Background: Interaction between DNA polymerase α-primase (Pol-prim) and the viral helicase (LTag) is critical for SV40 primosome activity, but the protein interfaces remain poorly characterized.
Results: The detailed LTag interface with the Pol-prim regulatory subunit is revealed by co-crystal structure and confirmed by mutational analysis.
Conclusion: Pol-prim/LTag interaction activates the SV40 primosome.
Significance: The LTag complex provides structural insight into a eukaryotic helicase-primase assembly.
Keywords: Crystallography, DNA Helicase, DNA Polymerase, DNA Primase, DNA Replication, SV40 T Antigen, Deformable Elastic Network Refinement, Primosome
Abstract
DNA polymerase α-primase (Pol-prim) plays an essential role in eukaryotic DNA replication, initiating synthesis of the leading strand and of each Okazaki fragment on the lagging strand. Pol-prim is composed of a primase heterodimer that synthesizes an RNA primer, a DNA polymerase subunit that extends the primer, and a regulatory B-subunit (p68) without apparent enzymatic activity. Pol-prim is thought to interact with eukaryotic replicative helicases, forming a dynamic multiprotein assembly that displays primosome activity. At least three subunits of Pol-prim interact physically with the hexameric replicative helicase SV40 large T antigen, constituting a simple primosome that is active in vitro. However, structural understanding of these interactions and their role in viral chromatin replication in vivo remains incomplete. Here, we report the detailed large T antigen-p68 interface, as revealed in a co-crystal structure and validated by site-directed mutagenesis, and we demonstrate its functional importance in activating the SV40 primosome in cell-free reactions with purified Pol-prim, as well as in monkey cells in vivo.
Introduction
All cells begin de novo DNA replication using RNA primers that are synthesized by a primase on single-stranded template DNA and then extended by a processive DNA polymerase. In prokaryotic cells, primer synthesis is coupled with unwinding of the parental DNA by ring-shaped hexameric replicative helicases, viral or cellular, that translocate with 5′ to 3′ polarity and a single-stranded DNA (ssDNA)6-binding protein. Dynamic physical and functional interactions among the primase, helicase, and ssDNA-binding proteins, which constitute a primosome, coordinate their activities at the replication fork (1–4). In eukaryotic cells, the primase activity is part of a DNA polymerase α-primase complex (Pol-prim), but understanding of how its activity is coordinated with that of the eukaryotic Cdc45-MCM-GINS replicative helicases and replication protein A (RPA), the major ssDNA-binding protein, remains incomplete (5–7).
Pol-prim consists of four subunits (8, 9). The catalytic primase subunit p48 associates with p58 to form a stable heterodimer that synthesizes an RNA primer of 8–12 nucleotides and then, through a so far elusive mechanism, transfers it internally to the associated p180 subunit (10–12). The p180 DNA polymerase subunit then elongates the RNA primers into RNA/DNA primers of about 30–35 nucleotides. The p68 or B-subunit is not essential for the enzymatic activities of Pol-prim, but it is essential in vivo and may regulate Pol-prim function in response to cell cycle signaling or at telomeres (13–17). Structure-function studies indicate that the C-terminal zinc domain of p180 anchors both the p68 subunit and the p58 subunit of the primase dimer in the complex (9, 18). Unexpectedly, these studies also revealed that several hundred residues at the N termini of the p180 and p68 subunits were dispensable for primase and polymerase activities (9, 17) and hence were presumed to be unstructured (18). Single particle electron microscopy reconstructions of Pol-prim domains and subcomplexes lacking the N-terminal regions suggest a modular architecture composed of globular domains joined by flexible linkers (18, 19). Crystal structures of the p58 C-terminal domain from human and yeast (20–22) and an NMR solution structure of the N-terminal domain of human p68 (23) have also recently become available. To our knowledge, no structures of Pol-prim subunits or domains in complex with a replicative helicase are currently available.
Here, we use SV40 DNA replication in a cell-free reaction as a model to study the role of human Pol-prim in a simple primosome. The purified hexameric replicative helicase SV40 large T antigen (LTag), human Pol-prim, human RPA, and any topoisomerase that can relax positively supercoiled DNA (Escherichia coli DNA gyrase and human topoisomerases I and II) are sufficient to initiate replication of supercoiled SV40 DNA (24–26). LTag is a modular protein with three structured domains joined by flexible peptides (Fig. 1A). LTag unwinds the origin DNA, recruits RPA to the template, and then orchestrates RPA displacement by human Pol-prim for primer synthesis and extension. In a simpler reaction utilizing RPA-coated ssDNA as the template, LTag and Pol-prim are sufficient to reconstitute SV40 primosome activity even in the absence of LTag helicase activity (27–30). These findings suggested that the key role of LTag in primosome activity may be to displace RPA and, in concert, position Pol-prim on the exposed ssDNA. As in prokaryotic primosomes, weak physical interactions among LTag, Pol-prim, and RPA are observed and have been implicated in primosome activity (14, 23, 27, 30–33). In solution, hexameric LTag binds to the Pol-prim heterotetramer with 1:1 stoichiometry, in which at least three subunits of Pol-prim contact LTag (Fig. 1B) (34, 35). However, the overall architecture of this primosome assembly, the functional interplay among the proteins that give rise to activity, and the path of the DNA through the assembly are not known.
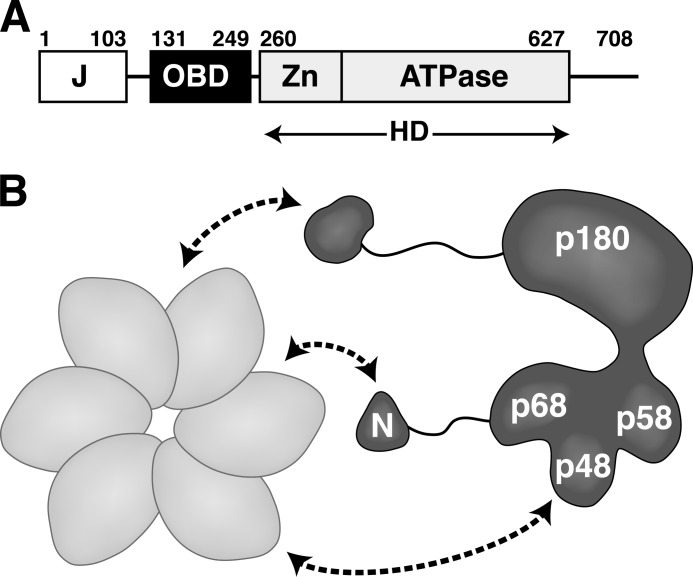
FIGURE 1.
Schematic diagram of the interactions of hexameric LTag with Pol-prim. A, modular architecture of SV40 LTag (27, 51). Each of the three structured domains is sufficient for its biochemical activity as follows: DnaJ chaperone domain (J), origin DNA binding domain (OBD), and helicase domain (HD), composed of the zinc (Zn) and ATPase subdomains, are boxed. Flexible linker regions are indicated as lines. B, hexameric helicase domain of LTag (light gray) contacts Pol-prim (dark gray) through at least three subunits (curved arrows). The N-terminal regions (N) of p180 and p68 are dispensable for enzymatic activity but interact physically with LTag (9, 17, 23, 60).
To develop a detailed mechanistic understanding of this simple eukaryotic primosome, we have begun to define the physical interactions among these three modular proteins and their potential functional roles in primosome activity. The previously unrecognized N-terminal domain of Pol-prim p68 (p68N, residues 1–78), a four-helix bundle, interacts with LTag helicase domain (residues 260–627). A p68N truncation in a mutant Pol-prim complex abolishes SV40 primosome activity in two different assays, implicating p68N-LTag interaction in primosome activity (23). On the surface of p68N, a small hydrophobic patch flanked by acidic charges docks on LTag and is needed for SV40 primosome activity (23). An initial screen for a corresponding docking interface on LTag identified Lys-425 as important for docking and for primosome activity (27). However, the absence of a hydrophobic patch adjacent to Lys-425 on the LTag that could complement the docking surface on p68N was puzzling (27), raising questions about the role of Lys-425 in the p68-LTag binding interface and in primosome activity.
In this study, we have re-examined the LTag-p68N interface using protein crystallography. We present a 5-Å crystal structure of the p68N domain of human Pol-prim bound to LTag hexameric helicase. We use newly developed refinement methods to show for the first time the detailed molecular interactions between p68 and LTag. Mutational analysis of residues within the interface confirms the importance of these sites for LTag-p68 interaction and for primosome activity, indicating that this site represents one of the docking sites critical for assembling the functional architecture of the Pol-prim·helicase complex of the SV40 primosome.
EXPERIMENTAL PROCEDURES
Expression and Purification of SV40 LTag and p68N
The LTag helicase domain (residues 260–627) was expressed in E. coli as a thrombin-cleavable GST-LTag fusion protein using the pGEX-2T vector as described (36). The fusion protein was isolated by glutathione affinity chromatography, and LTag released by thrombin cleavage was further purified by Superdex-200 gel filtration chromatography (GE Healthcare). The N-terminal domain (residues 1–78) of human p68 (p68N) was expressed in E. coli as a PreScission protease-cleavable His6 tag fusion protein and purified by Ni-NTA column chromatography as described (23). After on-column cleavage of the fusion protein, p68N was further purified by S75 size exclusion chromatography, and monomeric species were pooled.
Crystallization
Each protein was concentrated in crystallization buffer (25 mm Tris-HCl, pH 8.0, 250 mm NaCl, and 10 mm dithiothreitol). Concentrated LTag (15 mg ml−1) was mixed with p68N (5.3 mg ml−1) at an optimized molar ratio of 1:1.5 (LTag monomer/ p68N) and crystallized by the hanging-drop vapor diffusion method. Crystals were grown at 18 °C from 2-μl drops against 1 ml of well buffer (0.96 m sodium malonate, pH 6.0). Crystals were soaked stepwise in increasing concentrations of sodium malonate, pH 6.0 (1.25, 1.75, 2.25, and 2.75 m), for about 5 min at each step, and then flash-frozen in liquid nitrogen.
Data Collection, Structure Determination, and Refinement
Data were collected at Beamlines 8.3.1 at the Advanced Light Source (Berkeley, CA) and 23-IDD at Argonne National Laboratory and processed with HKL-2000 (37). To solve the structure, the previously determined LTag structures in various nucleotide states (36, 38, 39) were used as search models for molecular replacement. Knowing that one asymmetric unit may contain up to 12 LTag subunits that could form two hexamers in the crystal, we used monomeric and hexameric LTag models to do the molecular replacement search. Eventually, only the ATP-bound form (Protein Data Bank code 1SVM) (38) as a hexamer yielded a correct solution, with two hexamers in one asymmetric unit. The initial electron density map calculated with the molecular replacement solution model containing only 12 subunits of LTag already revealed some p68N density around the LTag hexamer (supplemental Fig. S1A). Subsequent 12-fold noncrystallographic symmetry averaging based on LTag subunits proved to be powerful for improving the density of p68N (supplemental Fig. S1B). Using the improved electron density map, the p68N NMR solution structure (Protein Data Bank code 2KEB) was used for docking into the non-model biased but reasonably defined density map. After the docking, it was clear that part of the main chain of the NMR structure of p68N needed to be adjusted to fit into the main-chain density. Successive rounds of model refinement and density modification further improved the phases and the density of p68Ns for further model adjustment. Even though the data completeness for the structural determination here is 75%, it should be noted that in previously reported structures, where multiple fold noncrystallographic symmetry averaging was available, data completeness around 50% (40, 41), and in one case as low as 20% (42), was sufficient for the structural determination and refinement. Given previous examples, with 12-fold noncrystallographic symmetry available in this structure, 75% data completeness should be sufficient.
Because of the relatively low resolution data (5.0 Å), refinement of the final model was further performed using the newly released deformable elastic network (DEN) refinement method that is specifically designed for structure refinement of low resolution x-ray diffraction data frequently encountered with large protein complexes (43). DEN refinement has been shown to work well for structure refinement of x-ray diffraction data of resolution around 5–6 Å (43–45). Briefly, DEN (implemented within CNS) refines structures by using previously solved high resolution structures of homologous proteins as reference structures to build springs between random pairs of atoms within the low resolution model being refined. These springs are deformable and updated with each round of DEN-assisted refinement, allowing for large differences between the reference structure and the refined structure while still applying local conserved conformational similarities that are supported by diffraction data. These springs provided structural information to supplement the atom placement supported solely from diffraction data. Strength of springs (wDEN) and the weight of the reference data (γ) are experimentally defined parameters determined by multiple refinement runs and assessed by improvements in Rfree. We created a script to test different values for wDEN and γ along a grid pattern. For each pair of values, we ran 10 refinements in our supercomputer, using different random numbers for the initial velocity assignments in refinement. Using the wDEN and γ values that gave the best average of Rfree, the best resultant structure revealed that DEN refinement changed most of the side-chain conformations at the interface between LTag and p68N to make much better inter-molecular interactions than those in the originally positioned model. The density of p68N was improved even further in the map calculated from the final model after DEN refinement (supplemental Fig. S1C), and the final refinement statistics and geometry are very good (Table 1). The coordinate file and structure factors (Protein Data Bank code 4E2I) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics.
TABLE 1.
Data collection and refinement statistics (molecular replacement)
| LTag-p68N | |
|---|---|
| Data collection statistics | |
| Space group | p41212 |
| Cell dimensions | |
| a, b, c | 249.1, 249.1, 387.0 Å |
| α, β, γ | 90, 90, 90° |
| Resolution | 50.01 to 5.00 Å (5.18 to 5.00 Å)a |
| Rsym or Rmerge | 9.3% (96%) |
| I/σI | 22.7 (1.6) |
| Completeness | 74.7% (78.5%) |
| Redundancy | 4.7 (4.7) |
| Refinement | |
| Resolution | 50.0-5.0 Å |
| No. of reflections | 35,234 |
| Rwork/Rfree | 30.47/31.39% |
| No. of atoms | |
| Protein | 41,874 |
| Root mean square deviations | |
| Bond lengths | 0.002597 Å |
| Bond angles | 0.65442° |
| Ramachandran plot | |
| Most favored regions | 81.7% |
| Additional allowed regions | 17.2% |
| Generously allowed regions | 0.7% |
| Disallowed regions | 0.3% |
| Protein Data Bank code | 4E2I |
a Values in parentheses are for highest resolution shell.
Preparation of Other Proteins
Mutations were introduced into the coding sequence of full-length LTag by standard site-directed mutagenesis, cloned into pFastBac1, and recombinant baculoviruses were generated by the Bac-to-Bac system in Sf9 cells (Invitrogen). Full-length LTag was expressed in Hi5 cells infected with recombinant baculoviruses (5 infectious units per cell) and purified by Pab101 immunoaffinity chromatography essentially as described (46) with the following modifications. Cells were lysed in lysis buffer (50 mm Tris-HCl, pH 8, 10 μm ZnCl2, 1 mm EDTA, pH 8, 100 mm NaCl, 0.5% Nonidet P-40, 1 μg/ml aprotinin, 1 μm leupeptin, 200 μm PMSF) using a Dounce homogenizer. Clarified lysates were rocked with Pab101-Sepharose beads for 2 h at 4 °C. The beads were washed extensively with wash buffer 1 (50 mm Tris-HCl, pH 8, 150 mm NaCl, 5 mm EDTA, pH 8, 10 μm ZnCl2, 0.5% Nonidet P-40) and wash buffer 2 (20 mm HEPES-KOH, pH 7.8, 5 mm NaCl, 0.1 mm EDTA, pH 8, 10 μm ZnCl2, 10% (v/v) glycerol). Protein was eluted with 20% (v/v) glycerol, 20 mm triethylamine and neutralized by addition of 0.5 m HEPES-KOH, pH 7.0, to a final pH of ∼7.5 and flash-frozen. The yield from 2 × 108 cells was ∼2 mg.
Pol-prim was expressed in Hi5 insect cells infected with four recombinant baculoviruses and purified by immunoaffinity chromatography as described previously (47). The human p180 subunit was purified identically, except that Hi5 cells were infected with only the p180 baculovirus. Human topoisomerase I was expressed in Hi5 cells using recombinant baculovirus and purified as described (48). Recombinant human RPA was expressed in E. coli and purified as described (49).
To prepare GST-p68N for pulldown assays of full-length LTag, residues 1–78 of p68 were cloned into pAT109, and the protein was expressed in E. coli by autoinduction and purified on glutathione-agarose. For full-length LTag pulldowns by primase, His6-FLAG×2-p58 was co-expressed with untagged p48 in E. coli and purified as described (50). The DBD of p53 (residues 92–292) was cloned into pAT109 to generate an N-terminal GST fusion, which was expressed by autoinduction and purified on glutathione-agarose. LT108 (LTag residues 108–627) was purified as described previously (51).
Affinity Pulldown Assays
Recombinant p68N-His6 fusion proteins (WT or mutant) and LTag helicase domain proteins (WT or mutant) were expressed in E. coli. All of the proteins were purified as described for crystallography except that the His6 tag was left intact on p68N, and the uncleaved fusion protein was eluted with lysis buffer containing high concentrations of imidazole (25 mm Tris-HCl, pH 8.0, 250 mm NaCl, and 150 mm). The LTag binding assay was carried out as follows. First, aliquots of 15 μl of Ni-NTA resin were incubated with 60 μg of His6-tagged p68N proteins in an Eppendorf tube for 30 min at 4 °C, followed by two washes with a total of 2 ml of buffer (25 mm Tris-HCl, pH 8.0, 250 mm NaCl). The resin was incubated with 100 μg of LTag in a 50-μl volume for another 30 min at 4 °C, followed by five washes with a total of 5 ml of buffer (25 mm Tris-HCl, pH 8.0, 250 mm NaCl, and 30 mm imidazole). As a negative control, 15 μl of Ni-NTA resin in the absence of His6-tagged p68N was incubated with 100 μg of LTag, followed by the same wash procedure as above. Washed resin was boiled in SDS loading buffer, and the dissociated proteins were separated by 10% SDS-PAGE and analyzed by Coomassie staining.
For full-length LTag pulldowns by GST-p68N, 50 pmol of GST or GST-p68N was bound to glutathione-agarose in binding buffer (40 mm HEPES-KOH, pH 7.9, 10 mm KCl, 7 mm magnesium acetate, 2% milk). For p180 or FLAG-primase pulldowns, 7 μg of p180 was pre-bound to SJK237-31 anti-p180 IgG (52) coupled to Sepharose, or 5 μg of His-FLAG-primase was pre-bound to FLAG M2 resin (Sigma). 1 μg of full-length LTag (or mutants), which had been preincubated for 30 min at room temperature in binding buffer containing 2 mm ATP, was added, and beads were incubated 1 h at 4 °C. Beads were washed extensively with wash buffer (40 mm HEPES-KOH, pH 7.9, 25 mm KCl, 7 mm magnesium acetate, 0.25% inositol, 0.01% Nonidet P-40). For pulldowns of LT108 with GST-p53 DBD, 50 pmol of GST or GST-p53 DBD was bound to glutathione-agarose in binding buffer (30 mm HEPES-KOH, pH 7.8, 10 mm KCl, 7 mm MgCl2, 10 μm ZnCl2, 2% milk). 2 μg of LT108 (or mutants) was added, and beads were incubated as above and then washed extensively with wash buffer (30 mm HEPES-KOH, pH 7.8, 25 mm KCl, 7 mm MgCl2, 10 μm ZnCl2, 0.25% inositol, 0.01% Nonidet P-40). Bound proteins were boiled in denaturing sample buffer, subjected to SDS-PAGE, and visualized by Western blot with monoclonal Pab101 for LTag (53), rabbit polyclonal anti-Tag for LT108 (54), monoclonal 2CT25 for p180 (55), monoclonal 8G10 for p48 (35), polyclonal anti-GST (Invitrogen), and chemiluminescence.
Helicase Assay
Partially complementary DNA oligonucleotides 5′-(dT)44GCTCGTGCAATGAGGCCGAGGCGGCCTCGGCCTCCGTGACCACG and 5′-CGTGGTCACGGAGGCCGAGGCCGCCTCGGCCTCATTGCACGAGC(dT)44 were annealed to generate a Y-shaped helicase substrate with 44-nucleotide single-stranded tails and a 44-nucleotide duplex, radiolabeled at the 5′ ends with 32P and purified by S200 gel filtration chromatography. Helicase assays were performed by incubating the substrate DNA with WT or mutant LT108 at 37 °C over a time course ranging from 30 s to 40 min in a 20-μl volume reaction containing 15 nm substrate, 500 nm (as monomer) of the indicated LTag proteins, 5 mm ATP, 1 mm DTT, and 0.1 mg ml−1 BSA in helicase buffer (10 mm MgCl2, 20 mm Tris-HCl, pH 7.5). The reaction was terminated by adding 5 μl of stop solution containing 100 mm EDTA, 0.5% SDS, 0.1% xylene cyanol, 0.1% bromphenol blue, and 50% glycerol. The reactions were electrophoresed on a 12% polyacrylamide gel in Tris borate/EDTA buffer for 90 min at 120 V. The gel was dried and then exposed on a phosphorimager.
Initiation of SV40 DNA Replication
Monopolymerase (29) assay mixtures (20 μl) consisted of 250 ng of supercoiled SV40 origin-containing pUC-HS plasmid DNA (2.8 kb), 300 ng of RPA, 300 ng of topoisomerase I, 125–500 ng of Pol-prim, and 300 ng of full-length LTag in initiation buffer (40 mm HEPES-KOH, pH 7.9, 10 mm magnesium acetate, 1 mm DTT, 4 mm ATP, 0.2 mm each of GTP, UTP, and CTP, 0.1 mm each of dGTP, dATP, and dTTP, 0.02 mm dCTP, 40 mm creatine phosphate, 40 μg/ml of creatine kinase) supplemented with 3 μCi of [α-32P]dCTP (3,000 Ci/mmol; PerkinElmer Life Sciences). Reactions were carried out and results evaluated as described (27).
SV40 DNA Replication in Monkey Cells
The pMini plasmid was first constructed as a minimal vector for cloning mutations into genomic SV40 DNA. The region of pBluescript-KS (Stratagene) containing the pMB1 origin and the β-lactamase resistance gene was amplified by PCR using pBS-KS-EcoRI-Forward (GCGGCGAATTCACGCAGGAAAGAACATGTGAGC) and pBS-KS-EcoRI-Reverse (GCGGCGAATTCGGGAAATGTGCGCGGAAC). The PCR product was digested with EcoRI and ligated, and then standard site-directed mutagenesis was performed to remove the remaining EarI site, generating pMini. WT SV40 genomic DNA was digested with EcoRI and cloned into pMini to generate pMini-SV40. Mutant LTag coding sequences were isolated from pFastBac1 clones (as EarI/BamHI fragments) and used to replace the WT LTag coding sequence in pMini-SV40. All clones were verified by DNA sequencing. BSC40 monkey kidney cells were grown as described (54). The pMini-SV40 DNAs were digested with EcoRI and purified by a PCR purification kit (Fermentas), and 2 μg of each linearized DNA was transfected per 35-mm dish of BSC40 cells using FuGENE HD (Roche Applied Science) according to the manufacturer's protocol.
Protein extracts were prepared at 24 h post-transfection by resuspending cells in lysis buffer (10 mm HEPES-KOH, pH 7.5, 250 mm NaCl, 5 mm EDTA, pH 8, 1% Nonidet P-40, 200 μm PMSF, 1 μg/ml aprotinin, 1 μm leupeptin), incubated on ice for 30 min, and clarified by centrifugation for 15 min at 4 °C and 17,500 × g. Samples were quantified by BCA (Pierce), and 4 μg of total protein were analyzed by SDS-PAGE and Western blot using anti-LTag Pab101 (53) or anti-actin (Santa Cruz Biotechnology, I-19) and chemiluminescence.
Southern Blot Analysis
DNA was extracted from 5 × 105 cells at 48 h post-transfection as described (56), digested with DpnI, separated in an agarose gel, and transferred to Zeta Probe membrane (Bio-Rad). Radiolabeled DNA probes were generated by random priming. The template for the SV40 probe was prepared from purified SV40 genomic DNA that was excised from pSVWT (57) with BamHI. To control for recovery of the DNA samples from transfected cells, a mitochondrial DNA probe was prepared by PCR of BSC40 DNA extracts using the oligonucleotides Mito-Forward (GGAGCTCTCCATGCATTTGGTATC) and Mito-Reverse (GGTGTGGATGTAAGTGGTGTCTTTG). Hybridized blots were visualized using a Typhoon Trio phosphorimager (GE Healthcare) and quantified using ImageQuant 5.2 (GE Healthcare). To correct for sample to sample variations in the recovery of replicated SV40 form I DNA, each SV40 signal was normalized to that of the respective mitochondrial DNA signal. To correct for nonspecific background, SV40 signal from the replication-dead D474N was then subtracted from that of WT and the other mutants. The corrected WT replication signal was set to 100%, and the mutant replication signals were expressed relative to that of WT.
RESULTS
Overall Structure of the LTag-p68N Complex Hexamer
To advance understanding of the molecular basis for replicative helicase interactions with Pol-prim in eukaryotes, we obtained a co-crystal of the LTag helicase domain (residues 260–627) and the N-terminal domain of the human Pol-prim regulatory subunit p68 (p68N, residues 1–78). The LTag·p68N complex crystallized in the space group P41212 (a = 249.0 Å, b = 249.0 Å, c = 387.0 Å, α = 90°, β = 90°, γ = 90°; Table 1). Using conventional refinement methods, the low resolution diffraction data obtained from the crystal would ordinarily be insufficient to build to a detailed structural model of the protein complex. However, this problem can be addressed using a recently developed extension of the structure refinement method known as a deformable elastic network, the basis of which is that the number of observable diffracted intensities at even 5 Å exceeds the number of torsion angle degrees of freedom of a macromolecule (43). DEN incorporates specific information from known high resolution structures of homologous proteins as reference models, together with stereochemical information, allowing for refinement using relatively low resolution diffraction data (see under “Experimental Procedures” for a detailed description). This approach allowed us to place most of the side chains in the structure at 5.0 Å and yielded good refinement statistics (Table 1).
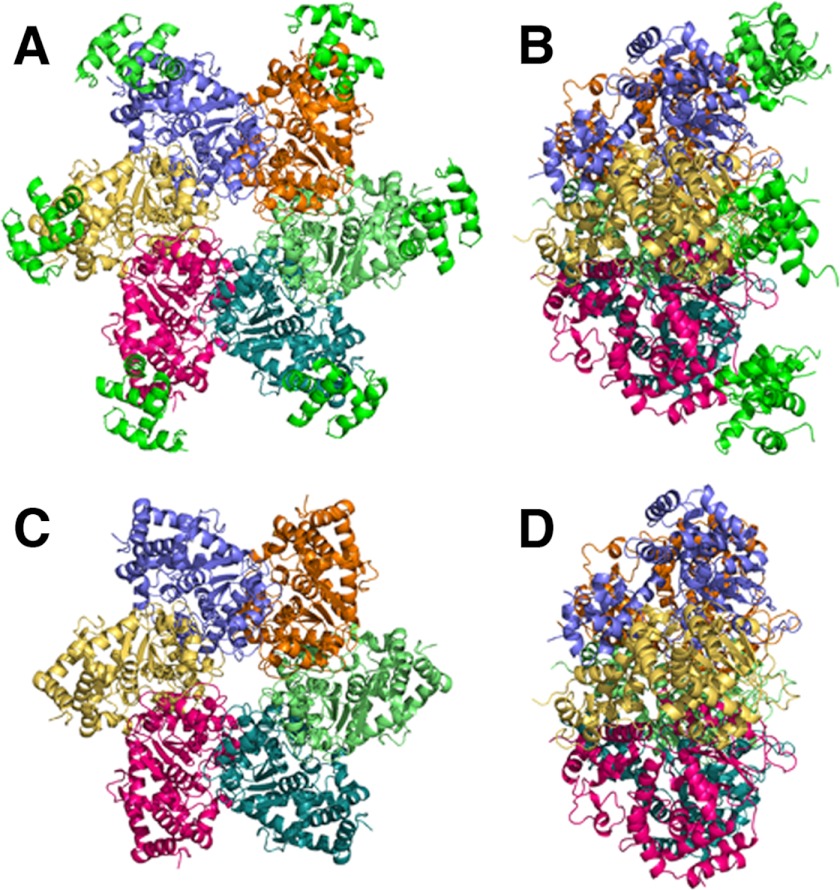
In the structure, one asymmetric unit contained 12 molecules of the LTag helicase domain and 11 molecules of p68N. The 12 LTag molecules assembled into two hexamers. In one hexamer, each individual LTag subunit binds to one molecule of p68N (Fig. 2, A and B), and the other LTag hexamer binds to only five p68N molecules, with the sixth p68N absent due to crystal packing. In the LTag·p68N complex, the LTag hexamer adopts a conformation very similar to that of the ATP-bound LTag crystal structure (38). The p68N domain adopts a compact four-helical bundle structure as in the previously reported NMR solution structure (23), but parts of the main chain display some differences from the NMR p68N structure (supplemental Fig. S1D). Each p68N molecule binds to a surface at the outermost tip of the LTag subunit, remote from the central channel formed by the hexamer (Fig. 2, C and D).
FIGURE 2.
Overall structure of the LTag-p68N complex. The C-terminal (A) and side (B) views of the LTag helicase domain hexamer in complex with p68N are shown. The six molecules of p68N are colored in green, and each LTag subunit is in a discrete color. For comparison, the C-terminal (C) and side (D) views of LTag hexamer alone are shown.
LTag-p68N Interface
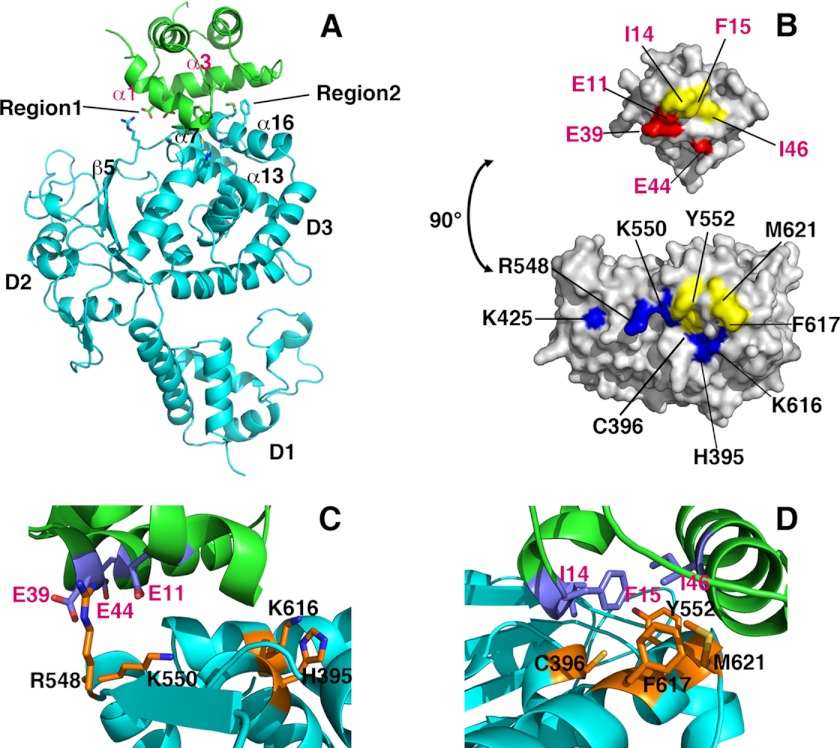
The buried surface area in each LTag-p68N interface is ∼858 Å2 (Fig. 3, A and B), consistent with the weak interaction measured in solution by isothermal titration calorimetry (Kd 6 ± 1 μm) (23). The p68N-docking surface of LTag was formed by elements of the α-helical D3 domain and faces away from the AAA+ motor domain (D2) and the Zn domain (D1). Specifically, the LTag α-helix 7 (α7), α13, the loop connecting α13 and β-strand 5, and α16 interact with p68N α1 and α3 (Fig. 3A). The interface on LTag consists of a hydrophobic patch centered on Tyr-552, Phe-617, and Met-621, flanked on one edge by a few positively charged residues. The LTag residue that had previously been implicated in p68N binding, Lys-425 (27), is not part of this interface (Fig. 3B and supplemental Fig. S2). On the p68N side, the interface also displays a hydrophobic patch, centered on Ile-14, Phe-15, and Ile-46 and flanked by negatively charged residues Glu-11, Glu-39, and Glu-44 (Fig. 3B), consistent with previously reported two-hybrid screening, pulldown assays, and NMR studies of p68N (23). Thus, the structural model reveals an LTag-p68N interface composed of hydrophobic and oppositely charged electrostatic surfaces that complement each other to form the complex (Fig. 3, C and D).
FIGURE 3.
Detailed LTag-p68N interface interactions. A, ribbon illustration of the complex structure of one LTag molecule (in cyan) binding to one p68N (in green). LTag domains D1, D2, and D3 are indicated. Secondary structures involved in the interaction of both proteins are labeled for LTag and p68N, respectively. Two regions featuring hydrophobic and electrostatic interactions are indicated. B, surface representation of LTag (bottom) and p68N (top), showing the interface areas on both proteins. The residues involved in the interface contacts are colored as follows: hydrophobic residues in yellow, positively charged residues in blue, and negatively charged residues in red. The LTag Lys-425 is not part of the p68N-binding residues, but it is located immediately next to the interface. C and D, close-up views of the detailed LTag-p68N interactions within region 1 (C) and region 2 (D), showing the charge-charge interactions in region 1 and the hydrophobic interactions in region 2, respectively.
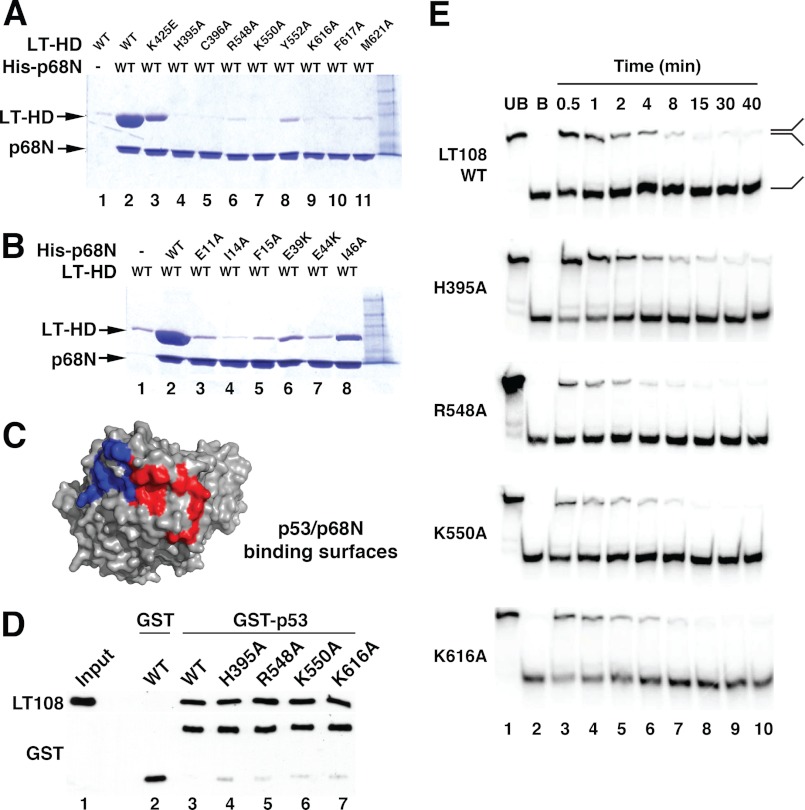
Although we were able to refine the model by placing most of the side chains in the final model at 5 Å and the final refinement statistics generated by DEN refinement are very good, the accuracy of the side chain positions is still a concern given the relatively low resolution diffraction data used for DEN refinement. As a result, a thorough biochemical validation of the interface and the functional specificity of the binding interaction is imperative. Toward this end, LTag and p68N residues at the observed interface were substituted by alanine using site-directed mutagenesis. Soluble purified wild type (WT) or mutant LTag helicase domain proteins were pulled down on His-tagged p68N beads, and bound proteins were visualized by denaturing gel electrophoresis and Coomassie staining (Fig. 4A). The results indicated that the background binding of LTag to the nickel resin alone was low (Fig. 4A, lane 1), but significant binding of LTag to the Ni resin pre-bound with His6-p68N was detected (lane 2). In contrast, with the exception of Lys-425E, which showed a reduction in binding relative to WT (Fig. 4A, lane 3), all LTag mutants had near background binding (lanes 4–11). This result was further confirmed by mutations on the p68N surface. Six mutant His-tagged p68N proteins bound poorly to WT LTag (Fig. 4B, compare lanes 3–7 with lane 2). These results demonstrate the critical role of each of these residues in the LTag-p68N interface identified in the structural model.
FIGURE 4.
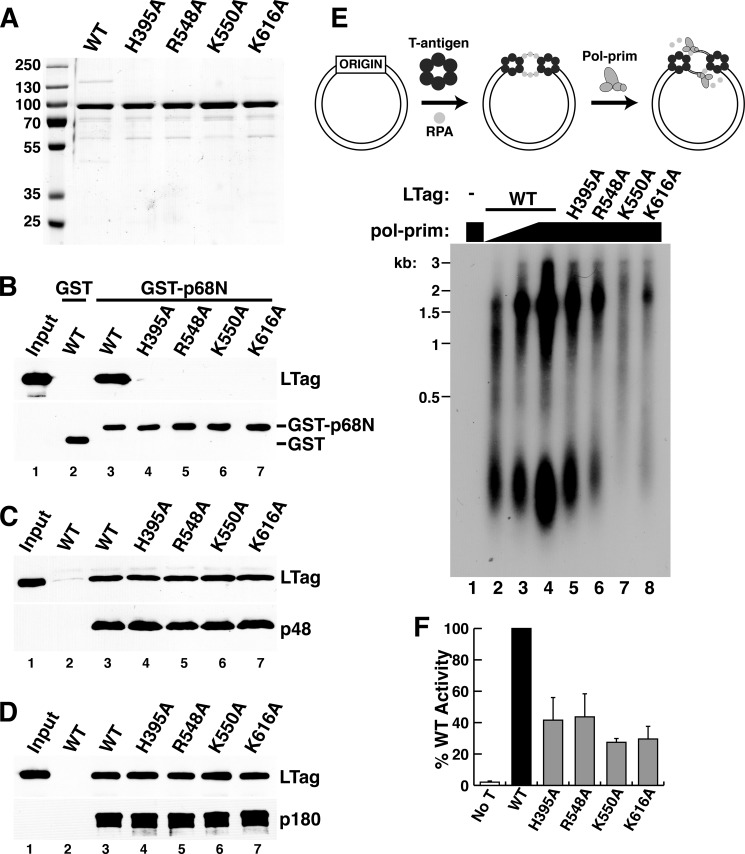
Mutational analysis of the LTag-p68N interface and functional validation. A, pulldown assays were performed to evaluate the effect of LTag mutations of the residues within the interface on binding to p68N. Lane 1, LTag helicase domain (LT-HD) retained on Ni resin in the absence of His6-p68N; lane 2, the LTag retained on His6-p68N-bound Ni resin; lanes 3–11, mutant LTags (as marked) retained on His6-p68N-bound Ni resin. B, pulldown assays were performed to evaluate the effect of p68N mutations of the residues within the interface on binding of LTag. Lane 1, LTag retained on Ni resin in the absence of His6-p68N; lane 2, LTag retained on His6-p68N-bound Ni resin; lanes 3–8, LTags retained on Ni resin bound to mutant His6-p68Ns (as marked). For the pulldown assays in A and B, the input LTag for initial incubation for each lane was 100 μg (see “Experimental Procedures”). C, binding interfaces on LTag for p53 (red) (39) and for p68 (blue) (Fig. 3) are adjacent but distinct. D, glutathione-agarose beads bound to either GST (lane 2) or GST-p53 DBD (lanes 3–7) were incubated with LT108 WT or substitution proteins as indicated. Retained proteins were visualized by Western blot with the indicated antibodies. Lane 1, 15% of the LT108 input amounts used in lanes 2–7. E, helicase activities of LT108 mutants that have disrupted p68N binding were assayed over a time course of 30 s to 40 min. Lanes 1 and 2 contain unboiled (UB) and boiled (B) DNA substrate.
Specificity of the LTag-p68N Binding Interface
As a first approach to validate the functional specificity of the LTag-p68 interface in the structural model, we examined the interaction of LTag with the human tumor suppressor protein p53. Like p68N, the DNA binding domain (DBD) of p53 binds to the helicase domain of LTag with a 1:1 stoichiometry (39). A high resolution co-crystal structure of six p53 DBDs with hexameric LTag helicase domain revealed a large binding interface that was fully validated by site-directed mutagenesis of the interface (39). Interestingly, the conformation of p53 DBD-bound LTag hexamer closely resembled that of ATP-bound LTag (39). Moreover, bound p53 inhibited LTag ATPase, DNA helicase, and hence replication activities and directly interfered with Pol-prim binding to LTag (58, 59). These observations suggested the possibility that p68N and p53 might dock on overlapping surfaces of LTag. However, an overlay of the previously defined p53 interface (39) with that of p68N on LTag in our structural model predicts adjacent, but nonoverlapping, binding interfaces (Fig. 4C).
To test this prediction, GST-p53 DBD pulldown assays were conducted with soluble LT108 WT and four of the alanine substitution proteins defective in binding to p68N. No significant differences between WT and mutant LT108 binding to p53 were detected (Fig. 4D, compare lanes 4–7 with lane 3). We conclude that p53 and p68N bind to adjacent but distinct surfaces on LTag, suggesting that the observed competition could arise through steric hindrance.
To further corroborate the specificity of the p68-docking interface in LTag, we monitored the DNA helicase activity of WT LT108 and four LT108 proteins with alanine substitutions that disrupt interaction with p68N. All four LT108 variants displayed helicase activity similar to that of WT (Fig. 4E, compare upper panel with lower panels), indicating that these substitutions do not indirectly compromise the enzymatic activity of the LTag helicase domain. Taken together, the data in Fig. 4 indicate that the substitutions in LTag that disrupt binding to p68N do not abrogate other known functions of the helicase domain.
Role for LTag-p68 Interaction in Primosome Activity
The SV40 primosome activity of Pol-prim containing a p68 mutation that either abolished or reduced LTag binding was reduced in proportion to its defect in binding to LTag (23), suggesting that p68N docking on LTag was crucial for this activity. The detailed LTag-p68N interactions revealed here now allow us to further test the importance of the LTag interface residues in SV40 primosome activity. Recombinant full-length WT and four mutant LTags were stable and purified in similar yields (Fig. 5A). Binding of p68N to purified full-length WT LTag was easily detected, but p68N bound poorly to the mutant proteins (Fig. 5B, compare lanes 4–7 with lane 3). In contrast, interactions of the mutant LTags with the primase and p180 subunits of Pol-prim were indistinguishable from those of WT LTag (Fig. 5, C and D). We conclude that the substitutions in LTag specifically weaken its interaction with p68, but not with other Pol-prim subunits.
FIGURE 5.
Specific role of LTag-p68 interaction in primosome activity. A, purified full-length WT and the indicated mutant LTags were separated by SDS-PAGE and stained with Coomassie Brilliant Blue. Marker proteins are shown at left. B, glutathione-agarose beads bound to either GST (lane 2) or GST-p68N (lanes 3–7) were incubated with full-length WT or the indicated point mutant LTags. Retained proteins were analyzed by Western blot with the indicated antibodies. Lane 1, 15% of the LTag input used for pulldowns in lanes 2–7. C and D, FLAG beads without (lane 2) or with bound p48/His6-FLAG×2-p58 heterodimer (C) or SJK237-31-Sepharose beads without (lane 2) or with bound p180 (D) were incubated with soluble, purified full-length WT or the point mutant LTags as indicated. Lane 1 shows 7.5% of the LTag input used for pulldowns in lanes 2–7. E, initiation of SV40 DNA replication initiation was assayed in monopolymerase reactions containing purified LTag, RPA, topoisomerase, and Pol-prim (diagram). Radiolabeled products of reactions lacking LTag (lane 1) or containing 300 ng of WT or the indicated mutant LTags (lanes 2–8) and varying amounts of Pol-prim (lane 2, 125 ng; lane 3, 250 ng; lanes 1 and 4–8, 500 ng) were analyzed by denaturing gel electrophoresis and phosphorimaging. DNA size markers are indicated (kb). F, initiation activity from three independent experiments as in E was quantified by phosphorimaging, and the activity of each mutant LTag was expressed relative to that of the WT LTag activity in each experiment. No T, as a negative control, LTag was omitted from the sample. Error bars indicate standard deviation.
To monitor the primosome activity of the full-length LTag mutant proteins, we used the SV40 monopolymerase assay, which measures primer synthesis coupled with unwinding of supercoiled DNA containing the SV40 origin (29). Although newly synthesized primers can be directly detected in this assay in the absence of deoxyribonucleotides, measurement of primer-dependent extension into RNA-DNA products in the presence of radiolabeled dCTP was used here to provide greater sensitivity (29). In this assay, products of two general size classes were formed as follows: a longer product, approximately one-half the size of the template, that primarily results from replication of the leading strand; and a shorter fragment, about 100–300 nucleotides in length, that represents synthesis of Okazaki fragments. In reactions containing DNA, WT LTag, RPA, and topoisomerase, radiolabeled products accumulated in proportion to the amount of purified Pol-prim present in the assay (Fig. 5E, lanes 2–4). No products were observed when LTag was omitted (Fig. 5E, lane 1). LTag substitutions H395A, R548A, K550A, and K616A significantly reduced primosome activity relative to that of an equal amount of WT LTag (Fig. 5, E, lanes 4–8, and F). The magnitude of this defect was very similar to the ∼60% drop in primosome activity observed in reactions conducted with Pol-prim containing the I14A-substituted p68, which disrupts binding to LTag (23). The results demonstrate that interaction of p68 with the LTag interface identified in our structural model is important for primosome activity in a reconstituted reaction.
Biological Role of LTag-p68 Interaction in SV40 DNA Replication in Vivo
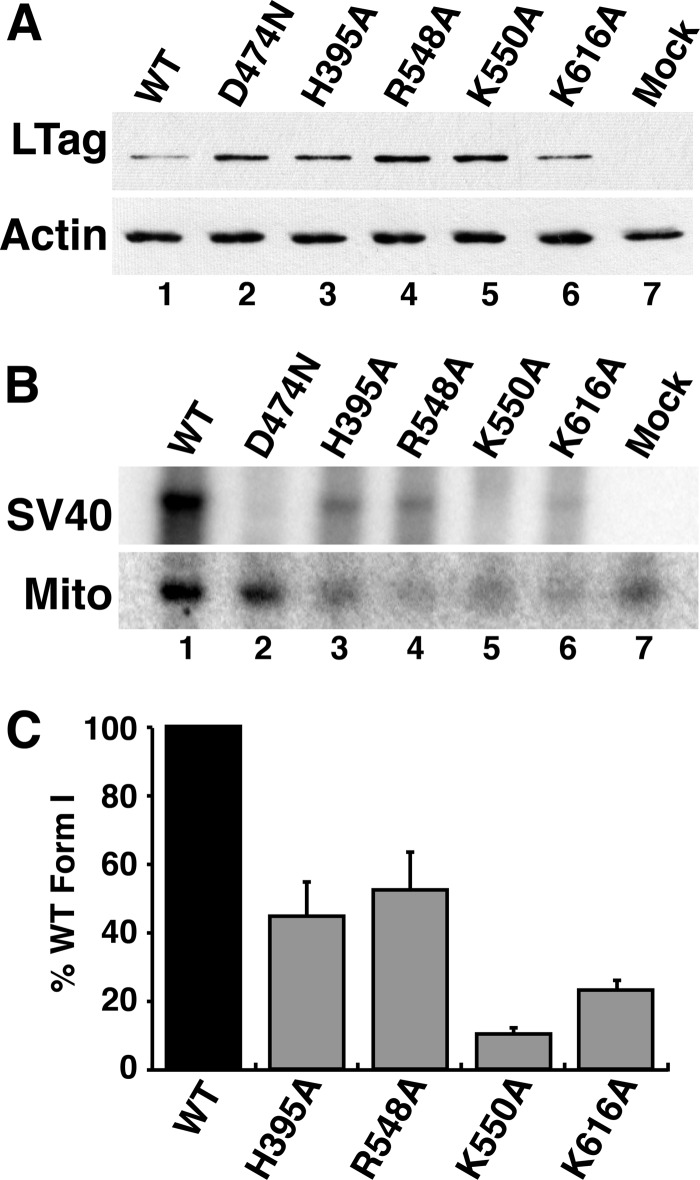
A cell-free primosome assay with purified DNA and proteins is of course highly simplified relative to the intracellular environment in which viral chromatin replicates. To determine whether this in vitro p68-LTag interaction contributes to viral mini-chromosome replication in the natural host monkey cells, we prepared bacterial plasmid DNAs that each contained a complete SV40 genome encoding the WT or one of the mutant H395A, R548A, K550A, or K616A LTags with defective primosome activity. An SV40 genome encoding the Walker B LTag substitution D474N, which disrupts the essential replicative helicase activity, was used as a negative control (27). We introduced this panel of SV40 genomic DNAs into monkey cells and monitored LTag expression and viral DNA replication products.
24 h after transfection of cells with WT and mutant SV40 DNA, whole cell extracts were prepared, and LTag expression was monitored by immunoblotting. WT and each of the mutant LTags accumulated at detectable levels, with some variation among samples (Fig. 6A). Immunofluorescence microscopy confirmed that the WT and mutant LTags accumulated in the nuclei of the transfected cells as expected (data not shown). To evaluate viral DNA replication at 48 h after transfection, low molecular weight DNA was isolated from cells expressing WT and mutant LTag, digested with DpnI to fragment residual input DNA, and analyzed by Southern blot and phosphorimaging. Daughter DNA was easily detected in the WT samples, and not in the Walker B mutant samples, as expected (Fig. 6B, lanes 1 and 2). The poor replication of the LTag mutant genomes H395A, R548A, K550A, and K616A in monkey cells mirrors their poor primosome activity in vitro (Fig. 6B, lanes 3–6). Quantification of results from two independent transfection experiments confirmed that a single substitution in the LTag-p68 interface was sufficient to significantly diminish replication activity in vivo (Fig. 6C). We conclude that viral mini-chromosome replication in mammalian cells depends on the p68-LTag primosome interface identified in our structural model.
FIGURE 6.
LTag-p68 interaction is required to replicate SV40 chromatin in monkey kidney cells. A, whole cell extracts were prepared from BSC40 cells transfected 24 h earlier with genomic SV40 DNA encoding WT LTag, the Walker B mutant LTag D474N, or the indicated mutant LTags with defects in p68 binding. A 4-μg sample of total protein from each extract was analyzed by SDS-PAGE and Western blotting to evaluate LTag expression. Actin served as control for equal protein loading. B, low molecular weight DNAs were extracted from a parallel experiment (A) at 48 h after transfection and analyzed by Southern blotting with a radiolabeled SV40 DNA probe and with a human mitochondrial (Mito) DNA probe as a recovery control. C, radiolabeled signals for SV40 form I DNA replication product and mitochondrial DNA in each lane were quantified using phosphorimaging. Replication signals were corrected for sample to sample recovery variations using mitochondrial DNA and for background using the helicase-dead D474N mutant and were compared with that of WT in the same experiment, as detailed under “Experimental Procedures.” The graph shows the average value from two independent transfection experiments. Error bars represent standard deviation.
DISCUSSION
The replisome is a multiprotein machine composed of various “moving components” that display a high degree of dynamic coordination. Although the key protein players and their interactions in prokaryotic primosomes and replisomes have been characterized biochemically and structurally, understanding of their eukaryotic counterparts is much less comprehensive, in part due to the lack of structural information. Here, we have applied a powerful new extension of DEN refinement methods to a low resolution co-crystal structure of a human Pol-prim domain (p68 N-terminal domain, residues 1–78) bound to the helicase domain (residues 260–627) of the replicative helicase LTag. The resulting structural model of the complex reveals in detail a network of complementary hydrophobic and electrostatic interactions at the protein interface that have been extensively validated by site-directed mutagenesis, highlighting the effectiveness of these new refinement methods. Using this panel of mutants, we show that the p68-LTag interface is not essential for LTag helicase activity or for binding to the p53 tumor suppressor protein, to the Pol-prim catalytic subunit p180, or to primase. In contrast, the integrity of the p68-LTag interface is vital for initiation of SV40 DNA replication in a cell-free reaction reconstituted with purified proteins, as well as in the natural monkey kidney host cells.
The identification of the LTag binding surface on p68N as a hydrophobic patch rimmed by acidic charge (23) led to the expectation that the p68N binding surface on LTag would be composed of a complementary hydrophobic patch bound by basic charge. A two-hybrid screen of charge-reverse substitutions in LTag surface residues for mutations that disrupted binding to p68N identified LTag Lys-425 as a potential interacting residue (27), but no hydrophobic patch could be identified in the immediate vicinity. The structural model reported here reveals that Lys-425 is not part of the interface (Fig. 3B), but rather it resides on the surface of LTag near both the P-loop of the AAA+ subdomain and Arg-548, which is positioned at the edge of the LTag-p68 interface (supplemental Fig. S2). It is likely that the K425E substitution perturbed the LTag surface sufficiently to weaken the interaction with p68N. In addition, disruption of the P-loop might explain the low ATPase activity displayed by K425E LTag (27).
Extensive biochemical studies have shown that specific interactions between LTag and Pol-prim are required for primosome activity (14, 23, 55, 60). Surface plasmon resonance measurements indicate that Pol-prim interacts with hexameric LTag with nanomolar affinity (35). However, the interaction of p68 with a single LTag subunit is weak (Kd 6 μm) when measured in solution by isothermal titration calorimetry (23). This difference in affinity suggests that additional interactions between Pol-prim and LTag must exist. LTag variants that fail to bind to p68 retain WT interaction with the p180 and primase subunits of Pol-prim (Fig. 5, B–D), implying that LTag has at least two more, likely different, interfaces that bind to Pol-prim and remain to be identified. Like the LTag-p68 interaction, each of these interactions is relatively weak but contributes to the overall affinity of Pol-prim for the LTag hexamer. Together, they are likely to play critical roles in primosome activity, perhaps by maintaining proper orientation of Pol-prim relative to the hexameric helicase or coordinating helicase activity with primer synthesis and extension. The p68-docking site on LTag is of course present on all six subunits of the hexamer, but given the 1:1 (Pol-prim/LTag hexamer) stoichiometry observed in solution (34) and the similar mass of hexameric LTag and Pol-prim (∼500 and 350 kDa), it seems likely that the 1:1 stoichiometry is the functional complex. Thus, the structural arrangement of Pol-prim bound to LTag is inherently asymmetric, which is likely to be functionally significant.
Conserved Polyomavirus Interface for Pol-prim Interaction?
Analysis of LTag protein sequences from other primate polyomaviruses revealed the LTag residues critical for binding p68 to be well conserved (supplemental Fig. S3A). Similarly, the extreme N-terminal end (residues ∼1–80), and specifically the residues implicated in LTag binding (supplemental Fig. S3B, boxed), are conserved among mammals. Thus, the p68-LTag binding interface and its function in polyomaviral primosome activity may be conserved. However, whether the p68N domain regulates primosome activity in chromosomal replication remains an open question (61, 62).
Emerging Eukaryotic Primosome Architecture?
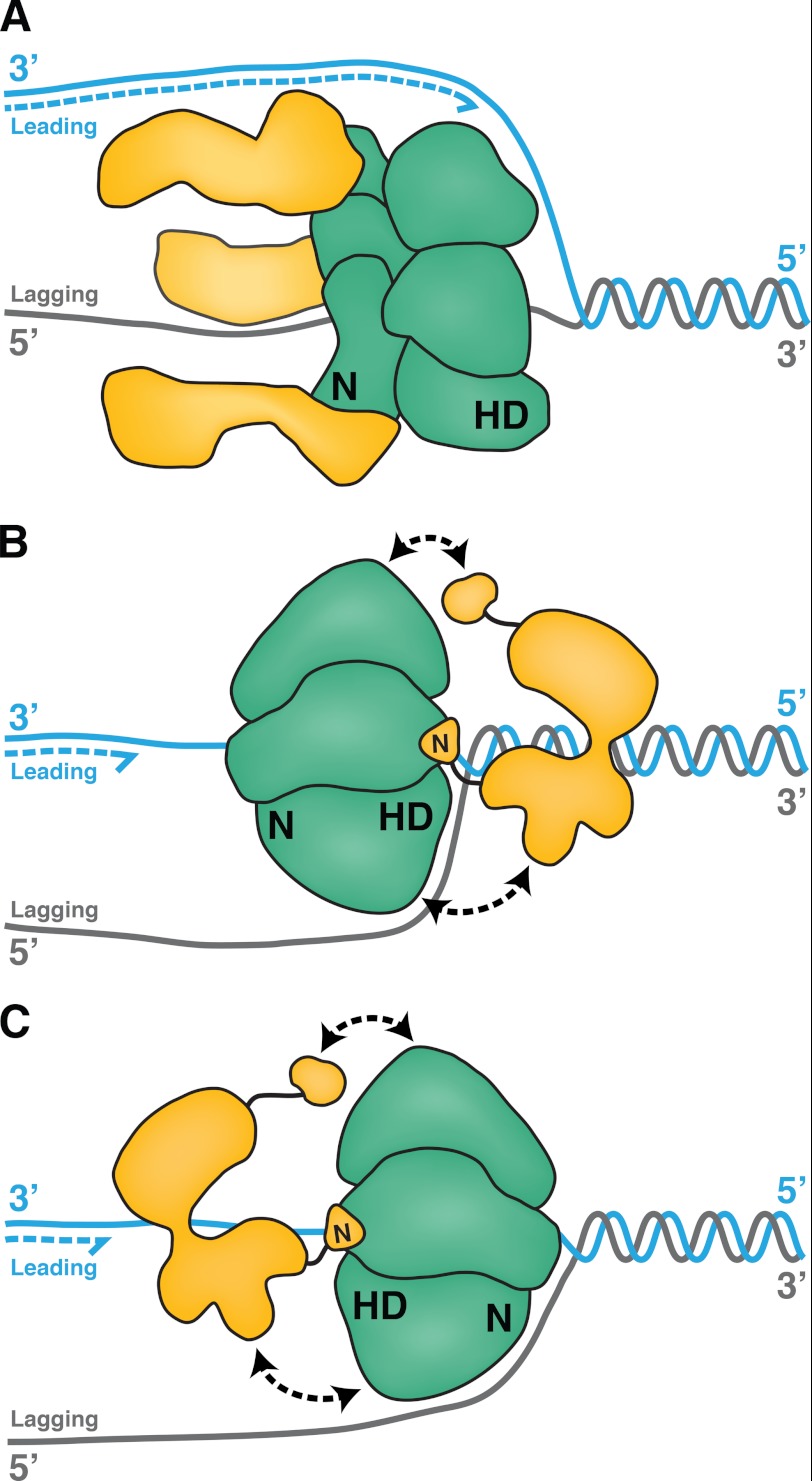
Our current understanding of the interactions of replicative helicases with DNA primases is based largely on prokaryotic replisomes that have been extensively characterized in multiple systems (1–4, 11). The replicative helicases of prokaryotes translocate with 5′ to 3′ polarity on the lagging strand template, displacing the leading strand template (Fig. 7A). FRET analysis revealed that the E. coli replicative helicase DnaB is oriented with its C-terminal motor domains facing toward the duplex DNA substrate (63, 64) and its primase-interacting regions following along behind (Fig. 7A) (65–67). The formation of a trombone loop in the lagging strand template is thought to enable primase, assisted by the χ/ψ subunits of the DNA polymerase III holoenzyme, to displace ssDNA-binding protein and generate RNA primers for Okazaki fragment synthesis (68 and references therein).
FIGURE 7.
Variations on primosome architecture. A, in a model prokaryotic primosome, the E. coli replicative helicase, DnaB (green), translocates along the lagging strand template with 5′ to 3′ polarity, its C-terminal helicase domains (HD) facing toward the duplex DNA (gray/cyan). Three molecules of primase, DnaG (yellow), interact with the N-terminal (N) domain of DnaB and trail behind. B, LTag hexamer (green) interacts through its C-terminal helicase domains (HD) with at least three subunits of Pol-prim (yellow). In one possible model of the SV40 primosome, the entire central channel of the hexameric helicase encircles one ssDNA, with the helicase domains facing toward the duplex as LTag translocates with 3′ to 5′ polarity. We suggest that Pol-prim docking on LTag (black double arrows) positions primase to bind the lagging strand template. C, in another possible model, the N-terminal domains of LTag could face the DNA duplex.
In contrast, the eukaryotic and archaeal replicative helicase assemblies translocate with 3′ to 5′ polarity on the leading strand template. Mini-chromosome maintenance (MCM) proteins constitute the core of the helicase and supply its motor function (5, 69–71). FRET analyses indicate that the archaeal MCM complex translocates 3′ to 5′ with its C-terminal motor domains oriented toward the duplex DNA as it displaces the complementary 5′ strand (72, 73). Accumulating evidence suggests that the excluded 5′ strand wraps the positively charged exterior surface of the MCM complex (6, 74, 75).
Although the orientation of the LTag helicase on a substrate DNA has not been experimentally determined, the 3′–5′ polarity of translocation on ssDNA and unwinding of DNA fork substrates, overall structural organization, and basic exterior surface of the LTag helicase domain resemble properties of MCM helicase, suggesting that an MCM-like orientation of LTag on DNA would be plausible (Fig. 7B). In this orientation, DNA polymerase δ would synthesize the leading strand behind the translocating LTag hexamer (30, 76, 77). The LTag helicase domains interact with the long flexible N-terminal extensions of the p180 and p68 subunits of Pol-prim, likely spatially positioning Pol-prim on the lagging strand template for primer synthesis (14, 23, 27, 28, 47, 60). Alternatively, LTag may be oriented with its N-terminal domains toward the duplex DNA and the motor domains following behind, as suggested by a crystal structure of the superfamily 3 bovine papillomaviral helicase E1 on ssDNA (78, 79). This orientation could also allow the LTag helicase domains to interact with the N-terminal extensions of p68 and p180 to bring the primase and lagging strand template together for Okazaki fragment synthesis (Fig. 7C).
In summary, this study reveals for the first time the molecular interactions of LTag with the Pol-prim regulatory subunit p68 and has important implications for understanding how interaction of p68 with LTag activates SV40 primosome activity. Despite the relatively small interface between LTag-p68, we have now shown that both sides of this interface are critical for primosome activity, suggesting that each of these weak interactions between Pol-prim and LTag is important for proper function of the highly dynamic complex machine (80). Future work to characterize the remaining Pol-prim-LTag interactions and their orientation at a fork will advance our understanding of this model eukaryotic primosome.
Acknowledgments
We thank the staff at Advanced Light Source Beamlines 8.3.1, 8.2.1, and 5.0.2, the Lawrence Berkeley Laboratory, and the staff at beamline 23-IDD, Argonne National Laboratory, for assistance in data collection. We thank the Biophysics Core Laboratory, University of Southern California, for their support; W. J. Chazin for a p68N expression vector and discussions; J. M. Pipas for thoughtful contributions and support for this study; J. Hurwitz and V. Bermudez for p58-FLAG baculovirus; E. Kremmer for anti-p48 antibody, and U. Dadwal for assistance with mutagenesis. We also thank Dr. A. Brunger for giving us access to the pre-release version of the program DEN, designed for refining low resolution structures. The Vanderbilt-Ingram Cancer Center, Vanderbilt University, was the recipient of National Institutes of Health Grant P30 CA068485.
This work was supported, in whole or in part, by National Institutes of Health Grants GM080338 and AI055926 (to X. S. C.) and GM52948 (to E. F.).

This article contains supplemental Figs. S1–S3.
- ssDNA
- single-stranded DNA
- AAA+
- ATPases associated with various cellular activities
- DBD
- DNA binding domain
- LTag
- large tumor antigen
- MCM
- mini-chromosome maintenance
- Pol-prim
- DNA polymerase α-primase
- RPA
- replication protein A
- Ni-NTA
- nickel-nitrilotriacetic acid
- DEN
- deformable elastic network
- wDEN
- weight for DEN.
REFERENCES
- 1. Hamdan S. M., Richardson C. C. (2009) Motors, switches, and contacts in the replisome. Annu. Rev. Biochem. 78, 205–243 [DOI] [PubMed] [Google Scholar]
- 2. Hamdan S. M., van Oijen A. M. (2010) Timing, coordination, and rhythm. Acrobatics at the DNA replication fork. J. Biol. Chem. 285, 18979–18983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Langston L. D., Indiani C., O'Donnell M. (2009) Whither the replisome. Emerging perspectives on the dynamic nature of the DNA replication machinery. Cell Cycle 8, 2686–2691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Patel S. S., Pandey M., Nandakumar D. (2011) Dynamic coupling between the motors of DNA replication. Hexameric helicase, DNA polymerase, and primase. Curr. Opin. Chem. Biol. 15, 595–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Costa A., Ilves I., Tamberg N., Petojevic T., Nogales E., Botchan M. R., Berger J. M. (2011) The structural basis for MCM2–7 helicase activation by GINS and Cdc45. Nat. Struct. Mol. Biol. 18, 471–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Costa A., Onesti S. (2009) Structural biology of MCM helicases. Crit. Rev. Biochem. Mol. Biol. 44, 326–342 [DOI] [PubMed] [Google Scholar]
- 7. Masai H., Matsumoto S., You Z., Yoshizawa-Sugata N., Oda M. (2010) Eukaryotic chromosome DNA replication. Where, when, and how? Annu. Rev. Biochem. 79, 89–130 [DOI] [PubMed] [Google Scholar]
- 8. Kunkel T. A., Burgers P. M. (2008) Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 18, 521–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mizuno T., Yamagishi K., Miyazawa H., Hanaoka F. (1999) Molecular architecture of the mouse DNA polymerase α-primase complex. Mol. Cell. Biol. 19, 7886–7896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Copeland W. C., Wang T. S. (1993) Enzymatic characterization of the individual mammalian primase subunits reveals a biphasic mechanism for initiation of DNA replication. J. Biol. Chem. 268, 26179–26189 [PubMed] [Google Scholar]
- 11. Kuchta R. D., Stengel G. (2010) Mechanism and evolution of DNA primases. Biochim. Biophys. Acta 1804, 1180–1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zerbe L. K., Kuchta R. D. (2002) The p58 subunit of human DNA primase is important for primer initiation, elongation, and counting. Biochemistry 41, 4891–4900 [DOI] [PubMed] [Google Scholar]
- 13. Grossi S., Puglisi A., Dmitriev P. V., Lopes M., Shore D. (2004) Pol12, the B-subunit of DNA polymerase α, functions in both telomere capping and length regulation. Genes Dev. 18, 992–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Collins K. L., Russo A. A., Tseng B. Y., Kelly T. J. (1993) The role of the 70-kDa subunit of human DNA polymerase α in DNA replication. EMBO J. 12, 4555–4566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Eichinger C. S., Mizuno T., Mizuno K., Miyake Y., Yanagi K., Imamoto N., Hanaoka F. (2009) Aberrant DNA polymerase α is excluded from the nucleus by defective import and degradation in the nucleus. J. Biol. Chem. 284, 30604–30614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Foiani M., Marini F., Gamba D., Lucchini G., Plevani P. (1994) The B-subunit of the DNA polymerase α-primase complex in Saccharomyces cerevisiae executes an essential function at the initial stage of DNA replication. Mol. Cell. Biol. 14, 923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mizuno T., Ito N., Yokoi M., Kobayashi A., Tamai K., Miyazawa H., Hanaoka F. (1998) The second-largest subunit of the mouse DNA polymerase α-primase complex facilitates both production and nuclear translocation of the catalytic subunit of DNA polymerase α. Mol. Cell. Biol. 18, 3552–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klinge S., Núñez-Ramírez R., Llorca O., Pellegrini L. (2009) Three-dimensional architecture of DNA pol α reveals the functional core of multisubunit replicative polymerases. EMBO J. 28, 1978–1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Núñez-Ramírez R., Klinge S., Sauguet L., Melero R., Recuero-Checa M. A., Kilkenny M., Perera R. L., García-Alvarez B., Hall R. J., Nogales E., Pellegrini L., Llorca O. (2011) Flexible tethering of primase and DNA pol α in the eukaryotic primosome. Nucleic Acids Res. 39, 8187–8199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Agarkar V. B., Babayeva N. D., Pavlov Y. I., Tahirov T. H. (2011) Crystal structure of the C-terminal domain of human DNA primase large subunit. Implications for the mechanism of the primase-polymerase α switch. Cell Cycle 10, 926–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sauguet L., Klinge S., Perera R. L., Maman J. D., Pellegrini L. (2010) Shared active site architecture between the large subunit of eukaryotic primase and DNA photolyase. PLoS One 5, e10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Vaithiyalingam S., Warren E. M., Eichman B. F., Chazin W. J. (2010) Insights into eukaryotic DNA priming from the structure and functional interactions of the 4Fe-4S cluster domain of human DNA primase. Proc. Natl. Acad. Sci. U.S.A. 107, 13684–13689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang H., Weiner B. E., Zhang H., Fuller B. E., Gao Y., Wile B. M., Zhao K., Arnett D. R., Chazin W. J., Fanning E. (2010) Structure of a DNA polymerase α-primase domain that docks on the SV40 helicase and activates the viral primosome. J. Biol. Chem. 285, 17112–17122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dean F. B., Bullock P., Murakami Y., Wobbe C. R., Weissbach L., Hurwitz J. (1987) Simian virus 40 (SV40) DNA replication. SV40 large T antigen unwinds DNA containing the SV40 origin of replication. Proc. Natl. Acad. Sci. U.S.A. 84, 16–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ishimi Y., Sugasawa K., Hanaoka F., Eki T., Hurwitz J. (1992) Topoisomerase II plays an essential role as a swivelase in the late stage of SV40 chromosome replication in vitro. J. Biol. Chem. 267, 462–466 [PubMed] [Google Scholar]
- 26. Yang L., Wold M. S., Li J. J., Kelly T. J., Liu L. F. (1987) Roles of DNA topoisomerases in simian virus 40 DNA replication in vitro. Proc. Natl. Acad. Sci. U.S.A. 84, 950–954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Huang H., Zhao K., Arnett D. R., Fanning E. (2010) A specific docking site for DNA polymerase α-primase on the SV40 helicase is required for viral primosome activity, but helicase activity is dispensable. J. Biol. Chem. 285, 33475–33484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Collins K. L., Kelly T. J. (1991) Effects of T antigen and replication protein A on the initiation of DNA synthesis by DNA polymerase α-primase. Mol. Cell. Biol. 11, 2108–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Matsumoto T., Eki T., Hurwitz J. (1990) Studies on the initiation and elongation reactions in the simian virus 40 DNA replication system. Proc. Natl. Acad. Sci. U.S.A. 87, 9712–9716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yuzhakov A., Kelman Z., Hurwitz J., O'Donnell M. (1999) Multiple competition reactions for RPA order the assembly of the DNA polymerase δ holoenzyme. EMBO J. 18, 6189–6199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Arunkumar A. I., Klimovich V., Jiang X., Ott R. D., Mizoue L., Fanning E., Chazin W. J. (2005) Insights into hRPA32 C-terminal domain-mediated assembly of the simian virus 40 replisome. Nat. Struct. Mol. Biol. 12, 332–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jiang X., Klimovich V., Arunkumar A. I., Hysinger E. B., Wang Y., Ott R. D., Guler G. D., Weiner B., Chazin W. J., Fanning E. (2006) Structural mechanism of RPA loading on DNA during activation of a simple pre-replication complex. EMBO J. 25, 5516–5526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Melendy T., Stillman B. (1993) An interaction between replication protein A and SV40 T antigen appears essential for primosome assembly during SV40 DNA replication. J. Biol. Chem. 268, 3389–3395 [PubMed] [Google Scholar]
- 34. Huang S. G., Weisshart K., Gilbert I., Fanning E. (1998) Stoichiometry and mechanism of assembly of SV40 T antigen complexes with the viral origin of DNA replication and DNA polymerase α-primase. Biochemistry 37, 15345–15352 [DOI] [PubMed] [Google Scholar]
- 35. Weisshart K., Förster H., Kremmer E., Schlott B., Grosse F., Nasheuer H. P. (2000) Protein-protein interactions of the primase subunits p58 and p48 with simian virus 40 T antigen are required for efficient primer synthesis in a cell-free system. J. Biol. Chem. 275, 17328–17337 [DOI] [PubMed] [Google Scholar]
- 36. Li D., Zhao R., Lilyestrom W., Gai D., Zhang R., DeCaprio J. A., Fanning E., Jochimiak A., Szakonyi G., Chen X. S. (2003) Structure of the replicative helicase of the oncoprotein SV40 large tumor antigen. Nature 423, 512–518 [DOI] [PubMed] [Google Scholar]
- 37. Otwinowski Z., Minor W. (1997) Macromolecular Crystallography, Part A. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 38. Gai D., Zhao R., Li D., Finkielstein C. V., Chen X. S. (2004) Mechanisms of conformational change for a replicative hexameric helicase of SV40 large tumor antigen. Cell 119, 47–60 [DOI] [PubMed] [Google Scholar]
- 39. Lilyestrom W., Klein M. G., Zhang R., Joachimiak A., Chen X. S. (2006) Crystal structure of SV40 large T-antigen bound to p53. Interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev. 20, 2373–2382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Grimes J. M., Burroughs J. N., Gouet P., Diprose J. M., Malby R., Ziéntara S., Mertens P. P., Stuart D. I. (1998) The atomic structure of the bluetongue virus core. Nature 395, 470–478 [DOI] [PubMed] [Google Scholar]
- 41. Simpson A. A., Chipman P. R., Baker T. S., Tijssen P., Rossmann M. G. (1998) The structure of an insect parvovirus (Galleria mellonella densovirus) at 3.7 Å resolution. Structure 6, 1355–1367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. McClain B., Settembre E., Temple B. R., Bellamy A. R., Harrison S. C. (2010) X-ray crystal structure of the rotavirus inner capsid particle at 3.8 Å resolution. J. Mol. Biol. 397, 587–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schröder G. F., Levitt M., Brunger A. T. (2010) Super-resolution biomolecular crystallography with low resolution data. Nature 464, 1218–1222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ataide S. F., Schmitz N., Shen K., Ke A., Shan S. O., Doudna J. A., Ban N. (2011) The crystal structure of the signal recognition particle in complex with its receptor. Science 331, 881–886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Brunger A. T., Das D., Deacon A. M., Grant J., Terwilliger T. C., Read R. J., Adams P. D., Levitt M., Schröder G. F. (2012) Application of DEN refinement and automated model building to a difficult case of molecular-replacement phasing. The structure of a putative succinyl-diaminopimelate desuccinylase from Corynebacterium glutamicum. Acta Crystallogr. D Biol. Crystallogr. 68, 391–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ott R. D., Wang Y., Fanning E. (2002) Mutational analysis of simian virus 40 T-antigen primosome activities in viral DNA replication. J. Virol. 76, 5121–5130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ott R. D., Rehfuess C., Podust V. N., Clark J. E., Fanning E. (2002) Role of the p68 subunit of human DNA polymerase α-primase in simian virus 40 DNA replication. Mol. Cell. Biol. 22, 5669–5678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Stewart L., Champoux J. J. (1999) Purification of baculovirus-expressed human DNA topoisomerase I. Methods Mol. Biol. 94, 223–234 [DOI] [PubMed] [Google Scholar]
- 49. Henricksen L. A., Umbricht C. B., Wold M. S. (1994) Recombinant replication protein A. Expression, complex formation, and functional characterization. J. Biol. Chem. 269, 11121–11132 [PubMed] [Google Scholar]
- 50. Copeland W. C. (1997) Expression, purification, and characterization of the two human primase subunits and truncated complexes from Escherichia coli. Protein Expr. Purif. 9, 1–9 [DOI] [PubMed] [Google Scholar]
- 51. Cuesta I., Núñez-Ramírez R., Scheres S. H., Gai D., Chen X. S., Fanning E., Carazo J. M. (2010) Conformational rearrangements of SV40 large T antigen during early replication events. J. Mol. Biol. 397, 1276–1286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tanaka S., Hu S. Z., Wang T. S., Korn D. (1982) Preparation and preliminary characterization of monoclonal antibodies against human DNA polymerase α. J. Biol. Chem. 257, 8386–8390 [PubMed] [Google Scholar]
- 53. Gurney E. G., Tamowski S., Deppert W. (1986) Antigenic binding sites of monoclonal antibodies specific for simian virus 40 large T antigen. J. Virol. 57, 1168–1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhao X., Madden-Fuentes R. J., Lou B. X., Pipas J. M., Gerhardt J., Rigell C. J., Fanning E. (2008) Ataxia telangiectasia-mutated damage-signaling kinase- and proteasome-dependent destruction of Mre11-Rad50-Nbs1 subunits in simian virus 40-infected primate cells. J. Virol. 82, 5316–5328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Dornreiter I., Höss A., Arthur A. K., Fanning E. (1990) SV40 T antigen binds directly to the large subunit of purified DNA polymerase α. EMBO J. 9, 3329–3336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hirt B. (1967) Selective extraction of polyoma DNA from infected mouse cell cultures. J. Mol. Biol. 26, 365–369 [DOI] [PubMed] [Google Scholar]
- 57. Fanning E., Westphal K. H., Brauer D., Cörlin D. (1982) Subclasses of simian virus 40 large T antigen. Differential binding of two subclasses of T antigen from productively infected cells to viral and cellular DNA. EMBO J. 1, 1023–1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Braithwaite A. W., Sturzbecher H. W., Addison C., Palmer C., Rudge K., Jenkins J. R. (1987) Mouse p53 inhibits SV40 origin-dependent DNA replication. Nature 329, 458–460 [DOI] [PubMed] [Google Scholar]
- 59. Gannon J. V., Lane D. P. (1987) p53 and DNA polymerase α compete for binding to SV40 T antigen. Nature 329, 456–458 [DOI] [PubMed] [Google Scholar]
- 60. Dornreiter I., Copeland W. C., Wang T. S. (1993) Initiation of simian virus 40 DNA replication requires the interaction of a specific domain of human DNA polymerase α with large T antigen. Mol. Cell. Biol. 13, 809–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Foiani M., Liberi G., Lucchini G., Plevani P. (1995) Cell cycle-dependent phosphorylation and dephosphorylation of the yeast DNA polymerase α-primase B-subunit. Mol. Cell. Biol. 15, 883–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Voitenleitner C., Rehfuess C., Hilmes M., O'Rear L., Liao P. C., Gage D. A., Ott R., Nasheuer H. P., Fanning E. (1999) Cell cycle-dependent regulation of human DNA polymerase α-primase activity by phosphorylation. Mol. Cell. Biol. 19, 646–656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Fang L., Davey M. J., O'Donnell M. (1999) Replisome assembly at oriC, the replication origin of E. coli, reveals an explanation for initiation sites outside an origin. Mol. Cell 4, 541–553 [DOI] [PubMed] [Google Scholar]
- 64. Jezewska M. J., Rajendran S., Bujalowski W. (1998) Complex of Escherichia coli primary replicative helicase DnaB protein with a replication fork. Recognition and structure. Biochemistry 37, 3116–3136 [DOI] [PubMed] [Google Scholar]
- 65. Corn J. E., Pelton J. G., Berger J. M. (2008) Identification of a DNA primase template tracking site redefines the geometry of primer synthesis. Nat. Struct. Mol. Biol. 15, 163–169 [DOI] [PubMed] [Google Scholar]
- 66. Bailey S., Eliason W. K., Steitz T. A. (2007) Structure of hexameric DnaB helicase and its complex with a domain of DnaG primase. Science 318, 459–463 [DOI] [PubMed] [Google Scholar]
- 67. Wang G., Klein M. G., Tokonzaba E., Zhang Y., Holden L. G., Chen X. S. (2008) The structure of a DnaB-family replicative helicase and its interactions with primase. Nat. Struct. Mol. Biol. 15, 94–100 [DOI] [PubMed] [Google Scholar]
- 68. Marceau A. H., Bahng S., Massoni S. C., George N. P., Sandler S. J., Marians K. J., Keck J. L. (2011) Structure of the SSB-DNA polymerase III interface and its role in DNA replication. EMBO J. 30, 4236–4247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bochman M. L., Schwacha A. (2008) The Mcm2–7 complex has in vitro helicase activity. Mol. Cell 31, 287–293 [DOI] [PubMed] [Google Scholar]
- 70. Moyer S. E., Lewis P. W., Botchan M. R. (2006) Isolation of the Cdc45·Mcm2–7·GINS (CMG) complex, a candidate for the eukaryotic DNA replication fork helicase. Proc. Natl. Acad. Sci. U.S.A. 103, 10236–10241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Fletcher R. J., Bishop B. E., Leon R. P., Sclafani R. A., Ogata C. M., Chen X. S. (2003) The structure and function of MCM from archaeal M. thermoautotrophicum. Nat. Struct. Biol. 10, 160–167 [DOI] [PubMed] [Google Scholar]
- 72. McGeoch A. T., Trakselis M. A., Laskey R. A., Bell S. D. (2005) Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism. Nat. Struct. Mol. Biol. 12, 756–762 [DOI] [PubMed] [Google Scholar]
- 73. Sanchez-Berrondo J., Mesa P., Ibarra A., Martínez-Jiménez M. I., Blanco L., Méndez J., Boskovic J., Montoya G. (2012) Molecular architecture of a multifunctional MCM complex. Nucleic Acids Res. 40, 1366–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Graham B. W., Schauer G. D., Leuba S. H., Trakselis M. A. (2011) Steric exclusion and wrapping of the excluded DNA strand occurs along discrete external binding paths during MCM helicase unwinding. Nucleic Acids Res. 39, 6585–6595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rothenberg E., Trakselis M. A., Bell S. D., Ha T. (2007) MCM forked substrate specificity involves dynamic interaction with the 5′-tail. J. Biol. Chem. 282, 34229–34234 [DOI] [PubMed] [Google Scholar]
- 76. Murakami Y., Hurwitz J. (1993) Functional interactions between SV40 T antigen and other replication proteins at the replication fork. J. Biol. Chem. 268, 11008–11017 [PubMed] [Google Scholar]
- 77. Waga S., Stillman B. (1998) The DNA replication fork in eukaryotic cells. Annu. Rev. Biochem. 67, 721–751 [DOI] [PubMed] [Google Scholar]
- 78. Duderstadt K. E., Berger J. M. (2008) AAA+ ATPases in the initiation of DNA replication. Crit. Rev. Biochem. Mol. Biol. 43, 163–187 [DOI] [PubMed] [Google Scholar]
- 79. Enemark E. J., Joshua-Tor L. (2006) Mechanism of DNA translocation in a replicative hexameric helicase. Nature 442, 270–275 [DOI] [PubMed] [Google Scholar]
- 80. Stauffer M. E., Chazin W. J. (2004) Structural mechanisms of DNA replication, repair, and recombination. J. Biol. Chem. 279, 30915–30918 [DOI] [PubMed] [Google Scholar]