Background: Vms1 is a recently isolated Cdc48 cofactor but has few degradation targets under normal growth conditions.
Results: Vms1 and Cdc48 promote Cdc13 turnover, which involves both the proteasome and autophagy.
Conclusion: Our study reveals novel functions of Vms1 and autophagy.
Significance: Cdc13 may be the first ubiquitylated yeast protein subject to both degradation pathways under normal growth conditions.
Keywords: Autophagy, Proteasome, Protein Degradation, Ubiquitin, Ubiquitylation
Abstract
Vms1 is a newly identified Cdc48-binding protein. The biological function of Vms1 remains obscure. Here, we show that both Cdc48 and Vms1, but not Cdc48 cofactors Ufd1 and Ufd2, are crucial for the degradation of Cdc13, a telomere regulator. Interestingly, both autophagy and the proteasome are involved in Cdc13 turnover. Toxicity associated with accumulation of large amounts of Cdc13 in vms1Δ or autophagy mutants underscores the significance of the proteolytic regulation of Cdc13. Because few ubiquitylated yeast proteins are known to be degraded by autophagy under non-stress conditions, the identification of Cdc13 as a target of autophagy provides a valuable tool to unravel the mechanism of autophagy-mediated selective protein degradation.
Introduction
CDC48 encodes an essential ATPase that regulates a myriad of cellular processes, including cell cycle progression, DNA repair, gene expression, membrane fusion, and proteolysis (1–3). In many cases, Cdc48 uses its ATPase activity to catalyze the disassembly of a protein complex and/or the extraction of specific proteins (1, 3, 4). Cdc48 fulfills its diverse cellular functions through its interaction with different cofactors (e.g. Ufd1–3, Png1, Otu1, Ubx1–7, SVIP, and gp78) (1, 5). A new member of the Cdc48 cofactor family is Vms1, an evolutionarily conserved protein that resides mainly in the cytosol (6, 7). Upon mitochondrial stress, a small population of Vms1 is relocalized to mitochondria and works with Cdc48 to promote mitochondrial protein turnover (e.g. Fzo1), which in turn protects cells from various insults (6). Moreover, yeast Vms1 was shown to play a moderate, albeit undefined role in endoplasmic reticulum (ER)2-associated degradation because human cystic fibrosis transmembrane conductance regulator protein, an ER-associated degradation substrate, was stabilized modestly in vms1Δ cells (7). Nevertheless, its physiological function remains poorly understood, as degradation of few substrates is significantly affected in vms1Δ cells under non-stress conditions.
Here, we found that Cdc48 and Vms1 promote the degradation of Cdc13, a telomere regulator in yeast. Cdc13 is a single-stranded DNA-binding protein that plays critical roles in telomerase regulation, telomere replication, and maintenance (8). Cdc13 was previously shown to be degraded (9). We found that Cdc13 turnover involves both the proteasome and autophagy. Importantly, accumulation of large amounts of Cdc13 in cells lacking VMS1 or specific autophagy regulators (e.g. ATG14 and ATG1) leads to growth inhibition, suggesting that tight control of Cdc13 levels is crucial for cell growth and survival. Autophagy has long been regarded as a stress response system to degrade proteins and organelles nonspecifically by the lysosome but has also emerged as a regulatory pathway that selectively eliminates specific ubiquitylated proteins (10–12). Genetic analysis in yeast has led the way in uncovering key components and critical mechanistic attributes of autophagy, which has recently been implicated in human diseases such as cancer, neurodegenerative disorders (e.g. Huntington and Parkinson), and diabetes (10). Despite the fact that studies in yeast have been instrumental in illuminating the elusive principles governing autophagy, only a handful of autophagy substrates have been found in yeast under normal growth conditions. Our study leads not only to novel insights into the biological function of the Vms1-Cdc48 pathway but also to the physiological role of the autophagy system.
EXPERIMENTAL PROCEDURES
Yeast Strains and Plasmids
Haploid strains lacking VMS1, UFD2, or specific ATG genes in the BY4741 background were purchased from Open Biosystems (Huntsville, AL). Strains KFY116 (cdc48-1), cim5-1, and ufd1-1 were obtained from Drs. K. Frolich (University of Tübingen), T. Suzuki (Osaka University), and A. Varshavsky (California Institute of Technology). The plasmid pEGH-Cdc13 expressing both His6- and GST-tagged Cdc13 from the GAL1 promoter, was obtained from Dr. Brenda Andrews (University of Toronto). VMS1 was amplified by PCR to incorporate the FLAG epitope and cloned into the yeast vector pESC-Leu (Agilent Technologies). For its expression in Escherichia coli, His6-tagged Vms1 was cloned into pET28a (Novagen) and expressed in BL21 cells. Cultures were grown in rich medium (yeast extract, peptone, and dextrose) or synthetic medium containing standard ingredients and 1) 2% glucose (SD medium), 2) 2% raffinose (SR medium), 3) 2% galactose (SG medium), or 4) 2% raffinose + 2% galactose.
Expression Shutoff Assay
Wild-type or mutant yeast cells carrying plasmid pEGH-Cdc13 were grown at 30 °C to A600 ∼ 1 in synthetic medium with auxotrophic supplements and 2% raffinose as the carbon source. Protein expression was induced with 2% galactose for 2 h and then repressed by the addition of 2% glucose. Cycloheximide (100 μg/ml) was also added to stop translation. Samples were withdrawn at the indicated time points. Proteins were extracted by the glass bead method using yeast lysis buffer (50 mm HEPES (pH 7.5), 5 mm EDTA, 0.15 m NaCl, and 1% Triton X-100), processed for immunoprecipitation with glutathione-Sepharose beads (GE Healthcare), and resolved by 8% SDS-PAGE. Immunoblots were probed with anti-His6 antibody (Sigma), followed by detection with goat anti-mouse HRP conjugate using ECL reagents (GE Healthcare). The stable protein Rpt5 was used as a loading control in the expression shutoff experiments.
Co-immunoprecipitation Assay
Protein-protein interactions were assayed by co-immunoprecipitation. Yeast cells carrying either an empty vector or pESC-Vms1-FLAG (bearing FLAG-tagged Vms1) and pRS314-PUFD2-Ufd2-Myc (expressing Myc-tagged Ufd2), pRS415G-RGS/His6-Cdc48 (expressing RGS/His6-tagged Cdc48) (4), or pGEH-Cdc13 (expressing GST-His6-tagged Cdc13), as indicated, were grown to log phase. Cells were lysed with glass beads and immunoprecipitated with beads coated with the indicated antibodies for 2 h at 4 °C. Immunoprecipitates were resolved by SDS-PAGE, transferred to PVDF membrane, and immunoblotted with the antibodies indicated.
RESULTS
Vms1 Directly Binds Cdc48
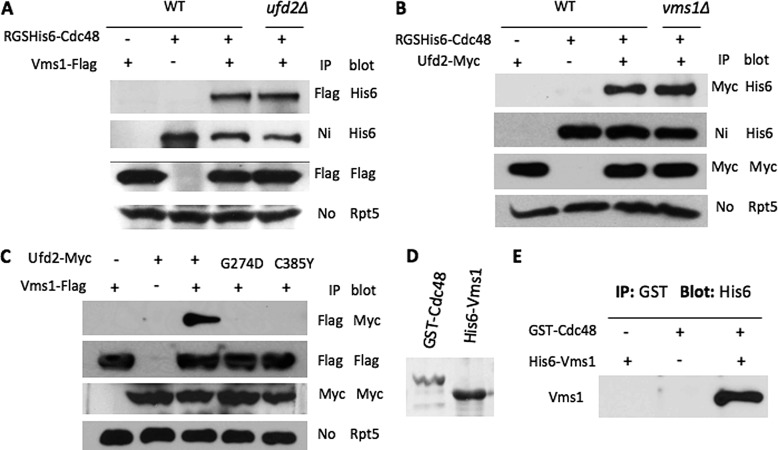
Several genome-wide studies previously revealed that Vms1 may interact with both Cdc48 and Ufd2 (13–15), which was validated by two recent reports (6, 7) while our work was in progress. To understand the interactions among them, we employed various mutants to evaluate whether these interactions (i.e. Ufd2-Cdc48, Vms1-Cdc48, and Vms1-Ufd2) are separate or interdependent. We carried out co-immunoprecipitation experiments in yeast and found that the in vivo binding of Cdc48 to Ufd2 or Vms1 was not affected by the absence of Vms1 or Ufd2, respectively (Fig. 1, A and B). Furthermore, Vms1 interacted with wild-type Ufd2, but not Ufd2 mutants (i.e. G274D and C385Y) defective for Cdc48 binding (Fig. 1C) (4), suggesting that the Vms1-Ufd2 interaction observed is mediated by their separate associations with Cdc48. Although Vms1 was shown to bind Cdc48 in yeast, it was unclear whether the Vms1-Cdc48 interaction is direct or facilitated by other proteins. To this end, we expressed Vms1 and Cdc48 in E. coli and further purified these two proteins (Fig. 1D). In vitro binding reactions using purified proteins indicated that Vms1 directly bound Cdc48 (Fig. 1E). Although many Cdc48 cofactors bind ubiquitin (Ub) chains, we failed to detect Vms1-Ub binding (data not shown).
FIGURE 1.
Interactions among Cdc48, Vms1, and Ufd2. A, co-immunoprecipitation analysis of in vivo interactions between yeast Cdc48 and Vms1 in the presence or absence of Ufd2. Proteins were extracted from yeast cells expressing RGS/His6-tagged Cdc48 and FLAG-tagged Vms1 and immunoprecipitated with beads coupled to various antibodies as indicated. Strains are indicated above the panels. The antibodies used for immunoprecipitation (IP) and Western blotting are indicated on the right of the panels. Equal amounts of protein extracts were used as determined by blotting with anti-Rpt5 antibody (lower panel). B, interactions between Cdc48 and Ufd2 in wild-type or vms1Δ cells. C, associations between Vms1 and Ufd2 derivatives in yeast. Ufd2 alleles defective for Cdc48 binding were described previously (4). D, purification of GST-tagged Cdc48 and His6-tagged Vms1 from E. coli. The Coomassie Blue-stained gel shows purified Cdc48 and Vms1. E, Vms1 directly binds Cdc48 in vitro. Purified GST-Cdc48 and His6-Vms1 were mixed in vitro in the presence of GST-Sepharose and analyzed by Western blotting using anti-His6 antibody.
Vms1 and Cdc48 Are Involved in Cdc13 Turnover
Vms1 was shown to promote mitochondrial protein degradation under stress conditions (6). Vms1 also regulates the degradation of misfolded secretory proteins (7), the effect of which is modest in single vms1Δ mutant cells but much more evident in double mutants harboring a deletion of VMS1 and several other known proteolytic factors (e.g. Ubx4, Ufd2, and Ubx1) involved in protein quality control. However, a specific degradation substrate exhibiting significantly compromised proteolysis in the single vms1Δ mutant is pivotal for the understanding of Vms1 function and has yet to be found under normal growth conditions.
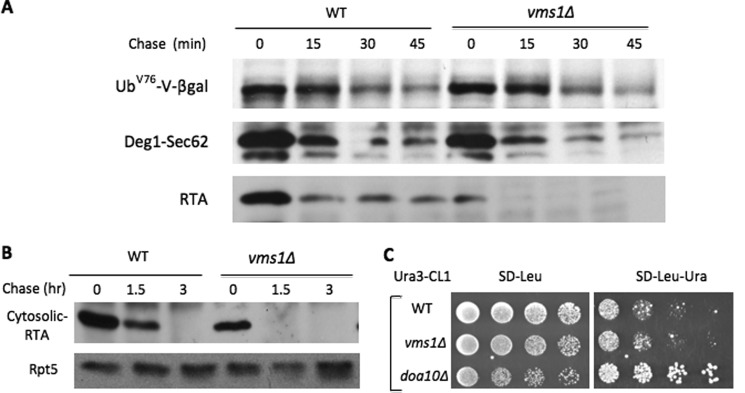
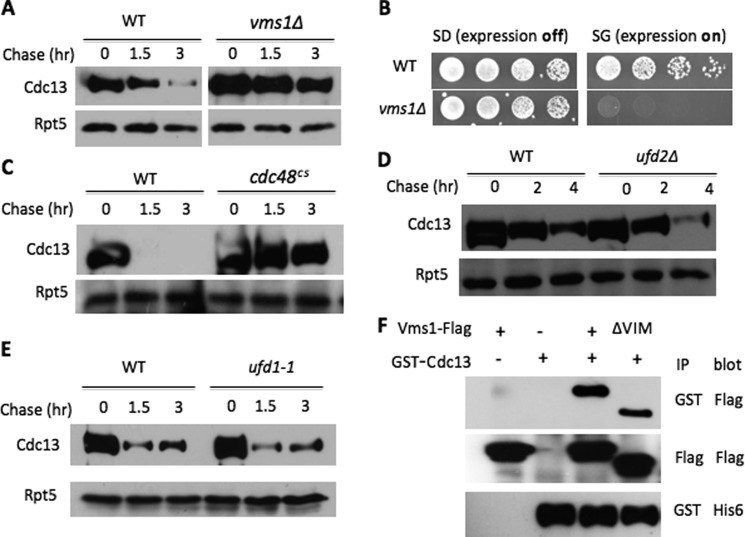
To understand the specific role of Vms1 in proteolysis, we evaluated the requirement of Vms1 for the degradation of a variety of proteasomal substrates localized at different compartments (e.g. nucleus, cytosol, and ER) and/or distinct Ub E3 ligases (e.g. anaphase-promoting complex (APC), SCF, Doa10, and Hrd1). Using pulse-chase or expression shutoff assays, we found that VMS1 deletion did not impair the turnover of a number of substrates tested, including the cytosolic Ub fusion protein UbV76-V-β-gal (Ufd4 E3) (Fig. 2A and supplemental Fig. S1); Deg1-Sec62 (Doa10 E3-regulated membrane protein) (Fig. 2A and supplemental Fig. S1); the misfolded secretory ricin A chain (RTA) allele (Hrd1 E3-regulated luminal protein) (Fig. 2A and supplemental Fig. S1) and a cytosolic version of misfolded RTA (Fig. 2B); cytosolic Ura3-CL1 (Doa10 E3) (Fig. 2C); the nuclear protein Mps1 kinase (APC/cyclosome E3) (data not shown); nuclear cell cycle regulators Clb2 (APC E3), Dbf4 (APC E3), Cdc6 (SCF E3), and Cln2 (SCF E3) (supplemental Fig. S2, A–D and G–J); and two transcription factors, Tfb4 and α1 (supplemental Fig. S2, E, F, K, and L). Interestingly, the degradation of Cdc13 (9), a nuclear telomere-binding protein, appeared markedly compromised in vms1Δ mutant cells (Fig. 3A and supplemental Fig. S3). We previously demonstrated that accumulation of large amounts of substrates can cause toxicity in cells compromised for proteolysis (16). Indeed, Cdc13 overexpression led to growth retardation in vms1Δ mutants (Fig. 3B), supporting the involvement of Vms1 in Cdc13 turnover.
FIGURE 2.
Degradation of many proteasomal substrates is not compromised in vms1Δ cells. A, pulse-chase analysis of various substrates in wild-type and vms1Δ cells. The protein stability of the indicated proteasomal substrates in wild-type or vms1Δ cells was determined as described previously (22). Briefly, cells were pulse-labeled with 35S for 10 min, followed by a chase with unlabeled methionine and cysteine. Samples were collected at various time points for immunoprecipitation and analyzed by autoradiography. B, degradation of cytosolic RTA is not impaired in vms1Δ cells. The signal sequence for the ER on the misfolded RTA allele was removed to construct a cytosolic version of RTA that is degraded by the proteasome. Wild-type and vms1Δ cells containing GAL1 promoter-driven cytosolic RTA were first grown in SR medium. Expression of FLAG-tagged cytosolic RTA was induced by the addition of galactose. Samples were taken after promoter shutoff at the time points indicated and analyzed by anti-FLAG Western blotting. Equal amounts of protein extracts were used and ascertained by blotting with anti-Rpt5 antibody (lower panel). The identities of the proteins are indicated on the left. C, Vms1 does not affect the Ura3-CL1 level. 5-Fold serial dilutions of wild-type, vms1Δ mutant, or doa10Δ mutant cells bearing Ura3-CL1 were spotted onto the indicated media. Plates were incubated for 2 days at 30 °C. The cytosolic Ura3-CL1 fusion protein was previously shown to be degraded by the Doa10 E3 pathway (24). The accumulation of Ura3-CL1 in doa10Δ cells allowed them to grow on medium lacking uracil (right panel).
FIGURE 3.
Vms1 and Cdc48 are required for Cdc13 degradation. A, degradation of Cdc13 in wild-type and vms1Δ cells. Cdc13 stability in the indicated strains was determined as described previously (18). In brief, expression of GST-His6-Cdc13 was induced with galactose. Samples were collected after expression shutoff by the addition of glucose and cycloheximide. Proteins were extracted and incubated with GST beads to enrich Cdc13, followed by Western blotting with anti-His6 antibody. Rpt5 served as a loading control. B, overexpression of Cdc13 causes growth retardation in vms1Δ cells. Wild-type and vms1Δ cells containing GAL1 promoter-regulated Cdc13 were grown to similar densities, and 5-fold serial dilutions were spotted onto SD medium (expression off) or SG medium (expression on). C–E, Cdc13 degradation requires Cdc48, but not Ufd1 and Ufd2. Cdc13 degradation was evaluated in wild-type, cdc48cs, ufd1-1, and ufd2Δ cells. F, Vms1 binds Cdc13. Proteins were extracted from yeast cells expressing GST-His6-tagged Cdc13 and FLAG-Vms1 alleles and immunoprecipitated (IP) with beads coated with the indicated antibodies, followed by immunoblotting with various antibodies.
Consistent with the protein-protein interaction analysis between Vms1 and Cdc48 or Ufd2 described above (Fig. 1), Cdc13 was stabilized in cdc48 mutants, but not ufd2Δ cells (Fig. 3, C and D). The Ub-binding protein Ufd1 is another Cdc48 cofactor that promotes the proteasome-mediated degradation of many Cdc48 targets. Interestingly, Ufd1 and Vms1 seem to bind Cdc48 separately (6, 7). In line with independent binding, Ufd1 was not involved in Cdc13 degradation (Fig. 3E), suggesting that Ufd1 and Vms1 define two distinct Cdc48-dependent degradation pathways.
We examined whether Vms1 interacts with Cdc13. As determined by co-immunoprecipitation analysis, Vms1 recognized Cdc13. Vms1 uses a VCP/Cdc48-interacting motif to bind Cdc48 (6). Interestingly, the VCP/Cdc48-interacting motif deletion of Vms1 retained the ability to bind Cdc13 (Fig. 3F), but not Cdc48, further suggesting that Vms1 may be a substrate recruitment factor for Cdc48.
Involvement of Ub and the Proteasome in Cdc13 Degradation
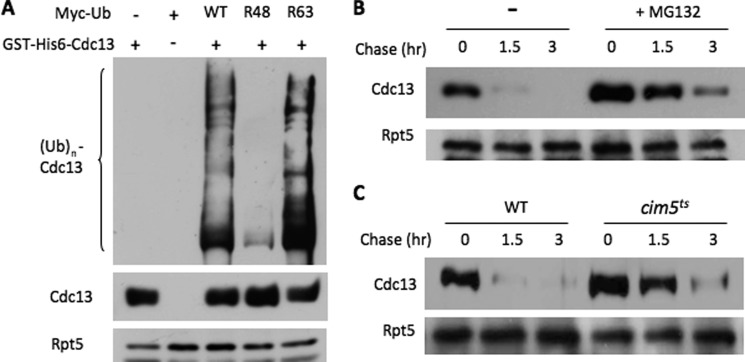
Cdc48 substrates are often marked with Ub molecules signaling for their destruction (1, 5). Cdc13 ubiquitylation was seen in wild-type yeast cells (Fig. 4A). We examined the Ub chain linkages decorated onto Cdc13 using the Ub mutants defective for Ub chain assembly. Mutation of Lys-48 drastically abated Cdc13 ubiquitylation (Fig. 4A). Next, we assessed the contribution of the proteasome in Cdc13 turnover. We found that Cdc13 was partially stabilized upon the addition of the proteasome inhibitor MG132 or in the proteasome mutant cim5 (Fig. 4, B and C, and supplemental Fig. S4), suggesting that Cdc13 is subjected in part to proteasome-mediated destruction.
FIGURE 4.
Cdc13 is ubiquitylated and partially degraded by the proteasome. A, Cdc13 ubiquitylation. GST-His6-Cdc13 was transformed with Myc-tagged Ub derivatives into wild-type cells. Cdc13 was precipitated with GST beads and blotted with anti-Myc antibody for ubiquitylated Cdc13 species (upper panel). The amounts of Cdc13 and Rpt5 in the extracts were also assessed by Western blotting (lower panels). B and C, compromised proteasomal activity partially stabilizes Cdc13. Cdc13 degradation was monitored as described for Fig. 3A in the presence or absence of the proteasome inhibitor MG132 or in cim5ts cells with compromised proteasome function.
Autophagy Plays a Key Role in Cdc13 Degradation
Because significant amounts of Cdc13 were ultimately destroyed in cells with compromised proteasomal activity (Fig. 4, B and C), the above results also indicate that another proteolytic system may be involved in Cdc13 regulation. Besides the proteasome system, autophagy has recently been tied to selective Ub-dependent protein degradation (17). Multiple types of autophagy exist and share the hallmark: cargoes in the cytoplasm are engulfed by a double-membrane vesicle termed the autophagosome and then delivered to the lysosome for destruction by resident hydrolases (10, 11, 17). In response to different environmental stimuli, various adaptations of the core autophagy machinery allow cells to accordingly modulate cellular contents, including organelles (peroxisomes, ribosomes, and mitochondria) and intracellular proteins (e.g. huntingtin and α-synuclein) (10, 11).
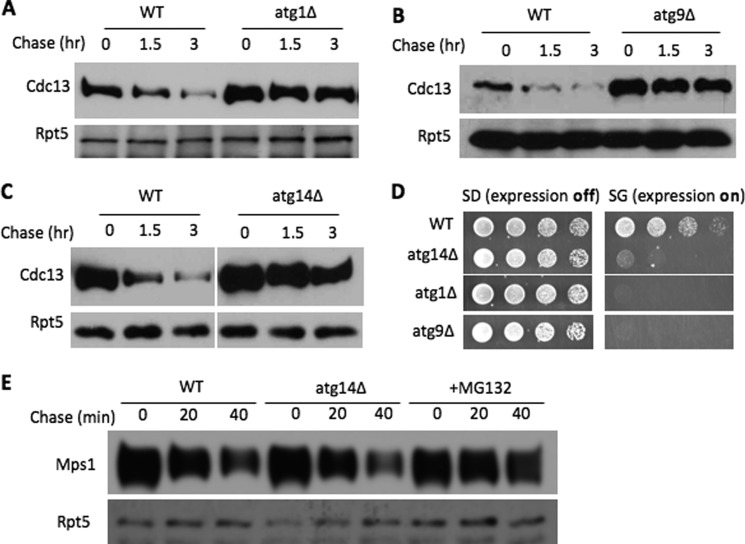
To determine the influence of autophagy on Cdc13 turnover, we first evaluated the degradation kinetics of Cdc13 in cells lacking one of the core components of the autophagy system (Fig. 5). We chose ATG1, ATG9, and ATG14, which are required for various steps of autophagy, including induction of autophagy and vesicle formation (10–12). ATG1 encodes a conserved Ser/Thr kinase critical for the activation of autophagy. Atg14 is a subunit of the phosphatidylinositol 3-kinase complex essential for establishing the pre-autophagosomal structure, a precursor to the autophagosome. Atg9 is a membrane protein proposed to be key in trafficking the membrane to the pre-autophagosomal structure. Interestingly, Cdc13 was efficiently degraded in wild-type cells but markedly stabilized in these autophagy mutants (Fig. 5, A–C, and supplemental Fig. S5), suggesting the involvement of autophagy in Cdc13 regulation. In contrast, the degradation of another nuclear protein, Mps1 (18), was minimally affected by ATG14 deletion but inhibited by the proteasome inhibitor (Fig. 5E), indicating the specific involvement of autophagy in Cdc13 degradation. Consistent with this idea, Cdc13 overexpression caused growth retardation in cells lacking these core autophagy genes (Fig. 5D).
FIGURE 5.
Autophagy promotes Cdc13 turnover. A–C, Cdc13 stability was determined in wild-type, atg1Δ, atg9Δ, or atg14Δ cells. D, Cdc13 overexpression inhibits growth of atg1Δ, atg9Δ, or atg14Δ cells. Serial dilutions of the indicated yeast cells bearing a plasmid for inducible Cdc13 expression were plated as described for Fig. 3B. E, the degradation of the nuclear protein kinase Mps1 requires the proteasome, but not autophagy. GST-His6-tagged Mps1 degradation was performed under conditions similar to those used for Cdc13 turnover and has been described previously (18).
To further understand the role of these genes in Cdc13 turnover, we monitored Cdc13 localization. In wild-type cells, Cdc13 appeared mainly in the nucleus (supplemental Fig. S6A), consistent with its nuclear function in telomere regulation. Two hours after the expression shutoff, the Cdc13 signal became faint, likely due to ongoing proteolysis. In contrast, a significant amount of Cdc13 in atg1Δ cells was observed outside the nucleus and formed a few foci (supplemental Fig. S6B), which were persistent after expression shutoff, suggesting that autophagy is likely involved in the cytosolic degradation of Cdc13. Interestingly, in vms1Δ mutants, Cdc13 was found in the cytosol as well, with a few foci present (supplemental Fig. S6C). In wild-type cells treated with the proteasome inhibitor, Cdc13 appeared mainly in the nucleus (supplemental Fig. S7), suggesting that the proteasome-mediated Cdc13 destruction likely operates in the nucleus.
Requirements of Various Autophagy Pathways in Cdc13 Regulation
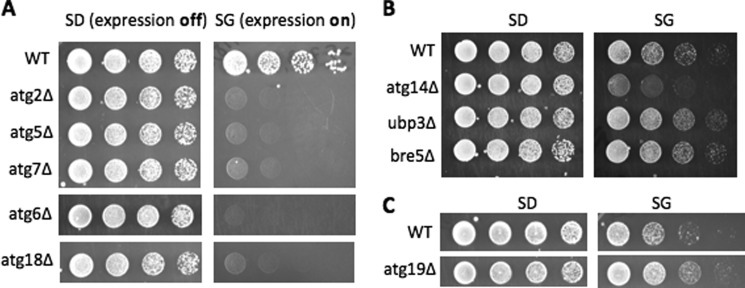
Different autophagy pathways are employed for distinct cargoes. Because Cdc13 overexpression triggered toxicity in vms1Δ mutants and ATG-deficient cells, this offers a convenient assay to assess the involvement of various autophagy pathways in Cdc13 degradation. First, to ascertain the involvement of autophagy in Cdc13 regulation, we assayed the effect of Cdc13 expression in cells lacking other core components of autophagy (Fig. 6A), including Atg5 and Atg7, which are part of the Ub-like conjugation system critical for the biogenesis of autophagosome-related membranes (10–12). As expected, Cdc13 overexpression caused growth inhibition in mutants lacking these factors (Fig. 6A).
FIGURE 6.
Cdc13 overexpression does not trigger toxicity in all autophagy mutants. A–C, yeast cells defective for different types of autophagy (e.g. ribophagy and Cvt) were transformed with plasmid pEGH-Cdc13. Serial dilutions of wild-type and the indicated mutant cells were spotted onto SD or SG plates.
We then assessed the requirement of the Ubp3-Bre5 ubiquitin protease complex, which is specifically involved in the autophagosome-dependent removal of excess ribosomes (a process termed ribophagy) but is not involved in other types of autophagy (19). Like wild-type cells, ubp3Δ and bre5Δ cells could tolerate Cdc13 overexpression (Fig. 6B), suggesting that Cdc13 degradation is likely ribophagy-independent. One of the best characterized autophagy pathways is the cytosol-to-vacuole transport (Cvt) pathway (11), which yeast cells employ to deliver two lysosomal hydrolases (e.g. Ape1 and Ams1) to the lysosome (vacuole in yeast). Atg19 is a cargo receptor only for the Cvt pathway, but not other types of autophagy (11). Interestingly, Cdc13 overexpression did not cause adverse growth effects in atg19Δ cells (Fig. 6C), suggesting Cdc13 does not require the Cvt pathway for its destruction. Cdc13 may employ a novel autophagy pathway. We will combine the toxicity and stability assays to identify relevant factors involved in Cdc13 regulation.
Rapamycin Promotes Cdc13 Degradation
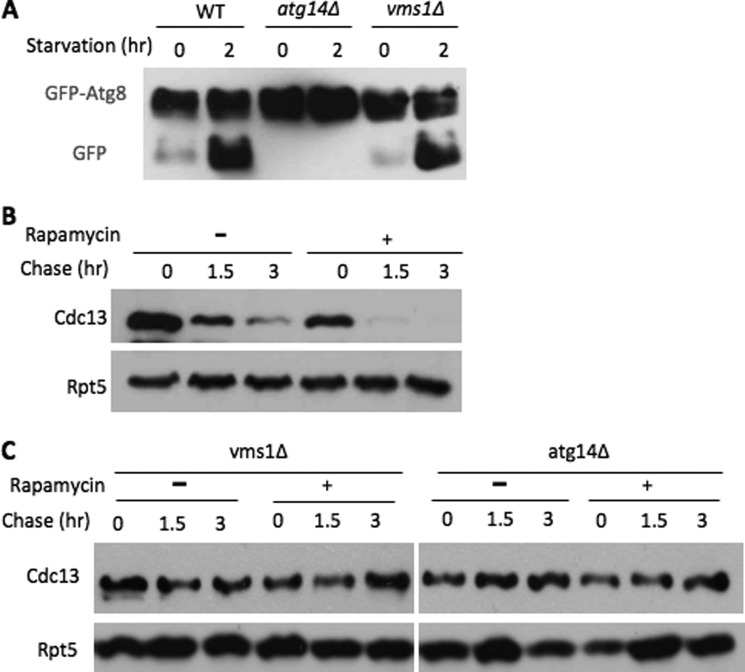
We next wanted to investigate the functional relationship between Vms1 and autophagy. Two classic autophagy pathways, nutrient starvation-induced autophagy and the Cvt pathway, were not compromised in cells lacking VMS1 (Fig. 7A) (7), suggesting that Vms1 is not a core component of the autophagy system. Three key autophagy regulators (i.e. Atg1, Atg5, and Atg8) were present at similar levels in wild-type and vms1Δ cells (supplemental Fig. S8), suggesting that there is no global alteration of expression levels of autophagy proteins in vms1Δ cells. The antibiotic rapamycin is a well known autophagy inducer through inhibition of Tor kinase (10, 12). Upon rapamycin addition, Cdc13 degradation was accelerated (Fig. 7B), indicating that enhanced autophagy activity stimulates Cdc13 destruction. In contrast, Cdc13 remained stable in cells lacking ATG14 or VMS1 upon rapamycin treatment (Fig. 7C), suggesting that the effect of rapamycin on Cdc13 requires the function of ATG14 and VMS1.
FIGURE 7.
Link between Vms1 and autophagy. A, nitrogen starvation-induced autophagy remains intact in vms1Δ cells. GFP-Atg8 was employed to monitor the nutrient deprivation-triggered autophagy (23). Briefly, wild-type, atg14Δ, and vms1Δ cells harboring GFP-Atg8 were grown in uracil-free SD medium to log phase. The cells were then shifted to starvation-inducing nitrogen-free SD medium. Samples were harvested at intervals and analyzed by Western blotting using anti-GFP antibody. Starvation-triggered autophagy leads to the cleavage of GFP-Atg8 (23), which is known to generate a free GFP moiety in wild-type cells, but not atg14Δ cells. B and C, Cdc13 stability was determined with or without rapamycin (2 μg/ml) in wild-type, atg14Δ, and vms1Δ cells.
DISCUSSION
Cdc48 plays important regulatory roles in diverse biological processes that are essential for cell survival and growth (1–3). The functional diversity of Cdc48 is achieved through different binding partners (e.g. Ufd1–3, SVIP, Png1, and Ubx1–7). A key to delineation of the specific role of Cdc48 in a particular process is to understand the cellular function of the individual Cdc48 cofactors involved (1, 2). One poorly understood Cdc48 cofactor is Vms1, a highly conserved eukaryotic protein (6, 7). Vms1 seems to exist in a Cdc48-Npl4 complex without Ufd1 (6, 7), a well characterized Cdc48-binding protein engaged in proteasome-mediated proteolysis (1, 2). We have demonstrated the direct binding between Vms1 and Cdc48. Vms1 promotes mitochondrial protein degradation under stress conditions (6). The majority of Vms1 remains in the cytosol (6). Vms1 collaborates with several proteolysis regulators (e.g. Ubx1 and Ufd2) for efficient elimination of a subset of misfolded ER-associated degradation proteins (7), although the specific mechanism is unclear. The identification of the telomere regulator Cdc13 as a Vms1 target expands the biological function of the Vms1-Cdc48 pathway. The toxicity associated with Cdc13 accumulation in vms1Δ cells further underscores the functional significance of Cdc13 regulation. Interestingly, Vms1 can bind Cdc13 without its VCP/Cdc48-interacting motif, suggesting that Vms1 may be a substrate recognition factor.
Our study also establishes Cdc13 as one of the few ubiquitylated proteins in yeast subject to autophagy-mediated degradation (10, 17). The renaissance of autophagy was initially propelled in part by the molecular dissection of the nitrogen starvation-induced autophagy process and the Cvt pathway in yeast and brought to the forefront in cell biology by its ties to human diseases, including cancer and neurodegenerative disorders (10, 12). Although autophagy was previously considered a nonselective degradation process, recent studies indicate that ubiquitylated proteins are specifically targeted for elimination by the autophagy system, although the underlying molecular mechanism remains obscure (17). Several Ub-binding proteins (e.g. p62, NBR1, Cdc48, and PLIC) have been implicated in bringing ubiquitylated substrates to the autophagy machinery in mammalian cells. Interestingly, mutations in human Cdc48 (also called VCP) that impair its function in autophagy lead to a degenerative disorder termed inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia (20, 21). It will be important to determine whether human Vms1 is involved in this disorder.
The participation of autophagy in a subset of regulated proteolysis, which is handled mainly by the proteasome, remains poorly understood. The identification and characterization of cis-elements and trans-factors critical for routing Cdc13 to specific degradation pathways should provide significant insight into the principles governing selective proteolysis on multiple fronts, including understanding the functioning of the Vms1 branch of the Cdc48 pathway, the mechanism underlying autophagy-mediated protein turnover, and the coordination between autophagy and the proteasome in modulating protein levels and various cellular events.
Acknowledgments
We are grateful to Drs. J. Laney, T. Suzuki, M. Hochstrasser, D. J. Klionsky, and Y. Ohsumi for plasmids. We thank C. Liu and V. Choe for helpful discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants GM078085 and P30 CA054174 and United States Department of Defense Grant W911NF-11-10466 (to H. R.).

This article contains supplemental Figs. S1–S8.
- ER
- endoplasmic reticulum
- Ub
- ubiquitin
- APC
- anaphase-promoting complex
- RTA
- ricin A chain
- Cvt
- cytosol-to-vacuole.
REFERENCES
- 1. Meyer H., Bug M., Bremer S. (2012) Emerging functions of the VCP/p97 AAA-ATPase in the ubiquitin system. Nat. Cell Biol. 14, 117–123 [DOI] [PubMed] [Google Scholar]
- 2. Stolz A., Hilt W., Buchberger A., Wolf D. H. (2011) Cdc48: a power machine in protein degradation. Trends Biochem. Sci. 36, 515–523 [DOI] [PubMed] [Google Scholar]
- 3. Ramanathan H. N., Ye Y. (2011) Revoking the cellular license to replicate: yet another AAA assignment. Mol. Cell 44, 3–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baek G. H., Kim I., Rao H. (2011) The Cdc48 ATPase modulates the interaction between two proteolytic factors, Ufd2 and Rad23. Proc. Natl. Acad. Sci. U.S.A. 108, 13558–13563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Buchberger A. (2010) Control of ubiquitin conjugation by Cdc48 and its cofactors. Subcell. Biochem. 54, 17–30 [DOI] [PubMed] [Google Scholar]
- 6. Heo J. M., Livnat-Levanon N., Taylor E. B., Jones K. T., Dephoure N., Ring J., Xie J., Brodsky J. L., Madeo F., Gygi S. P., Ashrafi K., Glickman M. H., Rutter J. (2010) A stress-responsive system for mitochondrial protein degradation. Mol. Cell 40, 465–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tran J. R., Tomsic L. R., Brodsky J. L. (2011) A Cdc48p-associated factor modulates endoplasmic reticulum-associated degradation, cell stress, and ubiquitinated protein homeostasis. J. Biol. Chem. 286, 5744–5755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pinto A. R., Li H., Nicholls C., Liu J. P. (2011) Telomere protein complexes and interactions with telomerase in telomere maintenance. Front. Biosci. 16, 187–207 [DOI] [PubMed] [Google Scholar]
- 9. Tseng S. F., Shen Z. J., Tsai H. J., Lin Y. H., Teng S. C. (2009) Rapid Cdc13 turnover and telomere length homeostasis are controlled by Cdk1-mediated phosphorylation of Cdc13. Nucleic Acids Res. 37, 3602–3611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang Z., Klionsky D. J. (2010) Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lynch-Day M. A., Klionsky D. J. (2010) The Cvt pathway as a model for selective autophagy. FEBS Lett. 584, 1359–1366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms: lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 [DOI] [PubMed] [Google Scholar]
- 13. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 14. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. (2002) Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 15. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrín-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 16. Liu C., van Dyk D., Xu P., Choe V., Pan H., Peng J., Andrews B., Rao H. (2010) Ubiquitin chain elongation enzyme Ufd2 regulates a subset of Doa10 substrates. J. Biol. Chem. 285, 10265–10272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kirkin V., Dikic I. (2011) Ubiquitin networks in cancer. Curr. Opin. Genet. Dev. 21, 21–28 [DOI] [PubMed] [Google Scholar]
- 18. Liu C., van Dyk D., Choe V., Yan J., Majumder S., Costanzo M., Bao X., Boone C., Huo K., Winey M., Fisk H., Andrews B., Rao H. (2011) Ubiquitin ligase Ufd2 is required for efficient degradation of Mps1 kinase. J. Biol. Chem. 286, 43660–43667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kraft C., Deplazes A., Sohrmann M., Peter M. (2008) Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10, 602–610 [DOI] [PubMed] [Google Scholar]
- 20. Watts G. D., Wymer J., Kovach M. J., Mehta S. G., Mumm S., Darvish D., Pestronk A., Whyte M. P., Kimonis V. E. (2004) Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 36, 377–381 [DOI] [PubMed] [Google Scholar]
- 21. Badadani M., Nalbandian A., Watts G. D., Vesa J., Kitazawa M., Su H., Tanaja J., Dec E., Wallace D. C., Mukherjee J., Caiozzo V., Warman M., Kimonis V. E. (2010) VCP-associated inclusion body myopathy and Paget disease of bone knock-in mouse model exhibits tissue pathology typical of human disease. PLoS ONE 5, e13183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kim I., Ahn J., Liu C., Tanabe K., Apodaca J., Suzuki T., Rao H. (2006) The Png1-Rad23 complex regulates glycoprotein turnover. J. Cell Biol. 172, 211–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nair U., Thumm M., Klionsky D. J., Krick R. (2011) GFP-Atg8 protease protection as a tool to monitor autophagosome biogenesis. Autophagy 7, 1546–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ravid T., Kreft S. G., Hochstrasser M. (2006) Membrane and soluble substrates of the Doa10 ubiquitin ligase are degraded by distinct pathways. EMBO J. 25, 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]