Background: The function of heme o synthase (Cox10) is linked to its oligomerization, a process coupled to Cox1 synthesis.
Results: The C-terminal segment of Cox1 and Coa2 mediates Cox10 oligomerization.
Conclusion: Coa2 is a key factor that mediates multimerization of both Cox10 and Cox15.
Significance: Novel insights on Cox10 function and mechanism of disease-causing dysfunction are provided.
Keywords: Cytochrome Oxidase, Heme, Mitochondria, Mitochondrial Diseases, Respiratory Chain, Coa1, Coa2, Cox10, Cox15
Abstract
The synthesis of the heme a cofactor used in cytochrome c oxidase (CcO) is dependent on the sequential action of heme o synthase (Cox10) and heme a synthase (Cox15). The active state of Cox10 appears to be a homo-oligomeric complex, and formation of this complex is dependent on the newly synthesized CcO subunit Cox1 and the presence of an early Cox1 assembly intermediate. Cox10 multimerization is triggered by progression of Cox1 from the early assembly intermediate to downstream intermediates. The CcO assembly factor Coa2 appears important in coupling the presence of newly synthesized Cox1 to Cox10 oligomerization. Cells lacking Coa2 are impaired in Cox10 complex formation as well as the formation of a high mass Cox15 complex. Increasing Cox1 synthesis in coa2Δ cells restores respiratory function if Cox10 protein levels are elevated. The C-terminal segment of Cox1 is important in triggering Cox10 oligomerization. Expression of the C-terminal 54 residues of Cox1 appended to a heterologous matrix protein leads to efficient Cox10 complex formation in coa2Δ cells, but it fails to induce Cox15 complex formation. The state of Cox10 was evaluated in mutants, which predispose human patients to CcO deficiency and the neurological disorder Leigh syndrome. The presence of the D336V mutation in the yeast Cox10 backbone results in a catalytically inactive enzyme that is fully competent to oligomerize. Thus, Cox10 oligomerization and catalytic activation are separate processes and can be uncoupled.
Introduction
The terminal oxidase in mitochondrial cytochrome c oxidase (CcO)3 reduces oxygen to water by electrons arising from the oxidation of NADH and FADH2. CcO contains four prosthetic groups involved in electron transfer, and these include a binuclear copper center designated CuA, an isolated heme a moiety, and a heterobimetallic copper-heme a center designated CuB-heme a3. Heme a is a modified protoheme cofactor uniquely used by CcO. Heme a is generated by two enzymes, heme o and heme a synthases, localized within the mitochondrial inner membrane (1). The heme o synthase, designated Cox10 in yeast, transfers a farnesyl diphosphate to a vinyl group of protoheme generating the hydroxyethyl-farnesyl heme o intermediate that is subsequently oxidized at a pyrrole ring methyl group to a formyl substituent by the heme a synthase, designated Cox15 (2). The two heme a moieties along with the CuB ion are inserted into the mitochondrially encoded Cox1 subunit at an early step in CcO biogenesis (3). The CuA site is formed in a second mitochondrially encoded subunit Cox2.
The synthesis of heme a is likely regulated to minimize excess free heme. The lack of any heme degradation pathway within mitochondria and the known toxicity of free heme suggest that heme a formation is coupled to CcO biogenesis. Cox10 is likely the rate-limiting enzyme in heme a formation, because Cox15 is present in excess (∼8-fold) over Cox10 in protein abundance (4).
Yeast Cox10 is a 46-kDa intrinsic membrane protein with 8–9 predicted transmembrane helices (5). We demonstrated previously that Cox10 assembles in a multimeric complex. Cells containing different epitope tags on distinct COX10 loci exhibit homotypic interaction suggesting that the multimeric complex may represent a homo-oligomer (6). The second enzyme in the heme a biosynthetic pathway, Cox15, forms a multimeric complex distinct from the Cox10 complex, and neither the Cox10 nor Cox15 multimer shows an apparent interaction with Cox1 (6).
Insights into the yeast Cox10 were gleaned by studies on CcO-deficient coa2Δ cells. These mutant cells exhibit a marked attenuation in Cox1; Cox1 is translated but is rapidly degraded yielding a Cox1 deficiency (7). The mutant phenotype is suppressed by a specific mutation in Cox10 resulting in a gain-of-function N196K substitution (6).
Cox10 oligomerization is impaired in coa2Δ cells, yet the steady-state level of Cox10 is unchanged. The presence of the N196K allele restores Cox10 multimerization and results in a marked enhancement in the abundance of the high mass complex relative to the WT Cox10 complex. Comparing amino acid substitutions at position 196, we showed that the suppressor activity of Cox10 correlates with the abundance of the multimeric Cox10 complex (6). Because suppressor activity is dependent on catalytic activity, the observations are consistent with the catalytically active state of Cox10 being the oligomeric complex.
A second mechanism by which respiratory competence is achieved in coa2Δ cells is through the depletion of the Oma1 metalloproteinase (6, 8). The restoration of respiratory growth in coa2Δ cells by either the presence of a gain-of-function Cox10 allele or impaired Cox1 degradation by Oma1 led us to postulate that Coa2 is an important assembly factor in Cox1 hemylation, most likely in the single low spin heme a subsite. Hemylation of Cox1 in coa2Δ cells may be achieved either by the presence of a more efficient Cox10 enzyme or impeding Cox1 degradation enabling more time for the inefficient Coa2-independent hemylation process.
The stability of the high mass Cox10 complex correlates with newly synthesized Cox1. In addition, formation of the initial Cox1 assembly intermediate consisting of Mss51, Coa3, Cox14, and Ssc1 (9–12) is important for Cox10 oligomerization (6). In these mutant cells, steady-state levels of Cox1 are markedly attenuated, but Cox1 synthesis proceeds normally.
A schematic of Cox1 maturation with respect to Coa2 and Cox10 is shown in Fig. 1. We postulated that progression of Cox1 from the Mss51 assembly intermediate mediates the oligomerization and activation of Cox10.
FIGURE 1.

Model for involvement of Coa2, Cox10, and Cox15 in Cox1 maturation. Newly synthesized Cox1 is initially trapped in the Mss51/Cox14/Coa3/Ssc1 intermediate. Coa1, presumably in concert with Coa2, triggers further progression of Cox1 to the state where it is competent to receive heme a moieties. Such progression is coupled to Cox10 oligomerization. Population of the heme a and a3 sites culminates with the involvement of Shy1 and addition of Cox5a and Cox6 subunits. The C-terminal segment of Cox1 used to trigger Cox10 oligomerization is shown as C-term.
The goal of this study was to deduce how Cox10 senses the availability or assembly state of Cox1 for its activation. To address this question, we used coa2Δ cells, which are compromised in Cox1 hemylation. We demonstrate that Cox10 oligomerization can be triggered in coa2Δ cells by either a combination of increased levels of WT Cox10 and enhanced Cox1 synthesis or the presence of an ectopic Cox1 domain consisting of the C-terminal 54 residues. However, only the combined enhanced Cox1 synthesis and elevated Cox10 condition restore respiratory growth. We also report on the functional effects of human Cox10 mutations that predispose human patients to CcO deficiency and the progressive neurological disorder Leigh syndrome.
MATERIALS AND METHODS
Strains and Growth Media
Saccharomyces cerevisiae strains used in this study are listed in Table 1. Yeast cells were cultured in either YP (1% yeast extract, 2% bactopeptone) or SC minimal media supplemented with appropriate nutrients. Either 1 or 2% glucose, 2% galactose, or glycerol/lactate were used as a sole carbon source. The chromosomal loci of the respective genes in yeast cells were either tagged with a 13× Myc epitope tag at the 3′ position or disrupted by homologous recombination as described previously (13). All generated strains were confirmed by PCR. Escherichia coli DH5α cells, used for cloning and plasmid propagation, were handled as described previously (14).
TABLE 1.
Yeast strains used in this work
Vectors and Constructs
To generate m-hSod1-C54, a 165-bp fragment encoding 54 C-terminal amino acids of Cox1 was PCR-amplified from total yeast DNA with 5′-GGTGTAATTGGGATCGCCCAAAACAATAAAGTTAATAATAAATCA-3′ and 5′-TGATTTATTATTAACTTTATTGTTTTGGGCGATCCCAATTACACC-3′ primers. Cloning of this segment of COX1 did not require any recoding for compatibility with cytosolic translation. Obtained fragment was appended to the 3′-end of matrix-targeted hSOD1, amplified from pRS423-m-hSOD1 plasmid (15) by overlap extension PCR with 5′-TATTTAGGATCCATGTTCGCGAAAACAGCAGCTGCTAATTTA-3′ and 5′-TATTTACTGCAGTTAAGATTGTACAGCTGGTGTATTAAATGA-3′. BamHI and PstI restriction sites were introduced at 5′- and 3′-ends of the chimera. The resulting 710-bp construct (pre-Sod2-hSod1-Cox1) was cloned into either pRS415 or pRS425 vectors under the control of the MET25 promoter and CYC1 terminator using the aforementioned restriction sites. The T188K, P217L, E328G, and E328V point substitutions in pRS416-COX10-13×Myc or pRS426-COX10-13×Myc (6) were generated by site-directed mutagenesis using the QuikChange kit (Stratagene). Successive rounds of site-specific mutagenesis created the T188K/P217L, T188K/N196K, and E328V/N196K double mutations. All constructs were verified by sequencing. The earlier described plasmids pRS426-MSS51, YEp352-MSS51, YEpLac112-MSS51, pRS426-SSC1, pRS425-COA2 (7, 10, 13), pRS426-COX10-13×Myc, pRS426-COX10-His6, and pRS416-COX10-His6 N196K (6) have also been used in this study. Plasmids were transformed into the yeast cells using lithium acetate procedure (16).
Mitochondrial Isolation and Assays
Intact mitochondria were isolated from yeast cells as described (17). Mitochondrial protein concentrations were quantified by the Bradford assay (18). Specific CcO enzymatic activity was determined as described (19), normalized to mitochondrial protein levels, and presented as a percentage of wild-type activity. Separation of the intact mitochondrial protein complexes by blue native-PAGE (BN-PAGE) was performed as before (3). Mitochondria were lysed in 1% digitonin, and solubilized protein complexes were resolved on a continuous 5–13% gradient gel, transferred onto polyvinylidene difluoride (PVDF) membrane, and analyzed by immunoblotting. For assessment of the steady-state protein levels, either entire mitochondria or clarified mitochondrial lysates were loaded onto a denaturing 12% polyacrylamide gel, subjected to SDS-PAGE, and transferred onto a nitrocellulose membrane.
In Vivo Labeling of the Mitochondrial Translation Products
Yeast cells were pre-cultured overnight in either complete or supplemented SC medium containing 2% galactose, back-diluted, and grown to an A600 of 0.8. The labeling, preparation, and separation of the samples by SDS-PAGE were done as described previously (20). The gel was dried, and separated radiolabeled proteins were visualized by autoradiography.
Immunoassays
Proteins were detected with indicated primary antibodies and visualized with horseradish peroxidase-conjugated secondary antibodies and ECL reagents (Millipore). Anti-Myc antibody was from Roche Diagnostics. Antibodies to the mitochondrial outer membrane porin were from Invitrogen; the Cox1, Cox2, and Cox3 subunits of CcO were from Mitosciences. Dr. Alex Tzagoloff kindly provided Atp2 (F1) antiserum. Anti-hSOD1 serum was purchased from Santa Cruz Biotechnology.
Miscellaneous
Sensitivities of the yeast strains and transformants to hydrogen peroxide were tested as described previously (21). Bioinformatic analysis of protein sequences was done using TMpred, MultAlin (22), and BoxShade software.
RESULTS
Effects of Cox1 Synthesis on Cox10 Oligomerization
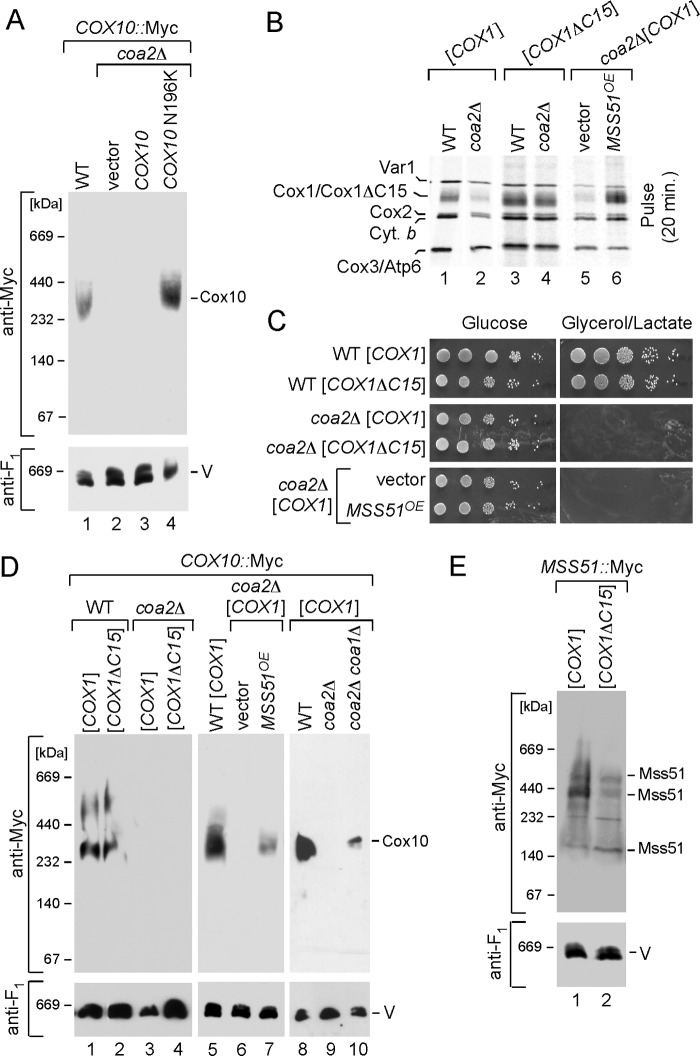
As mentioned, Cox10 oligomerization was markedly attenuated in coa2Δ cells (Fig. 2A) (6). The presence of the N196K mutant allele of Cox10 restores oligomerization in coa2Δ cells. The inability of Cox10 to form the complex in coa2Δ cells is not related to Cox10 protein stability, because steady-state levels of Cox10 are normal in the mutant cells (6). The facile degradation of Cox1 observed in coa2Δ cells suggested that the absence of Cox1 multimerization might be due to the low levels of Cox1. To address the role of Cox1 in the Cox10 oligomerization process, we tested the effects of enhanced Cox1 synthesis in coa2Δ cells.
FIGURE 2.
Cox10 oligomeric complex is attenuated in coa2Δ mitochondria and can be partially restored by increased Cox1 synthesis. A, mitochondria (30 μg) from the wild-type (WT) cells with an endogenously tagged 13×Myc COX10 gene or its Coa2-deficient derivative, expressing either Cox10 or Cox10 N196K mutant, were solubilized in the buffer containing 1% digitonin. Clarified lysates were loaded onto 5–13% continuous gradient gel, and protein complexes were separated under native conditions. Cox10-13×Myc complexes were visualized by immunoblotting with anti-Myc antibodies. The monomeric complex V detected with anti-Atp2 (anti-F1) antiserum served as a loading control. B, in vivo labeling of mitochondrial translation products. COX10::13×Myc WT or coa2Δ cells with either full-length or truncated Cox1 or coa2Δ cells expressing MSS51 were pulsed with [35S]methionine for 20 min at 30 °C. The samples were subjected to 12% SDS-PAGE, and the gel containing separated labeled polypeptides was dried and analyzed by autoradiography. C, respiratory growth of the strains described for B. Cells were pre-grown in synthetic supplemented medium containing 2% galactose and 0.1% glucose, serially diluted and spotted onto plates containing either 2% glucose or glycerol/lactate as a sole carbon source. Pictures were taken after 2 (for glucose plates) and 4 (for glycerol-lactate plates) days of incubation at 30 °C. D, BN-PAGE analysis of the strains described in B and C as well as WT, coa2Δ, coa1Δ, and coa2Δ coa1Δ cells with COX10::13×Myc chromosomal tag was performed as in A. E, mitochondria derived from MSS51::13×Myc WT strains with either full-length or truncated COX1 were solubilized in 1% digitonin and analyzed by native electrophoresis.
Many CcO assembly mutants exhibit reduced levels of Cox1 synthesis due to sequestration of the Mss51 translational activator of Cox1 in a stalled Cox1-containing complex containing Mss51, Cox14, Coa3, and Ssc1 (Fig. 1) (9–12). Sequestration of Mss51 within this early Cox1 assembly intermediate results in insufficient levels of free Mss51 to stimulate Cox1 translation. This attenuation in Cox1 synthesis can be overcome by one of four ways as follows: 1) overexpression of Mss51; 2) depletion of proteins that form the high mass Mss51 complex (Cox14 or Coa3) resulting in more Mss51 available for translational initiation of Cox1; 3) depletion of Coa1 that interacts with Cox1 downstream of Mss51, or 4) using a Cox1 mutant strain lacking its C-terminal 15 residues that destabilizes the Mss51-containing Cox1 complex (10, 12, 23).
Cox1 synthesis was monitored by an in vivo mitochondrial translation assay. As expected, the levels of newly synthesized Cox1 in coa2Δ cells are enhanced by either Mss51 overexpression or in cells containing the truncated Cox1 (Fig. 2B, compare lanes 4 and 6) (23). However, the increased levels of newly synthesized Cox1 failed to restore respiratory growth in coa2Δ cells (Fig. 2C). Likewise, deletion of COA1 in coa2Δ cells increases Cox1 synthesis, yet the mutant cells fail to respire (7).
To assess whether the enhanced levels of newly synthesized Cox1 were sufficient to trigger Cox10 oligomerization, coa2Δ cells harboring episomal MSS51, lacking Coa1, or containing the COX1 truncation mutant along with a Myc epitope-tagged Cox10 allele were isolated, and mitochondria derived from these cells were used in BN-PAGE. High levels of Mss51 or a deletion of COA1 led to a weak stabilization of oligomerized Cox10 (Fig. 2D, lane 7), although neither condition was sufficient to restore respiratory growth. However, no Cox10 complex formation was observed in coa2Δ with the C-terminal truncated Cox1 (Fig. 2D, lane 4).
The failure of the elevated Cox1 in the ΔC15-truncated Cox1 cells lacking Coa2 to stimulate Cox10 oligomerization, unlike enhanced Cox1 arising from overexpression of Mss51, may arise from the reported destabilization of the Mss51-containing Cox1 assembly intermediate (23). The Mss51-containing Cox1 early assembly intermediate is attenuated in abundance in Coa2-containing cells with the ΔC15-truncated Cox1 (Fig. 2E, lane 2), yet the Mss51 complex is of sufficient stability to contribute to Cox10 complex formation in these Coa2-containing cells (Fig. 2D, lane 2). The failure of the ΔC15-truncated Cox1 to stimulate Cox10 oligomerization in coa2Δ cells may occur if the C-terminal Cox1 segment had a role in triggering Cox10 multimerization in addition to mediating the Mss51/Cox14 interaction.
Ectopic Expression of Cox1 C-terminal Tail Restores Cox10 Complex in coa2Δ Cells
Yeast Cox1 consists of 12 TM helices and has a 54-residue-long C-terminal segment that packs against matrix-facing subunits especially Cox4 (24). We sought to test whether the C-terminal 54 residues of Cox1, when appended to a soluble matrix protein, would trigger Cox10 oligomerization. We postulated that enhanced Cox10 oligomerization might enhance hemylation of endogenous Cox1 thereby restoring respiratory growth of coa2Δ cells.
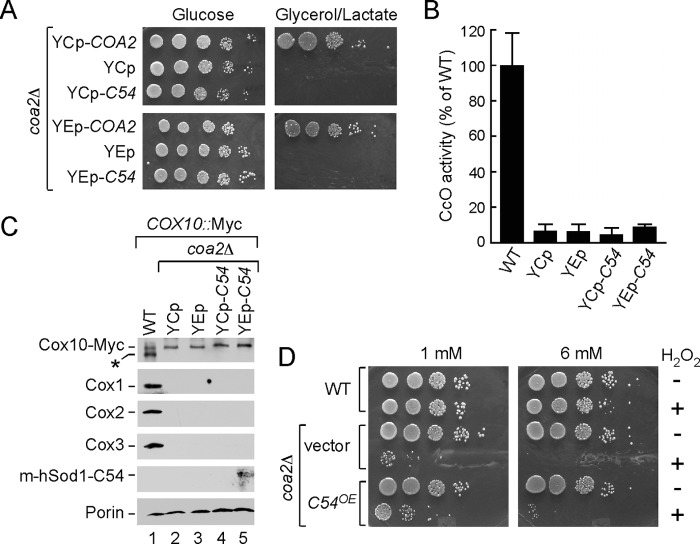
We attached the C-terminal 54 residues of Cox1 to human Sod1 expressed within the matrix. We had shown previously that a Sod2/hSod1 fusion protein consisting of the Sod2 mitochondrial target sequence fused to the human Sod1 molecule accumulated within the yeast mitochondrial matrix (25). The human Sod1 was used as a convenient, soluble passenger protein (Fig. 3A). The Sod1-Cox1 fusion protein was expressed to a lower level relative to the matrix-targeted Sod1 protein alone (Fig. 3A), and the chimera protein was localized to the matrix (data not shown). Despite the reduced levels of the fusion protein, expression of the chimeric protein in coa2Δ cells from low and high copy plasmids resulted in a concentration-dependent restoration of Cox10 oligomerization (Fig. 3B). Cells with the chimera expressed from a high copy YEp vector (YEp-C54) exhibited higher abundance Cox10 multimers relative to cells with low copy chimera (YCp-C54). Cells expressing matrix-targeted human Sod1 without the Cox1 appendage failed to induce Cox10 complex formation (Fig. 3C). The presence of the Cox1 C-terminal segment appeared more effective in promoting Cox10 complex formation, compared with elevating endogenous Cox1 levels by the overexpression of MSS51 (Fig. 3D).
FIGURE 3.
Ectopic expression of the 54 C-terminal residues of Cox1 in coa2Δ cells restores Cox10 oligomerization in a dose-dependent manner. A, schematic view of the mitochondrial matrix-targeted human Sod1 and its derivative containing C-terminal residues of Cox1 (left panel). Right panel shows immunoblot of mitochondria isolated from the COX10::13×Myc coa2Δ cells expressing respective chimeras. Purified mitochondria (15 or 30 μg) were separated by SDS-PAGE and analyzed by Western blot with antibodies against hSod1 and outer mitochondrial membrane protein porin (loading control). B, Cox10 oligomerization in COX10::13×Myc WT or COX10::13×Myc coa2Δ cells transformed with either centromeric (YCp) or episomal (YEp) vectors expressing m-hSod1-C54 was tested by native electrophoresis as in Fig. 2A. Protein complexes were visualized with anti-Myc and anti-Atp2 (anti-F1) antisera. C, BN-PAGE analysis of Cox10 oligomer in mitochondria derived from COX10::13×Myc coa2Δ cells expressing m-hSod1. D, assessment of Cox10 oligomerization in COX10::13×Myc coa2Δ cells overexpressing either m-hSod1-C54 or MSS51 was carried out as described above. E, distributions of Cox10–13×Myc in mitochondria (30 μg) derived from COX10::13×Myc WT or COX10::13×Myc cox14Δ and panel COX10::13×Myc mss51Δ cells expressing m-hSOD1-C54 were analyzed by native electrophoresis.
We tested whether the Mss51-containing Cox1 complex was required for the Sod1-Cox1 chimera-mediated Cox10 oligomerization by expressing the fusion protein in cells lacking Mss51 or Cox14. One key difference between the two strains is that no mitochondrial Cox1 synthesis is apparent in mss51Δ cells, whereas Cox1 is efficiently synthesized in cox14Δ cells but is unstable (10). As can be seen in Fig. 3E, the expression of episomal Sod1-Cox1 failed to induce Cox10 oligomerization in either mss51Δ or cox14Δ cells. Thus, chimera-induced Cox10 complex formation is dependent on the Mss51-containing Cox1 assembly intermediate shown in Fig. 1.
Cox10 Oligomerization Induced by Cox1 C-terminal End Expression Is Uncoupled from Respiratory Function
Mutant coa2Δ cells harboring the Sod1-Cox1 chimera were tested for respiratory function by plating cells in serial dilution on medium containing either glucose or glycerol/lactate. As can be seen in Fig. 4A, the presence of the Sod1-Cox1 chimera failed to restore growth on glycerol/lactate medium regardless of its expression level. Consistent with the lack of growth, no increase in CcO catalytic activity was observed in mitochondria from coa2Δ cells with the Sod1-Cox1 chimera (Fig. 4B). It is unlikely that the Sod1-Cox1-induced Cox10 oligomer is active, because no stabilization of CcO subunits, especially Cox1, was observed (Fig. 4C). This is in contrast to coa2Δ cells harboring the N196K mutant Cox10 that stabilizes Cox1 and enables CcO biogenesis to proceed (6).
FIGURE 4.
Expression of the m-hSod1-C54 does not restore respiratory function of coa2Δ cells. A, Coa2-deficient cells, transformed with either centromeric (YCp) or episomal (YEp) vectors expressing COA2 or m-hSOD1-C54, were grown and plated as described in Fig. 2C. The growth was assessed after 2 and 6 days of incubation at 30 °C for glucose and glycerol/lactate plates, respectively. B, CcO-specific activities of mitochondria derived from the strains listed in A. Activities are shown as a percentage of wild-type specific activity, and error bars indicate S.D. (n = 3). C, steady-state levels of CcO core subunits (Cox1, Cox2, and Cox3) and Cox10-13×Myc in 20 μg of mitochondria isolated from the aforementioned transformants were analyzed by immunoblotting with respective antibodies. Anti-hSod1 was used to visualize m-hSod1-C54; porin levels served as a loading control and were detected with the respective antibody. Asterisk indicates nonspecific band. D, coa2Δ cells transformed with either empty vector or YEp m-hSod1-C54 and wild-type (WT) cells were grown to mid-exponential phase and incubated with (+) or without (−) indicated concentrations of H2O2 for 2 h at 30 °C. Following the incubation, serial dilutions were made and plated onto solid 2% glucose-containing media. The growth was assessed after 36–48 h of incubation at 30 °C.
A second assay of Cox10 catalytic function that we exploited is the Cox10-dependent sensitivity of several CcO assembly mutant cells to hydrogen peroxide (7). Mutant cells stalled in Cox1 maturation at a stage in which the heme a sites are occupied exhibit peroxide sensitivity likely due to the reactive heme a3 subsite. Cells lacking Coa2 are susceptible to growth arrest by pretreatment with hydrogen peroxide prior to subsequent replating (Fig. 4D). Deletion of COX10 in coa2Δ cells abrogates this peroxide sensitivity, likely through blocking hemylation of Cox1. Although Cox1 levels are low in coa2Δ cells, sufficient levels of either hemylated Cox1 or free heme a exist to generate this sensitivity (7). The presence of the Sod1-Cox1 fusion did not induce enhanced peroxide sensitivity as would be expected if Cox10 was activated and generated excess heme o. These studies suggest that although the Sod1-Cox1 chimera induced Cox10 multimerization, the complex was inactive catalytically.
Cox1 C-terminal Domain Triggers Formation of the Coa1-containing Cox1 Complex
Because the Sod1-Cox1 chimera failed to restore respiratory growth in coa2Δ cells, we sought to assess which step in Cox1 maturation was impaired. Initially, we addressed the state of Cox1 assembly intermediates in mutant cells containing the chimera. As mentioned, the initial assembly intermediate consists of Cox1 associated with Mss51, Cox14, Coa3, and Ssc1 (see Fig. 1) (9–12). Newly synthesized Cox1 transitions to later assembly intermediates, including complexes with assembly factors Coa1 and Shy1 (3, 20, 26). Hemylation of Cox1 and formation of the CuB center occurs in the later Cox1 intermediate containing Shy1 (3). In the absence of Cox1 hemylation, e.g. in cox10Δ cells, only the Shy1-containing Cox1 complex is perturbed (3).
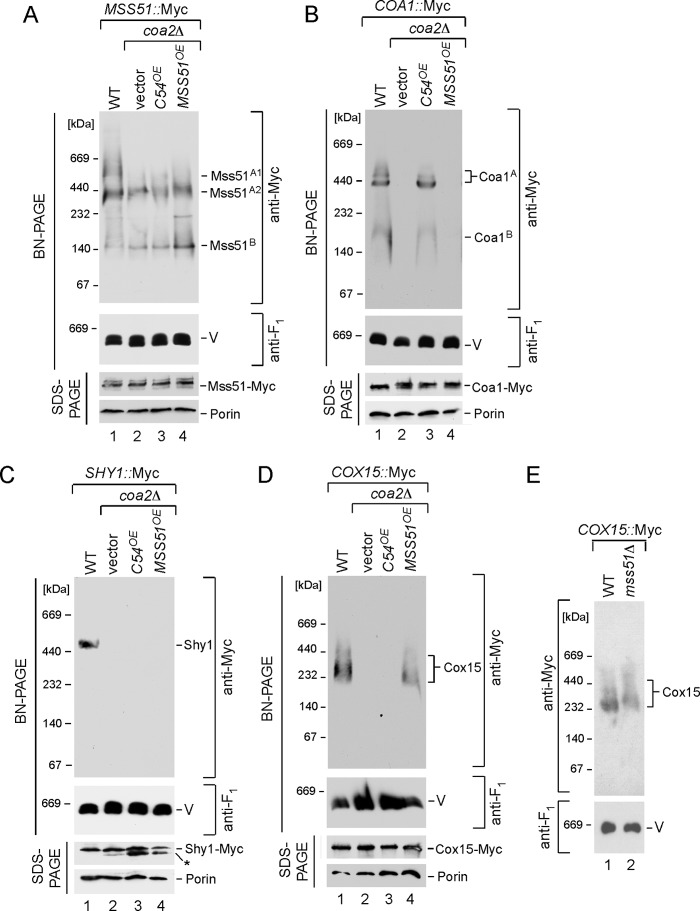
Cells lacking Coa2 contain normal levels of the early Mss51-containing Cox1 complex but lack downstream Coa1-containing and Shy1-containing Cox1 complexes (Fig. 5, A–C) (Shy1 complex is shown in Fig. 1) (3). To assess whether the Sod1-Cox1 fusion protein would restore any Cox1 assembly intermediates, we expressed the fusion protein in cells harboring chromosomal Myc epitope tags on Mss51, Coa1, and Shy1. The presence of the C-terminal 54 residues of Cox1 restored the Coa1-containing Cox1 complex (Fig. 5B) but not the Shy1-containing Cox1 complex (Fig. 5C). Although the Coa1 complex contains Cox1, we were unable to see any appreciable accumulation of Cox1 by steady-state immunoblotting likely due to instability. The presence of the Sod1-Cox1 fusion did not affect steady-state levels of Mss51, Coa1, or Shy1 (Fig. 5, B and C, SDS-PAGE lanes). In contrast, overexpression of MSS51 failed to induce formation of the high mass Coa1 complex in coa2Δ cells (Fig. 5B, lane 4). The Sod1-Cox1 fusion protein may facilitate release of Cox1 from the Mss51-containing complex.
FIGURE 5.
Expression of the m-hSOD1-C54 and MSS51 exerts different effects on Cox1 assembly intermediates in coa2Δ cells. A–D, mitochondria from MSS51::13×Myc WT or MSS51::13×Myc coa2Δ (A), COA1::13×Myc WT or COA1::13×Myc coa2Δ (B), SHY1::13×Myc WT or SHY1::13×Myc coa2Δ (C), and COX15::13×Myc WT or COX15::13×Myc coa2Δ (D) cells overexpressing m-hSOD1-C54 or MSS51 were analyzed by BN-PAGE as in Fig. 2A. 13×Myc tag-containing complexes were visualized by immunoblotting with anti-Myc antibodies. F1β subunit of monomeric complex V was detected with anti-Atp2 (anti-F1). The bottom of each panel shows steady-state levels of the respective proteins from the same strains analyzed by denaturing SDS-PAGE and Western blotting with antibodies against the Myc epitope and porin (loading control). 30 μg of purified mitochondria were used for BN-PAGE analyses in each case except for SHY1::13×Myc set, where 50 μg of mitochondria were tested. 10 μg of isolated organelles were used for SDS-PAGE. The asterisk in C denotes degradation product. E, BN-PAGE analysis of Cox15 oligomeric complex in mitochondria derived from the WT and mss51Δ cells bearing the COX15::13×Myc endogenous tag.
Cox1 C-terminal Domain Induces Formation of Cox10 Oligomers but Not Cox15 Complex Formation
Hemylation of Cox1 requires both Cox10 and Cox15. Heme o produced by Cox10 is oxidized to heme a by the Cox15 heme a synthase. As mentioned, Cox15 forms a high mass multimer that is distinct from the Cox10 complex (6). The Cox15 complex is also attenuated in coa2Δ cells, yet steady-state levels of Cox15 are wild-type (Fig. 5D). The presence of the Cox1 C-terminal 54-residue segment failed to induce Cox15 complex formation in coa2Δ cells, although overexpression of MSS51 induces Cox15 complex formation. Cox15 oligomerization is distinct from that of Cox10 in that Cox15 complex formation is not totally dependent on newly synthesized Cox1. Cells lacking Mss51 retain limited quantities of high mass Cox15 (Fig. 5E). Thus, the trigger for Cox10 and Cox15 multimerization has unique aspects for each.
The lack of respiratory function in coa2Δ cells by the presence of the Sod1-Cox1 fusion may relate to the inability of the Cox1 C-terminal segment to stabilize the Shy1-containing Cox1 assembly intermediate or induce Cox15 oligomerization. However, no definitive information is available on whether the oligomerized Cox15 is required for its catalytic function.
Analysis of Cox10 Patient Mutations in Yeast Protein
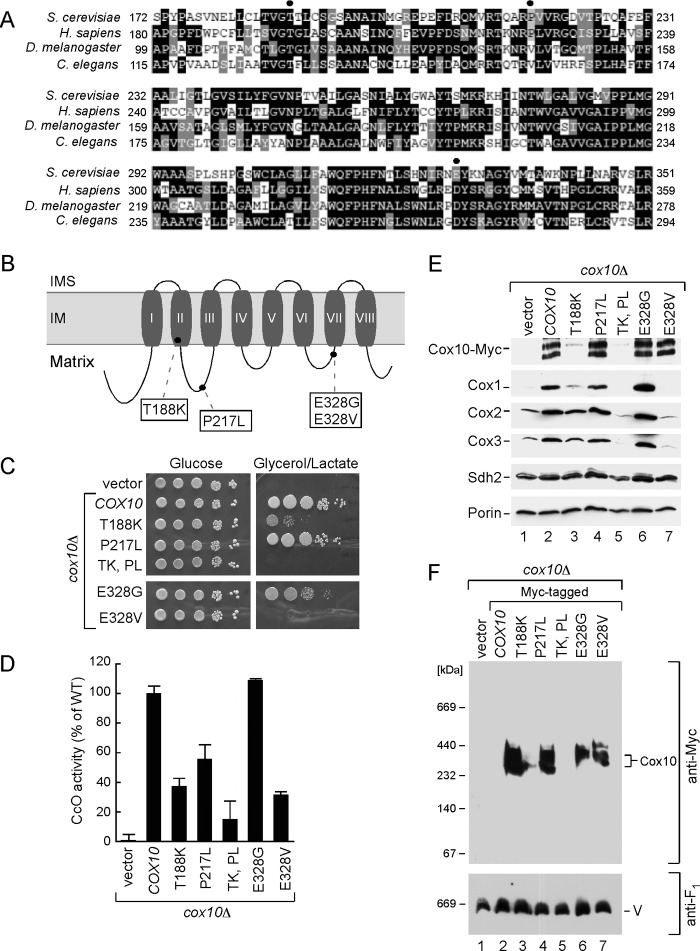
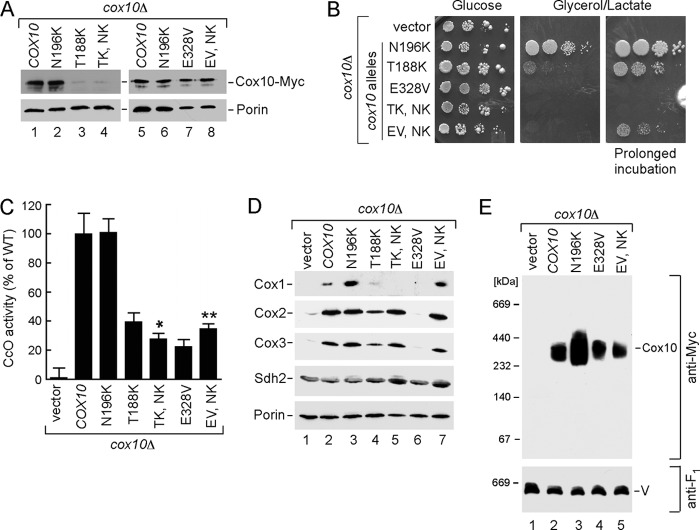
To evaluate the significance of Cox10 oligomerization in relation to human disease, we evaluated mutations reported in CcO-deficient patients who presented with a range of phenotypes from Leigh syndrome to fatal infantile hypertrophic cardiomyopathy or lactic acidosis (27, 28). The Cox10 mutations reported to date include N204K, D336G, or D336V and a double T196K/P225L substitution. Surprisingly, the N204K mutation that was identified in a patient with encephalopathy and CcO deficiency is the corresponding residue and mutation as the N196K gain-of-function mutation we isolated in yeast Cox10. The N204K is an attenuating mutation in human Cox10, because expression of WT human Cox10 but not the N204K mutant showed reduced function when expressed in a yeast cox10Δ strain (27). Human patient cells harboring the double T196K/P225L Cox10 mutant had 40% of WT CcO activity, whereas cells with the D336G or D336V substitution had ∼18% of WT activity (28). To assess the consequences of mutations at the corresponding residues in yeast Cox10 (Thr-188, Pro-217, and Glu-328) (Fig. 6A), mutations were introduced into yeast COX10 and tested in cox10Δ cells. Thr-188 is located in the middle of the second TM motif; Pro-217 is present in the matrix loop connecting TM2 and TM3 adjacent to other residues important for enzyme function. Glu-328 is located in another matrix-facing loop connecting TM6 and TM7 (Fig. 6B). T188K and P217L substitutions were tested singly and as a double mutation, and both E328G and E328V substitutions were engineered.
FIGURE 6.
Analysis of Cox10 human pathogenic mutations in the yeast model. A, multiple sequence alignment of the Cox10 conserved region from different species. Sequences were aligned using the MultAlin and BoxShade programs. Identical amino acid residues are shown in black; conserved residues are in dark gray, and the similar ones are in light gray. The circles indicate amino acid residues mutated in Leigh syndrome patients. B, schematic view of yeast Cox10. Predicted transmembrane domains (I–VIII) are indicated. Marked are the substitutions corresponding to pathogenic mutations in human protein. C, respiratory growth of wild-type (WT) and cox10Δ cells of BY4743 background expressing WT Cox10 or its T188K, P217W, T188K/P217W, E328G, and E328V mutant forms. Cells were handled and tested as in Fig. 2C, except that glycerol/lactate plates were incubated up to 6 days at 30 °C. D, mitochondria derived from the cells described in C were used to assess CcO activity. Enzymatic activities are shown as a percentage of WT-specific activity. The data represent an average of three independent measurements; the error bars indicate S.D. E, steady-state levels of Cox10-13Myc and its mutant forms as well as core CcO subunits in the respective mitochondria (10 μg) were analyzed by immunoblot with indicated antibodies. Subunit Sdh2 of succinate dehydrogenase and outer membrane protein porin were detected with respective antibodies and served as controls. F, BN-PAGE analysis of the aforementioned mitochondria. Samples were handled and analyzed as in Fig. 2.
Cells harboring T188K Cox10 were partially compromised in respiratory growth, whereas the P217L Cox10 variant supported glycerol/lactate growth (Fig. 6C), but CcO activity was slightly impaired (Fig. 6D). The double T188K/P217L mutant was unable to support respiratory growth (Fig. 6C), and CcO activity in the mutant cells was markedly attenuated (Fig. 6D). The E328G Cox10 mutant supported respiratory growth and contributed to appreciable CcO activity, whereas cells containing the E328V Cox10 were impaired in respiration and CcO activity. The T188K mutant Cox10 protein was unstable, and the double T188K/P217L protein was even more compromised in stability, such that steady-state levels of CcO subunits were dramatically reduced, thus explaining the impaired respiratory growth (Fig. 6E). Whereas no useful information could be gleaned from these mutants due to protein instability, the E328G and E328V mutant proteins were expressed stably (Fig. 6E). Cells harboring E328V Cox10 showed markedly reduced steady-state levels of Cox1-Cox3. Despite the reduced Cox1 levels, BN-PAGE of mitochondria from E328V cells showed the normal Cox10 oligomeric complex (Fig. 6F).
Previously, we demonstrated that a H317A mutant Cox10 was unstable, but the addition of an N196K substitution imparted protein stability (6). We tested whether the introduction of an N196K substitution would stabilize the T188K mutant protein or activate the E328V mutant. A double T188K/N196K Cox10 mutant exhibited no enhanced protein stability (Fig. 7A, TK,NK), and these cells were more compromised in glycerol/lactate growth (Fig. 7B) and CcO activity (Fig. 7C), compared with the single T188K mutant. In contrast, the introduction of the N196K allele in the E328V Cox10 backbone resulted in a modest improvement in both glycerol/lactate growth and CcO activity (Fig. 7, B and C). As expected from the enhanced CcO activity, steady-state levels of CcO subunits also increased (Fig. 7D).
FIGURE 7.
E328V mutant phenotype can be partially rescued by cis-effect of N196K substitution without changes to Cox10 oligomerization. A, Western blot analysis of the mitochondria from the BY4743 cox10Δ cells expressing either WT Cox10-13×Myc or indicated mutant forms of the protein. 10 μg of mitochondrial proteins were separated by SDS-PAGE and detected with antibodies against Myc epitope or porin. B, respiratory growth of the BY4743 cox10Δ cells transformed with indicated constructs. Transformants were handled as in Fig. 2C. Pictures of the plates were taken after 2 (glucose) or 4 and 8 (glycerol lactate) days of incubation at 30 °C. C, CcO-specific activities of mitochondria isolated from indicated transformants were determined as described in Fig. 6D. D, immunoblot analysis of the steady-state levels of Cox1, Cox2, Cox3, Sdh2, and porin analyzed in 10 μg of transformant-derived mitochondria. E, oligomeric state of Cox10-13×Myc and its mutant forms was analyzed by BN-PAGE as described above.
The increased abundance of the Cox10 oligomer seen with the N196K Cox10 variant, relative to WT Cox10, was not apparent when the N196K was present together with the E328V substitution (Fig. 7E, compare lanes 4 with 5). The enhanced CcO activity in the N196K/E328V double mutant without a change in oligomeric Cox10 highlights the gain-of-function activity of the N196K substitution.
Role of WT Cox10 as a Suppressor of coa2Δ Cells
This study reveals that the respiratory deficiency of coa2Δ cells may arise from impaired oligomerization of Cox10 and Cox15, and these events are coupled to the levels of newly synthesized Cox1. Stimulation of Cox1 synthesis in coa2Δ cells is sufficient to stimulate Cox15 multimerization but is only partially efficient in stimulating Cox10 complex formation (Fig. 2D and Fig. 5D).
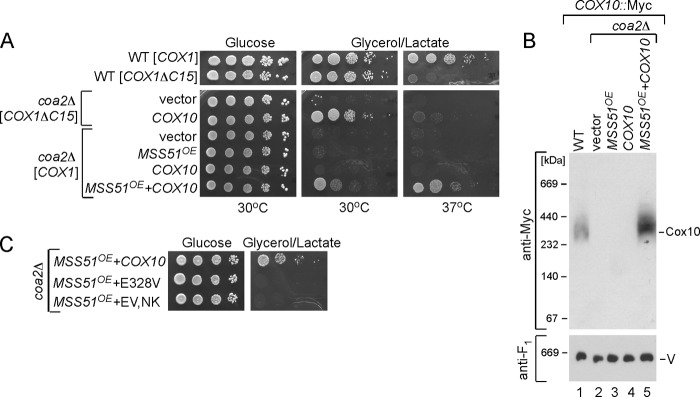
We tested whether a combination of enhanced Cox1 synthesis and elevated levels of WT Cox10 would restore respiratory growth of coa2Δ cells (Fig. 8A). We tested the combination of WT Cox10 in addition to overexpression of MSS51 or the presence of the Cox1ΔC15 allele. Whereas neither overexpression of MSS51 nor the presence of the Cox1ΔC15 allele in coa2Δ cells supported respiratory growth (Fig. 2C), the addition of YCp COX10 restored limited respiratory growth in both cases (Fig. 8A). The synergistic effect of elevated levels of both COX10 and COX1 synthesis resulted in a marked increase in the abundance of the Cox10 oligomeric complex, a situation not seen merely by expression of WT Cox10 (Fig. 8B).
FIGURE 8.
Synergistic effect of increased Cox1 synthesis and Cox10 expression restores respiration in coa2Δ cells. A, respiratory growth of the WT cells with either full-length or truncated Cox1 or respective coa2Δ cells bearing YCp-Cox10 (low copy), YEp-MSS51 (high copy), or co-expressing both constructs. Cells were pre-cultured and tested as described in Fig. 2C except that the plates were incubated at either 30 or 37 °C. B, BN-PAGE analysis of mitochondria derived from coa2Δ transformants described in A. The exposure shown does not reveal the low abundance of the Cox10 oligomer with overexpression of MSS51. C, growth of coa2Δ cells co-expressing YEp-MSS51 and YCp-COX10 or its mutant alleles was tested as in A.
The synergistic effect of COX10 and MSS51 is dependent on a functional Cox10 molecule. Co-expression of MSS51 and the COX10 mutant allele containing the E328V substitution failed to show any restoration of respiratory growth (Fig. 8C). Likewise, no respiratory growth was restored in coa2Δ cells expressing MSS51 and the Sod1-Cox1 chimera (data not shown). Thus, a bypass of Coa2 can occur upon increasing Cox1 synthesis and either having elevated levels of WT Cox10 or an allele (e.g. N196K) that stabilizes the oligomer form of the enzyme.
DISCUSSION
The Cox10 farnesyltransferase is an essential enzyme for heme a formation. Mutations in human Cox10 have been reported in CcO-deficient patients with leukodystrophy, Leigh syndrome, and fatal infantile hypertrophic cardiomyopathy (27–29).
Yeast Cox10 exists within a multimeric unit, and this complex is dependent on the presence of newly synthesized Cox1 (6). We sought to deduce how Cox10 senses the availability or assembly state of Cox1 for its activation. We show that an increase in newly synthesized Cox1 through overexpression of its translation activator Mss51 or deletion of COA1 in coa2Δ cells modestly restores Cox10 multimerization, but this process is enhanced when elevated levels of the WT enzyme exist. A clear link exists between the levels of newly synthesized Cox1 and the oligomeric state of Cox10. We demonstrate that the C-terminal segment of Cox1 is important in triggering Cox10 oligomerization. First, a Cox1 mutant lacking the C-terminal 15 residues is compromised in triggering Cox10 complex formation in coa2Δ cells. Second, we demonstrated that expression of the C-terminal 54 residues of Cox1 appended to a heterologous matrix protein leads to efficient Cox10 complex formation in coa2Δ cells; however, that was insufficient to restore respiratory function in the mutant cells.
Cox10 oligomerization is dependent on the presence of the early Mss51-containing Cox1 assembly intermediate. Although the high mass Mss51 complex is present in coa2Δ cells, Cox10 oligomerization is impaired. Cox10 multimerization is likely triggered by progression of Cox1 from the Mss51 complex to a downstream intermediate (see Fig. 1). Coa1 appears to be important in the release of Cox1 from the Mss51 complex to downstream maturation (Fig. 8).
The release of Cox1 from the Mss51 complex may expose the Cox1 C-terminal segment to trigger Cox10 oligomerization and activation. Cox10 is predicted to have matrix-facing loops connecting TM domains that contain catalytically important residues. The C-terminal 54 residues of Cox1 may induce Cox10 complex formation through a transient interaction with Cox10. The mSod1-Cox1 chimera may mimic the effect of the endogenous Cox1 in promoting Cox10 oligomerization. However, no stable interaction was observed between Cox10 and the mSod1-Cox1 chimera in coimmunoprecipitation studies (data not shown). Alternatively, the mSod1-Cox1 chimera may induce Cox10 oligomerization indirectly through enhancing formation of the Coa1-containing Cox1 assembly intermediate that is downstream of the Mss51-containing Cox1 complex (3). The chimera may trigger the release of Cox1 from the Mss51-containing Cox1 complex. A third scenario for the effect of the chimera on Cox10 complex formation is that the Cox1 54-residue segment may compete with endogenous newly synthesized Cox1 for proteolytic degradation. This scenario is unlikely as the Sod1-Cox1 chimera stimulates only Cox10, but not Cox15, multimerization. Elevated Cox1 synthesis through overexpression of Mss51 restores Cox15 complex formation in coa2Δ cells. The lack of respiratory growth in coa2Δ cells containing the Sod1-Cox1 chimera may arise from the impairment in Cox15 and not Cox10. The high mass Cox15 complex is distinct from the Cox10 oligomeric complex. Unlike Cox10, Cox15 complex formation is not strictly linked to newly synthesized Cox1. The attenuation of the Cox15 high mass complex in coa2Δ cells but not mss51Δ cells suggests that Coa2 may have a role in mediating Cox15 multimerization also. Yet the trigger for Cox10 and Cox15 high mass complex formation differs. At present, no clear information exists on whether this high mass Cox15 complex is critical for its function.
Cox10 oligomerization is not impaired in downstream mutants such as sco1Δ cells (6). Cox1 progression from the Mss51-containing complex proceeds normally in sco1Δ cells and thus may account for the normal Cox10 multimerization.
Additional insights into Cox10 emerged from analyses of patient mutations in human Cox10. The most revealing mutants were the D336G and D336V substitutions that correspond to Asp-328 in the yeast protein. Asp-328 is not an essential catalytic residue as the E328G variant is functional; however, the E328V mutant is compromised in function. This mutant is impaired in catalytic activity but not in the ability to oligomerize. Cox10 oligomerization and catalytic activity are not necessarily linked processes. The Val substitution may have a negative steric effect on the catalytic function. The nearby His-317 residue is proposed to be an axial heme ligand stabilizing the protoheme substrate in the active site (30).
This study reveals novel insights into Coa2. The rapid turnover of Cox1 in coa2Δ cells appears to arise from the impaired hemylation of Cox1 or the instability of Cox1 prior to the hemylation step. The N196K Cox10 gain-of-function mutant restores respiratory growth in coa2Δ cells likely by a combination of its enhanced stabilization of the oligomeric complex, as well as catalytic efficiency. In the absence of the stabilizing N196K substitution in Cox10, elevated levels of newly synthesized Cox1 in combination with high levels of WT Cox10 restore respiratory function in coa2Δ cells. Coa2 appears to facilitate the coupling of newly synthesized Cox1 to Cox10 oligomerization and activation. We showed previously that restoration of respiratory function in coa2Δ cells occurs through the depletion of Oma1. The ablation of this protease may increase the abundance or stability of downstream Cox1 assembly intermediates that can undergo inefficient hemylation in the absence of Coa2. The depletion of Oma1 also restores Cox10 oligomerization.
We reported previously that Coa2 forms a transient interaction with the Shy1-containing Cox1 assembly intermediate. The Shy1-containing assembly intermediate is the likely complex in which the CuB-heme a3 bimetallic center is formed (3). Coa2 may also function as a chaperone stabilizing formation of the heme a subsite. Formation of this heme a center is predicted to stabilize the Cox1 helical bundle. The fully extended farnesyl group of this heme center packs within an α-helical bundle formed by helices 1, 11, and 12 of Cox1 (31). The rapid turnover of Cox1 in coa2Δ cells may arise from an inefficient population of the heme a subsite leading to a misfolded Cox1 conformer.
Because Coa2 has an apparent role in coupling Cox1 maturation to Cox10 oligomerization, a role in Cox15 complex formation, and a role in chaperoning the heme a subsite formation, an important unresolved question is whether these are independent functions of Coa2 or whether these processes are linked. Resolution of this question will require new strategies and experimental approaches.
Acknowledgment
We thank Dr. Antonio Barrientos for the YEplac112-MSS51 plasmid.
This work was supported, in whole or in part, by National Institutes of Health Grant ES03817 from the NIEHS (to D. R. W.) and Training Grant T32 DK007115 (to H. K.). This work was also supported by American Heart Association Grant 10POST4300044 (to O. K.) and Programa de Apoyo a Proyectos de Investigacion e Innovacion Tecnologica, Universidad Nacional Autonoma de Mexico Grant IN82505 (to X. P.-M.).
- CcO
- cytochrome c oxidase
- BN-PAGE
- blue native-PAGE
- TM
- transmembrane.
REFERENCES
- 1. Moraes C. T., Diaz F., Barrientos A. (2004) Defects in the biosynthesis of mitochondrial heme c and heme a in yeast and mammals. Biochim. Biophys. Acta 1659, 153–159 [DOI] [PubMed] [Google Scholar]
- 2. Saiki K., Mogi T., Anraku Y. (1992) Heme O biosynthesis in Escherichia coli. The cyoE gene in the cytochrome bo operon encodes a protoheme IX farnesyltransferase. Biochem. Biophys. Res. Commun. 189, 1491–1497 [DOI] [PubMed] [Google Scholar]
- 3. Khalimonchuk O., Bestwick M., Meunier B., Watts T. C., Winge D. R. (2010) Formation of the redox cofactor centers during Cox1 maturation in yeast cytochrome oxidase. Mol. Cell. Biol. 30, 1004–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang Z., Wang Y., Hegg E. L. (2009) Regulation of the heme A biosynthetic pathway. Differential regulation of heme A synthase and heme O synthase in Saccharomyces cerevisiae. J. Biol. Chem. 284, 839–847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nobrega M. P., Nobrega F. G., Tzagoloff A. (1990) COX10 codes for a protein homologous to the ORF1 product of Paracoccus denitrificans and is required for the synthesis of yeast cytochrome oxidase. J. Biol. Chem. 265, 14220–14226 [PubMed] [Google Scholar]
- 6. Bestwick M., Khalimonchuk O., Pierrel F., Winge D. R. (2010) The role of Coa2 in hemylation of yeast Cox1 revealed by its genetic interaction with Cox10. Mol. Cell. Biol. 30, 172–185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pierrel F., Khalimonchuk O., Cobine P. A., Bestwick M., Winge D. R. (2008) Coa2 is an assembly factor for yeast cytochrome c oxidase biogenesis facilitating the maturation of Cox1. Mol. Cell. Biol. 28, 4927–4939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Khalimonchuk O., Jeong M. Y., Watts T., Ferris E., Winge D. R. (2012) Selective Oma1-mediated proteolysis of the Cox1 subunit of cytochrome oxidase in assembly mutants. J. Biol. Chem. 287, 7289–7300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perez-Martinez X., Broadley S. A., Fox T. D. (2003) Mss51p promotes mitochondrial Cox1p synthesis and interacts with newly synthesized Cox1p. EMBO J. 22, 5951–5961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barrientos A., Zambrano A., Tzagoloff A. (2004) Mss51p and Cox14p jointly regulate mitochondrial Cox1p expression in Saccharomyces cerevisiae. EMBO J. 23, 3472–3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fontanesi F., Soto I. C., Horn D., Barrientos A. (2010) Mss51 and Ssc1 facilitate translational regulation of cytochrome c oxidase biogenesis. Mol. Cell. Biol. 30, 245–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mick D. U., Vukotic M., Piechura H., Meyer H. E., Warscheid B., Deckers M., Rehling P. (2010) Coa3 and Cox14 are essential for negative feedback regulation of COX1 translation in mitochondria. J. Cell Biol. 191, 141–154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Pierrel F., Bestwick M. L., Cobine P. A., Khalimonchuk O., Cricco J. A., Winge D. R. (2007) Coa1 links the Mss51 post-translational function to Cox1 cofactor insertion in cytochrome c oxidase assembly. EMBO J. 26, 4335–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sambrook J., Fritsch E. F., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 15. Cobine P. A., Ojeda L. D., Rigby K. M., Winge D. R. (2004) Yeast contain a nonproteinaceous pool of copper in the mitochondrial matrix. J. Biol. Chem. 279, 14447–14455 [DOI] [PubMed] [Google Scholar]
- 16. Schiestl R. H., Gietz R. D. (1989) High efficiency transformation of intact yeast cells using single-stranded nucleic acid as a carrier. Curr. Genet. 16, 339–346 [DOI] [PubMed] [Google Scholar]
- 17. Daum G., Böhni P. C., Schatz G. (1982) Import of proteins into mitochondria, cytochrome b2, and cytochrome c peroxidase is located in the intermembrane space of yeast mitochondria. J. Biol. Chem. 257, 13028–13033 [PubMed] [Google Scholar]
- 18. Bradford M. M. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254 [DOI] [PubMed] [Google Scholar]
- 19. Taanman J. W., Capaldi R. A. (1992) Purification of yeast cytochrome c oxidase with a subunit composition resembling the mammalian enzyme. J. Biol. Chem. 267, 22481–22485 [PubMed] [Google Scholar]
- 20. Barrientos A., Korr D., Tzagoloff A. (2002) Shy1p is necessary for full expression of mitochondrial COX1 in the yeast model of Leigh syndrome. EMBO J. 21, 43–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Khalimonchuk O., Bird A., Winge D. R. (2007) Evidence for a pro-oxidant intermediate in the assembly of cytochrome oxidase. J. Biol. Chem. 282, 17442–17449 [DOI] [PubMed] [Google Scholar]
- 22. Corpet F. (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16, 10881–10890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Shingú-Vázquez M., Camacho-Villasana Y., Sandoval-Romero L., Butler C. A., Fox T. D., Pérez-Martínez X. (2010) The carboxyl-terminal end of Cox1 is required for feedback assembly regulation of Cox1 synthesis in Saccharomyces cerevisiae mitochondria. J. Biol. Chem. 285, 34382–34389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tsukihara T., Aoyama H., Yamashita E., Tomizaki T., Yamaguchi H., Shinzawa-Itoh K., Nakashima R., Yaono R., Yoshikawa S. (1996) The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science 272, 1136–1144 [DOI] [PubMed] [Google Scholar]
- 25. Cobine P. A., Pierrel F., Bestwick M. L., Winge D. R. (2006) Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase. J. Biol. Chem. 281, 36552–36559 [DOI] [PubMed] [Google Scholar]
- 26. Mick D. U., Wagner K., van der Laan M., Frazier A. E., Perschil I., Pawlas M., Meyer H. E., Warscheid B., Rehling P. (2007) Shy1 couples Cox1 translational regulation to cytochrome c oxidase assembly. EMBO J. 26, 4347–4358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Valnot I., von Kleist-Retzow J. C., Barrientos A., Gorbatyuk M., Taanman J. W., Mehaye B., Rustin P., Tzagoloff A., Munnich A., Rötig A. (2000) A mutation in the human heme A:farnesyltransferase gene (COX10) causes cytochrome c oxidase deficiency. Hum. Mol. Genet. 9, 1245–1249 [DOI] [PubMed] [Google Scholar]
- 28. Antonicka H., Leary S. C., Guercin G. H., Agar J. N., Horvath R., Kennaway N. G., Harding C. O., Jaksch M., Shoubridge E. A. (2003) Mutations in COX10 result in a defect in mitochondrial heme A biosynthesis and account for multiple early onset clinical phenotypes associated with isolated COX deficiency. Hum. Mol. Genet. 12, 2693–2702 [DOI] [PubMed] [Google Scholar]
- 29. Vempati U. D., Torraco A., Moraes C. T. (2008) Mouse models of oxidative phosphorylation dysfunction and disease. Methods 46, 241–247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mogi T. (2009) Overexpression and characterization of Bacillus subtilis heme O synthase. J. Biochem. 145, 669–675 [DOI] [PubMed] [Google Scholar]
- 31. Yoshikawa S., Shinzawa-Itoh K., Nakashima R., Yaono R., Yamashita E., Inoue N., Yao M., Fei M. J., Libeu C. P., Mizushima T., Yamaguchi H., Tomizaki T., Tsukihara T. (1998) Redox-coupled crystal structural changes in bovine heart cytochrome c oxidase. Science 280, 1723–1729 [DOI] [PubMed] [Google Scholar]