Background: PML is a tumor suppressor involved in response to viral and genotoxic stress.
Results: Depletion of IL6 siRNA-mediated knockdown of STAT3 or NEMO suppresses PML gene expression.
Conclusion: PML is regulated via IL-6-dependent JAK-STAT3 and PI3K-NFκB signaling pathways in an autocrine/paracrine manner.
Significance: Paracrine regulation of PML gene expression is a part of tissue adaptation to local stress.
Keywords: Cancer Tumor Promoter, DNA-binding Protein, Protein Phosphorylation, Protein Translocation, Tyrosine Protein Kinase (Tyrosine Kinase), JAK/STAT3 and NFκB Pathway, NEMO, PML Gene Expression, Autocrine/Paracrine Signaling, Interleukin 6
Abstract
Tumor suppressor PML is induced under viral and genotoxic stresses by interferons and JAK-STAT signaling. However, the mechanism responsible for its cell type-specific regulation under non-stimulated conditions is poorly understood. To analyze the variation of PML expression, we utilized three human cell types, BJ fibroblasts and HeLa and U2OS cell lines, each with a distinct PML expression pattern. Analysis of JAK-STAT signaling in the three cell lines revealed differences in levels of activated STAT3 but not STAT1 correlating with PML mRNA and protein levels. RNAi-mediated knockdown of STAT3 decreased PML expression; both STAT3 level/activity and PML expression relied on IL6 secreted into culture media. We mapped the IL6-responsive sequence to an ISRE(−595/−628) element of the PML promoter. The PI3K/Akt/NFκB branch of IL6 signaling showed also cell-type dependence, being highest in BJ, intermediate in HeLa, and lowest in U2OS cells and correlated with IL6 secretion. RNAi-mediated knockdown of NEMO (NF-κ-B essential modulator), a key component of NFκB activation, suppressed NFκB targets LMP2 and IRF1 together with STAT3 and PML. Combined knockdown of STAT3 and NEMO did not further promote PML suppression, and it can be bypassed by exogenous IL6, indicating the NF-κB pathway acts upstream of JAK-STAT3 through induction of IL6. Our results indicate that the cell type-specific activity of IL6 signaling pathways governs PML expression under unperturbed growth conditions. As IL6 is induced in response to various viral and genotoxic stresses, this cytokine may regulate autocrine/paracrine induction of PML under these pathophysiological states as part of tissue adaptation to local stress.
Introduction
Promyelocytic (PML)4 nuclear bodies (NBs) are complex structures of mammalian nuclei comprising more than 100 proteins of various function including important cell cycle and cell fate regulators and factors involved in response to DNA damage (1). PML, a scaffold protein required for structural maintenance of PML NBs, is considered a tumor suppressor (for review, see e.g. Refs. 2 and 3), as loss of PML gene integrity by chromosomal translocation and gene fusion is linked to pathogenesis of acute promyelocytic leukemia (4) and mice with ablation of PML are tumor-prone (5). Importantly, PML expression is frequently deregulated in many human solid tumors (6–8). Early stages of epithelial tumors feature higher levels of PML than normal tissue cells, whereas advanced invasive tumor stages are associated with down-regulation of PML. Interestingly, Koken et al. (8) reported a high abundance of PML in tumor stroma regardless of tumor stage, especially in the vascular component. These expression patterns indicate decreased expression or loss of PML during acquisition of the invasive phenotype and involvement of a paracrine mechanism in PML induction (8). The latter notion is consistent with several studies showing that PML is inducible by cytokines, namely type I and type II interferons (9–12).
The mechanism of tumor-suppressive function of PML is not completely understood. In general, PML plays a role in cellular senescence and apoptosis (for reviews, see e.g. Refs. 13 and 14). Some effects of PML can be linked directly to PML protein itself, whereas others can be attributed to the function of PML nuclear compartment, which is built with the aid of PML tetramers. Soluble PML can bind to p53, a transcription factor mediating DNA damage response, senescence, and apoptosis, and facilitates acetylation, stabilization, and phosphorylation-mediated activation of p53 (15, 16). Moreover, as a direct transcriptional target of p53, PML is implicated in a positive feedback loop controlling p53 activity (17). Furthermore, PML and PML NBs cooperate with pRb in formation of chromatin-dense nuclear structures known as senescence-associated heterochromatin foci (18–21) observed in some forms of cellular senescence (22). Multiplication of PML NBs is observed in almost all types of cellular senescence (13, 15, 23–26). The elevation of PML in senescent cells is mediated at the transcription level (25) via activated Janus kinase/signal transducer and transcription activator (JAK/STAT) signaling (27) and/or post-translationally in some cell types (28).
Several groups including ours reported that various genotoxic stresses leading to activation of DNA damage response induce expression of a complex cytokine network (for reviews, see Refs. 29–31), which can also include type I and type II interferons (32). Activated JAK/STAT signaling accompanying lasting DNA damage response during drug-induced premature senescence contributes to multiplication of nuclear PML compartment through modulation of PML transcript level (33). Notably, PML NBs were found to quickly reassemble after DNA damage and to co-associate with persistent DNA lesions (33–35), implicating PML NBs in metabolism of damaged DNA (36). Importantly, persistent DNA damage response activity and development of cellular senescence is a feature characteristic for early stages of human tumorigenesis (37–41). Collectively, these findings suggest that under conditions of genotoxic stress the PML compartment is regulated at least in part in an autocrine/paracrine manner via secreted cytokines activating the JAK/STAT signaling pathway. Although we reported previously that JAK/STAT signaling (33) directly modulates PML transcription, the key cytokine responsible for PML activation was not determined due to a wide spectrum of cytokines produced by senescent cells.
In contrast to our understanding of PML gene induction during genotoxic stress, regulation of PML transcription under unstressed conditions is currently unclear. Despite that it has been known for almost two decades that various normal and malignant human cell types both in vitro and in vivo harbor variable numbers of PML nuclear bodies (42), the underlying mechanism responsible for such differences is unknown. In addition, the number of PML NBs noticeably differs even among individual cells in a given cell population (43) partly reflecting cell cycle dependence (8) or proliferative age (see e.g. Ref. 23).
In this study we address some of the open questions about PML regulation and show that expression of PML under unperturbed cell culture conditions is partially dependent on IL6, whose level of secretion is cell type-dependent. In general, IL6 is a functionally pleiotropic cytokine produced by many cell types in response to injury, inflammation, and infection. IL6 is an important regulator of cell proliferation and survival, and it is involved in regenerative and inflammatory processes (for review, see Ref. 44). Such functions of IL6 are mediated via its binding to membrane-bound or -soluble α-subunit IL6 receptor. IL6-IL6 receptor complexes then associate at the cellular membrane with two molecules of gp130 subunit followed by gp130 homodimer formation, which results in activation of kinases JAK1, JAK2, and TYK2. These events trigger engagement of phosphatase Src homology domains containing tyrosine phosphatase-2 (SHP-2) and subsequent initiation of signaling cascades including JAK/STAT-3, phosphoinositide 3-kinase (PI3K)-protein kinase B/Akt (PKB/Akt) and ras/raf/mitogen-activated protein kinase pathway (MAPK) (for review, see e.g. Refs. 44 and 45). Our present results also show that activities of both the JAK/STAT and PI3K/Akt pathways correlate with PML expression, which in turn is IL6-inducible. Furthermore, down-regulation of key components of either of the two signaling pathways leads to suppression of PML levels. We conclude that IL6 signaling represents an important determinant of regulation of the PML nuclear compartment.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used: mouse monoclonal antibody PG-M3 against PML (for immunofluorescence), rabbit polyclonal antibodies against PML (for immunoblots), STAT5, phosphotyrosine 694 of STAT5, STAT3 (clone C-20), NEMO, and p53 (all from Santa Cruz Biotechnology (Santa Cruz, CA)); mouse monoclonal antibody against phosphotyrosine 705 of STAT3, rabbit polyclonal antibodies against phosphotyrosine 701 of STAT1, phosphoserine 15 of p53, rabbit monoclonal antibodies against phosphoserine 473 of Akt and against total Akt (all from Cell Signaling Technology (Danvers, MA); mouse monoclonal antibody against GAPDH (GeneTEX, Irvine, CA); mouse monoclonal antibody against phosphoserine 139 of histone H2AX (Millipore, Billerica, MA); mouse monoclonal antibody against total STAT1 (SM2 clone, Exbio, Vestec, Czech Republic). For immunofluorescence, secondary antibodies anti-mouse IgG antibody conjugated with Cy3 (Jackson ImmunoResearch Laboratories, West Grove, PA) and anti-rabbit IgG antibody Alexa 488 (Invitrogen) were used.
Cell Cultures
Human cancer cell lines HeLa (cervix carcinoma), U2OS (osteosarcoma), and BJ normal human fibroblasts (at population doublings between 30 and 45) were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Cells were kept at 37 °C under 5% CO2 atmosphere and 95% humidity.
Indirect Immunofluorescence
Cells grown on glass coverslips were fixed by 4% formaldehyde and permeabilized by 0.1% Triton X-100 in two consecutive steps, each for 15 min at room temperature. After washing with PBS, cells were incubates in 10% FBS (diluted in PBS) for 30 min to block unspecific signal. After this step cells were incubated with diluted primary antibodies for 1 h at room temperature and then extensively washed with PBS, 0.1% Tween 20. The incubation with secondary antibodies was performed for 1 h at room temperature. To counterstain nuclei, coverslips were mounted in Mowiol containing 4′,6-diamidino-2-phenylindole (Sigma) and viewed by a fluorescence microscope (Leica DMRXA).
Quantitative Real Time RT-PCR (qRT-PCR)
Total RNA samples were isolated using the RNeasy Mini kit (Qiagen, MD) according to the manufacturer's protocol. First strand cDNA was synthesized from 200 ng of total RNA with random hexamer primers using TaqMan Reverse Transcription Reagents (Applied Biosystems). qRT-PCR was performed in ABI Prism 7300 (Applied Biosystems) using SYBR Green I Master Mix (Applied Biosystems) with the following set of primers: PML (designed to encompass PML exons common to all isoforms), 5′-CCG CAA GAC CAA CAA CAT CTT-3′, 5′-CAG CGG CTT GGA ACA TCC T-3′; actin, 5′-AGG CAC CAG GGC GTG AT-3′, 5′-TCG CCC ACA TAG GAA TCC TT-3′; STAT3, 5′-CTT TGA GAC CGA GGT GTA TCA CC-3′, 5′-GGT CAG CAT GTT GTA CCA CAG G-3′; NEMO, 5′-GGT GGA GCA CCT GAA GAG AT-3′, 5′-CAG AGC CTG GCA TTC CTT AG-3′; IL6, 5′-AGC CCT GAG AAA GGA GAC ATG TA-3′, 5′-TCT GCC AGT GCC TCT TTG C-3′; IRF1, 5′- AAA AGG AGC CAG ATC CCA AGA-3′, 5′- CAT CCG GTA CAC TCG CAC AG-3′; LMP2, 5′-TGT GCA CTC TCT GGT TCA GC-3′, 5′-GGA GGT TCC TCC AGT TCT ATC C-3′; CTNNB1, 5′-CAC AAG CAG AGT GCT GAA GGT G-3′, 5′-GAT TCC TGA GAG TCC AAA GAC AG-3′. The relative quantity of cDNA was estimated by ΔΔCt, and data were normalized to β-actin. Samples were measured in triplicate.
SDS-PAGE and Immunoblotting
Cells were harvested into Laemmli SDS sample lysis buffer, sonicated, and centrifuged at 16,000 × g for 10 min. Proteins concentration was estimated by the BCA method (Pierce). 100 mm DTT and 0.01% bromphenol was added to lysates before separation in polyacrylamide gels by SDS-PAGE (9% gels were used). The same protein amount (25 μg for BJ cells, 35 μg for other cell lines; in experiments with cell line comparison, 25 μg of protein was used) was loaded into each well. Proteins were electrotransferred onto a nitrocellulose membrane using wet transfer and detected by specific antibodies combined with horseradish peroxidase-conjugated secondary antibodies (goat anti-rabbit, goat anti-mouse; Bio-Rad). Peroxidase activity was detected by ECL (Pierce). GAPDH was used as a marker of equal loading.
Estimation of PML Promoter Activity
The Bluescript II SK+ plasmid containing the PML promoter fragment (−809/+633) in front of a luciferase reporter gene (PML 1.44-Luc) was a gift from H. de Thé (27). ΔISRE PML-Luc was made by deletion of a 34-bp fragment (−595/−628) containing the ISRE element using adjacent BglII and NcoI restriction sites. To measure luciferase activity, cells were seeded at 6 × 104 per well in 12-well plates 1 day before transfection. 450 ng of PML 1.44-Luc or ΔISRE PML-Luc with 50 ng of vector containing Renilla luciferase under thymidine kinase promoter (pRL-TK) were transfected to each well using FuGENE 6 (Roche Diagnostics). 24 h after transfection cell lysates were harvested according to the manufacturer's protocol. For the dual luciferase assay, luciferase activities were quantified with a luminometer (GloMax®-Multi Microplate Multimode Reader, Turner Biosystems, CA) using the Dual Luciferase Reporter Assay System (Promega, Madison, WI). The values are given as ratios of PML-Luc and pRL-TK luminescence (arbitrary units).
Determination of Cytokines in Cultivation Media
The conditioned medium from cells was collected 24 h after fresh medium was changed, and the numbers of cells per each dish were counted. The concentrations of IL6 and IL1β were estimated by a FACS bead array using FlowCytomix Human Simplex kit (IL6-BMS8213FF, IL1β-BMS8224FF; Bender MedSystems, Wien, Austria) on flow cytometer LSRII (BD Biosciences) according to the manufacturer's protocol.
Estimation of IL6 Biological Activity
To test the effectiveness of IL6 depletion mediated via IL6 antibody (2 μg/ml; goat polyclonal antibody; R&D Systems, Inc., Minneapolis, MN), growth dependence of mouse hybridoma B9 cells on the presence of IL6 was utilized (46). The conditioned media from BJ and HeLa cells incubated for 2 and 4 days, respectively, with IL6 antibody were transferred in a 1:1 dilution with fresh medium to mouse hybridoma B9 cells seeded in triplicate at a density of 25,000 cells/ml on 24-well plates. As positive or negative controls, B9 cells were cultivated with or without the addition of recombinant IL6 (100 pg/ml, Peprotech, NJ), respectively. 50-μl aliquots of B9 cell cultures were removed after 3 days, and cell growth and viability were measured after staining with Hoechst 33258 (Invitrogen) by flow cytometer (BD LSRII, BD Biosciences). To test for antibody toxicity in B9 cells, goat polyclonal antibody against HP1γ (Santa Cruz) was used at a concentration 2 μg/ml.
siRNA-mediated Gene Knockdown
Specific siRNAs were introduced into cells using Lipofectamine™ RNAiMAX (Invitrogen). Non-sense siRNA sequences were used as negative control siRNAs. siRNA was used are shown (non-sense siRNA sequences and siSTAT3 (Ambion); si NEMO (Dharmacon)): siSTAT3, #1 (5′-GGCUGGACAAUAUCAUUGAtt-3′), #2 (5′-GCCUCAAGAUUGACCUAGAtt-3′), #3 (5′-GCACCUUCCUGCUAAGAUUtt-3′); siNEMO, a mix of four siRNAs (no sequence available).
Chromatin Immunoprecipitation
Chromatin immunoprecipitation was performed as described previously (47, 48). Briefly, untreated BJ, HeLa, U2OS, and IL6-treated HeLa (5 ng/ml for 0, 0.5, 1, and 6 h) and U2OS (5 ng/ml for 48 h) were fixed (5 × 106 cells/sample) with 1% formaldehyde for 15 min, and the reaction was stopped with 0.125 m glycine. Cells were lysed in radioimmune precipitation cell lysis buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mm Tris, pH 8, 5 mm EDTA) for 10 min on ice. All buffers were supplemented with protease inhibitors (Complete EDTA-free, Roche Diagnostics). Samples were sonicated 33 times for 10 s with 50-s pause at 23% amplitude (Digital Sonifier 450, Branson Ultrasonics Corp.) in an ice bath. Protein concentration was adjusted to 1 mg/ml with radioimmune precipitation assay buffer. A part of each lysate was saved as a control reaction input. Lysates were precleared with protein A/G UltraLink Resin beads (Thermo Scientific) pre-equilibrated in radioimmune precipitation assay buffer. Precleared samples were then incubated with individual antibodies (STAT3, clone C-20, and STAT5; Santa Cruz) overnight at 4 °C followed by incubation with A/G UltraLink Resin beads for 3 h at 4 °C. The same total protein amount was used for each reaction. Immunocomplexes bound on beads were washed twice with radioimmune precipitation assay buffer, four times with LiCl buffer (100 mm Tris-HCl, pH 8.0, 1 mm EDTA, 500 mm LiCl, 0.5% Nonidet P-40, 0.5% sodium deoxycholate), and twice with TE buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA). Protein-DNA complexes were eluted with 0.1 m Tris-HCl, pH 8.0, 1 mm EDTA, and 1% SDS, de-cross-linked in the presence of 200 mm NaCl for 5 h at 65 °C, and then treated with proteinase K (20 μg/sample) for 30 min at 45 °C. DNA was extracted with phenol/chloroform, precipitated, and PCR-amplified. The following primers encompassing ISRE element in PML promoter were used: 5-TCAAGGGACTCAGCCAACTGG-3 and 5-GAGGCATGGTGGGCTCCT-3.
Statistical Analysis
Data were expressed as the means ± S.E. Analysis of variance was used for statistical evaluation of the data. p value<0.05 was expressed as significant difference.
RESULTS
PML NBs, PML Protein, and Transcript Level Are Modulated in a Paracrine Manner
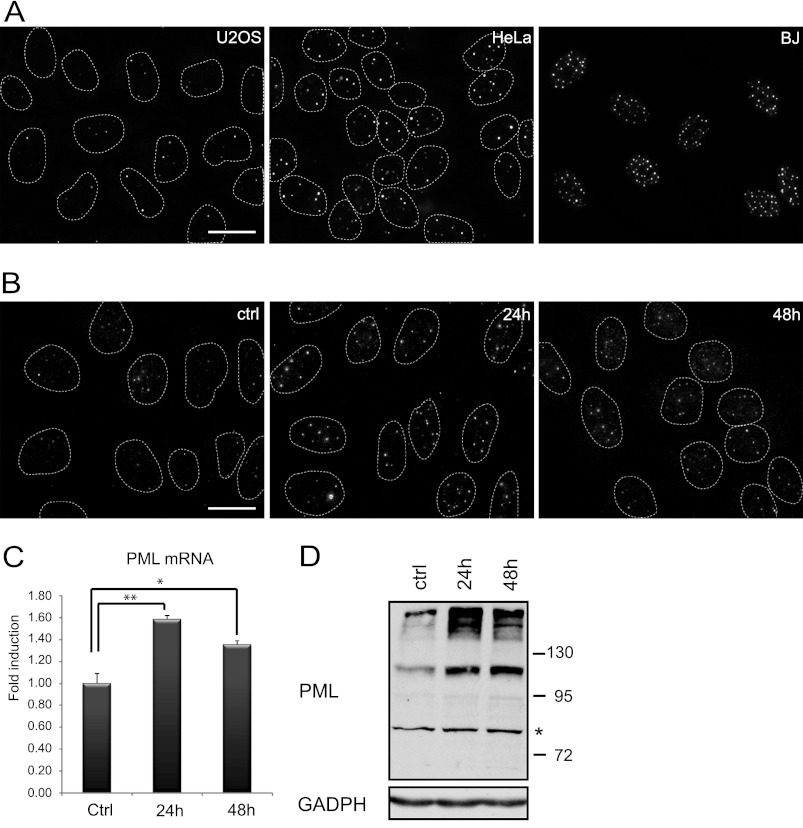
In our previous work we showed that PML gene transcription is regulated in an autocrine manner via cytokine-activated JAK/STAT signaling pathways in human normal and cancer cell lines exposed to various genotoxic compounds (33). To provide further insight into PML gene regulation, we asked whether autocrine/paracrine signaling determines PML levels also in naïve cells grown under unperturbed conditions in vitro. First, we selected three human cell types that feature low (U2OS cells, 3–4 PML NBs), medium (HeLa cells, 7–8 PML NBs), and high (BJ cells, PD 35, >30 PML NBs) numbers of PML NBs per nucleus, respectively, when grown under standard conditions (see Fig. 1A for PML NBs immunofluorescence staining). Next, we exposed U2OS cells with low content of PML NB to culture medium conditioned by BJ fibroblasts (population doubling 38) for 1 and 2 days and examined the number of PML NBs (Fig. 1B). We found an increase of the number of PML NBs in U2OS cells exposed to BJ-conditioned media. This intriguing result was further corroborated by the elevated PML mRNA examined by real time RT-qPCR (Fig. 1C) and elevated PML protein determined by immunoblotting (Fig. 1D). Both protein and mRNA levels of PML significantly increased in U2OS cells exposed to preconditioned media already after 24 h. These data indicate that the variation of basal PML gene expression seen in various cell types under normal cell culture conditions depends on signaling molecules secreted into culture media, consistent with the hypothesis that autocrine/paracrine mechanisms contribute to regulation of the PML compartment.
FIGURE 1.
PML and PML NBs are controlled by paracrine signaling. A, immunofluorescence detection of PML NBs in untreated U2OS, HeLa, and BJ cells cultured under standard conditions is shown. Immunofluorescence detection of PML NBs (B) and mRNA levels of PML (C) were quantified by qRT-PCR in U2OS cells treated 24 and 48 h with conditioned medium from BJ cells (diluted 1:1 with fresh medium). The values represent the average of two independent experiments performed in triplicate and are given as -fold induction of PML mRNA levels relative to control U2OS cells treated with conditioned medium from U2OS cells (diluted 1:1 with fresh medium); error bars represent S.E. Asterisks (*) and (**) represent p values <0.05 and <0.01, respectively. β-Actin was used as a reference gene. Bar, 15 μm. D, shown is immunoblot detection of PML in U2OS cells after 24 and 48 h of treatment with conditioned medium from BJ cells (diluted 1:1 with fresh medium). Bands with lower mobility than 130 kDa represent unmodified PML isoforms. A nonspecific band is marked by asterisk. GAPDH was used as a loading control.
Activity of IL6-JAK/STAT Signaling Is Cell Type-dependent and Correlates with PML Expression
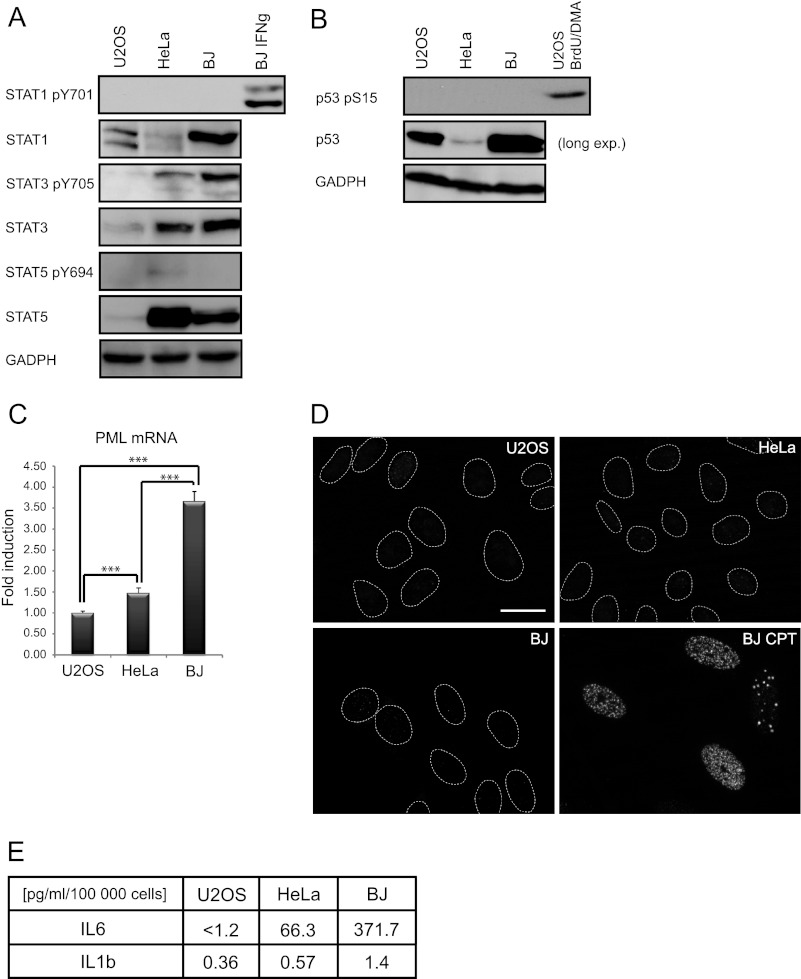
As reported previously, PML transcription is regulated by interferon-stimulated JAK/STAT signaling (27, 33). Therefore, we next compared the basal activity of JAK/STAT1/3/5 signaling in U2OS, HeLa, and BJ cells. The level of activated STAT1 (detected as STAT1 phosphorylated at tyrosine 701) was undetectable in all three cell types, yet it was inducible upon IFNγ treatment in all three cell types (Fig. 2A, only BJ is shown). The level of activated STAT5 (detected as STAT5-phosphorylated at tyrosine 694) was also almost undetectable in all three cell lines (Fig. 2A). In contrast, basal levels of activated STAT3 (phosphorylated at tyrosine 705) were detectable, highest in BJ, and lowest in U2OS cells (Fig. 2A), correlating with PML mRNA levels (Fig. 2C) and the number of PML NBs (Fig. 1A) in these cell types. Because p53 was reported as a candidate inducer of PML (17) and endogenous stress can activate p53, we assessed the basal level of the activated form of p53 (detected with antibody against p53 phosphorylated at serine 15). Serine 15-phosphorylated p53 was very low-to-undetectable in all three cell types (see Fig. 2B) consistent with low levels of endogenous DNA damage assessed by nuclear foci of serine 139-phosphorylated histone H2AX (γH2AX), a surrogate marker of DNA damage signaling (49), in contrast to elevated γH2AX in positive control cells treated by camptothecin (Fig. 2D). Interestingly, the concentration of the inductor of the JAK/STAT3 pathway, IL6, when measured in conditioned media of each of the three cell types was highest in BJ, lower in HeLa, and below the detection limit (1.2 pg/ml of assay used; see “Experimental Procedures”) in U2OS cells, correlating again with levels of activated STAT3 and the number of PML NBs (Fig. 2E). Intriguingly, IL6 levels correlated with IL1β, which was reported to induce IL6 (Ref. 50; Fig. 2E). Thus the positive correlation between expression of PML NBs and levels of secreted IL6 and phosphorylated STAT3 in different cell lines suggested a role of activated IL6-JAK/STAT3 signaling in regulation of basal PML gene expression.
FIGURE 2.
Cell type-dependent differences in IL6-STAT3 activity and expression of PML. A, shown is immunoblot detection of total STAT1, STAT3, STAT5, tyrosine 701-phosphorylated STAT1, tyrosine 705-phosphorylated STAT3, and tyrosine 694-phosphorylated STAT5 in U2OS, HeLa, and BJ cells cultivated at standard conditions and BJ exposed to IFNγ (10 IU/ml) for 24 h (included as a positive control of STAT1 activation); GAPDH was used as a loading control. B, shown is immunoblot detection of total and p53 serine 15-phosphorylated p53 in U2OS, HeLa, and BJ cultivated at standard conditions; U2OS cell treated with a combination of 10 μm BrdU and 10 μm distamycin A (BrdU/DMA) are included as positive control; GAPDH was used as a loading control. C, PML mRNA levels were detected by real time qRT-PCR in U2OS, HeLa, and BJ cells cultivated at standard conditions. The values represent the average of two independent experiments performed in triplicate and are given as the -fold induction of PML mRNA levels relative to U2OS; β-catenin (CTNNB1) was used as a reference gene. Asterisks (***) represent a p value <0.005. D, shown is immunofluorescence detection of endogenous level of γH2AX, a marker of DNA damage response, in U2OS, HeLa, and BJ cells. BJ treated for 4 days with camptothecin (BJ CPT) were used as a positive control of γH2AX formation. Bar, 15 μm. E, shown is an estimation of IL6 and IL1β in medium of U2OS, HeLa, and BJ cells measured by FACS beads.
Down-regulation of IL6/STAT3 Signaling Results in Suppression of PML Gene Expression
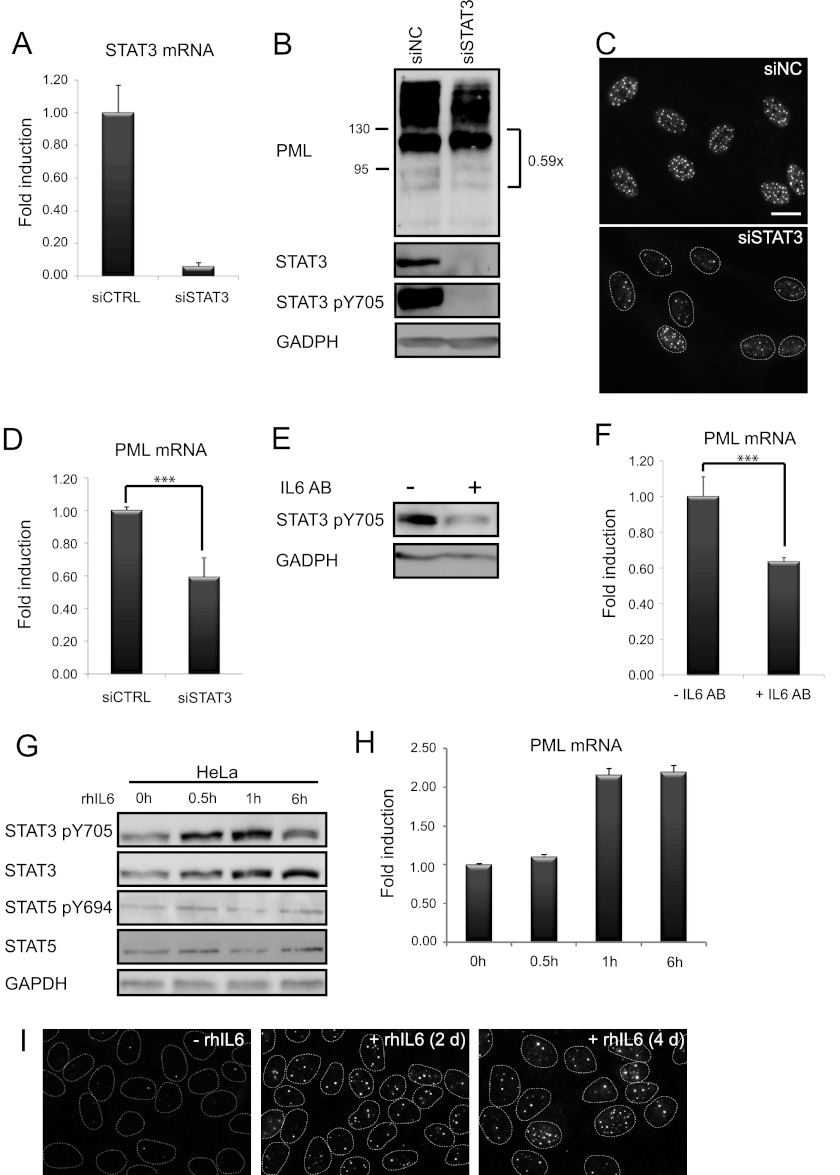
To test directly whether IL6-JAK/STAT3 signaling is involved in PML gene regulation, siRNA-mediated knockdown of STAT3 was performed in BJ cells, and the effect of such STAT3 depletion on PML expression was estimated. Three independent specific STAT3 siRNAs were tested (see supplemental Fig. 1A), and siSTAT3 #2 was selected for further experiments. STAT3 siRNA transfection resulted in almost complete knockdown of STAT3 at mRNA (>90%) and protein levels by 48 h after transfection (Fig. 3, A and B). This was accompanied by a substantial decrease of numbers of PML NBs (Fig. 3C) and PML protein levels (Fig. 3B) and suppression of PML mRNA (Fig. 3D). To analyze whether the effect of activated STAT3 on PML gene expression is mediated via secreted IL6, BJ-conditioned medium was depleted for IL6 with a specific antibody (51). The effectiveness of IL6 depletion was verified utilizing growth dependence of murine hybridoma B9 cells on human IL6 (46). Both recombinant human IL6 (rhIL6)-supplemented (100 pg/ml) and BJ-conditioned media stimulated proliferation of B9 cells, whereas IL6-depleted BJ-conditioned or rhIL6-negative (control) medium had no effect on growth of B9 cells (supplemental Fig. 1B). Substitution of IL6 antibody by a control-unrelated antibody (to HP1) had no effect on the ability of BJ-conditioned media to support growth of B9 cells, thereby excluding any potential cytostatic effects of neutralizing antibodies (supplemental Fig. 1B). In BJ cells, the level of activated STAT3 (Fig. 3E) as well as PML mRNA (Fig. 3F) was decreased after 2 days of cultivation in medium depleted of IL6. A similar effect of IL6 neutralization on PML level was reproduced in HeLa cells (see supplemental Fig. 1C). Likewise, HeLa conditioned medium but not its IL6-depleted form supported growth of B9 cells (supplemental Fig. 1D). Furthermore, stimulation of HeLa with exogenous rhIL6 (5000 pg/ml) led to increase of activated and total STAT3 levels after 30 min post-stimulation and persisted for at least additional 6 h (Fig. 3G). This was accompanied by elevation of PML transcript, the increased level of which was clearly observed after 1 h of IL6 treatment (Fig. 3H). Notably, the same treatment of U2OS cells led to a delayed response; the increase of activated and total STAT3 was not observable until 48 h of IL6 exposure (supplemental Fig. 1E) followed by elevation of PML mRNA transcript (supplemental Fig. 1F). Stimulation of U2OS and HeLa cells with IL6 also increased the number of PML NBs (Fig. 3I and data not shown). Altogether, these findings indicate that IL6 and STAT3 are involved in regulation of basal PML gene expression.
FIGURE 3.
IL6-STAT3 pathway modulates basal PML gene expression. A, shown is efficiency of STAT3 knockdown by specific siRNA detected on mRNA given as the -fold induction of PML mRNA levels relative to BJ transfected with control nonspecific siRNA (β-actin was used as a reference gene). B, shown is protein level by immunoblot detection of PML, total, and activated STAT3 in BJ cells 2 days after transfection. Ratios of major unmodified PML isoforms and GAPDH, estimated by densitometric analysis, are given on the right side of the PML immunoblot. Shown is down-regulation of PML after STAT3 knockdown, estimated at protein levels by immunofluorescence detection of PML NBs (C) and on mRNA levels by qRT-PCR in BJ cells (D) 2 days after transfection. The values represent the average of three independent experiments performed in triplicate and are given as -fold induction of PML mRNA levels relative to BJ transfected with control nonspecific siRNA. Bar, 15 μm. E, shown is down-regulation of STAT3 activity measured as tyrosine 705-phosphorylated STAT3 2 days after depletion of IL6 from medium by IL6 antibody (IL6 AB, 2 μg/ml ; antibody was at all times in the medium) in BJ cells; GAPDH was used as a loading control. F, down-regulation of PML mRNA levels was estimated by real time qRT-PCR in BJ cells 2 days after depletion of IL6 from medium by IL6 antibody (AB). The values represent the average of three independent experiments performed in triplicate and are given as -fold induction of PML mRNA levels relative to BJ without IL6 depletion; β-actin was used as a reference gene. Asterisks (***) represent p values < 0.005. G, time-course response of total and activated STAT3 and STAT5 levels was detected by immunoblotting as tyrosine 705-phosphorylated STAT3 and tyrosine 694-phosphorylated STAT5 in HeLa cells treated with IL6 (5000 pg/ml). H, time course response of PML mRNA levels in HeLa cells treated with IL6 (5000 pg/ml) estimated by RT-qPCR. The values represent the average of two independent experiments performed in triplicate and are given as -fold induction of PML mRNA levels relative to untreated HeLa cells. I, induction of PML protein levels was detected as PML NBs by indirect immunofluorescence in U2OS cells after a 2- and 4-day exposure to recombinant human IL6 (rhIL6; 1000 pg/ml). Bar, 15 μm.
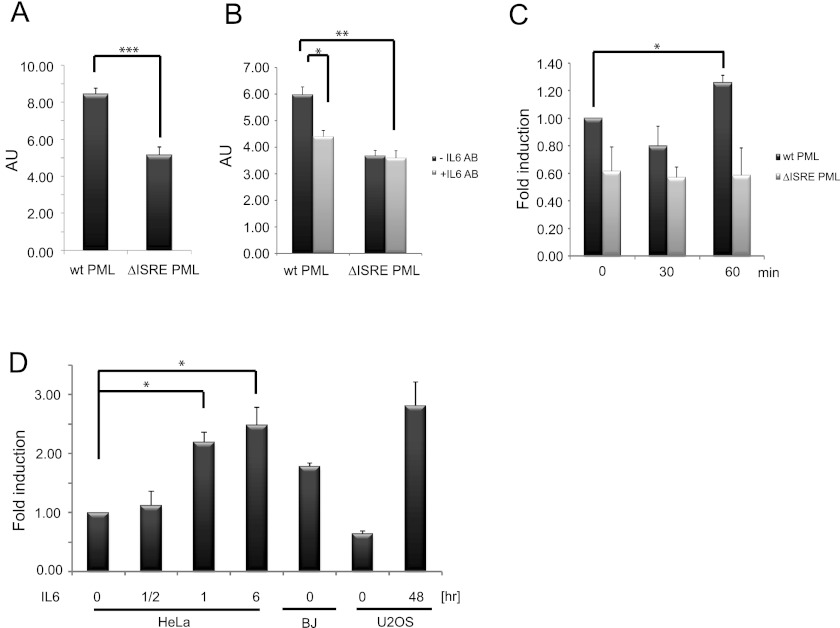
PML Gene Induction by IL6-STAT3 Is Regulated via an ISRE Element in the PML Gene Promoter
Previously, the main IFN-JAK/STAT signaling-responsive regulatory site of PML gene promoter has been mapped to a DNA binding element of the PML proximal promoter, ISRE (+605/+618; numbered relative to transcription start; Ref. 27). We found that deletion of the ISRE element within the 1.44-kbp HindIII fragment of the proximal PML gene promoter regions driving a luciferase reporter led to a significant decrease of basal reporter gene activity (Fig. 4A). Moreover, depletion of culture medium with the IL6 neutralizing antibody resulted in a decrease of luciferase reporter gene activity driven with promoter containing wild type ISRE element, whereas the reporter gene activity of construct with deleted ISRE element remained unchanged irrespective of IL6 depletion (Fig. 4B). On the other hand, the increase of luciferase activity in HeLa cells treated with IL6 was observed after 1 h for wild type ISRE but not for PML promoter with deleted ISRE (Fig. 4C). These results further support the role of the ISRE DNA binding site in response to IL6. To prove direct binding of STAT3 to human PML ISRE binding site in vivo, we performed chromatin immunoprecipitation (ChIP). As shown in Fig. 4D, treatment of HeLa (for 0.5, 1, and 6 h) and U2OS cells (48 h) with IL6 resulted in increased binding of STAT3 to the PML gene regulatory region-containing ISRE sequence in comparison to the non-stimulated state. No increase of STAT5 binding to PML ISRE was observed in HeLa cells treated with IL6 (supplemental Fig. 1, G and H). Note that STAT3 binding in unperturbed BJ, HeLa, and U2OS correlated with basal levels of PML expression in these cell types.
FIGURE 4.
ISRE element of PML promoter gene is involved in basal transcription of PML gene. Shown is luciferase reporter gene activity under the control of PML gene promoter (−809 bp/+633 bp relative to transcription start) in HeLa cells with an intact or deleted (−595 bp/−628 bp) ISRE DNA binding site (cells were harvested 24 h after transfection; the average values representing three independent experiments are given as arbitrary units (AU); error bars represent S.E.) (A), with or without IL6-depleting antibody (IL6 AB; 2 μg/ml; average values representing two independent experiments are given as arbitrary units; error bars represent S.E.) (B), and with or without IL6 treatment (5000 pg/ml; average values representing two independent experiments are given as -fold induction; error bars represent S.E.) (C). Asterisks (*, **, and ***) represent p values <0.05, <0.01, and <0.005, respectively. D, shown is evaluation of STAT3 binding to PML ISRE element in untreated BJ, HeLa, and U2OS cells and in HeLa and U2OS cells treated with IL6 for 0.5, 1, and 6 h in HeLa and 48 h in U2OS cells, respectively, quantified with RT-qPCR after chromatin immunoprecipitation. The values quantified with RT-qPCR represent average of two independent experiments performed in triplicate and are expressed as -fold induction relative to HeLa untreated cells.
Comparison of the PML ISRE sequence with the sequence of STAT3 consensus binding site derived from previously identified STAT3 binding sequences on STAT3-regulated genes, as listed in Table 3 in Ehret et al. (52), showed a good match (supplemental Fig. 1I). Collectively, these findings indicated that PML gene expression is directly regulated by an IL6-activated signaling pathway and STAT3 as transcription activator directly binding to the PML gene promoter.
NFκB Signaling Contributes to IL6-STAT3-mediated PML Expression
Besides the JAK/STAT3 pathway, IL6 stimulates the PI3K/Akt-PKB/ κB kinase/NFκB signaling cascade through activation of JAK kinases (53). Therefore, to investigate the mechanism controlling both PML gene expression and activity of the IL6-STAT3 pathway, we focused on NFκB signaling, as IL6 can regulate its own expression through a positive feedback loop involving NFκB (see e.g. Ref. 54). Notably, the levels of the active serine 473-phosphorylated form of Akt kinase were proportional to levels of IL6, activated STAT3, and PML in the three cell types tested (supplemental Fig. 2A). RNAi-mediated down-regulation of NEMO, the γ-subunit of IκB kinase required for kinase activity of the complex and activation of NFκB (55), depleted (>90%) NEMO mRNA (see Fig. 5, A and G, and supplemental Fig. 2E) and protein (Fig. 5B) in BJ cells 2 days after siRNA transfection. To assess the impact of NEMO knockdown, we estimated mRNA levels of known NFκB targets LMP2 and IRF1, both of which were decreased (supplemental Fig. 2, B and C). Importantly, we found that the levels of PML mRNA (Fig. 5C) and PML protein (Fig. 5B) were also decreased in such NEMO-depleted cells. The resulting decrease of PML protein on immunoblot was relatively modest, as expected from the long half-life of PML (>24 h; see Ref. 33). NEMO knockdown was also accompanied by a decrease of total and activated STAT3 (Fig. 5B). As IL6 is one of the transcriptional targets of NFκB, we estimated its transcript and protein levels after NEMO depletion. As expected, both IL6 mRNA (Fig. 5D) and protein (supplemental Fig. 2F) were decreased, consistent with reduction of NFκB activity and explaining the concomitant decrease of STAT3 activation. The effect of NEMO down-regulation on STAT3 activation and PML mRNA levels was bypassed by the addition of rhIL6 into culture medium during siRNA treatment (see “Experimental Procedures” for details; Fig. 5, E and F). However, the addition of rhIL6 had no significant effect on mRNA levels of NEMO itself and the known NFκB target IRF1 (Fig. 5, G and H). Thus, to test whether NFκB signaling acts upstream of or in parallel to IL6-STAT3 signaling to control PML gene expression, the double knockdown of STAT3 and NEMO in BJ cells was performed and analyzed. We observed no additive or synergistic effects of the combined STAT3/NEMO knockdown on the number of PML NBs and PML mRNA (Fig. 5, I and J; for knockdown efficiency, see supplemental Fig. 2, D and E), indicating that the NFκB pathway acts upstream of the IL6-STAT3 signaling module. We interpret these data as evidence for a role of NFκB signaling in control of a previously unrecognized IL6-dependent autocrine/paracrine loop that maintains activity of the IL6-STAT3 pathway and PML level.
FIGURE 5.
NFκB pathway controls PML expression through regulation of IL6 level. A, shown is efficiency of NEMO knockdown by specific siRNA detected on mRNA level 2 days after transfection in BJ. B, down-regulation of PML, NEMO, and the activated and total form of STAT3 in BJ detected on immunoblot 2 and 4 days after transfection of NEMO siRNA; GAPDH was used as loading control. Down-regulation of PML (D) and IL6 mRNA (D) levels quantified by qRT-PCR in BJ cells 2 days after NEMO knockdown is shown. The values represent the average of three independent experiments and are given as the -fold induction relative to BJ cells transfected with control nonspecific siRNA; error bars represent S.E. β-Actin was used as a reference gene. E, shown is immunoblot detection of the STAT3-activated form after NEMO knockdown and NEMO knockdown with the addition of recombinant human IL6 protein (5000 pg/ml) into medium of BJ cells. Shown is down-regulation of PML (F), NEMO (G), and IRF1 (H) mRNA levels 2 days after NEMO knockdown and NEMO knockdown with the addition of recombinant human IL6 protein (5000 pg/ml) quantified by real time qRT-PCR in BJ cells. The values represent the average of two independent experiments and are shown as -fold induction relative to BJ cells transfected with control nonspecific siRNA; error bars represent S.E.; β-actin was used as a reference gene. Shown is down-regulation of PML mRNA (I) and protein levels detected as PML NBs (J) 2 days after NEMO, STAT3, or combined NEMO and STAT3 knockdown by specific siRNAs in BJ cells. The mRNA values represent the average of two independent experiments and are shown as -fold induction relative to BJ cells transfected with control nonspecific siRNA; error bars represent S.E. β-Actin was used as a reference gene. Bar, 15 μm. Asterisks (*, **, and ***) represent p value <0.05, <0.01, and <0.005, respectively.
DISCUSSION
PML is a multifunctional protein that belongs to the family of interferon-stimulated genes. Expression of PML is stimulated by interferons of type I and II (9–12), which are produced during cellular responses to viral infection (for review, see Ref. 56) and some genotoxic insults (32, 33). Response of PML to both types of interferons is mediated via transcription factors STAT1 and STAT2 (27), and it is suppressed by inhibitors of histone deacetylases (47). Besides STAT1 and STAT2, overexpression of STAT5a led to elevation of PML NBs (57). In addition to this mode of transcriptional regulation, Shtutman et al. (58) showed that overexpression of β-catenin induces PML, indicating that Wnt signaling can participate in regulation of PML gene expression. Furthermore, De Stanchina et al. (17) reported that induction of PML during oncogenic Ras signaling depends on intact Arf and p53 and that PML is a direct target of p53. None of these studies, however, has addressed the mechanism of constitutive expression of this important antiviral and tumor suppressor protein. Here we show that the variation of PML content in different cell types propagated under standard cell culture conditions relies on constitutive activity of IL6-dependent signaling and thus is regulated in autocrine/paracrine manner. Analysis of two IL6-dependent signaling pathways, JAK/STAT3 and Akt/NFκB, by experimental manipulation of key components revealed responses of PML transcript and protein levels. Our data also indicate a functional cross-talk of the two pathways through a positive regulatory loop consisting of self-regulated expression of IL6 via Akt/NFκB (see the scheme supplemental Fig. 2G).
Although the role of IL6 as a signaling molecule that mobilizes organism-protective systems including innate and acquired immunity is well established, the precise functions of PML and PML NBs in cellular stress responses are not well understood. There is accumulating evidence that PML is involved in response to 1) viral infections and 2) genotoxic stress (for review, see Ref. 59). Importantly, both types of response result in induction of IL6, suggesting a potential physiologically significant link between IL6 signaling and PML.
PML and Viral Infections
Interferons are produced in virus-infected cells and regulate cellular antiviral responses via controlling expression of a large number of interferon-stimulated genes, which then interfere with critical processes of viral replication cycle. Importantly, overexpression of PML can establish resistance to infection by some viruses (such as RNA vesicular stomatitis virus and influenza virus) through preventing their replication by interference with viral mRNA and protein synthesis (60). The role of PML in protection against viral infection is indirectly supported also by the existence of some viruses that developed mechanisms to bypass (or even take advantage of) PML function (for review, e.g. see Ref. 61). Remarkably, infection of cells with certain (mainly DNA) viruses including herpes simplex virus type-1 (HSV-1) and human cytomegalovirus (HCMV) results in disruption of PML NBs (62), which is mediated by virally encoded proteins ICP0 of HSV-1 (63) and IE1 of HCMV (Ref. 64; for review, see Ref. 65), respectively. Both viral proteins ICP0 and IE1 reduce SUMOylation of PML, which is a prerequisite for PML NBs formation (66). A nucleus entering viral capsid proteins can sequester PML before disaggregation (67), and PML NBs disruption is linked to formation of viral DNA replication compartments containing PML liberated from PML NBs (68–71). Moreover, HSV-1 infection is accompanied by proteasome-dependent degradation of PML (72). Importantly, oncogenic viruses, such as hepatitis C virus HCV inactivate the PML tumor-suppressive pathway and thus block activation of p53 (73).
PML and Genotoxic Stress
Intriguingly, the common theme of response to viral infections and DNA damage is dynamic redistribution of PML NBs and associated proteins to sites of viral genomes and DNA damage, respectively (for review, see Ref. 59). The first indirect evidence of PML involvement in response to genotoxic stress came from the findings that NBS1 and MRE11, components of the Mre11-Rad50-NBS1 complex that senses, signals, and partly processes DNA double strand breaks, are present in PML NBs (74, 75). Lately, Carbone et al. (34) showed the increase of PML and PML NBs after ionizing irradiation accompanied by delayed but gradual colocalization of PML NBs with sites of persistent DNA breaks. The colocalization of PML NBs with late DNA damage foci was confirmed in other studies utilizing genotoxic drugs as DNA-damaging agents (33, 35) or bacterial toxins (26).5 As mentioned above, the common denominator of these two types of stresses (viral and genotoxic) is activation of a complex cytokine network including induction and secretion of IL6 (26, 32, 76–78). In our previous work we showed that both JAK/STAT1/2 and JAK/STAT3 signaling is involved in stress-induced PML gene transcription (32, 33). One of the key questions raised by those studies was which other cytokines in addition to interferons induce PML (13). Until now, however, the critical ligand activating the STAT3 pathway remained unidentified due to a wide spectrum of genotoxic stress-induced cytokines.
The IL6-STAT3 Pathway Controls Basal PML Gene Expression
With the goal to identify the mechanism of PML transcription under unperturbed cell culture conditions, we employed three cell types (BJ, HeLa, and U2OS) that differ in constitutive numbers of PML NBs, PML mRNA, and protein levels. Remarkably, medium conditioned with PML NB high content BJ cells induced PML mRNA, protein, and PML NBs in U2OS with low content PML NBs, indicating autocrine/paracrine signaling as a main mode of regulation of PML “basal” transcription. As we observed no differences in endogenous DNA damage response (determined as the activation of p53 and presence of γH2AX/53BP1 foci) or activity in stress signaling (as judged from the T180/Y182 activated form of p38MAPK; data not shown) under our culture conditions, we excluded the effect of activated p53 on PML transcription. However, when analyzing the activity of JAK/STAT1/2, JAK/STAT5, and JAK/STAT3 pathways, we noticed cell type-specific differences in the level of activated STAT3 but not STAT1 and STAT5. Moreover, we found marked cell type-dependent variation in the amount of STAT3-activating ligand IL6 secreted into culture media, which correlated well not only with activated STAT3 but also with numbers of PML NBs, PML protein, and mRNA levels. Importantly, both media supplemented with human recombinant IL6- or BJ cells-conditioned media containing IL6 resulted in induction of PML and PML NBs in U2OS or HeLa cells. In addition, both depletion of IL6 in culture medium with IL6 neutralizing antibody and siRNA-mediated knockdown of STAT3 led to a decrease of STAT3 active form, PML mRNA and PML NBs in BJ cells.
The responsiveness to IL6 can be modulated by the level of expression of its cognate receptor IL6 receptor and other components of this signaling pathway. This is supported by a kinetic study of STAT3 activation in HeLa and U2OS cells by IL6 that was accompanied by a proportional response of STAT3-dependent PML transcription in these two types of cells. Although the response of HeLa cells to exogenous IL6 was fast, as is expected for this type of signaling, there was a significant delay in STAT3 tyrosine 705 phosphorylation and related STAT3 and PML gene induction in U2OS cells. We suggest that the basis for such a difference is the initial “constitutive” level of STAT3 at the time of exogenous IL6 addition, being relatively high in HeLa but low in U2OS cells. Such effect can reflect the existence of positive feedback (see below), which contributes to accumulation of components of the signaling pathway thus progressively amplifying the cell response to an ongoing stimulus.
As shown previously, deletion of PML ISRE element led to a decrease of luciferase reporter gene activity in cells exposed to type I interferons (27). Our findings summarized on Fig. 4, including chromatin immunoprecipitation to prove direct binding of STAT3 to the PML gene regulatory region in vivo, indicate that IL6-mediated STAT3 signaling controls PML transcription through the same DNA binding element, which comprises a nucleotide sequence similar to the common consensus binding site of STAT transcription factors (52). In fact, the PML ISRE element resembles STAT3 consensus better than that of STAT1 (79) (see supplemental Fig. 1E). Altogether, our data indicate that IL6 controls PML gene expression under unperturbed cell culture conditions via JAK/STAT3 signaling and direct binding of STAT3 to the PML regulatory region.
Role of Akt-NFκB Signaling in IL6-mediated Induction of PML
The activation of IL6 receptor transmits signal to three pathways: (i) JAK/STAT3, (ii) PKB/Akt, and (iii) ras/raf/MAPK. Because IL6 gene expression is directly controlled via NFκB (80), we hypothesized that IL6 transcription and production is under IL6 self-control in a positive feedback loop via activation of the Akt-NFκB pathway. Indeed, the active form of kinase Akt correlated with secreted IL6 in the three cell types tested here, being highest in BJ and undetectable in U2OS. siRNA mediated knockdown of a key component of NFκB activation, the γ-subunit of IκB kinase NEMO, resulted in suppression of IL6 protein and mRNA levels and activated STAT3 and PML mRNA and protein levels. We cannot exclude the possibility that PML transcription is at least in part directly regulated by NFκB, as several putative NFκB binding sites are present in the proximal PML promoter. However, the fact that knockdown of NEMO and down-regulation of NFκB signaling (confirmed with decreased expression of known NFκB targets IRF1 and LMP2; (81, 82) can be bypassed by the addition of recombinant IL6 suggests that NFκB signaling is not acting in parallel but rather upstream of STAT3 signaling via modulation of IL6 production. Indeed, the combined knockdown of NEMO and STAT3 did not result in either additive or synergistic effects, thereby further supporting our conclusion that both modules operate in succession along the same signaling pathway.
The cell type-specific nature of variation in constitutive IL6 signaling and resulting PML expression has not been addressed here except for the observed correlations of the analyzed parameters. This is an interesting question that deserves further research to identify key component(s) of a negative feedback, which sets the basal level of the IL6 signaling circuitry. Whether this putative negative feedback acts on regulation of IL1β signaling (50) as a potential upstream stimulus of IL6 induction under normal conditions should be further investigated.
Conclusion
In this study we demonstrated that interleukin 6 (also called interferon β2) regulates basal PML transcription via JAK/STAT3 and Akt/NFκB signaling, thus broadening the spectrum of cytokines involved in PML gene expression. We anticipate that this mechanism participates in part in PML gene regulation also in scenarios when cells in tissues encounter viral or genotoxic stress, which are accompanied by IL6 induction. This can be the underlying mechanism of elevated PML observed in initial stages of several human tumors (8). Notably, initial but not advanced tumor stages contain senescent cells (38), and there is evidence that all forms of cellular senescence are accompanied by production of IL6 (26, 32, 76–78, 83). The paracrine effects of IL6 can also explain the elevation of PML and PML NBs in tumor stroma and tumor endothelial cells (8). Given the involvement of PML in cellular stress responses, we propose that such paracrine component of PML gene induction is a part of tissue adaptation to local stress, aiming to alarm and prepare the surrounding intact cells to face the challenge of incoming damage to avoid further spreading of injury.
This work was supported by Grant Agency of the Czech Republic (Project 204/08/1418), Institutional Grant (Project RVO 68378050), the Danish Cancer Society, the Danish National Research Foundation, the Czech Ministry of Education (MSM6198959216), Novo Nordisk (R153-A12997), and the European Commission 7th Framework Programme (Projects Infla-Care, Biomedreg and TRIREME).

This article contains supplemental Figs. 1 and 2.
S. Hubackova, K. Krejcikova, J. Bartek, and Z. Hodny, unpublished results.
- PML
- promyelocytic leukemia protein
- NBs
- nuclear body
- ISRE
- interferon-stimulated response element
- NFκB
- nuclear factor κB
- qRT-PCR
- quantitative real time RT-PCR
- rhIL6
- recombinant human IL6
- NEMO
- NF-κ-B essential modulator.
REFERENCES
- 1. Dellaire G., Bazett-Jones D. P. (2007) Beyond repair foci. Subnuclear domains and the cellular response to DNA damage. Cell Cycle 6, 1864–1872 [DOI] [PubMed] [Google Scholar]
- 2. Ruggero D., Wang Z. G., Pandolfi P. P. (2000) The puzzling multiple lives of PML and its role in the genesis of cancer. Bioessays 22, 827–835 [DOI] [PubMed] [Google Scholar]
- 3. Bernardi R., Pandolfi P. P. (2007) Structure, dynamics, and functions of promyelocytic leukaemia nuclear bodies. Nat. Rev. Mol. Cell Biol. 8, 1006–1016 [DOI] [PubMed] [Google Scholar]
- 4. de Thé H., Chomienne C., Lanotte M., Degos L., Dejean A. (1990) The t(15;17) translocation of acute promyelocytic leukaemia fuses the retinoic acid receptor α gene to a novel transcribed locus. Nature 347, 558–561 [DOI] [PubMed] [Google Scholar]
- 5. Wang Z. G., Delva L., Gaboli M., Rivi R., Giorgio M., Cordon-Cardo C., Grosveld F., Pandolfi P. P. (1998) Role of PML in cell growth and the retinoic acid pathway. Science 279, 1547–1551 [DOI] [PubMed] [Google Scholar]
- 6. Gurrieri C., Capodieci P., Bernardi R., Scaglioni P. P., Nafa K., Rush L. J., Verbel D. A., Cordon-Cardo C., Pandolfi P. P. (2004) Loss of the tumor suppressor PML in human cancers of multiple histologic origins J. Natl. Cancer Inst. 96, 269–279 [DOI] [PubMed] [Google Scholar]
- 7. Gambacorta M., Flenghi L., Fagioli M., Pileri S., Leoncini L., Bigerna B., Pacini R., Tanci L. N., Pasqualucci L., Ascani S., Mencarelli A., Liso A., Pelicci P. G., Falini B. (1996) Heterogeneous nuclear expression of the promyelocytic leukemia (PML) protein in normal and neoplastic human tissues. Am. J. Pathol. 149, 2023–2035 [PMC free article] [PubMed] [Google Scholar]
- 8. Koken M. H., Linares-Cruz G., Quignon F., Viron A., Chelbi-Alix M. K., Sobczak-Thépot J., Juhlin L., Degos L., Calvo F., de Thé H. (1995) The PML growth-suppressor has an altered expression in human oncogenesis. Oncogene 10, 1315–1324 [PubMed] [Google Scholar]
- 9. Lavau C., Marchio A., Fagioli M., Jansen J., Falini B., Lebon P., Grosveld F., Pandolfi P. P., Pelicci P. G., Dejean A. (1995) The acute promyelocytic leukaemia-associated PML gene is induced by interferon. Oncogene 11, 871–876 [PubMed] [Google Scholar]
- 10. Korioth F., Gieffers C., Maul G. G., Frey J. (1995) Molecular characterization of NDP52, a novel protein of the nuclear domain 10, which is redistributed upon virus infection and interferon treatment. J. Cell Biol. 130, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Der S. D., Zhou A., Williams B. R., Silverman R. H. (1998) Identification of genes differentially regulated by interferon α, β, or γ using oligonucleotide arrays. Proc. Natl. Acad. Sci. U.S.A. 95, 15623–15628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chelbi-Alix M. K., Pelicano L., Quignon F., Koken M. H., Venturini L., Stadler M., Pavlovic J., Degos L., de Thé H. (1995) Induction of the PML protein by interferons in normal and APL cells. Leukemia 9, 2027–2033 [PubMed] [Google Scholar]
- 13. Bourdeau V., Baudry D., Ferbeyre G. (2009) PML links aberrant cytokine signaling and oncogenic stress to cellular senescence Front. Biosci. 14, 475–485 [DOI] [PubMed] [Google Scholar]
- 14. Krieghoff-Henning E., Hofmann T. G. (2008) Role of nuclear bodies in apoptosis signaling. Biochim. Biophys. Acta 1783, 2185–2194 [DOI] [PubMed] [Google Scholar]
- 15. Pearson M., Carbone R., Sebastiani C., Cioce M., Fagioli M., Saito S., Higashimoto Y., Appella E., Minucci S., Pandolfi P. P., Pelicci P. G. (2000) PML regulates p53 acetylation and premature senescence induced by oncogenic Ras. Nature 406, 207–210 [DOI] [PubMed] [Google Scholar]
- 16. Louria-Hayon I., Grossman T., Sionov R. V., Alsheich O., Pandolfi P. P., Haupt Y. (2003) The promyelocytic leukemia protein protects p53 from Mdm2-mediated inhibition and degradation. J. Biol. Chem. 278, 33134–33141 [DOI] [PubMed] [Google Scholar]
- 17. de Stanchina E., Querido E., Narita M., Davuluri R. V., Pandolfi P. P., Ferbeyre G., Lowe S. W. (2004) PML is a direct p53 target that modulates p53 effector functions. Mol. Cell 13, 523–535 [DOI] [PubMed] [Google Scholar]
- 18. Narita M., Nũnez S., Heard E., Narita M., Lin A. W., Hearn S. A., Spector D. L., Hannon G. J., Lowe S. W. (2003) Rb-mediated heterochromatin formation and silencing of E2F target genes during cellular senescence. Cell 113, 703–716 [DOI] [PubMed] [Google Scholar]
- 19. Zhang R., Poustovoitov M. V., Ye X., Santos H. A., Chen W., Daganzo S. M., Erzberger J. P., Serebriiskii I. G., Canutescu A. A., Dunbrack R. L., Pehrson J. R., Berger J. M., Kaufman P. D., Adams P. D. (2005) Formation of MacroH2A-containing senescence-associated heterochromatin foci and senescence driven by ASF1a and HIRA. Dev. Cell 8, 19–30 [DOI] [PubMed] [Google Scholar]
- 20. Zhang R., Chen W., Adams P. D. (2007) Molecular dissection of formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27, 2343–2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ye X., Zerlanko B., Zhang R., Somaiah N., Lipinski M., Salomoni P., Adams P. D. (2007) Definition of pRB- and p53-dependent and -independent steps in HIRA/ASF1a-mediated formation of senescence-associated heterochromatin foci. Mol. Cell. Biol. 27, 2452–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kosar M., Bartkova J., Hubackova S., Hodny Z., Lukas J., Bartek J. (2011) Senescence-associated heterochromatin foci are dispensable for cellular senescence, occur in a cell type- and insult-dependent manner and follow expression of p16(ink4a). Cell Cycle 10, 457–468 [DOI] [PubMed] [Google Scholar]
- 23. Janderová-Rossmeislová L., Nováková Z., Vlasáková J., Philimonenko V., Hozák P., Hodný Z. (2007) PML protein association with specific nucleolar structures differs in normal, tumor and senescent human cells. J. Struct. Biol. 159, 56–70 [DOI] [PubMed] [Google Scholar]
- 24. Jiang W. Q., Ringertz N. (1997) Altered distribution of the promyelocytic leukemia-associated protein is associated with cellular senescence. Cell Growth Differ. 8, 513–522 [PubMed] [Google Scholar]
- 25. Ferbeyre G., de Stanchina E., Querido E., Baptiste N., Prives C., Lowe S. W. (2000) PML is induced by oncogenic ras and promotes premature senescence. Genes Dev. 14, 2015–2027 [PMC free article] [PubMed] [Google Scholar]
- 26. Blazkova H., Krejcikova K., Moudry P., Frisan T., Hodny Z., Bartek J. (2010) Bacterial intoxication evokes cellular senescence with persistent DNA damage and cytokine signaling. J. Cell. Mol. Med. 14, 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Stadler M., Chelbi-Alix M. K., Koken M. H., Venturini L., Lee C., Saïb A., Quignon F., Pelicano L., Guillemin M. C., Schindler C., (1995) Transcriptional induction of the PML growth suppressor gene by interferons is mediated through an ISRE and a GAS element. Oncogene 11, 2565–2573 [PubMed] [Google Scholar]
- 28. Scaglioni P. P., Yung T. M., Cai L. F., Erdjument-Bromage H., Kaufman A. J., Singh B., Teruya-Feldstein J., Tempst P., Pandolfi P. P. (2006) A CK2-dependent mechanism for degradation of the PML tumor suppressor. Cell 126, 269–283 [DOI] [PubMed] [Google Scholar]
- 29. Coppé J. P., Desprez P. Y., Krtolica A., Campisi J. (2010) The senescence-associated secretory phenotype. The dark side of tumor suppression. Annu. Rev. Pathol. 5, 99–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kuilman T., Peeper D. S. (2009) Senescence-messaging secretome, SMS-ing cellular stress. Nat. Rev. Cancer 9, 81–94 [DOI] [PubMed] [Google Scholar]
- 31. Young A. R., Narita M. (2009) SASP reflects senescence. EMBO Rep. 10, 228–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Novakova Z., Hubackova S., Kosar M., Janderova-Rossmeislova L., Dobrovolna J., Vasicova P., Vancurova M., Horejsi Z., Hozak P., Bartek J., Hodny Z. (2010) Cytokine expression and signaling in drug-induced cellular senescence. Oncogene 29, 273–284 [DOI] [PubMed] [Google Scholar]
- 33. Hubackova S., Novakova Z., Krejcikova K., Kosar M., Dobrovolna J., Duskova P., Hanzlikova H., Vancurova M., Barath P., Bartek J., Hodny Z. (2010) Regulation of the PML tumor suppressor in drug-induced senescence of human normal and cancer cells by JAK/STAT-mediated signaling. Cell Cycle 9, 3085–3099 [DOI] [PubMed] [Google Scholar]
- 34. Carbone R., Pearson M., Minucci S., Pelicci P. G. (2002) PML NBs associate with the hMre11 complex and p53 at sites of irradiation induced DNA damage. Oncogene 21, 1633–1640 [DOI] [PubMed] [Google Scholar]
- 35. Dellaire G., Kepkay R., Bazett-Jones D. P. (2009) High resolution imaging of changes in the structure and spatial organization of chromatin, γ-H2A.X, and the MRN complex within etoposide-induced DNA repair foci. Cell Cycle 8, 3750–3769 [DOI] [PubMed] [Google Scholar]
- 36. Dellaire G., Bazett-Jones D. P. (2004) PML nuclear bodies: dynamic sensors of DNA damage and cellular stress. Bioessays 26, 963–977 [DOI] [PubMed] [Google Scholar]
- 37. Bartkova J., Horejsí Z., Koed K., Krämer A., Tort F., Zieger K., Guldberg P., Sehested M., Nesland J. M., Lukas C., Ørntoft T., Lukas J., Bartek J. (2005) DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 [DOI] [PubMed] [Google Scholar]
- 38. Bartkova J., Rezaei N., Liontos M., Karakaidos P., Kletsas D., Issaeva N., Vassiliou L. V., Kolettas E., Niforou K., Zoumpourlis V. C., Takaoka M., Nakagawa H., Tort F., Fugger K., Johansson F., Sehested M., Andersen C. L., Dyrskjot L., Ørntoft T., Lukas J., Kittas C., Helleday T., Halazonetis T. D., Bartek J., Gorgoulis V. G. (2006) Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature 444, 633–637 [DOI] [PubMed] [Google Scholar]
- 39. Di Micco R., Fumagalli M., Cicalese A., Piccinin S., Gasparini P., Luise C., Schurra C., Garre' M., Nuciforo P. G., Bensimon A., Maestro R., Pelicci P. G., d'Adda di Fagagna F. (2006) Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature 444, 638–642 [DOI] [PubMed] [Google Scholar]
- 40. Gorgoulis V. G., Vassiliou L. V., Karakaidos P., Zacharatos P., Kotsinas A., Liloglou T., Venere M., Ditullio R. A., Jr., Kastrinakis N. G., Levy B., Kletsas D., Yoneta A., Herlyn M., Kittas C., Halazonetis T. D. (2005) Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature 434, 907–913 [DOI] [PubMed] [Google Scholar]
- 41. Bartek J., Lukas J., Bartkova J. (2007) DNA damage response as an anti-cancer barrier. Damage threshold and the concept of “conditional haploinsufficiency.” Cell Cycle 6, 2344–2347 [DOI] [PubMed] [Google Scholar]
- 42. Stuurman N., de Graaf A., Floore A., Josso A., Humbel B., de Jong L., van Driel R. (1992) A monoclonal antibody recognizing nuclear matrix-associated nuclear bodies. J. Cell Sci. 101, 773–784 [DOI] [PubMed] [Google Scholar]
- 43. Koken M. H., Puvion-Dutilleul F., Guillemin M. C., Viron A., Linares-Cruz G., Stuurman N., de Jong L., Szostecki C., Calvo F., Chomienne C. (1994) The t(15;17) translocation alters a nuclear body in a retinoic acid-reversible fashion. EMBO J. 13, 1073–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Scheller J., Chalaris A., Schmidt-Arras D., Rose-John S. (2011) The pro- and anti-inflammatory properties of the cytokine interleukin-6. Biochim. Biophys. Acta 1813, 878–888 [DOI] [PubMed] [Google Scholar]
- 45. Heinrich P. C., Behrmann I., Haan S., Hermanns H. M., Müller-Newen G., Schaper F. (2003) Principles of interleukin (IL)-6-type cytokine signaling and its regulation. Biochem. J. 374, 1–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Aarden L. A. (1989) Hybridoma growth factor. Ann. N.Y. Acad. Sci. 557, 192–198, discussion 198–199 [DOI] [PubMed] [Google Scholar]
- 47. Vlasáková J., Nováková Z., Rossmeislová L., Kahle M., Hozák P., Hodny Z. (2007) Histone deacetylase inhibitors suppress IFNα-induced up-regulation of promyelocytic leukemia protein. Blood 109, 1373–1380 [DOI] [PubMed] [Google Scholar]
- 48. Carey M., Peterson C. L., Smale S. T. (2008) Transcriptional Regulation in Eukaryotes: Concepts, Strategies, and Techniques, 2 Ed., pp. 132–139, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 49. Rogakou E. P., Pilch D. R., Orr A. H., Ivanova V. S., Bonner W. M. (1998) DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273, 5858–5868 [DOI] [PubMed] [Google Scholar]
- 50. Eda H., Burnette B. L., Shimada H., Hope H. R., Monahan J. B. (2011) Interleukin-1β-induced interleukin-6 production in A549 cells is mediated by both phosphatidylinositol 3-kinase and interleukin-1receptor-associated kinase-4. Cell Biol. Int. 35, 355–358 [DOI] [PubMed] [Google Scholar]
- 51. Walter M., Liang S., Ghosh S., Hornsby P. J., Li R. (2009) Interleukin 6 secreted from adipose stromal cells promotes migration and invasion of breast cancer cells. Oncogene 28, 2745–2755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Ehret G. B., Reichenbach P., Schindler U., Horvath C. M., Fritz S., Nabholz M., Bucher P. (2001) DNA binding specificity of different STAT proteins. Comparison of in vitro specificity with natural target sites. J. Biol. Chem. 276, 6675–6688 [DOI] [PubMed] [Google Scholar]
- 53. Takahashi-Tezuka M., Hibi M., Fujitani Y., Fukada T., Yamaguchi T., Hirano T. (1997) Tec tyrosine kinase links the cytokine receptors to PI 3-kinase probably through JAK. Oncogene 14, 2273–2282 [DOI] [PubMed] [Google Scholar]
- 54. Matsusaka T., Fujikawa K., Nishio Y., Mukaida N., Matsushima K., Kishimoto T., Akira S. (1993) Transcription factors NF-IL6 and NF-κB synergistically activate transcription of the inflammatory cytokines, interleukin 6, and interleukin 8. Proc. Natl. Acad. Sci. U.S.A. 90, 10193–10197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Tegethoff S., Behlke J., Scheidereit C. (2003) Tetrameric oligomerization of IκB kinase γ (IKKγ) is obligatory for IKK complex activity and NF-κB activation. Mol. Cell. Biol. 23, 2029–2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Biron C. A. (1998) Role of early cytokines, including α, and β interferons (IFN-α/β) in innate and adaptive immune responses to viral infections. Semin. Immunol. 10, 383–390 [DOI] [PubMed] [Google Scholar]
- 57. Mallette F. A., Gaumont-Leclerc M. F., Huot G., Ferbeyre G. (2007) Myc down-regulation as a mechanism to activate the Rb pathway in STAT5A-induced senescence. J. Biol. Chem. 282, 34938–34944 [DOI] [PubMed] [Google Scholar]
- 58. Shtutman M., Zhurinsky J., Oren M., Levina E., Ben-Ze'ev A. (2002) PML is a target gene of β-catenin and plakoglobin and coactivates β-catenin-mediated transcription. Cancer Res. 62, 5947–5954 [PubMed] [Google Scholar]
- 59. Everett R. D. (2006) Interactions between DNA viruses, ND10 and the DNA damage response. Cell. Microbiol. 8, 365–374 [DOI] [PubMed] [Google Scholar]
- 60. Chelbi-Alix M. K., Quignon F., Pelicano L., Koken M. H., de Thé H. (1998) Resistance to virus infection conferred by the interferon-induced promyelocytic leukemia protein. J. Virol. 72, 1043–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Regad T., Chelbi-Alix M. K. (2001) Role and fate of PML nuclear bodies in response to interferon and viral infections. Oncogene 20, 7274–7286 [DOI] [PubMed] [Google Scholar]
- 62. Maul G. G., Guldner H. H., Spivack J. G. (1993) Modification of discrete nuclear domains induced by herpes simplex virus type 1 immediate early gene 1 product (ICP0) J. Gen. Virol. 74, 2679–2690 [DOI] [PubMed] [Google Scholar]
- 63. Everett R. D., Maul G. G. (1994) HSV-1 IE protein Vmw110 causes redistribution of PML. EMBO J. 13, 5062–5069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Wilkinson G. W., Kelly C., Sinclair J. H., Rickards C. (1998) Disruption of PML-associated nuclear bodies mediated by the human cytomegalovirus major immediate early gene product. J. Gen. Virol. 79, 1233–1245 [DOI] [PubMed] [Google Scholar]
- 65. Sternsdorf T., Grötzinger T., Jensen K., Will H. (1997) Nuclear dots. Actors on many stages. Immunobiology 198, 307–331 [DOI] [PubMed] [Google Scholar]
- 66. Müller S., Dejean A. (1999) Viral immediate-early proteins abrogate the modification by SUMO-1 of PML and Sp100 proteins, correlating with nuclear body disruption. J. Virol. 73, 5137–5143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Puvion-Dutilleul F., Venturini L., Guillemin M. C., de Thé H., Puvion E. (1995) Sequestration of PML and Sp100 proteins in an intranuclear viral structure during herpes simplex virus type 1 infection. Exp. Cell Res. 221, 448–461 [DOI] [PubMed] [Google Scholar]
- 68. Burkham J., Coen D. M., Weller S. K. (1998) ND10 protein PML is recruited to herpes simplex virus type 1 prereplicative sites and replication compartments in the presence of viral DNA polymerase. J. Virol. 72, 10100–10107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Burkham J., Coen D. M., Hwang C. B., Weller S. K. (2001) Interactions of herpes simplex virus type 1 with ND10 and recruitment of PML to replication compartments. J. Virol. 75, 2353–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Everett R. D., Murray J. (2005) ND10 components relocate to sites associated with herpes simplex virus type 1 nucleoprotein complexes during virus infection. J. Virol. 79, 5078–5089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Everett R. D., Rechter S., Papior P., Tavalai N., Stamminger T., Orr A. (2006) PML contributes to a cellular mechanism of repression of herpes simplex virus type 1 infection that is inactivated by ICP0. J. Virol. 80, 7995–8005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Chelbi-Alix M. K., de Thé H. (1999) Herpes virus induced proteasome-dependent degradation of the nuclear body-associated PML and Sp100 proteins. Oncogene 18, 935–941 [DOI] [PubMed] [Google Scholar]
- 73. Herzer K., Weyer S., Krammer P. H., Galle P. R., Hofmann T. G. (2005) Hepatitis C virus core protein inhibits tumor suppressor protein promyelocytic leukemia function in human hepatoma cells. Cancer Res. 65, 10830–10837 [DOI] [PubMed] [Google Scholar]
- 74. Lombard D. B., Guarente L. (2000) Nijmegen breakage syndrome disease protein and MRE11 at PML nuclear bodies and meiotic telomeres. Cancer Res. 60, 2331–2334 [PubMed] [Google Scholar]
- 75. Mirzoeva O. K., Petrini J. H. (2001) DNA damage-dependent nuclear dynamics of the Mre11 complex. Mol. Cell. Biol. 21, 281–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Kuilman T., Michaloglou C., Vredeveld L. C., Douma S., van Doorn R., Desmet C. J., Aarden L. A., Mooi W. J., Peeper D. S. (2008) Oncogene-induced senescence relayed by an interleukin-dependent inflammatory network. Cell 133, 1019–1031 [DOI] [PubMed] [Google Scholar]
- 77. Acosta J. C., O'Loghlen A., Banito A., Guijarro M. V., Augert A., Raguz S., Fumagalli M., Da Costa M., Brown C., Popov N., Takatsu Y., Melamed J., d'Adda di Fagagna F., Bernard D., Hernando E., Gil J. (2008) Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell 133, 1006–1018 [DOI] [PubMed] [Google Scholar]
- 78. Rodier F., Coppé J. P., Patil C. K., Hoeijmakers W. A., Muñoz D. P., Raza S. R., Freund A., Campeau E., Davalos A. R., Campisi J. (2009) Persistent DNA damage signaling triggers senescence-associated inflammatory cytokine secretion. Nat. Cell Biol. 11, 973–979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Horvath C. M., Wen Z., Darnell J. E., Jr. (1995) A STAT protein domain that determines DNA sequence recognition suggests a novel DNA-binding domain. Genes Dev. 9, 984–994 [DOI] [PubMed] [Google Scholar]
- 80. Libermann T. A., Baltimore D. (1990) Activation of interleukin-6 gene expression through the NF-κB transcription factor. Mol. Cell. Biol. 10, 2327–2334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rein T., Müller M., Zorbas H. (1994) In vivo footprinting of the IRF-1 promoter. Inducible occupation of a GAS element next to a persistent structural alteration of the DNA. Nucleic Acids Res. 22, 3033–3037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Wright K. L., White L. C., Kelly A., Beck S., Trowsdale J., Ting J. P. (1995) Coordinate regulation of the human TAP1 and LMP2 genes from a shared bidirectional promoter. J. Exp. Med. 181, 1459–1471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Minamino T., Yoshida T., Tateno K., Miyauchi H., Zou Y., Toko H., Komuro I. (2003) Ras induces vascular smooth muscle cell senescence and inflammation in human atherosclerosis. Circulation 108, 2264–2269 [DOI] [PubMed] [Google Scholar]