [PubMed] [Google Scholar]
- 44. Ashkenazi A., Shai Y. (2011) Insights into the mechanism of HIV-1 envelope-induced membrane fusion as revealed by its inhibitory peptides. Eur. Biophys. J. 40, 349–357 [DOI] [PubMed] [Google Scholar]
- 45. Wild C., Oas T., McDanal C., Bolognesi D., Matthews T. (1992) A synthetic peptide inhibitor of human immunodeficiency virus replication: correlation between solution structure and viral inhibition. Proc. Natl. Acad. Sci. U.S.A. 89, 10537–10541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Jiang S., Lin K., Strick N., Neurath A. R. (1993) HIV-1 inhibition by a peptide. Nature 365, 113. [DOI] [PubMed] [Google Scholar]
- 47. Wild C. T., Shugars D. C., Greenwell T. K., McDanal C. B., Matthews T. J. (1994) Peptides corresponding to a predictive alpha-helical domain of human immunodeficiency virus type 1 gp41 are potent inhibitors of virus infection. Proc. Natl. Acad. Sci. U.S.A. 91, 9770–9774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shimura K., Nameki D., Kajiwara K., Watanabe K., Sakagami Y., Oishi S., Fujii N., Matsuoka M., Sarafianos S. G., Kodama E. N. (2010) Resistance profiles of novel electrostatically constrained HIV-1 fusion inhibitors. J. Biol. Chem. 285, 39471–39480 [DOI] [PMC free article] [PubMed] [Google Scholar]