Background: Innate immune protein surfactant protein D (SP-D) helps to protect against influenza A virus (IAV) infection.
Results: In pigs, this protein exhibits a unique sugar-binding site in its carbohydrate recognition domain.
Conclusion: Distinct interactions with viral mannose-rich glycans contribute to the profound antiviral activity of porcine SP-D.
Significance: These findings could help explain why pigs serve as mixing vessels for IAV.
Keywords: Host Defense, Influenza Virus, Lectin, Molecular Dynamics, Surfactant Protein D, X-ray Crystallography, Carbohydrate Recognition, Collectin, Viral Glycan
Abstract
Pigs can act as intermediate hosts by which reassorted influenza A virus (IAV) strains can be transmitted to humans and cause pandemic influenza outbreaks. The innate host defense component surfactant protein D (SP-D) interacts with glycans on the hemagglutinin of IAV and contributes to protection against IAV infection in mammals. This study shows that a recombinant trimeric neck lectin fragment derived from porcine SP-D (pSP-D) exhibits profound inhibitory activity against IAV, in contrast to comparable fragments derived from human SP-D. Crystallographic analysis of the pSP-D fragment complexed with a viral sugar component shows that a unique tripeptide loop alters the lectin site conformation of pSP-D. Molecular dynamics simulations highlight the role of this flexible loop, which adopts a more stable conformation upon sugar binding and may facilitate binding to viral glycans through contact with distal portions of the branched mannoside. The combined data demonstrate that porcine-specific structural features of SP-D contribute significantly to its distinct anti-IAV activity. These findings could help explain why pigs serve as important reservoirs for newly emerging pathogenic IAV strains.
Introduction
Influenza is a highly contagious respiratory disease, estimated to cause between 250,000 and 500,000 deaths worldwide each year. Its potential to cause pandemic influenza outbreaks in humans was illustrated recently by the swine-origin pandemic flu of 2009 (1), which emphasized the importance of pigs as animal reservoirs from which novel reassorted influenza A virus (IAV)4 strains can be introduced into the human population. Before IAV infection of the host is established, the virus encounters various lines of innate defenses, including an important class of mammalian innate immune proteins, the collagenous C-type lectins known as collectins. Lung collectins are involved in the early pulmonary response to limit infection and spread of IAVs and other pathogens in the airways. By now, it is well established that among these collectins, surfactant protein D (SP-D) appears particularly important in the context of IAV. Apart from the serum bovine collectin conglutinin, native SP-D exhibits the strongest anti-IAV activity compared with other collectins as indicated by various in vitro studies (2, 3), although its protective role is underscored in studies with SP-D knock-out mice (4, 5). SP-D-mediated protection is primarily established by reducing the number of infectious particles via collectin-mediated aggregation of viral particles, which prevents attachment of virus to the host respiratory epithelium and induces phagocytic responses resulting in enhanced viral clearance (opsonization). In addition, SP-D is involved in control of pulmonary inflammation in early stages of IAV infection (4–6), prevents deactivation of neutrophils (7), and could also bridge the innate and adaptive immune responses by modulating the function of dendritic cells and T cells (8).
SP-D is mainly secreted as a dodecameric four-armed structure (9, 10) in which each arm represents a trimeric oligomer, the basic structural unit common for the collectin family members. Within each subunit, four major domains can be distinguished as follows: an N-terminal cross-linking domain; a collagen-like triple helical domain; a neck region, and a carbohydrate recognition domain (CRD). The neck region, a short stretch of 33 amino acids, mediates formation of a triply wound α-helical coiled-coil domain (11). The three globular C-type lectin domains are clustered at the C terminus of each subunit to facilitate multivalent high affinity interactions between the ligand-binding sites of SP-D with patterns of glycoconjugates expressed on the surface of pathogens. Prior in vitro and in vivo studies have shown that the trimeric arrangement and carbohydrate-binding characteristics of the CRD are important structural requirements that confer the strong antiviral activity of SP-D against IAV (4, 5, 12–14). Correspondingly, neck and CRD domains (NCRDs) possess some measure of the antiviral activity inherent in the full-length, more highly oligomerized SP-D.
The CRDs of SP-D recognize high mannose glycans present on the hemagglutinin (HA) and/or neuraminidase of IAV (15, 16). The sensitivity of various IAV strains to SP-D-mediated inhibition therefore depends largely on the degree of viral glycosylation (15, 17, 18). This was also illustrated by a recent study with pandemic 2009 H1N1 IAV that, in contrast to seasonal H1N1 strains, appears to be poorly glycosylated and therefore exhibits resistance against the inhibitory activity of innate immune proteins like hSP-D (19).
Other modes of interaction can occur between SP-D and IAV, independent of viral glycosylation. Our investigations into the primary structure and biochemical properties of porcine SP-D (pSP-D) have revealed that pSP-D is unique in that it is the only SP-D species known to be equipped with an N-linked complex sialic acid-rich glycan in its CRD (10, 20). These sialic acids are likely to interact with the conserved sialic acid receptor, located on the HA of all IAVs, that mediates binding of the virus to sialoglycans expressed on the respiratory epithelium, initiating IAV infection of the lung. In vitro neutralization studies with IAV revealed that the N-linked oligosaccharide in the CRD of pSP-D isolated from bronchoalveolar lavage fluid adds to the lectin-mediated interactions between pSP-D and glycans present on the surface proteins of IAV (21). As a result, this N-linked glycan not only enhances the overall antiviral activity of pSP-D but also broadens the range of viral strains that can be inhibited by pSP-D, including strains that are resistant to inhibition by hSP-D (21–23). We speculate that this distinct pSP-D activity against IAV might be involved in the process of interspecies transmittance of IAV strains with birds and humans that readily occurs in pigs (24, 25).
To investigate the respective contributions of the CRD lectin-mediated IAV interactions versus the CRD N-linked glycosylation interactions, we initiated studies of the recombinant pSP-D NCRD either retaining (RpNCRD) or lacking (RpNCRD-dNG) the N-linked glycosylation site through site-directed mutation (N303Q). Previous studies performed with enzymatically N-deglycosylated pSP-D had established that the CRD-mediated interactions between pSP-D and IAV glycans are significantly stronger as compared with human and rat SP-D (23); however, the mechanistic basis of this distinction remained to be determined. Crystallographic studies provide the three-dimensional structure of RpNCRD-dNG complexed with d-mannose. The thermodynamics of pig and human SP-D complexed with or without oligomannose were characterized through molecular dynamics simulations. The combined data suggest that specific features of the pig SP-D lectin site region facilitate recognition of highly branched mannose-rich oligosaccharides expressed on the IAV surface.
EXPERIMENTAL PROCEDURES
Construction of pSP-D-NCRD cDNA
A full-length pSP-D cDNA clone in pCR4-TOPO vector (26) was used as a template in a standard 50-μl PCR using proofstart DNA polymerase to amplify the NCRD region of pSP-D starting at the codon coding for Gly-223 (numbers according to Ref. 20) and ending 142 nucleotides downstream from the stop codon (total length, 613 bp). For subcloning purposes, the forward primer included a BsmbI/BamHI restriction site (PDNCRDFOR, gcgtctcggatcc GGC ATC ACT GCT CTG AGG C; all DNA sequences described in this study are 5′ to 3′ with gene-specific sequences in capitals), and the reverse primer contained a NotI sequence (PSPDREV, 5′-gcggccgc TGA GGG AGG CGT TCC ATA GGC-3′). The amplified DNA fragment was gel-purified, and 3′-A overhangs were added by incubating 10 min at 72 °C in the presence of 1 unit of TaqDNA polymerase and 0.4 mm dATP. The gel-purified product was cloned into pCR4-TOPO sequencing vector using a TOPO T/A cloning kit (Invitrogen). Insert-positive clones were isolated and sequenced to rule out any errors. The resulting pSP-DNCRD construct was digested with BsmBI/NotI, gel-extracted, and ligated into pUPE 105.03 expression vector (U-Protein Express B.V., Utrecht, The Netherlands), a modified version of the pTT3 expression vector (27) containing an artificial cystatin signal peptide (28). No tags were used. Subclones containing the cDNA were identified by BamHI/NotI restriction analysis.
Site-directed Mutagenesis
Site-directed mutagenesis was performed on the pSP-DNCRD cDNA clone in pCR4-TOPO vector (described above) by PCR using the Quikchange II site-directed mutagenesis kit. To generate an N323Q (Asn-303 in hSP-D sequence) mutation to produce the pSP-DNCRDNKO mutant having the Asn-323–Phe-324–Thr-325 glycosylation motif changed into a nonglycosylating sequence in its CRD, the following primers were used: PSPDKOFOR, GAC TGA CAT CAA GAC GGA GGG CCA GTT CAC CTA CCC CAC GGG GGA with PSPDKOREV, CTC CCC CGT GGG GTA GGT GAA CTG GCC CTC CGT CTT GAT GTC AGT C (mutation codon underlined). After sequence verification, the pCR4-TOPO insert was digested with BsmBI/NotI, gel-purified, and transferred to pUPE105.03 as described for the pSP-DNCRD cDNA clone.
Recombinant Expression of Porcine and Human SP-D Trimeric NCRDs
Recombinant collectins were produced using HEK293 cells stably expressing Epstein-Barr virus nuclear antigen 1 (EBNA1). HEK293E cells were transiently transfected as described previously (29). The pSP-D sequence-containing pUPE105.03 expression plasmids were purified using an endotoxin-free plasmid maxi prep kit. Expression media containing RpNCRD and RpNCRD-dNG, which has the N303Q substitution that results in a deleted N-glycosylation site in its CRD, were harvested by centrifugation 6 days post-transfection. Large scale expression media were concentrated 10-fold and diafiltrated into 5 mm Hepes, 0.9% NaCl, 5 mm EDTA, pH 7.4, using the Quixstand Hollow Fiber System (GE Healthcare) using a cartridge with a 10-kDa cutoff. RhNCRD was produced in Escherichia coli as described previously (30).
Purification of Recombinant Porcine and Human SP-D-NCRDs
RpNCRD and RpNCRD-dNG were isolated from HEK293E cell-free diafiltrated medium as described previously for the isolation of SP-D from porcine bronchoalveolar lavage (10) with the exception of Tris-HCl, which was replaced by Hepes (5 mm, pH 7.4) in all buffers. Instead of a single overnight incubation at 4 °C, the medium was subjected to a batchwise overnight incubation in the presence of 5 mm calcium chloride and mannan-agarose (5-ml bed volume/liter of medium). This procedure was repeated twice to increase yield. Purification was performed in the absence of Tween 80 throughout the procedure. Mannan-bound RpNCRD and RpNCRD-dNG were eluted with 5 mm Hepes, 0.9% NaCl, 5 mm EDTA, pH 7.4, filtrated through a 5-μm filter and concentrated to ∼5 ml via ultrafiltration with Amicon Ultracel centrifugal filters (30-kDa cutoff; Millipore). Gel filtration chromatography was performed using an ÄKTAPurifier10 system that was equipped with a Hiload 16/60 Superdex200 PREP grade column. Fractions were analyzed by SDS-PAGE analysis and Western blot analysis. Pooled fractions were further concentrated by Ultracel centrifugation and analyzed for protein content. Endotoxin levels were determined with the ToxinSensor LAL assay kit (ranging between 10 and 100 pg/μg SP-D). RhNCRD was purified by maltosyl-agarose chromatography followed by gel filtration chromatography and characterized as described previously (30). Endotoxin values of RhNCRD preparations were below 0.1 pg/μg protein. Recombinant full-length porcine SP-D (RpSP-D) was produced and purified as described (26).
Biochemical Characterization of RpNCRDs
SDS-PAGE analysis, Western blot analysis, and N-glycanase treatment to determine the presence of N-linked oligosaccharides on RpNCRD preparations were performed as described in detail for the characterization of natural pSP-D (10). Chemical cross-linking of RpNCRD and RpNCRD-dNG was achieved with a BS3 cross-linker as described previously (26).
Hemagglutination Inhibition of IAV by SP-D
Inhibition of HA activity (HAI) was performed on IAVs grown in chicken eggs and purified on sucrose gradients as described (31) and included the following strains: A/Philippines/82(H3N2) (Phil) provided by Dr. E. Margot Anders (Department of Microbiology and Immunology, University of Melbourne, Melbourne, Australia); A2/Aichi68/H3N2 (Aichi) provided by American Type Culture Collection; and A/Puerto Rico/8/34(H1N1) (PR-8) provided by Dr. Jon Abramson (Department of Pediatrics, Wake Forest University, NC). The hemagglutination titer of each virus preparation was determined by titration of virus samples in PBS++ (PBS with 1 mm calcium chloride and 0.5 mm magnesium chloride) with thoroughly washed human type O, Rh− red blood cells as described (32). HAI was measured by serially diluting SP-D preparations (25 μl per well) in round-bottom 96-well plates using PBS++ as diluent. After adding 25 μl of IAV solution (final concentration, 40 HA units/ml), the IAV/SP-D mixture was preincubated for 15 min, followed by addition of 50 μl of human type O erythrocyte suspension in PBS++. The minimal concentration of a collectin, required to fully inhibit HA caused by IAV, was indicated by the formation of a pellet of red blood cells after incubation for 2 h. The highest collectin concentration tested was 40 μg/ml. The entire procedure was carried out at room temperature. All measurements were Ca2+-dependent because addition of EDTA reduced all HAI activity to 0; HAI was measured in triplicate for all preparations. Statistical comparisons were performed by Student's paired, two-tailed t test.
Collectin-mediated Protection of MDCK Cells against Infection by IAV
Fluorescent focus assays were performed to assess the inhibitory activity of RpSP-D, RpNCRD, RpNCRD-dNG, and RhNCRD against the Phil strain of IAV on infectivity of MDCK cells. A fixed amount of virus was preincubated with increasing concentrations of collectin (0–40 μg) for 30 min at 37 °C, added to confluent monolayers of MDCK cells, and incubated for 45 min at 37 °C. After washing, the monolayers were incubated for 7 h at 37 °C in Dulbecco's modified Eagle's medium, washed, and fixed with 80% (v/v) acetone at 4 °C. Thereafter, the monolayers were labeled with monoclonal antibody against IAV viral nucleoprotein (monoclonal antibody A-3; a kind gift of Nancy Cox, Centers for Disease Control and Prevention, Influenza Branch, Atlanta, GA) for 30 min at 4 °C. After three washes, the MDCK cells were incubated with FITC-labeled goat anti-mouse IgG. The numbers of infectious foci were counted by fluorescence microscopy and expressed as a percentage of the number of infectious foci formed in the control incubation (IAV added to monolayer in absence of collectin). Statistical comparisons were made using Student's paired, two-tailed t test.
Crystallization Procedure and Data Collection
Crystals of RpNCRD-dNG were grown in hanging drops by mixing protein (12.3 mg/ml in 0.9% (w/v) sodium chloride, 5 mm EDTA, 15 mm calcium chloride, and 20 mm d-mannose) with an equal amount of reservoir solution (0.1 m BisTris, pH 6.5, 20% (w/v) PEG 3350, 200 mm magnesium chloride, and 15% v/v glycerol) on a siliconized coverslip, then inverting over a well of a 0.5-ml reservoir solution on a VDX plate, and equilibrating at 17 °C. Crystals appeared in 3–4 days. Crystals were then soaked in a 30% (w/v) d-mannose solution, which is cryoprotective. Crystals were cooled to 80–100 K in a vapor nitrogen stream prior to data collection. Diffraction data were collected on a RAXIS-IV image plate system mounted on a Rigaku RU-300 rotating anode. Data were indexed and processed using DENZO and SCALEPACK.
Structure Solution and Refinement
Solution of the RpNCRD-dNG structure was performed using molecular replacement as implemented in Phenix (33). The search model was the B chain from the structure of RhNCRD complexed with maltotriose (PDB code 2GGU) (34) that had been treated with the Sculptor program within Phenix, removing the waters, ligands, and one of the three calcium ions. The top solution was then submitted to AutoBuild (35) in Phenix for composite-omit map rebuilding and density modification of the molecular replacement solution. Iterative rounds of manual rebuilding and refinement were then carried out in Phenix and Coot (36), and translation libration screw-motion (TLS) parameters were used for B factor refinement (37, 38). The protein was defined as two zones for TLS refinement, one corresponding to the neck region (residues 203–235) and the other corresponding to the CRD (residues 236–355). These final refined coordinates, less the water and glycerol molecules, were then used as the starting model for a difference Fourier solution of the RpNCRD-dNG·d-mannose complex data. Data collection and refinement statistics are shown in Table 1. Figures were prepared using PyMOL, Swiss PDBViewer (39), and Povray (40). Alignments were performed in a Swiss PDBViewer.
TABLE 1.
Data collection and refinement statistics of pSP-DNCRD crystal structure
The number in parentheses is the value for reflections in the outermost shell. Ligands are d-mannose and calcium ions. The average B factor for protein atoms is calculated with the isotropic contribution to the B factor only, ignoring the contribution of the TLS terms.
| Parameter | Value |
|---|---|
| Data collection | |
| Space group | P63 |
| Unit cell dimensions | a = b = 66.024, c = 66.018 Å |
| Resolution | 15 to 2.2 Å (2.28 to 2.20 Å) |
| Number of reflections | 8361 (840) |
| Data cutoff | I ≤3σ |
| I/σ(I) | 41.9 (7.1) |
| Redundancy | 10.5 (9.1) |
| Completeness | 99.9% (99.4%) |
| Rmerge | 0.045 (0.416) |
| Refinement | |
| Rwork/Rfree | 0.1989/0.2373 |
| No. of protein atoms | 1206 |
| No. of water atoms | 36 |
| No. of ligand atoms | 14 |
| Average B factors | |
| Protein atoms | 60.2 |
| Waters | 48.3 |
| Ligand atoms | 85.0 |
| r.m.s.d. from ideal values | |
| Bond lengths | 0.002 Å |
| Bond angles | 0.542° |
| Chiral volume | 0.038 Å3 |
| Ramachandran | |
| Favored | 93.1% |
| Allowed | 6.9% |
| Disallowed | 0.0% |
Docking Calculations
Oligomannose models were obtained starting from the crystal structure coordinates of Fab 2G12 bound to Man9GlcNAc2 (PDB code 1OP5 (41)). This crystal structure has nine mannoses; for the docking, one of the nonreducing end mannoses was removed a priori to be accommodated by Glide as described under “Results.” A small number of starting conformations was generated by a Monte Carlo torsion/low mode sampling conformational search in Macromodel (Schrödinger, Inc.). The search was performed in an OPLS-2005 force field using the extended search option. 1000 searches were carried out, and redundant conformations were eliminated using a 1-Å all atom r.m.s.d. cutoff.
The protein part of the RpNCRD-dNG·mannose complex was prepared for docking calculations using the Protein Preparation Wizard in Maestro (Schrödinger, Inc., New York), which adds hydrogens, assigns charges, removes solvent molecules, optimizes the hydrogen bond network, and minimizes the starting structure.
The grid for docking was calculated using the prepared protein as described above. The grid was centered on a point about 7 Å above the plane of the side chain of Phe-335. This grid covers the entire top of the NCRD. Because it was assumed that the mannose bound in the lectin site would have a conserved structure whether as a monomer, as in the crystal structure, or as part of the oligomannose chain, positional constraints were added to the grid in the form of 1-Å spheres centered at the positions of the C3, O3, C4, and O4 atoms of the bound mannose in the crystal structure. During the docking, poses that do not position the C3, O3, C4, and O4 atoms of a specified mannose within the corresponding spheres were discarded. This enforced a given mannose to the lectin site, where it was docked in a conformation very similar to the crystal structure.
Dockings were run in Glide using the OPLS-2005 force field on each of the generated conformations for each oligomannose species tested. The parameters for the docking were altered from the defaults as follows: 50,000 poses were kept for the initial phase of docking; the scoring window was increased to 200; the expanded sampling mode was used; the best 1000 poses were kept for energy minimization; the maximum number of minimization steps was set to 500; and up to 10 poses were selected for post-docking minimization and output.
After docking, the poses were clustered using XCluster (Schrödinger Inc., New York). The top scoring clusters from the dockings were then analyzed to determine whether the poses were consistent with the true substrate N-linked glycoprotein binding to pSP-D. For the docking, only the mannose portion of oligomannose was used; the protein and the first two GlcNAcs of the N-linked glycosylation from the glycoprotein substrate of RpNCRD-dNG were not included in the calculation. Therefore, poses where addition of the substrate protein or the remaining sugars of the glycosylation site to their expected position on the docked mannoses would create severe steric clashes with the pSP-D protein were eliminated from further consideration.
Molecular Dynamics Simulations
Five systems, namely RpNCRD-dNG, RpNCRD-dNG·Man8 complex, RhNCRD, RhNCRD·Man8 complex, and RpNCRD-dNG·d-mannose complex, were studied using molecular dynamics simulation.
Topology files of protein and sugar were generated using the psfgen plugin of VMD (42). For systems of the RpNCRD-dNG, RpNCRD-dNG·Man8 complex, RhNCRD, RhNCRD·Man complex, and RpNCRD-dNG·d-mannose complex, each initial structure was placed in a TIP3P water box at 100 mm NaCl and neutralized using VMD (42). The simulated systems of RpNCRD-dNG and the RpNCRD-dNG·Man8 complex contain 100,782 and 100,804 atoms, respectively; the simulated systems of unliganded RhNCRD and the RhNCRD·Man8 complex contain 98,524 and 103,027 atoms, respectively.
Simulations were performed using the program NAMD 2.8 (43) with the CHARMM22 force field with CMAP corrections (44) for the protein and ions, the CHARMM36 force field for the sugars (45, 46), and the TIP3P water mode (47). Periodic boundary conditions were assumed, and the particle mesh Ewald summation method was employed for evaluating Coulomb forces. The van der Waals energy was calculated using a cutoff of 12 Å; the integration time step was 1 fs; the temperature was kept at 295 K by applying Langevin forces with a damping constant of 0.1 ps−1 (48) only to the oxygen atoms of water molecules. The methods mentioned were described by Phillips and co-workers (43).
Each simulated system was first subjected to 8000 steps of conjugate gradient minimization and then heated to 295 K in 4 ps, followed by a 500-ps equilibration with harmonic restraints applied to all protein heavy atoms, and by a 2-ns equilibration with no restraints under NPT ensemble conditions using Nosé-Hoover Langevin piston pressure control (48). The resulting equilibrated structure was used for a 65-ns production molecular dynamics simulation under NVT ensemble conditions. An analysis of molecular dynamics simulations and snapshots of the molecular structures were made with VMD (42). Loop region r.m.s.d. values were calculated using the heavy atoms of residues 324–329 for both proteins.
The atomic coordinates of the RpNCRD-dNG·d-mannose complex were taken from the corresponding crystal structure in this study. The complex was solvated into a TIP3 (47) water box of dimension 82 × 64 × 104 Å and was neutralized with 10 mm NaCl using VMD (42). The final simulated system contained 2375 atoms for RpNCRD-dNG·mannose and 48,530 atoms for water and NaCl. The simulation was performed using NAMD 2.8 (43). The system was energy-minimized for 2000 steps, followed by a 0.5-ns equilibration with RpNCRD-dNG·d-mannose complex atoms fixed and another 0.5 ns equilibration with the RpNCRD-dNG loop region (residues Asn-324 to Asn-330) released under NPT ensemble. Finally, a 30-ns production simulation was performed. The temperature and pressure were maintained at 300 K and 1 atm using Langevin thermostat with a damping constant of 5 ps−1 and Nosé-Hoover Langevin piston methods (48), respectively. Periodic boundary conditions and particle-mesh Ewald summation were used for effective full electrostatic interaction calculation. The integration time step was 1 fs.
RESULTS
Biochemical Characterization of Recombinant SP-D NCRD Preparations
The purification of RpNCRD and RpNCRD-dNG from HEK293E cell medium transiently transfected with the pSP-D NCRD sequence with or without the N323Q mutation, respectively, was performed as described previously using calcium-dependent mannan affinity chromatography followed by gel filtration chromatography (10). Chemical cross-linking followed by SDS-PAGE analysis showed major bands of ∼65 and 55 kDa for RpNCRD and RpNCRD-dNG, respectively (data not shown). Digestion of RpNCRD with N-glycanase resulted in a mobility shift toward the position of RpNCRD-dNG, indicating that the difference in apparent molecular mass between RpNCRD and RpNCRD-dNG was due to N-glycosylation of the CRD of RpNCRD. This indicated that the N-glycosylation sequon was successfully mutated by site-directed mutagenesis, yielding RpNCRD-dNG, the N-deglycosylated analog of RpNCRD.
The RhSP-D-derived NCRD fragment (RhNCRD) was purified and characterized as described previously (30). This preparation was shown to consist almost exclusively of trimers and bound mannosyl- or maltosyl-agarose in a Ca2+-dependent manner.
Comparison of the Antiviral Activity of RSP-Ds against IAV
The HAI activities of full-length pig SP-D (RpSP-D) or pig or human NCRDs (RpNCRD, RpNCRD-dNG, and RhNCRD) were measured against IAV strains Phil82(H3N2), Aichi(H3N2), and PR-8(H1N1) (Table 2). The susceptibility to inhibition by RpSP-D (Phil82 > PR-8 > Aichi) was generally in agreement with what was previously found in SP-D-mediated HAI studies on these IAV strains (23), although the PR-8 strain tended to be less resistant (similar to Aichi strain) for inhibition by RpSP-D compared with SP-D from porcine lung. Although RpSP-D was shown to be highly effective against all three strains (<1 μg/ml required to fully inhibit 40 HA units of virus), substantial inhibition was also mediated by RpNCRD. The activity of RpNCRD was shown to be dependent to a significant extent on the presence of the N-linked oligosaccharide, as illustrated by the significantly lower inhibitory activity measured for RpNCRD-dNG. It is important to note, however, that RpNCRD-dNG did have measurable HA inhibitory activity against the Phil82 and Aichi strains, whereas RhNCRD had no inhibitory activity for any IAV strain with the highest concentration tested (40 μg/ml). The lack of HA inhibitory activity of RpNCRD-dNG for the PR-8 strain is consistent with the total lack of N-linked glycans on the head domain of the HA of PR-8. Collectin-mediated protection of MDCK cells against infection by IAV was measured using the Phil strain (Fig. 1), and the outcome of these measurements supported the results obtained by HA inhibition measurements. Again, the relative neutralization activity against IAV between the four preparations was RpSP-D > RpNCRD > RpNCRD-dNG > RhNCRD. Of note, although the N-glycan of RpNCRD plays an important role in contributing to the neutralization activity against Phil82(H3N2), the N-deglycosylated variant RpNCRD-dNG still exhibited significant inhibitory activity against IAV albeit at higher concentrations (>5 μg/ml) compared with RpNCRD. In contrast, no significant neutralization activity could be measured for the human-derived analog (RhNCRD) for all concentrations tested (0–10 μg/ml).
TABLE 2.
HA inhibition of IAV by recombinant porcine and human SP-Ds
Values are mean ± S.E. of at least three experiments. ND means not detectable (no inhibition with highest collectin concentration tested, 40 μg/ml).
| Collectin preparation | Phila | Aichia | PR-8a |
|---|---|---|---|
| RpSP-D | 0.030 ± 0.004 | 0.61 ± 0.2 | 0.42 ± 0.08 |
| RpNCRD | 0.66 ± 0.2 | 3.3 ± 0.9 | 7.4 ± 3 |
| RpNCRD-dNG | 6.8 ± 0.7b | 19.8 ± 3c | ND |
| RhNCRD | ND | ND | ND |
a Strain designations are as follows: Phil, A/Philippines/82(H3N2); Aichi, A2/Aichi/68 (H3N2); PR-8, A/Puerto Rico/34 (H1N1).
b p < 0.005 compared with RpNCRD.
c p < 0.05 compared with RpNCRD. Statistical comparisons were by Student's paired, two-tailed t test.
FIGURE 1.

Neutralization of IAV infectivity by RpSP-D, RpNCRD, RpNCRD-dNG, and RhNCRD. Infectivity of IAV for MDCK cells was assessed by fluorescent focus formation (details under “Experimental Procedures”). The inhibitory activity of increasing concentrations of RpSP-D (full-length, closed squares) and the three neck-CRD preparations from pig (RpNCRD (closed triangles) and RpNCRD-dNG (open triangles) and human (RhNCRD, open squares) was assessed against the Phil strain. A fixed amount of virus was preincubated with increasing concentrations of collectin for 30 min at 37 °C, added to monolayers of MDCK cells and incubated for 45 min at 37 °C. After washing and fixation, the monolayers were labeled with monoclonal antibody to IAV nucleoprotein and FITC-labeled goat anti-mouse IgG. The numbers of infectious foci were counted and expressed as a percentage of the number of infectious foci formed in the control incubation (IAV added to monolayer in absence of collectin). Values are means ± S.E. of five experiments. *, p < 0.02 versus RhNCRD; **, p < 0.02 versus RhNCRD or RpSP-D-dNG. Statistical analysis was performed by two-tailed Student's t test.
Crystal Structure Analysis of RpNCRD-dNG
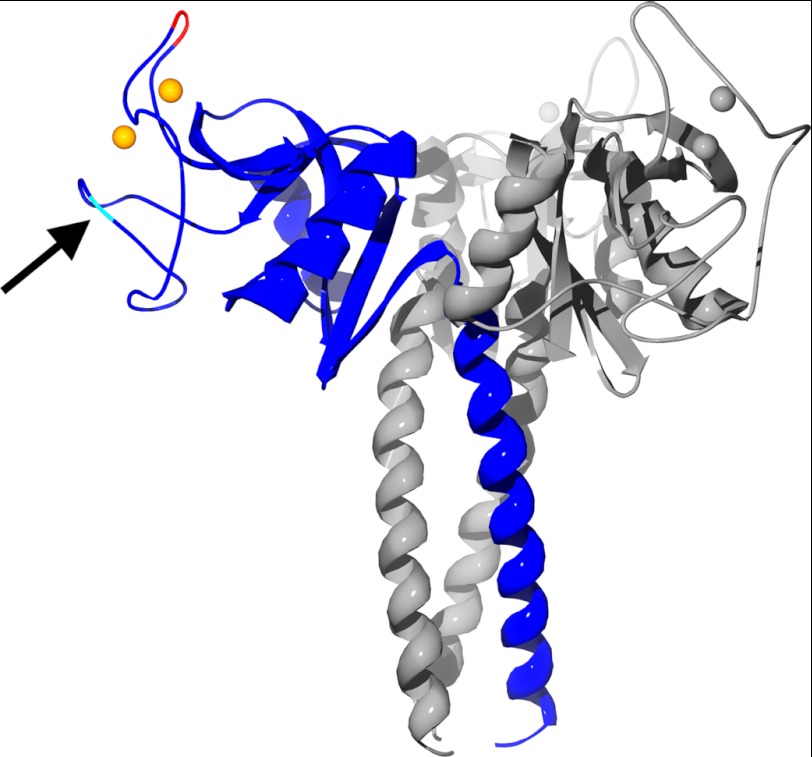
The tertiary structure of RpNCRD-dNG neck (residues 203–235) and CRD (residues 236–355) is very similar overall to that of RhNCRD as well as the two closely related collectins surfactant protein-A (SP-A) (49) and mannan-binding lectin (50). The RpNCRD-dNG crystal asymmetric unit is a monomer; however, the biological trimer is defined through crystallographic symmetry, where the helices that compose the neck domain from three molecules form a coiled-coil structure burying 6750 Å2 in the interface. The packing is analogous to that found in rat SP-A NCRD crystals, which also crystallize in space group P63 albeit with different unit cell dimensions (49). The RpNCRD-dNG CRD consists of a core with seven β-sheet regions flanked by two α-helices. Two loops on the protein, termed (as in mannan-binding lectin (51)) as the short loop (residues 297–303 in pSP-D) and the long loop (residues 307–331 in pSP-D), located in the CRD distal to the trimer axis, are involved in binding the two calcium ions observed in the structure. As shown in Fig. 2, the long loop contains an insertion in the pig sequence compared with that of the human. Fig. 2 also indicates the position of Asn-303, the site of the N-linked glycosylation. The pSP-D numbering is adjusted to account for these inserted residues between 326 and 327 to match the hSP-D numbering for all other residues.
FIGURE 2.
Ribbon diagram of the RpNCRD-dNG trimer. One monomer is highlighted in blue, and the residues of the insertion at residue 326 is highlighted in red. Calcium ions are shown as orange or gray spheres. The location of the N-glycosylation site is indicated by an arrow.
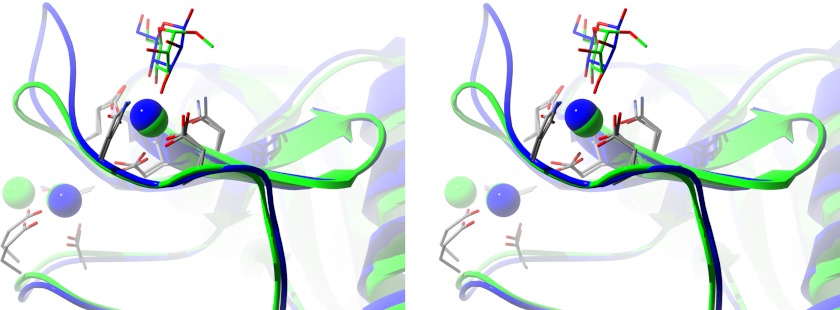
The lectin calcium site in RpNCRD-dNG is formed from residues on the long loop as well as residues from a nearby β-sheet region (residues 339–343) (Fig. 3). The calcium ion is coordinated by six protein oxygen atoms from five amino acids and two oxygen atoms from the ligands, creating a square antiprism. Mannose binds via the O3 and O4 oxygen atoms that form the canonical vicinal equatorial diol recognized by this type of lectin. Two of the amino acids, Glu-321 and Asn-323, are from a consensus sequence EPN found in most C-type lectins that favor binding of mannose. Two other amino acids, Asn-341 and Asp-342, are part of a consensus WND sequence found in C-type lectins. The other amino acid is Glu-329, which coordinates this calcium through its side chain.
FIGURE 3.
The d-mannose-liganded pNCRD-dNG lectin site structure. Wall-eyed stereo diagram showing the ribbon diagram of the pNCRD-dNG·d-mannose complex lectin site structure (blue) superposed onto the hSP-D·α-methyl-d-mannose complex structure (green) (PDB 3G81) (53). The calcium ions are rendered as spheres of appropriate color. Protein side chains that coordinate calcium are also shown. The three-residue insertion in pSP-D creates an extended binding site as can be seen.
A second nonlectin calcium is bound in the site between the two loops and is thought to play a structural role. This calcium is coordinated by five protein oxygen atoms from four protein side chains and one backbone oxygen atom, making a square pyramidal coordination sphere. Two of these side chains are from the short loop Asp-297 and Glu-301, the latter of which makes a bidentate interaction with calcium through its side chain. Other coordination interactions are formed with residues from the long loop, i.e. the side chains of Asn-324 and Asn-330 and the backbone oxygen atom of Glu-329.
There was clear electron density in the complex structure for d-mannose, which contains the required equatorial vicinal diol formed by the O3 and O4 atoms. Both of these atoms are within coordination distance of the lectin site calcium in the crystal structure. These oxygen atoms also make hydrogen bonds to protein side chains, O3 with the side chain of Asn-323 and Glu-321 and O4 with the side chain of Glu-329. The C4 and C6 atoms of the mannose ring are 3.5 Å away from the backbone carbonyl group of Asn-325, making van der Waal contacts. However, electron density in this region is relatively weak, consistent with the lack of H-bonds between the protein and this portion of the ring.
Docking Calculations
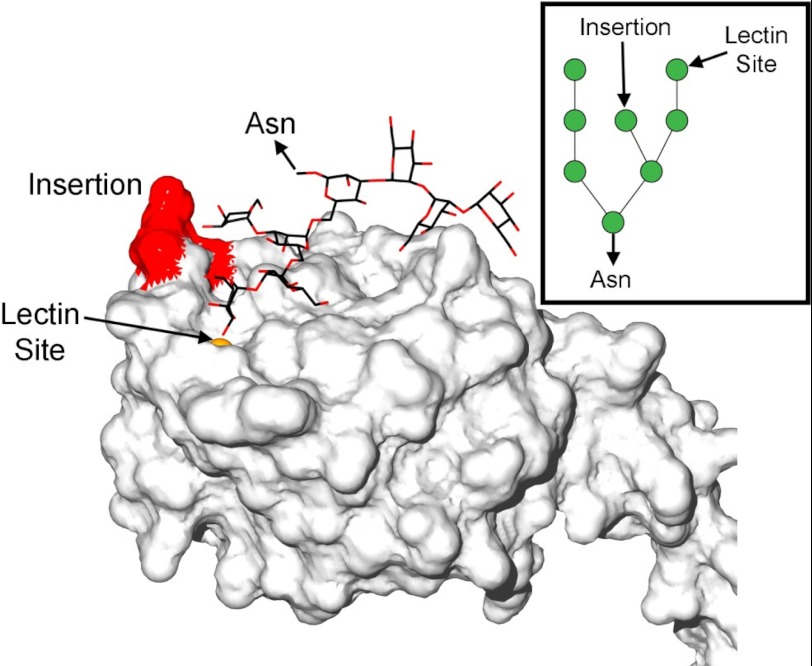
To better delineate the biologically relevant complex of RpNCRD-dNG bound to a glycoprotein, docking studies were employed to extend from the mannose present in the crystal structure to an RpNCRD-dNG·oligomannose complex. The highly branched octamannose (Man8), the largest oligomannose that can be accommodated by the docking program Glide (Schrödinger Inc., New York), was used in the calculations. Consistent with the crystallographic data, RpNCRD-dNG was positioned to bind to one of the nonreducing end terminal sugars through interactions with a vicinal equatorial diol formed by the O3 and O4 atoms of mannose, making similar interactions in the lectin site as observed in the crystal structure. The resulting docked structure from Glide is shown in Fig. 4. Man8 has three arms, and the terminal sugars from all three make contacts to RpNCRD-dNG (Fig. 4, inset). The docked structure presented here was the only terminal site where Man8 was successfully docked. This specific form of Man8 corresponds to that shown in an NMR study of ribonuclease B to be the major Man8 glycoform (52), supporting its biological relevance. Docking runs that constrained the terminal mannose from one of the other arms of Man8 in the lectin site failed to produce any conformations with favorable docking scores. The docked structure makes all the expected protein-sugar interactions in the lectin site, contacting residues Glu-321, Asn-323, Asn-325, Glu-329, Asn-341, Glu-342, and Lys-343. The other two arms make several other contacts to RpNCRD-dNG through mannoses farther from the lectin site. One arm contacts the protein making several favorable interactions with residues Glu-251, Gln-287, Asn-288, and Lys-289. These residues form a bulge in the surface of RpNCRD-dNG that the sugar chain wraps around. The last arm was observed to make contacts to residues Gly-326 and the porcine-specific tripeptide insertion Gly-326A, Ser-326B, and Ser-326C (GSS-loop) which is, upon sequence alignment, located between residues Gly-326 and Gly-327 of the hSP-D sequence. These contacts include potential hydrogen bonds to the O3 and O4 hydroxyls of the mannose ring; the stated residues are part of the long loop and include the three-residue insertion that distinguishes pSP-D from other SP-Ds. The residues form another bulge on the surface of the RpNCRD-dNG CRD, and the sugar chain wraps back around it to abut the mannose against the bulge.
FIGURE 4.
The docked complex of pNCRD-dNG and octamannose. The protein is shown in surface representation. Residues of the GSS-loop are colored in red to highlight the inserted residues in pSP-D as compared with hSP-D. The lectin site and the site of attachment of octamannose to the Asn residue in a polypeptide chain through two N-acetylglucosamines are indicated. The inset contains a schematic representation of octamannose, showing the connectivity of each mannose residue (green circles) and how mannose interacts with the lectin site and inserts into the docked structure.
Molecular Dynamics Simulations
In the crystal structure of the RpNCRD-dNG·d-mannose complex, the large loop conformation is stabilized by crystal lattice contacts from an adjacent trimer, leaving open the possibility that the loop conformation is different in the noncrystalline environment. Therefore, molecular dynamics simulations were performed to probe the range of loop conformations, as well as to compare the relative flexibility of the loops of RpNCRD-dNG and RhNCRD and to determine the effect of sugar binding on the stability of the loop regions.
Three sets of simulations were run for RpNCRD-dNG as follows: the RpNCRD-dNG·d-mannose crystallographic complex, the same structure with the mannose removed, and the docked Man8 complex. For RhNCRD, simulations started with coordinates from RhNCRD (PDB code 3G83 (53), with the ligand removed), with or without docked Man8. After 65-ns simulations, the average root mean square deviations (r.m.s.d.) of the loop regions of RpNCRD-dNG and RhNCRD are 2.69 and 2.11 Å, respectively, showing that the long loop of RpNCRD-dNG is indeed more flexible than that of RhNCRD. The simulations also suggest that the long loop of RpNCRD-dNG could adopt a conformation similar to that observed in the crystal structure, perhaps stabilized by sugar binding.
To determine the stabilization effect of sugar on the long loop of RpNCRD-dNG, comparisons of dynamics of the long loop were made with simulations with or without Man8. Fig. 5 shows that the r.m.s.d. of the long loop of the RpNCRD-dNG·Man8 complex (average r.m.s.d. = 1.79 Å) is smaller than that of RpNCRD-dNG in the absence of the sugar (average r.m.s.d. = 2.69 Å). The fluctuation of r.m.s.d. also becomes smaller when RpNCRD-dNG binds to Man8, indicating that the loop of RpNCRD-dNG exhibits reduced flexibility upon sugar binding. Simulations of the RpNCRD-dNG·d-mannose complex gave similar results as for RpNCRD-dNG in the absence of sugar, indicating that the larger, branched sugar is more effective than the monosaccharide at stabilizing the SP-D loop conformation. We also compared the flexibility of the loop of RhNCRD (average r.m.s.d. = 2.11 Å) and the loop of the RhNCRD·Man8 complex (average r.m.s.d. = 0.79 Å), showing that sugar binding also stabilizes the long loop of human SP-D (see supplemental Fig. S1 and supplemental Movies S1 and S2).
FIGURE 5.
Analysis of RpNCRD-dNG-loop by molecular dynamics simulation. Shown is the time evolution of the r.m.s.d. of the loop region of RpNCRD-dNG (green) and the RpNCRD-dNG·Man8 complex (pink) obtained from molecular dynamics simulations. Snapshots of the RpNCRD-dNG·Man8 complex and of RpNCRD-dNG are presented at bottom left and right, respectively. Calcium ions are shown as orange spheres. Two movies comparing the fluctuation of the loop of RpNCRD-dNG (supplemental Movie S1) and RpNCRD-dNG·Man8 complex (supplemental Movie S2) are provided.
The 30-ns trajectory of the RpNCRD-dNG loop region (residue Asn-324 to Asn-330) was analyzed with Cluster from Amber Tools (54). Similar coordinate frames from the trajectory based on r.m.s.d. of backbone atoms were grouped together, and three clusters are found with r.m.s.d. values of representative structures about 0.7, 1.4, and 1.7 Å compared with the starting model. Their percentages in the trajectory are 38.9, 39.4, and 15.7%. The lack of predominant structure in the RpNCRD-dNG·mannose complex is underscored by the loop r.m.s.d., which is fluctuating within the range of 2.5 Å. These data demonstrate that the monosaccharide binding to the RpNCRD-dNG lectin site undergoes no significant interactions with the loop.
DISCUSSION
The emergence of the swine origin H1N1/2009 in humans has revitalized the hypothesis that pigs are important facilitators of reassortment among influenza viruses, occasionally giving rise to human pandemics (55). Both adaptive and innate immune responses play an important role in the control of infectious disease caused by IAV and contribute to limiting the spread of these zoonotic pathogens from animal reservoirs to humans. The mechanisms by which viruses evolve in one host, cause zoonotic infection, and adapt to a new host species remain unclear, but it is speculated that the innate immune system, given its crucial role in early containment of IAV infection (56), could play an important role in avian-to-swine and swine-to-human host adaptation of IAVs in pigs.
SP-D is an important component of the early host defense mechanism against IAV, and our previous studies have shown that in pigs this collectin exhibits distinctly strong neutralization activity against a broad range of IAV strains (23). In this study, we describe the structure of a recombinant trimeric fragment of pSP-D that exhibits inhibitory activity against IAV. We characterized several porcine-specific structural features of pSP-D CRD that account for its overall high affinity binding of viral glycans and strong neutralization activity against IAV in pigs.
To investigate the mechanisms of interaction between the pSP-D NCRD and IAV, we cloned and expressed two recombinant truncated (NCRD) versions of pSP-D in which the Asn-303-linked glycan was present (RpNCRD) or deleted by site-directed mutagenesis (RpNCRD-dNG). For this purpose, we applied the HEK293 expression system that was previously used successfully for the production of full-length wild-type pSP-D, wild-type hSP-D, and various h/pSP-D derivatives (26). Biochemical characterization showed that both RpNCRD and RpNCRD-dNG were expressed as trimeric proteins, and N-glycanase digestions confirmed the absence of N-linked glycans in the RpNCRD-dNG preparation. Importantly, both preparations were extracted by Ca2+-dependent mannan-affinity chromatography, indicating that the two expressed porcine NCRDs can be assigned as properly folded, functionally active collectins. This expectation was confirmed for RpNCRD-dNG by formation of the crystallographic complex with d-mannose reported here.
The antiviral activity of both RpNCRDs was determined by HA inhibition measured against three different IAV strains representing two subtypes of IAV (H3N2 and H1N1, Table 2) with varying degrees of susceptibility to SP-D-mediated neutralization. For all strains, however, it was clearly demonstrated that RpNCRD exhibits HA inhibitory activity, in contrast to RhNCRD for which no activity could be measured in this assay. Dodecameric oligomerization thus appeared to be essential for native hSP-D to express its HA inhibitory activity, as determined previously (57, 58), driven primarily by the aggregation properties of the fully assembled hSP-D molecule.
The neutralization activity observed for both RpNCRDs is attributed to the relatively strong affinity expressed by their trimeric CRDs for viral (mannose-rich) glycans. The observed differences in antiviral activity as measured by HA inhibition were underlined by a neutralization of IAV (Phil strain) infectivity assay (Fig. 1). A relatively strong protective effect was shown not only for RpNCRD but also for RpNCRD-dNG, although RhNCRD was not able to prevent infection of MDCK cells by IAV. We therefore sought to compare the structural features of the N-deglycosylated NCRD of pSP-D with the NCRD of hSP-D to elucidate the basis for the functional enhancement of lectin activity that is associated with the CRD of pSP-D.
In cloning the cDNA of pSP-D (20), we found that, other than the N-linked glycosylation, its CRD is unique among SP-Ds in that it contains an insertion of three amino acids, Gly-Ser-Ser, immediately after Gly-326 (hSP-D sequence numbering), denoted as 326-GSS or the GSS-loop. By sequence alignment, this insertion is shown to be on the long loop of the RpNCRD-dNG crystal structure. Interestingly, the bovine collectin CL-43 contains a similar insertion of three amino acid residues (Arg-Ala-Lys; RAK) near the carbohydrate-binding site, and this insertion motif contributes to enhanced binding affinity for mannose-rich viral glycoconjugates (14, 59). Considering the proximal location of the GSS-loop toward the lectin-binding site in the CRD, it is hypothesized that this aberrant loop region could have an impact on the glycan binding characteristics of pSP-D. Consequently, interactions between pSP-D and specific glycans (e.g. viral sugars) could be enhanced compared with hSP-D, as observed in the study mentioned above for RpNCRD-dNG and similar to what was observed for the CL-43 derived RAK insertion into the hSP-D CRD (14).
The major differences in the RpNCRD-dNG molecular structure compared with that of RhNCRD are consequences of the greater length of the long loop in RpNCRD-dNG (Fig. 3). Crystallographic comparison of the RpNCRD-dNG (this study) and RhNCRD (53) complexes with d-mannose or α-methyl-d-mannose, respectively, shows that the CRDs overall are very similar, with r.m.s.d. fitting of 0.47–0.53 Å for corresponding residues. Calcium-binding residues in C-type lectin sites are strictly conserved in all SP-Ds characterized thus far. Superimposition of the lectin-binding sites of RpNCRD-dNG and RhNCRD calculated using all atoms from calcium-coordinating residues Glu-321, Asn-323, Glu-329, Asn-341, and Asp-342 and the lectin site calcium results in an r.m.s.d. of 0.31 Å. However, the mannose rings do not overlay well in the superimposed structures, consistent with slightly longer bond distances between the sugar and lectin site atoms in RpNCRD-dNG. The three additional amino acids Gly-326A–Ser-326B–Ser-326C form a loop that protrudes away from the lectin site. The broken electron density and high temperature factors for this loop suggest a high degree of conformational flexibility. In the crystal structure, residues Gly-326A–Ser-326B–Ser-326C (GSS-loop), Gly-327, and Ala-328 of this loop make close contacts to residues 239–242 from a neighboring molecule. Therefore, the position of the loop in the crystal structure may be affected by crystal contacts. To determine whether such contacts might occlude a more favorable conformation in a noncrystalline environment, molecular dynamics simulations were run on either of the crystallographic d-mannose complexes with the sugar either present or removed. In all cases, an ensemble of conformations was found from these simulations, supporting the idea that the loop is inherently flexible and therefore free to adopt the conformation observed in the crystal structure.
To better understand how pSP-D might interact with branched high mannose viral glycans compared with monosaccharides, we initiated docking studies with an appropriate Man8 (Fig. 4), for which starting crystallographic coordinates were available (41). Man8 dockings were performed such that the terminal mannoside bound in the lectin site overlaid the d-mannose in the crystallographic RpNCRD-dNG complex, fixing the starting position. Invariably, the docking showed potential for contacts between the Man8 branched arms and the flexible RpNCRD-dNG loop in the vicinity of the insertion. The docked Man8 structures showed the potential for favorable hydrogen bonds between residues Gly-326A, Ser-326B, and Ser-326C of pSP-D and the O3 and O4 hydroxyls of one of the terminal mannose sugars (Fig. 4). Overlay of the docked RpNCRD-dNG structure and RhNCRD demonstrates that hSP-D would be unable to make the interactions with Man8 as pSP-D does; the long loop is too short in the RhNCRD structure and lacks the bulge seen in RpNCRD-dNG. Amino acid substitutions also may play a role in the increased affinity of pSP-D for viral glycans. In addition to residues Gly-326, Gly-326A, Ser-326B, and Ser-326C, there are residues Glu-251, Gln-287, and Lys-289. In the hSP-D, these residues are Val-251, Lys-287, and Glu-289. Thus, the potential for increased hydrogen bonds and van der Waal contacts to the oligomannose substrate for pSP-D from these residues could explain the observed increased affinity of pSP-D for viral glycans compared with hSP-D.
The additional intermolecular contacts observed in the docking experiments led us to speculate that binding to the larger highly branched viral glycans might stabilize the flexible pSP-D loop. Therefore, we initiated molecular dynamics simulations on the docked RpNCRD-dNG·Man8 complex and compared the results with that of the unliganded RpNCRD-dNG and the crystallographic RpNCRD-dNG·d-α-mannose complex structures. Simulations demonstrated that the loop structure, relative to its unliganded or monosaccharide-bound counterparts, exhibited increased conformational stability. In contrast, further simulations run on unliganded RhNCRD compared with the corresponding docked Man8 complex showed no such effects because the hSP-D loop is intrinsically less flexible to begin with. It is possible that in vivo, the longer, more flexible loop of pSP-D allows it to make contacts with the glycan on HA that hSP-D cannot make, resulting in higher inhibitory activity against IAV.
In RhNCRD structures, four calcium ion sites have been defined as follows: one in the lectin site, one in the neck region close to the trimer axis, and two structural calcium ions bound in between the long and short loops of the CRD (60). In the RpNCRD-dNG structure, however, only two calcium ions were observed, one in the lectin site and one structural calcium ion sandwiched between the long and short loops (Fig. 3). No electron density was found in the electron density maps of RpNCRD-dNG for either the second structural calcium or the neck region calcium. In the case of the second structural calcium site observed in RhNCRD, but not RpNCRD-dNG, the site in the latter structure is occupied by the side chain of Arg-220 from a neighboring molecule in the crystal lattice. The side chain of Arg-220 makes a salt bridge to the side chain of Glu-301 as well as making hydrogen bonds to the backbone oxygen atoms of Asp-297 and Ile-298. We speculate that the lack of this second structural calcium in RpNCRD-dNG may be a factor in allowing the more flexible pSP-D loop to adopt an optimal conformation for interaction with IAV.
The proper folding of the long loop in the conformation observed in the RpNCRD-dNG crystal structure may be critical for recognition of oligomannose substrates by pSP-D. In previous work performed with full-length recombinant hSP-D derivatives, a Ser-Gly-Ala or “SGA”-loop (two residues shifted toward the C terminus as regards the 326-GSS loop) was inserted by site-directed mutagenesis at a predicted identical location in the CRD of hSP-D based upon sequence alignment with pSP-D. It was anticipated that this insertion would mimic the pSP-D loop bulge and enhance antiviral activity in hSP-D. However, in vitro IAV neutralization assays with these mutants demonstrated that insertion of the SGA-loop completely inactivated the antiviral activity of the RhSP-D mutant. Evaluation of this insertion strategy using the crystallographic data showed that the site of the insertion in RhNCRD did not mimic the corresponding tertiary structure of the RpNCRD-dNG loop because the structural alignment appears to differ slightly from the sequence alignment. In the crystal structures, superposition of the RhNCRD and RpNCRD-dNG backbones showed that human and porcine NCRD loop structures diverge distal to the inserted residues between residues Asn-325 and Ala-328, i.e. where Ala-328 of pSP-D aligns structurally with Ser-328 of hSP-D. Therefore, on a tertiary structure level, the insertion location is better described as residues Gly-Ser-Ser in-between Gly-326 and Gly-327 of the hSP-D sequence (326-GSS). Taken together, the absence of antiviral activity observed with the SGA-containing RhSP-D mutant may result from a structural alignment error but with the tripeptide situated closer to the N-terminal portion of the loop; it may be in a better position to interact with viral mannosides on the IAV, as suggested by the docking studies.
In summary, we have explored the mechanisms underlying antiviral activity of pSP-D and compared it with that of hSP-D. Porcine SP-D has two distinct features in its CRD that contribute to a better anti-IAV activity. These two features are the N-linked glycan at position Asn-303 and the long loop in the carbohydrate-binding site. Future work is needed to determine the structural features of the N-glycosylated pNCRD and possible synergy with the lectin site.
The strong antiviral activity of SP-D in pigs against IAV is intriguing, because pigs serve as a reservoir of H1, H3, H5, H7, and H9 subtype IAVs (61). Moreover, co-circulation of different IAVs and persistence of infection readily occur in these animals. Given these contradicting observations, it is speculated that a highly effective innate immune-driven clearance of IAV in pigs might delay or interfere with the adaptive immune response that is known to be crucial for elimination of the virus once an infection has been established. Within this context, it is of interest to note that pigs transiently infected with avian IAVs do not always generate detectable antibody responses (62, 63), and it is known that subclinical infections frequently occur in pigs (64). However, we cannot exclude the possibility that the distinct interactions between pSP-D and an exceptionally broad range of IAV strains, including poorly N-glycosylated strains that are not recognized by SP-D in humans as demonstrated in vitro (22), may favor reassortment conditions in which simultaneous uptake of these different viral strains by a single host cell is established more easily in pigs. Taken together, it remains to be investigated how and to what extent pSP-D contributes to these phenomena and whether pSP-D affects the process of transmission and/or adaptation of IAV in pigs, an animal species that is considered as an important intermediate host for reassortment of IAVs of avian and human origin (65).
Acknowledgments
M. V. E. and H. P. H. thank Sanne Raadsveld for technical support. Supercomputer time was provided by Texas Advanced Computing Center via TeraGrid Resource Allocation Committee Grant MCA93S028.
This work was supported, in whole or in part, by National Institutes of Health Grants AI083222 (to B. A. S.) and HL069031 (to K. L. H.). This work was also supported by European Union FP6 Grant Specific Targeted Research Project Contract LSHM-CT-2004-512093 (to H. P. H.) and Human Frontier Science Program Grant RGP0016/2009-C (to H. P. H.)

This article contains supplemental Fig. 1 and Movies S1 and S2.
The atomic coordinates and structure factors (code 4DN8) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- IAV
- influenza A virus
- CRD
- carbohydrate recognition domain
- HAI
- hemagglutination inhibition
- hSP-D
- human SP-D
- MDCK
- Madin-Darby canine kidney
- NCRD
- neck region plus CRD
- pSP-D
- porcine SP-D
- RhNCRD
- recombinant human SP-D-derived trimeric neck-CRD
- RpNCRD
- recombinant porcine SP-D-derived trimeric neck-CRD
- RpNCRD-dNG
- RpNCRD with deleted N-glycosylation site
- RpSP-D
- recombinant porcine SP-D
- GSS-loop
- pSP-D-specific insertion of Gly-Ser-Ser adjacent to residue Gly-326 of the SP-D consensus sequence
- SP-A
- surfactant protein A
- SP-D
- surfactant protein D
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- r.m.s.d.
- root mean square deviation
- PDB
- Protein Data Bank
- VMD
- visual molecular dynamics.
REFERENCES
- 1. Novel Swine-Origin Influenza A (H1N1) Virus Investigation Team, Dawood F. S., Jain S., Finelli L., Shaw M. W., Lindstrom S., Garten R. J., Gubareva L. V., Xu X., Bridges C. B., Uyeki T. M. (2009) Emergence of a novel swine-origin influenza A (H1N1) virus in humans. N. Engl. J. Med. 360, 2605–2615 [DOI] [PubMed] [Google Scholar]
- 2. Hartshorn K. L., Crouch E. C., White M. R., Eggleton P., Tauber A. I., Chang D., Sastry K. (1994) Evidence for a protective role of pulmonary surfactant protein D (SP-D) against influenza A viruses. J. Clin. Invest. 94, 311–319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hartshorn K. L., White M. R., Shepherd V., Reid K., Jensenius J. C., Crouch E. C. (1997) Mechanisms of anti-influenza activity of surfactant proteins A and D. Comparison with serum collectins. Am. J. Physiol. 273, L1156–L1166 [DOI] [PubMed] [Google Scholar]
- 4. Hawgood S., Brown C., Edmondson J., Stumbaugh A., Allen L., Goerke J., Clark H., Poulain F. (2004) Pulmonary collectins modulate strain-specific influenza a virus infection and host responses. J. Virol. 78, 8565–8572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. LeVine A. M., Whitsett J. A., Hartshorn K. L., Crouch E. C., Korfhagen T. R. (2001) Surfactant protein D enhances clearance of influenza A virus from the lung in vivo. J. Immunol. 167, 5868–5873 [DOI] [PubMed] [Google Scholar]
- 6. White M. R., Crouch E., Vesona J., Tacken P. J., Batenburg J. J., Leth-Larsen R., Holmskov U., Hartshorn K. L. (2005) Respiratory innate immune proteins differentially modulate the neutrophil respiratory burst response to influenza A virus. Am. J. Physiol. Lung Cell Mol. Physiol. 289, L606–L616 [DOI] [PubMed] [Google Scholar]
- 7. Hartshorn K. L., Reid K. B., White M. R., Jensenius J. C., Morris S. M., Tauber A. I., Crouch E. (1996) Neutrophil deactivation by influenza A viruses/ Mechanisms of protection after viral opsonization with collectins and hemagglutination-inhibiting antibodies. Blood 87, 3450–3461 [PubMed] [Google Scholar]
- 8. Pastva A. M., Wright J. R., Williams K. L. (2007) Immunomodulatory roles of surfactant proteins A and D. Implications in lung disease. Proc. Am. Thorac. Soc. 4, 252–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crouch E., Persson A., Chang D., Heuser J. (1994) Molecular structure of pulmonary surfactant protein D (SP-D). J. Biol. Chem. 269, 17311–17319 [PubMed] [Google Scholar]
- 10. van Eijk M., van de Lest C. H., Batenburg J. J., Vaandrager A. B., Meschi J., Hartshorn K. L., van Golde L. M., Haagsman H. P. (2002) Porcine surfactant protein D is N-glycosylated in its carbohydrate recognition domain and is assembled into differently charged oligomers. Am. J. Respir. Cell Mol. Biol. 26, 739–747 [DOI] [PubMed] [Google Scholar]
- 11. Hoppe H. J., Barlow P. N., Reid K. B. (1994) A parallel three-stranded α-helical bundle at the nucleation site of collagen triple-helix formation. FEBS Lett. 344, 191–195 [DOI] [PubMed] [Google Scholar]
- 12. Hartshorn K., Chang D., Rust K., White M., Heuser J., Crouch E. (1996) Interactions of recombinant human pulmonary surfactant protein D and SP-D multimers with influenza A. Am. J. Physiol. 271, L753–L762 [DOI] [PubMed] [Google Scholar]
- 13. Hartshorn K. L., White M. R., Tecle T., Tornoe I., Sorensen G. L., Crouch E. C., Holmskov U. (2007) Reduced influenza viral neutralizing activity of natural human trimers of surfactant protein D. Respir. Res. 8, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Crouch E., Tu Y., Briner D., McDonald B., Smith K., Holmskov U., Hartshorn K. (2005) Ligand specificity of human surfactant protein D. Expression of a mutant trimeric collectin that shows enhanced interactions with influenza A virus. J. Biol. Chem. 280, 17046–17056 [DOI] [PubMed] [Google Scholar]
- 15. Hartshorn K. L., White M. R., Voelker D. R., Coburn J., Zaner K., Crouch E. C. (2000) Mechanism of binding of surfactant protein D to influenza A viruses. Importance of binding hemagglutinin to antiviral activity. Biochem. J. 351, 449–458 [PMC free article] [PubMed] [Google Scholar]
- 16. Crouch E., Nikolaidis N., McCormack F. X., McDonald B., Allen K., Rynkiewicz M. J., Cafarella T. M., White M., Lewnard K., Leymarie N., Zaia J., Seaton B. A., Hartshorn K. L. (2011) Mutagenesis of surfactant protein D informed by evolution and x-ray crystallography enhances defenses against influenza A virus in vivo. J. Biol. Chem. 286, 40681–40692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hartshorn K. L., Webby R., White M. R., Tecle T., Pan C., Boucher S., Moreland R. J., Crouch E. C., Scheule R. K. (2008) Role of viral hemagglutinin glycosylation in anti-influenza activities of recombinant surfactant protein D. Respir. Res. 9, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Reading P. C., Pickett D. L., Tate M. D., Whitney P. G., Job E. R., Brooks A. G. (2009) Loss of a single N-linked glycan from the hemagglutinin of influenza virus is associated with resistance to collectins and increased virulence in mice. Respir. Res. 10, 117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Job E. R., Deng Y. M., Tate M. D., Bottazzi B., Crouch E. C., Dean M. M., Mantovani A., Brooks A. G., Reading P. C. (2010) Pandemic H1N1 influenza A viruses are resistant to the antiviral activities of innate immune proteins of the collectin and pentraxin superfamilies. J. Immunol. 185, 4284–4291 [DOI] [PubMed] [Google Scholar]
- 20. van Eijk M., Haagsman H. P., Skinner T., Archibald A., Reid K. B., Lawson P. R. (2000) Porcine lung surfactant protein D. Complementary DNA cloning, chromosomal localization, and tissue distribution. J. Immunol. 164, 1442–1450 [DOI] [PubMed] [Google Scholar]
- 21. van Eijk M., White M. R., Batenburg J. J., Vaandrager A. B., van Golde L. M., Haagsman H. P., Hartshorn K. L. (2004) Interactions of influenza A virus with sialic acids present on porcine surfactant protein D. Am. J. Respir. Cell Mol. Biol. 30, 871–879 [DOI] [PubMed] [Google Scholar]
- 22. Hillaire M. L., van Eijk M., van Trierum S. E., van Riel D., Saelens X., Romijn R. A., Hemrika W., Fouchier R. A., Kuiken T., Osterhaus A. D., Haagsman H. P., Rimmelzwaan G. F. (2011) Assessment of the antiviral properties of recombinant porcine SP-D against various influenza A viruses in vitro. PLoS One 6, e25005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. van Eijk M., White M. R., Crouch E. C., Batenburg J. J., Vaandrager A. B., Van Golde L. M., Haagsman H. P., Hartshorn K. L. (2003) Porcine pulmonary collectins show distinct interactions with influenza A viruses; role of the N-linked oligosaccharides in the carbohydrate recognition domain. J. Immunol. 171, 1431–1440 [DOI] [PubMed] [Google Scholar]
- 24. Webster R. G. (1997) Influenza virus. Transmission between species and relevance to emergence of the next human pandemic. Arch. Virol. Suppl. 13, 105–113 [DOI] [PubMed] [Google Scholar]
- 25. Neumann G., Noda T., Kawaoka Y. (2009) Emergence and pandemic potential of swine-origin H1N1 influenza virus. Nature 459, 931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. van Eijk M., Bruinsma L., Hartshorn K. L., White M. R., Rynkiewicz M. J., Seaton B. A., Hemrika W., Romijn R. A., van Balkom B. W., Haagsman H. P. (2011) Introduction of N-linked glycans in the lectin domain of surfactant protein D. Impact on interactions with influenza A viruses. J. Biol. Chem. 286, 20137–20151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Durocher Y., Perret S., Kamen A. (2002) High level and high throughput recombinant protein production by transient transfection of suspension-growing human 293-EBNA1 cells. Nucleic Acids Res. 30, E9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Barash S., Wang W., Shi Y. (2002) Human secretory signal peptide description by hidden Markov model and generation of a strong artificial signal peptide for secreted protein expression. Biochem. Biophys. Res. Commun. 294, 835–842 [DOI] [PubMed] [Google Scholar]
- 29. Morlot C., Hemrika W., Romijn R. A., Gros P., Cusack S., McCarthy A. A. (2007) Production of Slit2 LRR domains in mammalian cells for structural studies and the structure of human Slit2 domain 3. Acta Crystallogr. D Biol. Crystallogr. 63, 961–968 [DOI] [PubMed] [Google Scholar]
- 30. Wang H., Head J., Kosma P., Brade H., Müller-Loennies S., Sheikh S., McDonald B., Smith K., Cafarella T., Seaton B., Crouch E. (2008) Recognition of heptoses and the inner core of bacterial lipopolysaccharides by surfactant protein D. Biochemistry 47, 710–720 [DOI] [PubMed] [Google Scholar]
- 31. Hartshorn K. L., Ligtenberg A., White M. R., Van Eijk M., Hartshorn M., Pemberton L., Holmskov U., Crouch E. (2006) Salivary agglutinin and lung scavenger receptor cysteine-rich glycoprotein 340 have broad anti-influenza activities and interactions with surfactant protein D that vary according to donor source and sialylation. Biochem. J. 393, 545–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hartshorn K. L., Sastry K., White M. R., Anders E. M., Super M., Ezekowitz R. A., Tauber A. I. (1993) Human mannose-binding protein functions as an opsonin for influenza A viruses. J. Clin. Invest. 91, 1414–1420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Crouch E., McDonald B., Smith K., Cafarella T., Seaton B., Head J. (2006) Contributions of phenylalanine 335 to ligand recognition by human surfactant protein D. Ring interactions with SP-D ligands. J. Biol. Chem. 281, 18008–18014 [DOI] [PubMed] [Google Scholar]
- 35. Terwilliger T. C., Grosse-Kunstleve R. W., Afonine P. V., Moriarty N. W., Zwart P. H., Hung L. W., Read R. J., Adams P. D. (2008) Iterative model building, structure refinement, and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64, 61–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 37. Painter J., Merritt E. A. (2006) Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D Biol. Crystallogr. 62, 439–450 [DOI] [PubMed] [Google Scholar]
- 38. Painter J., Merritt E. A. (2006) TLSMD web server for the generation of multigroup TLS models. J. Appl. Crystallogr. 39, 109–111 [Google Scholar]
- 39. Guex N., Peitsch M. C. (1997) SWISS-MODEL and the Swiss-PdbViewer. An environment for comparative protein modeling. Electrophoresis 18, 2714–2723 [DOI] [PubMed] [Google Scholar]
- 40. Cason C., Froehlich T., Kopp N., Parker R. (2005) Persistence of Vision Raytracer. Pty. Ltd., Version 3.6, Williamstown, Victoria, Australia [Google Scholar]
- 41. Calarese D. A., Scanlan C. N., Zwick M. B., Deechongkit S., Mimura Y., Kunert R., Zhu P., Wormald M. R., Stanfield R. L., Roux K. H., Kelly J. W., Rudd P. M., Dwek R. A., Katinger H., Burton D. R., Wilson I. A. (2003) Antibody domain exchange is an immunological solution to carbohydrate cluster recognition. Science 300, 2065–2071 [DOI] [PubMed] [Google Scholar]
- 42. Humphrey W., Dalke A., Schulten K. (1996) VMD. Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 [DOI] [PubMed] [Google Scholar]
- 43. Phillips J. C., Braun R., Wang W., Gumbart J., Tajkhorshid E., Villa E., Chipot C., Skeel R. D., Kalé L., Schulten K. (2005) Scalable molecular dynamics with NAMD. J. Comput. Chem. 26, 1781–1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mackerell A. D., Jr., Feig M., Brooks C. L., 3rd (2004) Extending the treatment of backbone energetics in protein force fields. Limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J. Comput. Chem. 25, 1400–1415 [DOI] [PubMed] [Google Scholar]
- 45. Guvench O., Greene S. N., Kamath G., Brady J. W., Venable R. M., Pastor R. W., Mackerell A. D., Jr. (2008) Additive empirical force field for hexopyranose monosaccharides. J. Comput. Chem. 29, 2543–2564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Guvench O., Hatcher E. R., Venable R. M., Pastor R. W., Mackerell A. D. (2009) CHARMM additive all-atom force field for glycosidic linkages between hexopyranoses. J. Chem. Theory Comput. 5, 2353–2370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 [Google Scholar]
- 48. Martyna G. J., Tobias D. J., Klein M. L. (1994) Constant pressure molecular dynamics algorithms. J. Chem. Phys. 101, 4177–4189 [Google Scholar]
- 49. Head J. F., Mealy T. R., McCormack F. X., Seaton B. A. (2003) Crystal structure of trimeric carbohydrate recognition and neck domains of surfactant protein A. J. Biol. Chem. 278, 43254–43260 [DOI] [PubMed] [Google Scholar]
- 50. Weis W. I., Drickamer K. (1994) Trimeric structure of a C-type mannose-binding protein. Structure 2, 1227–1240 [DOI] [PubMed] [Google Scholar]
- 51. Weis W. I., Drickamer K., Hendrickson W. A. (1992) Structure of a C-type mannose-binding protein complexed with an oligosaccharide. Nature 360, 127–134 [DOI] [PubMed] [Google Scholar]
- 52. González L., Bruix M., Díaz-Mauriño T., Feizi T., Rico M., Solís D., Jiménez-Barbero J. (2000) Conformational studies of the Man8 oligosaccharide on native ribonuclease B and on the reduced and denatured protein. Arch. Biochem. Biophys. 383, 17–27 [DOI] [PubMed] [Google Scholar]
- 53. Crouch E., Hartshorn K., Horlacher T., McDonald B., Smith K., Cafarella T., Seaton B., Seeberger P. H., Head J. (2009) Recognition of mannosylated ligands and influenza A virus by human surfactant protein D: contributions of an extended site and residue 343. Biochemistry 48, 3335–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Case D. A., Cheatham T. E., 3rd, Darden T., Gohlke H., Luo R., Merz K. M., Jr., Onufriev A., Simmerling C., Wang B., Woods R. J. (2005) The Amber biomolecular simulation programs. J. Comput. Chem. 26, 1668–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Vijaykrishna D., Smith G. J., Pybus O. G., Zhu H., Bhatt S., Poon L. L., Riley S., Bahl J., Ma S. K., Cheung C. L., Perera R. A., Chen H., Shortridge K. F., Webby R. J., Webster R. G., Guan Y., Peiris J. S. (2011) Long term evolution and transmission dynamics of swine influenza A virus. Nature 473, 519–522 [DOI] [PubMed] [Google Scholar]
- 56. White M. R., Doss M., Boland P., Tecle T., Hartshorn K. L. (2008) Innate immunity to influenza virus: implications for future therapy. Expert Rev. Clin. Immunol. 4, 497–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. White M., Kingma P., Tecle T., Kacak N., Linders B., Heuser J., Crouch E., Hartshorn K. (2008) Multimerization of surfactant protein D, but not its collagen domain, is required for antiviral and opsonic activities related to influenza virus. J. Immunol. 181, 7936–7943 [DOI] [PubMed] [Google Scholar]
- 58. Tecle T., White M. R., Sorensen G., Gantz D., Kacak N., Holmskov U., Smith K., Crouch E. C., Hartshorn K. L. (2008) Critical role for cross-linking of trimeric lectin domains of surfactant protein D in antiviral activity against influenza A virus. Biochem. J. 412, 323–329 [DOI] [PubMed] [Google Scholar]
- 59. Hartshorn K. L., White M. R., Smith K., Sorensen G., Kuroki Y., Holmskov U., Head J., Crouch E. C. (2010) Increasing antiviral activity of surfactant protein D trimers by introducing residues from bovine serum collectins. Dissociation of mannan- binding and antiviral activity. Scand. J. Immunol. 72, 22–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shrive A. K., Tharia H. A., Strong P., Kishore U., Burns I., Rizkallah P. J., Reid K. B., Greenhough T. J. (2003) High resolution structural insights into ligand binding and immune cell recognition by human lung surfactant protein D. J. Mol. Biol. 331, 509–523 [DOI] [PubMed] [Google Scholar]
- 61. Liu W., Wei M. T., Tong Y., Tang F., Zhang L., Fang L., Yang H., Cao W. C. (2011) Seroprevalence and genetic characteristics of five subtypes of influenza A viruses in the Chinese pig population. A pooled data analysis. Vet. J. 187, 200–206 [DOI] [PubMed] [Google Scholar]
- 62. Hinshaw V. S., Webster R. G., Easterday B. C., Bean W. J., Jr. (1981) Replication of avian influenza A viruses in mammals. Infect. Immun. 34, 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Kida H., Ito T., Yasuda J., Shimizu Y., Itakura C., Shortridge K. F., Kawaoka Y., Webster R. G. (1994) Potential for transmission of avian influenza viruses to pigs. J. Gen. Virol. 75, 2183–2188 [DOI] [PubMed] [Google Scholar]
- 64. Van Reeth K. (2007) Avian and swine influenza viruses. Our current understanding of the zoonotic risk. Vet. Res. 38, 243–260 [DOI] [PubMed] [Google Scholar]
- 65. Brown I. H. (2000) The epidemiology and evolution of influenza viruses in pigs. Vet. Microbiol. 74, 29–46 [DOI] [PubMed] [Google Scholar]