Abstract
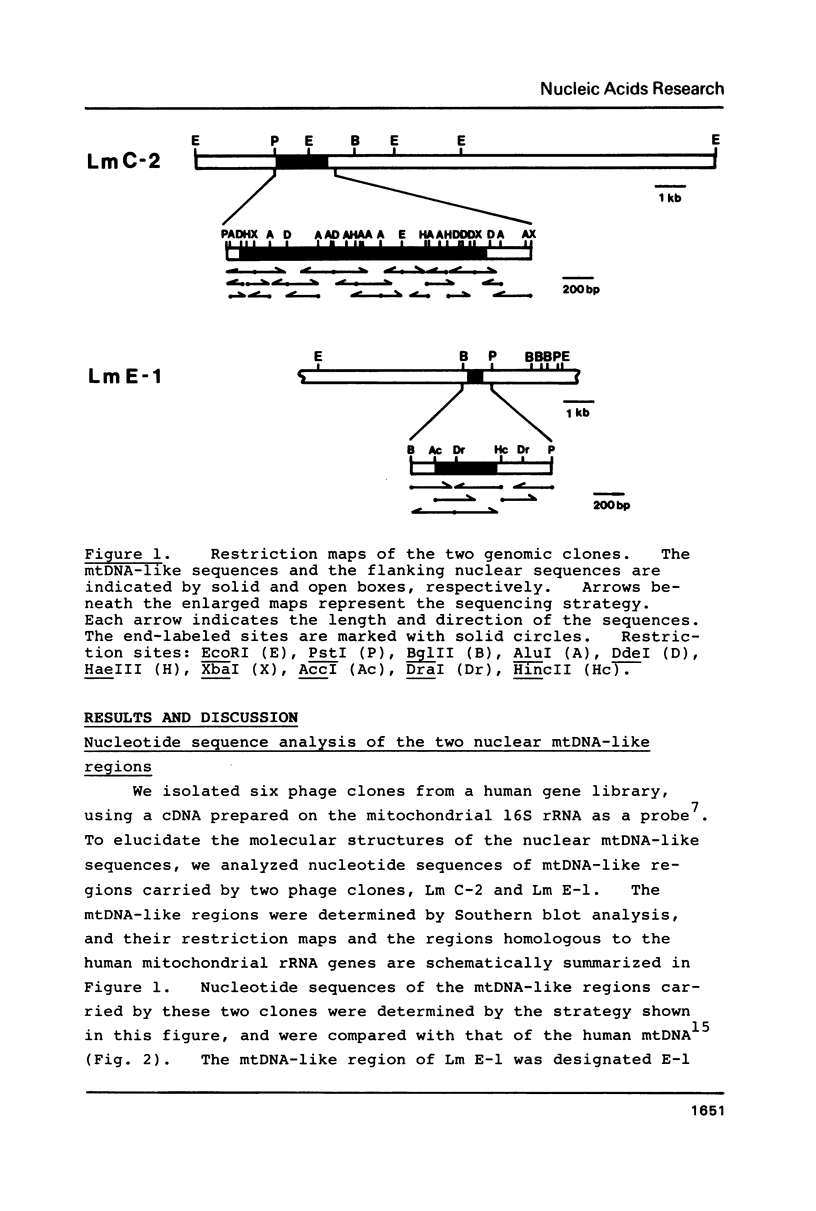
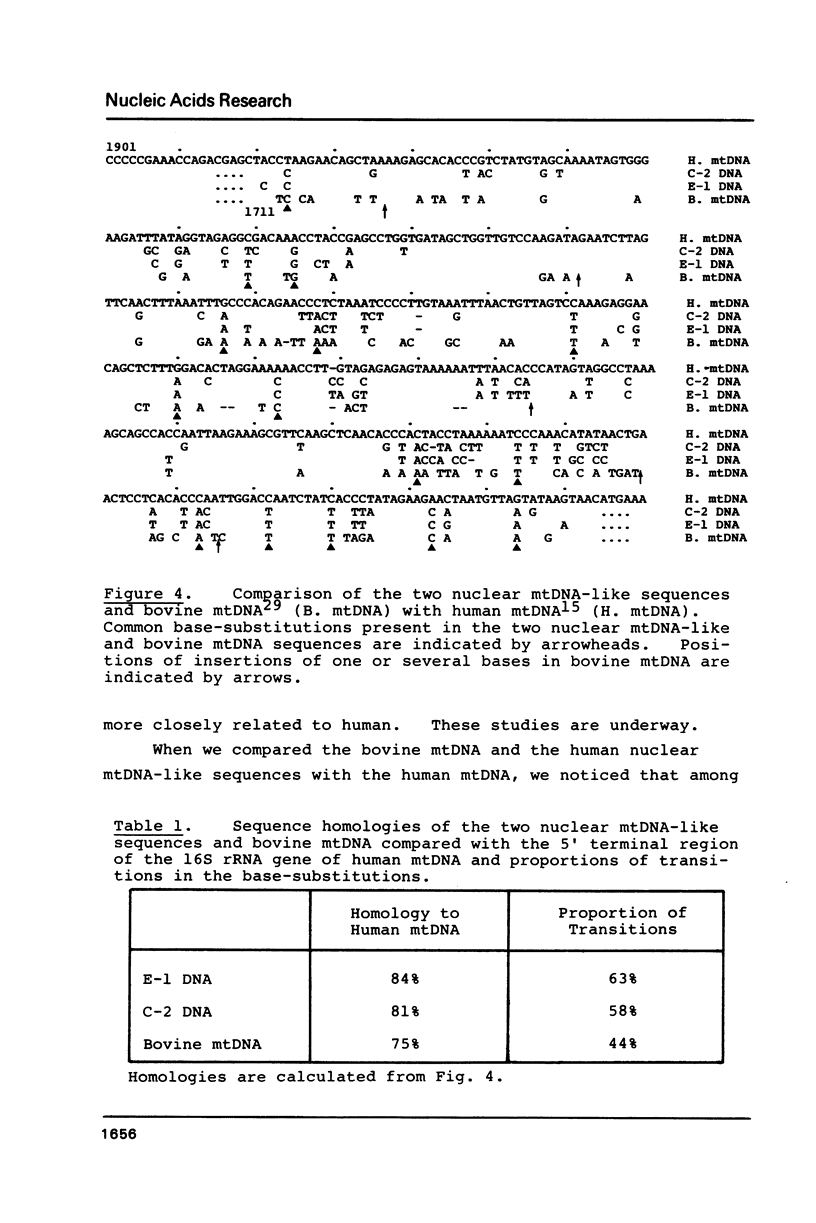
Two lambda phage clones carrying mitochondrial-DNA-like (mtDNA-like) sequences isolated from a human gene library were named Lm E-1 and Lm C-2, and their DNA structures were characterized. Lm E-1 contains about 0.4 kb DNA homologous to the 5' portion of the mitochondrial 16S ribosomal RNA (rRNA) gene and Lm C-2, a 1.6 kb DNA homologous to the 3' portion of the 12S rRNA gene and to almost all of the 16S rRNA gene. Comparisons of their nucleotide sequences with those of the corresponding regions of the human mtDNA revealed no detectable DNA rearrangement and their homologies to the human mtDNA are 84% and 80%, respectively. There are neither terminal repeats in the nuclear mtDNA-like sequences nor duplications of the nuclear DNAs flanking the mtDNA-like sequences. Evolutionary relationship between these two human nuclear mtDNA-like sequences and the human and bovine mtDNAs is discussed.
Full text
PDF







Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anderson S., Bankier A. T., Barrell B. G., de Bruijn M. H., Coulson A. R., Drouin J., Eperon I. C., Nierlich D. P., Roe B. A., Sanger F. Sequence and organization of the human mitochondrial genome. Nature. 1981 Apr 9;290(5806):457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- Anderson S., de Bruijn M. H., Coulson A. R., Eperon I. C., Sanger F., Young I. G. Complete sequence of bovine mitochondrial DNA. Conserved features of the mammalian mitochondrial genome. J Mol Biol. 1982 Apr 25;156(4):683–717. doi: 10.1016/0022-2836(82)90137-1. [DOI] [PubMed] [Google Scholar]
- Bloom G. D. A nucleus with cytoplasmic features. J Cell Biol. 1967 Oct;35(1):266–268. doi: 10.1083/jcb.35.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandes D., Schofield B. H., Anton E. Nuclear mitochondria? Science. 1965 Sep 17;149(3690):1373–1374. doi: 10.1126/science.149.3690.1373. [DOI] [PubMed] [Google Scholar]
- Brown W. M., Prager E. M., Wang A., Wilson A. C. Mitochondrial DNA sequences of primates: tempo and mode of evolution. J Mol Evol. 1982;18(4):225–239. doi: 10.1007/BF01734101. [DOI] [PubMed] [Google Scholar]
- Bullock P., Forrester W., Botchan M. DNA sequence studies of simian virus 40 chromosomal excision and integration in rat cells. J Mol Biol. 1984 Mar 25;174(1):55–84. doi: 10.1016/0022-2836(84)90365-6. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Wright R. M. DNA sequence of the excision sites of a mitochondrial plasmid from senescent Podospora anserina. Nucleic Acids Res. 1983 Apr 11;11(7):2111–2119. doi: 10.1093/nar/11.7.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrelly F., Butow R. A. Rearranged mitochondrial genes in the yeast nuclear genome. Nature. 1983 Jan 27;301(5898):296–301. doi: 10.1038/301296a0. [DOI] [PubMed] [Google Scholar]
- Gellissen G., Bradfield J. Y., White B. N., Wyatt G. R. Mitochondrial DNA sequences in the nuclear genome of a locust. Nature. 1983 Feb 17;301(5901):631–634. doi: 10.1038/301631a0. [DOI] [PubMed] [Google Scholar]
- Hadler H. I., Dimitrijevic B., Mahalingam R. Mitochondrial DNA and nuclear DNA from normal rat liver have a common sequence. Proc Natl Acad Sci U S A. 1983 Nov;80(21):6495–6499. doi: 10.1073/pnas.80.21.6495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs H. T., Posakony J. W., Grula J. W., Roberts J. W., Xin J. H., Britten R. J., Davidson E. H. Mitochondrial DNA sequences in the nuclear genome of Strongylocentrotus purpuratus. J Mol Biol. 1983 Apr 25;165(4):609–632. doi: 10.1016/s0022-2836(83)80270-8. [DOI] [PubMed] [Google Scholar]
- Klug H. Zum Vorkommen von Mitochondrien im Zellkern. Naturwissenschaften. 1966 Jul;53(13):339–339. doi: 10.1007/BF00631212. [DOI] [PubMed] [Google Scholar]
- Kuhara S., Matsuo F., Futamura S., Fujita A., Shinohara T., Takagi T., Sakaki Y. GENAS: a database system for nucleic acid sequence analysis. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):89–99. doi: 10.1093/nar/12.1part1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Miyata T., Yasunaga T. Rapidly evolving mouse alpha-globin-related pseudo gene and its evolutionary history. Proc Natl Acad Sci U S A. 1981 Jan;78(1):450–453. doi: 10.1073/pnas.78.1.450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mounts P., Kelly T. J., Jr Rearrangements of host and viral DNA in mouse cells transformed by simian virus 40. J Mol Biol. 1984 Aug 15;177(3):431–460. doi: 10.1016/0022-2836(84)90294-8. [DOI] [PubMed] [Google Scholar]
- Nomiyama H., Tsuzuki T., Wakasugi S., Fukuda M., Shimada K. Interruption of a human nuclear sequence homologous to mitochondrial DNA by a member of the KpnI 1.8 kb family. Nucleic Acids Res. 1984 Jul 11;12(13):5225–5234. doi: 10.1093/nar/12.13.5225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva H., Valle A., Flores L. D., Rivas M. C. Intranuclear mitochondriae in Hodgkin's disease. Virchows Arch B Cell Pathol. 1973 Jan 31;12(2):189–194. doi: 10.1007/BF02893997. [DOI] [PubMed] [Google Scholar]
- Savageau M. A., Metter R., Brockman W. W. Statistical significance of partial base-pairing potential: implications for recombination of SV40 DNA in eukaryotic cells. Nucleic Acids Res. 1983 Sep 24;11(18):6559–6570. doi: 10.1093/nar/11.18.6559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz M., Doerfler W. Deletion of cellular DNA at site of viral DNA insertion in the adenovirus type 12-induced mouse tumor CBA-12-1-T. Nucleic Acids Res. 1984 Jun 25;12(12):4959–4976. doi: 10.1093/nar/12.12.4959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stringer J. R. DNA sequence homology and chromosomal deletion at a site of SV40 DNA integration. Nature. 1982 Mar 25;296(5855):363–366. doi: 10.1038/296363a0. [DOI] [PubMed] [Google Scholar]
- Stringer J. R. Integrated simian virus 40 DNA: nucleotide sequences at cell-virus recombinant junctions. J Virol. 1981 May;38(2):671–679. doi: 10.1128/jvi.38.2.671-679.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuzuki T., Nomiyama H., Setoyama C., Maeda S., Shimada K., Pestka S. The majority of cDNA clones with strong positive signals for the interferon-induction-specific sequences resemble mitochondrial ribosomal RNA genes. Biochem Biophys Res Commun. 1983 Jul 29;114(2):670–676. doi: 10.1016/0006-291x(83)90833-1. [DOI] [PubMed] [Google Scholar]
- Tsuzuki T., Nomiyama H., Setoyama C., Maeda S., Shimada K. Presence of mitochondrial-DNA-like sequences in the human nuclear DNA. Gene. 1983 Nov;25(2-3):223–229. doi: 10.1016/0378-1119(83)90226-3. [DOI] [PubMed] [Google Scholar]
- Woodworth-Gutai M. Recombination in SV40-infected cells: nucleotide sequences at viral-viral recombinant joints in naturally arising variants. Virology. 1981 Mar;109(2):344–352. doi: 10.1016/0042-6822(81)90505-5. [DOI] [PubMed] [Google Scholar]
- Wright R. M., Cummings D. J. Integration of mitochondrial gene sequences within the nuclear genome during senescence in a fungus. Nature. 1983 Mar 3;302(5903):86–88. doi: 10.1038/302086a0. [DOI] [PubMed] [Google Scholar]
- Wright R. M., Horrum M. A., Cummings D. J. Are mitochondrial structural genes selectively amplified during senescence in Podospora anserina? Cell. 1982 Jun;29(2):505–515. doi: 10.1016/0092-8674(82)90167-2. [DOI] [PubMed] [Google Scholar]
- van den Boogaart P., Samallo J., Agsteribbe E. Similar genes for a mitochondrial ATPase subunit in the nuclear and mitochondrial genomes of Neurospora crassa. Nature. 1982 Jul 8;298(5870):187–189. doi: 10.1038/298187a0. [DOI] [PubMed] [Google Scholar]



