Background: Pseudomonas aeruginosa employs Tse1 to kill rival cells and Tsi1 to inactivate Tse1 for self-protection.
Results: Tse1 features a conserved catalytic site for murein hydrolysis, and Tsi1 specifically occupies the substrate-binding sites of Tse1.
Conclusion: Tse1 acts as a murein peptidase, and Tsi1 blocks its substrate binding.
Significance: This work builds a novel understanding of niche competition among bacteria.
Keywords: Hydrolases, Protein Complexes, Protein Structure, Pseudomonas aeruginosa, Virulence Factors, Three-dimensional Structures of Tse1/Tse1-Tsi1, T6SS, Immune Protein Tsi1, Virulence Effector Tse1
Abstract
Recently, it was identified that Pseudomonas aeruginosa competes with rival cells to gain a growth advantage using a novel mechanism that includes two interrelated processes as follows: employing type VI secretion system (T6SS) virulence effectors to lyse other bacteria, and at the same time producing specialized immunity proteins to inactivate their cognate effectors for self-protection against mutual toxicity. To explore the structural basis of these processes in the context of functional performance, the crystal structures of the T6SS virulence effector Tse1 and its complex with the corresponding immunity protein Tsi1 were determined, which, in association with mutagenesis and Biacore analyses, provided a molecular platform to resolve the relevant structural questions. The results indicated that Tse1 features a papain-like structure and conserved catalytic site with distinct substrate-binding sites to hydrolyze its murein peptide substrate. The immunity protein Tsi1 interacts with Tse1 via a unique interactive recognition mode to shield Tse1 from its physiological substrate. These findings reveal both the structural mechanisms for bacteriolysis and the self-protection against the T6SS effector Tse1. These mechanisms are significant not only by contributing to a novel understanding of niche competition among bacteria but also in providing a structural basis for antibacterial agent design and the development of new strategies to fight P. aeruginosa.
Introduction
Competitions for survival occur everywhere between living beings. Niche competition among bacteria in natural environments is fierce and often influences the pathogenesis and progress of certain human diseases during polymicrobial infections (1–4). In recent years, studies have identified that the type VI secretion system (T6SS)3 of Gram-negative bacteria not only serves as a virulence factor that directly attacks host cells, but it also participates in competitive interbacterial interactions by translocating effector proteins into rival cells (5–9). The important role of T6SS in interbacterial interactions in association with its presence in many disease cases caused by bacterial pathogens implies that it may be crucial in certain human polymicrobial infections.
It is now known that T6SS is a specialized protein secretory apparatus evolved by Gram-negative bacteria to deliver effector proteins into target cells in a cell contact-dependent manner. Two conserved T6SS components, hemolysin co-regulated protein and valine-glycine repeat protein G, have been structurally characterized and have remarkable homology to bacteriophage tail proteins (10–12). These findings prompt the speculation that T6SS may assemble as an inverted phage tail across the envelope of bacteria, targeting bacterial cells by a puncturing manner analogous to bacteriophage entry (13, 14). However, the mechanism underlying effector delivery through this secretory apparatus and the functions of the effectors in recipient cells remain unclear.
Most recently, it was demonstrated that the hemolysin co-regulated protein secretion island I-encoded T6SS (H1-T6SS) of Pseudomonas aeruginosa plays an important role in P. aeruginosa competition with other Gram-negative bacteria (15, 16). The system delivers two novel effector proteins, Tse1 and Tse3, into target bacterial cells, which breach the barrier of the Gram-negative outer membrane by virtue of the T6SS and act as lytic enzymes that hydrolyze the murein in the periplasm of recipient cells (16). Interestingly, P. aeruginosa cannot distinguish between its siblings and other bacteria, and thus the bacteria export these effectors into each other through the T6SS. To protect against mutual toxicity, P. aeruginosa produces two immunity proteins (Tsi1 and Tsi3) that specifically inactivate their cognate effectors (16). These findings indicate an ingenious bacterial niche competition strategy. Exploring the molecular mechanism of this strategy using a three-dimensional structural basis of the effectors and their specific immune proteins is of great significance for understanding interbacterial competitive interactions, and it also provides a potential application value in the development of effective antibacterial agents and approaches.
Tse1 has been functionally characterized as a dl-endopeptidase that specifically hydrolyzes the γ-d-glutamyl-l-meso-diaminopimelic acid (γ-d-glutamyl-DAP) linkage in the murein peptide bridge (16). However, the underlying mechanism for the recognition and cleaving of the murein peptide substrate by Tse1 has remained elusive because of the low primary sequence homology with known dl-endopeptidases and the lack of a solved structure for this T6SS effector. The immunity protein Tsi1, which is only detected in P. aeruginosa, also bears no significant sequence homology to characterized proteins. Therefore, the question of how Tsi1 specifically inactivates Tse1 to protect P. aeruginosa against mutual toxicity has remained open. In this study, we report on the crystal structures of Tse1 and its complex with Tsi1. These findings build a structural framework for the understanding of the bacteriolysis mechanisms underlying the T6SS effectors and self-protection via corresponding immunity proteins. In addition, these findings provide a structural basis for antibacterial agent design and the development of new strategies to fight P. aeruginosa.
EXPERIMENTAL PROCEDURES
Protein Expression and Purification
The gene sequences encoding the full-length Tse1 and truncated Tsi1 without the N-terminal 19-residue signal peptide were synthesized with proper optimization to reduce the high GC contents (Sangon Biotech, Shanghai, China). Then both genes were subcloned into a pET-28a vector (Novagen, Darmstadt, Germany) between the NdeI and XhoI sites, which resulted in an N-terminal fusion sequence (MGSSHHHHHHSSGLVPRGSHM). The mutants were generated by site-directed mutagenesis using the wild-type (WT) vector as the template and confirmed by DNA sequencing. The proteins were overexpressed in Escherichia coli strain BL21 (DE3) (Novagen, Darmstadt, Germany) grown in LB medium and induced with 0.2 mm isopropyl β-d-1-thiogalactopyranoside overnight at 289 K. Bacterial pellets were harvested by centrifugation at 4,000 × g and then resuspended in lysis buffer (300 mm NaCl, 50 mm NaH2PO4/Na2HPO4, pH 7.5, and 10 mm imidazole) with 0.1 mm PMSF and lysed by sonication in an ice bath. After centrifugation, the soluble proteins were first purified using Ni2+-resin (Novagen, Darmstadt, Germany) and then diluted with buffer A (100 mm NaCl, 50 mm Tris, pH 8.0, and 1% v/v glycerol), followed by a thrombin digestion of the His6 tag. A second nickel chromatography step was performed, and the flow-through proteins were collected and concentrated.
Tse1 was further purified by cation-exchange (HiTrap SP HP column, GE Healthcare) and gel filtration (Hiload 16/60 Superdex 75 column, GE Healthcare) chromatography. Selenomethionine-substituted (SeMet) Tse1 was produced by expression in the E. coli methionine auxotrophic strain B834 (DE3). The purification procedure for the SeMet derivative was the same as that of the native protein. Tsi1 was further purified by anion-exchange (HiTrap Q HP column, GE Healthcare) and gel filtration (Hiload 16/60 Superdex 75 column, GE Healthcare) chromatography. All the Tsi1 mutants were expressed and purified using the same protocol. The Tse1-Tsi1 complex was obtained by mixing two proteins with excess Tse1 followed by gel filtration (Hiload 16/60 Superdex 75 column, GE Healthcare) chromatography. All purified proteins were stored in buffer A (5 mm dithiothreitol and 0.2 mm EDTA added for the SeMet protein) at 193 K.
Crystallization and Data Collection
The crystallization experiments were performed using the hanging-drop vapor diffusion method at 293 K with 2-μl drops containing a 1-μl protein solution and 1-μl reservoir solution equilibrated over a 0.5-ml reservoir solution. Initial crystallization hits were found using an Index Kit (Hampton Research, Aliso Viejo, CA). The Tse1 crystals and SeMet-labeled Tse1 were grown in 1.8 m sodium formate and 50 mm MES, pH 6.2. The Tse1-Tsi1 complex crystals were grown from a solution of 20% (v/v) PEG4000, 100 mm sodium citrate, pH 5.4, and 20% (v/v) isopropyl alcohol.
The single wavelength anomalous diffraction data set for SeMet-labeled Tse1 was collected at beamline 3W1A of the Beijing Synchrotron Radiation Facility (Beijing, China). The high resolution data set of Tse1 and the Tse1-Tsi1 complex data set were collected at beamline 17A of the Photon Factory (KEK, Tsukuba, Japan). Prior to data collection, all of the crystals were transferred to a well solution supplied with 10% (v/v) ethylene glycol as a cryoprotectant.
Structure Determination and Refinement
All the data sets were processed with MOSFLM and scaled with SCALA from the CCP4 program suite (17). Phase determination and automatic model building of Tse1 were performed with PHENIX (18). The rest of the model was manually built with COOT (19). The refinement was carried out with PHENIX. The structure of the Tse1-Tsi1 complex was solved by molecular replacement with PHASER of the CCP4 program suite using the apo-Tse1 structure as a search model. Model building and structural refinement were performed with COOT and PHENIX, respectively. The statistics of data collection and refinement are summarized in Table 1. The quality of the final model was checked by MolProbity (20). Sequence alignment was generated by ClustalW (21). Our sequence alignment figure was produced by ESPript (22). All other figures were rendered in PyMOL.
TABLE 1.
Data collection and refinement statistics
| Se-Tse1 | Tse1 | Tse1-Tsi1 complex | |
|---|---|---|---|
| Data collection | |||
| Space group | P212121 | P212121 | P3221 |
| Cell dimensions | |||
| a, b, c | 39.26, 65.03, 75.39 Å | 38.45, 63.64, 73.74 Å | 97.00, 97.00, 292.17 Å |
| α, β, γ | 90, 90, 90° | 90, 90, 90° | 90, 90, 120° |
| Peak | |||
| Wavelength | 0.9795 Å | 1.0000 Å | 0.9788 Å |
| Resolutiona | 29.85 to 2.10 Å (2.21 to 2.10 Å) | 26.61 to 1.50 Å (1.58 to 1.50 Å) | 63.61 to 2.50 Å (2.64 to 2.50 Å) |
| Unique reflections | 12,563 | 29,544 | 56,166 |
| Rmergeb | 9.9% (24.0%) | 8.2% (26.6%) | 6.7 (38.7) |
| I/σI | 22.2 (13.4) | 14.6 (4.7) | 21.6 (5.6) |
| Completeness | 99.8% (99.8%) | 99.4% (96.0%) | 99.8% (99.4%) |
| Redundancy | 14.4 (14.7) | 6.7 (4.6) | 9.2 (8.9) |
| Refinement | |||
| Resolution | 26.61 to 1.50 Å | 63.61 to 2.50 Å | |
| No. of reflections | 29,040 | 55,504 | |
| Rwork/Rfree | 16.74/18.61% | 20.71/25.71% | |
| No. of atoms | |||
| Protein | 1140 | 8905 | |
| Formate− | 27 | ||
| Water | 264 | 341 | |
| B-factors | |||
| Protein | 12.05 Å2 | 53.23 Å2 | |
| Formate | 23.98 Å2 | ||
| Water | 25.04 Å2 | 43.47 Å2 | |
| Root mean square deviations | |||
| Bond lengths | 0.0066 Å | 0.0064 Å | |
| Bond angles | 1.014° | 1.055° | |
a Values in parentheses are for the data in the highest resolution shell.
b Rmerge = Σ|Ii − Im|/Σ Ii, where Ii is the intensity of the measured reflection, and Im is the mean intensity of all the symmetry-related reflections.
E. coli Growth Curves
The tse1 gene was also subcloned into a pET-22b vector (Novagen, Darmstadt, Germany) between the NcoI and XhoI sites for expression in the periplasm by fusion with an N-terminal signal peptide. The vector also added a C-terminal hexahistidine tag to the expressed protein, allowing for Western blot analysis of expression. The catalytic residue mutants were generated by site-directed mutagenesis using the WT-Tse1 as the template. BL21 (DE3) pLysS cells harboring WT-Tse1 expression plasmids and mutant Tse1 were grown overnight in LB medium at 310 K and subinoculated to a starting absorbance of ∼0.06 at 600 nm (A600) in LB medium. The cultures were grown to an A600 0.26–0.27 and induced with 0.1 mm isopropyl β-d-1-thiogalactopyranoside. The A600 values of the cultures were recorded for growth curves. Protein expression levels were detected at 40 min after induction by Western blot analysis against the anti-His antibody (Abmart, Shanghai, China) and the anti-β-lactamase antibody (Millipore, Billerica, MA).
Tse1 and Tsi1 Kinetic Assay
Protein kinetics were investigated using a surface plasmon resonance Biacore T100 machine (GE Healthcare) at 298 K. The running buffer (50 mm HEPES, pH 7.5, 100 mm NaCl, and 0.005% (v/v) Tween 20) was prepared, vacuum filtered, and degassed immediately prior to use. One CM5 sensor chip flow cell was activated for 7 min with a 1:1 mixture of 0.1 m N-hydroxysuccinimide and 0.1 m N-ethyl-N′-(3-diethylaminopropyl)-carbodiimide at a flow rate of 10 μl/min. Tse1, at a concentration of 2 μg/ml in 10 mm sodium acetate pH 5.0, was immobilized to 300 response units on the flow cell. The remaining binding sites were blocked by 1 m ethanolamine, pH 8.5.
The WT-Tsi1 and mutant Tsi1 proteins were injected at different concentrations over the Tse1 surface and blank flow cell for 2 min at a flow rate of 30 μl/min. After 2–4 min at dissociation phase, the bound protein was removed with a 30-s wash with 10 mm glycine-HCl, pH 2.0. The resulting data were analyzed with Biacore T100 evaluation software by fitting to a 1:1 Langmuir binding model.
RESULTS
Overall Structure of Tse1
Although Tse1 has been defined as a novel dl-endopeptidase that belongs to the superfamily of papain-like cysteine peptidases, determining the structure of Tse1 with available structures of other dl-endopeptidases as search models was unsuccessful. We solved the crystal structure of apo-Tse1 by selenium single-wavelength anomalous diffraction at a −1.5-Å resolution. The structure of apo-Tse1 contains one Tse1 molecule in an asymmetric unit, which is consistent with the observation that Tse1 exists as a monomer in solution (Fig. 1A). The final model includes residues 3–154 of Tse1. The first two residues are not included in the final model due to the lack of interpretable electron density. The details of the crystallographic analysis are summarized in Table 1.
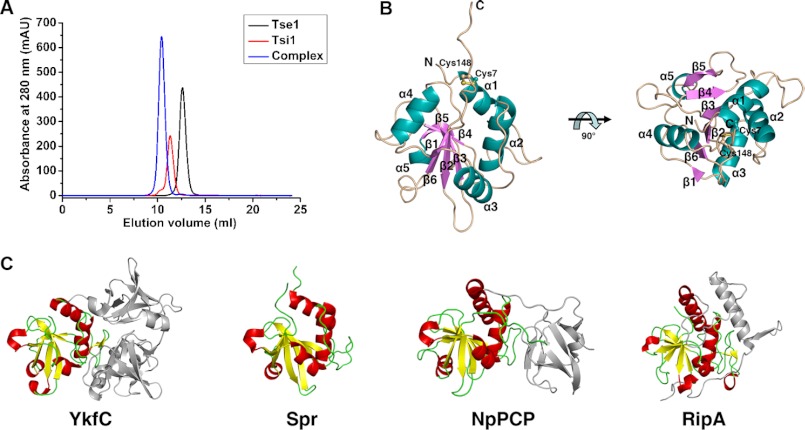
FIGURE 1.
Overall structure of Tse1. A, elution profiles of apo-Tse1, apo-Tsi1, and Tse1-Tsi1 complexes by size-exclusion chromatography. The single peaks of the Tse1, Tsi1, and Tse1-Tsi1 complexes correspond to apparent molecular masses of 16.7, 17.2, and 34.9 kDa, respectively. B, structure of apo-Tse1 shown from the side (left) and the top (right) views. The secondary structure elements are labeled and in different colors. The disulfide bonds formed between Cys-7 and Cys-148 are shown as ball and stick models. C, structural comparison between Tse1 and the NlpC/P60 domains of other dl-endopeptidases. The structures of other dl-endopeptidases are shown in the same superimposed orientation as Tse1 from the side view in B. The NlpC/P60 domains are highlighted.
Tse1 adopts a compact papain-like α+β fold that consists of six β-strands (β1-β6) and five helices (α1-α5). The six β-strands constitute a curved antiparallel β-sheet in the center of the structure, and the five helices flank each side of the central β-sheet (Fig. 1B). Following the last β6 is a long flexible loop that is fixed by a disulfide bond between Cys-148 within the loop and Cys-7 located at the N terminus of α1 (Fig. 1B). Extensive side-chain hydrophobic interactions play important roles in maintaining the integrated structure. Structural homology searches by DALI (23) reveal that Tse1 shares a notable structural similarity with characterized NlpC/P60 domains of other dl-endopeptidases. The significant hits retrieved by DALI include dl-endopeptidase YkfC from Bacillus cereus (Z = 10.0, PDB code 3H41) (24), membrane-anchored cell-wall hydrolase Spr from E. coli (Z = 8.8, PDB code 2K1G) (25), dl-endopeptidases NpPCP and AvPCP from cyanobacteria Nostoc punctiforme and Anabaena variabilis (Z = 8.6 and 8.5, PDB codes 2EVR and 2HBW) (26), and the peptidoglycan endopeptidases RipA and RipB from Mycobacterium tuberculosis (Z = 7.1 and 7.2, PDB code 3NE0 and 3PBI) (Fig. 1C) (27, 28). Structural comparisons between Tse1 and the NlpC/P60 domains of these dl-endopeptidases yield a moderate root mean square deviation (r.m.s.d.) of 2.7–3.8 Å for 112 aligned Cα atoms, indicative of notable differences between them, particularly in the flexible loop connecting the conserved second structure elements.
Tse1 Catalytic Site
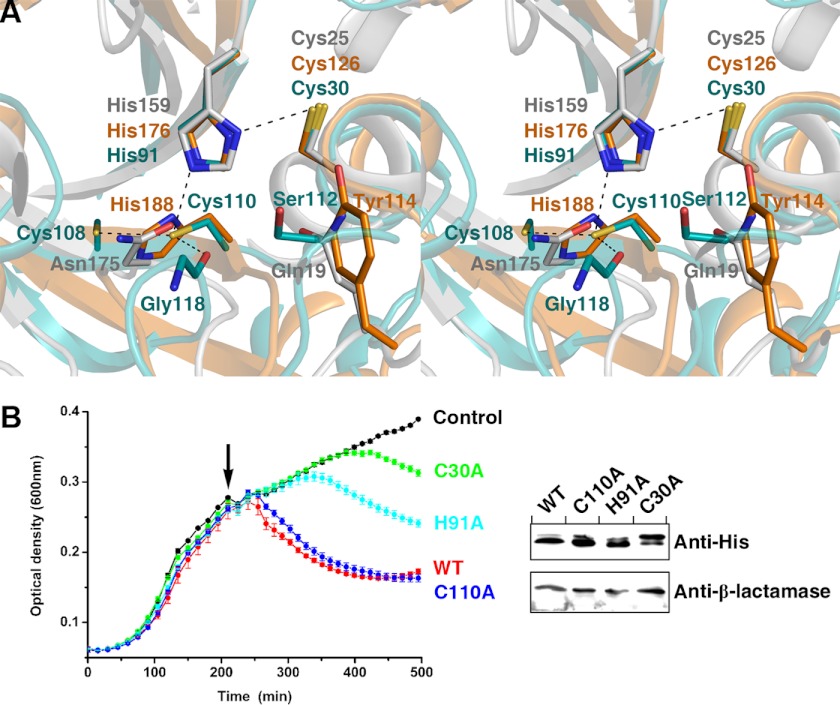
As a branch of papain-like cysteine peptidases widespread in bacteria, NlpC/P60 dl-endopeptidases possess the highly conserved catalytic triad Cys-His-Xaa (where Xaa is a polar residue) in which the catalytic Cys-His pair hydrolyzes the substrate and the third polar residue orients the second His to the first Cys (29). Sequence alignment indicates that despite bearing a low sequence homology with other NlpC/P60 dl-endopeptidases, Tse1 retains the crucial catalytic Cys-His pair (supplemental Fig. 1). In the apo-Tse1 structure, the reactive Cys-30 is located in the N terminus of α2, spatially adjacent to the catalytic His-91 located on the N terminus of β3. Both catalytic residues could be well superimposed to their corresponding counterparts in other NlpC/P60 dl-endopeptidases (Fig. 2A). Upon superimposition of the catalytic Cys-His pairs, there is a variation of the third residue. In other NlpC/P60 dl-endopeptidases, this position is often occupied by another histidine (24–26). Other polar residues, including Asn, Glu, Gln, and Asp, also occur at this position, although with low frequency (27, 28). Close inspection confirms that the third residue in the catalytic site of Tse1 is a cysteine (Cys-110) located in the C terminus of β4. Such an arrangement has not been previously observed in papain-like cysteine peptidases (Fig. 2A). Compared with the more commonly observed histidine, Cys-110 directs its sulfhydryl group farther away from the imidazole group of the second histidine, with the distance between the Sγ of Cys-110 and the Nδ1 of His-91 being 3.51 Å. Cys-110 is further stabilized by van der Waals interactions with the main-chain carbonyl group of Gly-118 and the sulfhydryl group of the proximal Cys-108 (Fig. 2A). In addition, another conserved tyrosine, which helps to form an “oxyanion hole” near its hydroxyl group in other NlpC/P60 dl-endopeptidases, has no counterpart but a serine (Ser-112) at the corresponding Tse1 position (Fig. 2A). Ser-112 is located on the flexible loop following Cys-110, spatially adjacent to the reactive Cys-30, and is probably responsible for the formation of the oxyanion hole.
FIGURE 2.
Catalytic site of Tse1 and mutagenesis of catalytic residues. A, stereoview presenting the superimpositions of the catalytic sites of Tse1, dl-endopeptidase NpPCP, and papain. The catalytic residues are shown as stick models with those of Tse1 in cyan, of NpPCP in orange, and of papain in gray. B, growth curves of E. coli expressing WT-Tse1 and catalytic residue mutants in the periplasm. Cultures were induced at the indicated time (arrow). Error bars indicate ± S.D. (n = 3). Western blot analysis of total protein levels of WT-Tse1 and mutant Tse1 for strains used in the experiments are presented in B. β-lactamase was used as a loading control.
To further confirm the contributions of the catalytic residues of Tse1, we expressed the mutants of the catalytic residues in the periplasm and tested their capacities to lyse bacterial cells. The results demonstrate that mutation of either catalytic Cys-30 or His-91 to alanine greatly reduced the bacteriolytic activities of Tse1, whereas mutation of Cys-110 to alanine had no obvious effect on bacteriolytic activity (Fig 2B). These observations confirmed that Tse1 acts as a papain-like cysteine peptidase. The conserved catalytic Cys-30–His-91 pair plays an important role in the catalytic activities of Tse1, whereas Cys-110 contributes little to the catalytic activities of Tse1.
Potential Substrate-binding Sites
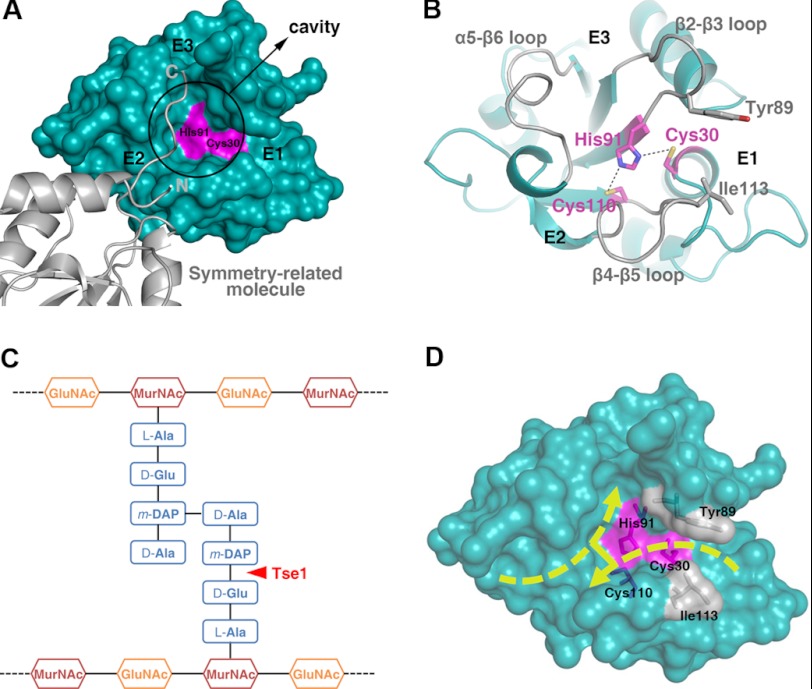
Surface inspection indicates that on top of the catalytic site of Tse1 is a large cavity with three independent entrances (E1, E2, and E3) related by a pseudo 3-fold axis (Fig. 3A). Three protruding loops (β2-β3, β4-β5, and α5-β6 loop) surround the cavity with two of them each binding an entrance (Fig. 3B). The distinct geometry of these areas indicates that they likely serve as the substrate-binding sites of Tse1 and recognize the cross-linked peptides of cell wall murein (Fig. 3C). Interestingly, in the structure of apo-Tse1, the C terminus of a symmetry-related molecule runs across E2, makes a 90° turn in the cavity, and flaps to E3 (Fig. 3A). In this way, the C-terminal flexible loop partially occupies the extensive substrate-binding sites, which, to a certain extent, mimics a portion of the Tse1 cross-linked murein peptide binding. In fact, the architecture of the substrate-binding sites could accommodate the cross-linked murein peptides well. Upon recognizing the cell wall peptidoglycan by Tse1, the acceptor murein tetrapeptide enters E2 with the extended glycan backbone out of the cavity, lying on the open surface of the protein. This tetrapeptide makes a fine turn in the cavity and directs the last residue to E3. The donor murein tetrapeptide enters E1 from the opposite direction. The scissile γ-d-glutamyl-DAP bond is clamped by Tyr-89 in the β2-β3 loop and Ile-113 in the β4-β5 loop, exposing it to the catalytic Cys-30 for specific cleavage. The last residue stretches into the cavity and cross-links with the third residue of the other tetrapeptide (Fig. 3D).
FIGURE 3.
Substrate-binding sites of Tse1. A, surface representation presenting the substrate-binding sites of Tse1. The catalytic Cys-30 and His-91 are colored in pink. The cavity is highlighted, and the entrances are labeled. The symmetry-related molecule is shown as a ribbon diagram. B, ribbon diagram of the substrate-binding sites of Tse1. The three protruding loops surrounding the cavity are labeled. The catalytic residues are shown as stick models and colored in pink. Tyr-89 and Ile-113, which clamp the scissile bond for hydrolysis, are also shown as stick models. C, schematic view of the cross-linked cell wall murein substrate. The scissile bond cleaved by Tse1 is highlighted. D, surface view showing the possible binding fashion of the cross-linked murein peptide substrate.
Overall Structure of Tse1-Tsi1 Complex
To elucidate the protection mechanism of P. aeruginosa against mutual toxicity by Tsi1, the crystal structure of Tse1 in complex with Tsi1 was determined at a 2.5-Å resolution by molecular replacement with the structure of apo-Tse1 as the search model. Each asymmetric unit contains four Tse1-Tsi1 complexes arranged in a D2 point group. Only a few interactions are observed between these four complexes. In addition, the Tse1-Tsi1 complex indicates an apparent molecular mass of 34.9 kDa by analytical size-exclusion chromatography (Fig. 1A). These observations suggest that Tse1 and Tsi1 form a complex with 1:1 stoichiometry. The four Tse1-Tsi1 complexes in the asymmetric unit are likely the result of crystal packing and represent no significant biological relevance. The structures of Tse1 and Tsi1 in the four complexes are essentially identical, with Cα r.m.s.d. of ∼0.22 to 0.36 and ∼0.29 to 0.48 Å for Tse1 and Tsi1, respectively. Thus, we hereafter confine our analyses and discussions to one Tse1-Tsi1 complex (composed of chain A for Tse1 and chain E for Tsi1). The final model of the Tse1-Tsi1 complex contains residues 3–148 of Tse1 and residues 21–168 of Tsi1. Bioinformatic analyses predict that the Tsi1 residues 1–19 compose a periplasmically localized signal peptide sequence and were truncated when we constructed the recombinant Tsi1 protein. Other terminal residues, comprising residues 1–2 and 149–154 of Tse1 and residues 20 and 169–172 of Tsi1, are omitted in the complex structure due to the lack of interpretable electron density. The details of the crystallographic analysis are summarized in Table 1.
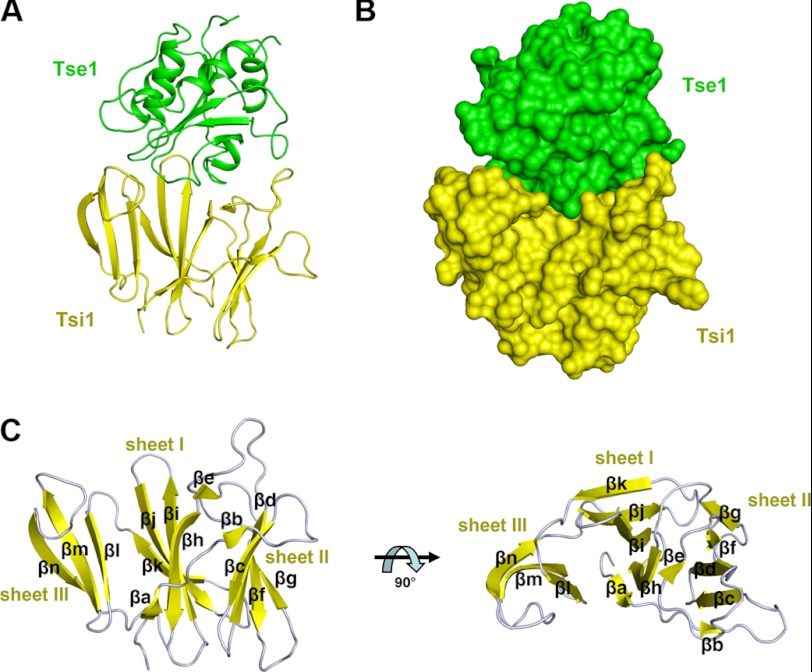
The structure of the Tse1-Tsi1 complex indicates that both proteins tightly interact with each other (Fig. 4, A and B). The complex buries a total of 3,951 Å2 of solvent-accessible surface area, with 1,869 Å2 contributed by Tse1 and 2082 Å2 contributed by Tsi1. Superposing the structure of Tse1 in the complex with that of apo-Tse1 yields an r.m.s.d. of 0.51 Å for 146 Cα atoms, indicative of no significant structural changes of Tse1 upon interaction with Tsi1 (supplemental Fig. 2). In the complex, Tsi1 adopts an all β fold consisting of 14 β-strands (βa-βn), which constitute three antiparallel β-sheets (termed as sheet I, II, and III) (Fig. 4C). Sheet I consists of βa, βh, βi, βj, and βk, located in the middle. Sheet II (composed of βb, βc, βd, βf, and βg) and sheet III (composed of βl, βm, and βn) are packed against each side of sheet I. The remaining short βe flanks the N terminus of βh at the base. All three sheets are curved and shaped like three blades (Fig. 4C). DALI searches (23) with the structure of Tsi1 as the bait obtained several structures containing a typical β-propeller fold, such as dipeptidyl aminopeptidase IV from Stenotrophomonas maltophilia (Z = 7.2, PDB code 2ECF) (30). Although the structure of Tsi1 can be roughly superimposed with part of the characterized β-propeller fold (supplemental Fig. 3), there are remarkable differences between them. The classic β-propeller is characterized by 4–8 blade-shaped β-sheets arranged toroidally around a central axis. Each sheet typically has four antiparallel β-strands. However, Tsi1 only has three blade-shaped β-sheets that do not form an integrated circle. Additionally, the three β-sheets have unequal antiparallel β-strands unlike those in a β-propeller.
FIGURE 4.
Overall structure of the Tse1-Tsi1 complex. A, ribbon representation of the complex structure. Tse1 is in green. Tsi1 is in yellow. B, surface view of the complex with the same colors as in A. C, ribbon diagram of Tsi1 from the side (left) and the top (right) views. The protein secondary structure elements are labeled.
Tsi1-Tse1 Interaction
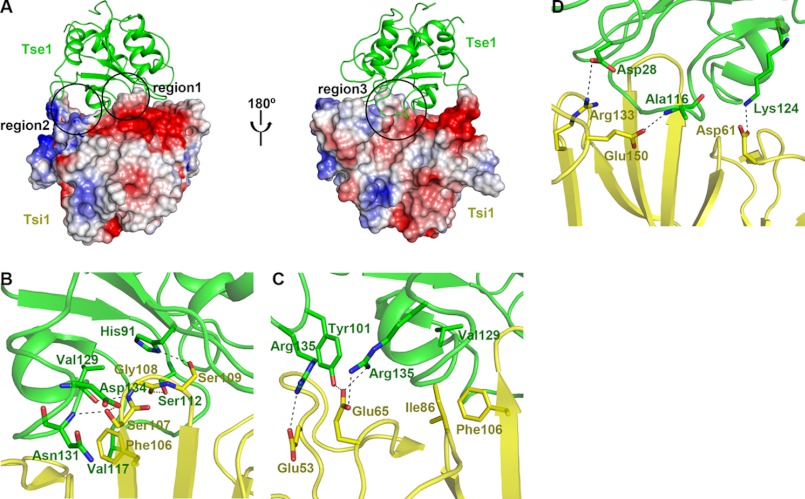
At the interface of the Tse1-Tsi1 complex, Tsi1 has a unique topography characterized by protruding rigid βi-βj turns in the center surrounded by a circle of low-lying areas. This structural feature provides a perfect fit for binding of the substrate-binding sites of Tse1. In other words, Tsi1 adopts a unique interactive mode to specifically recognize Tsi1, which gives rise to extensive interactions between these two proteins (Fig. 5A).
FIGURE 5.
Tsi1-Tse1 interaction. A, two opposite views of the interface between Tse1 and Tsi1. Tse1 is shown as a ribbon diagram in green. The electrostatic potential of Tsi1 is depicted with surface coloration from red to blue for negatively to positively charged regions. B–D, close-up view of the three contacting regions. All of the interacting residues are labeled and shown as stick models (Tse1 in green and Tsi1 in yellow). Hydrogen bonds and salt bridges are shown as black dashed lines.
The extensive interactions can be divided into three principal regions. 1) The rigid Tsi1 βi-βj turn and insert into the cavity of Tse1, contacting the floor and interacting with the surrounding Tse1 β2-β3 loop, β4-β5 loop, and α5-β6 loop (Fig. 5, A and B). This central region involves five hydrogen bonds. Among the interacting residues, Tsi1 Ser-107 forms two strong hydrogen bonds with the main chain of Tse1 Asn-131 and the side chain of Tse1 Asp-134, supplemented by van der Waals interactions with the side chain of Tse1 Asn-131. Tsi1 Ser-109 forms a hydrogen bond with the catalytic residue His-91 of Tse1 and fixes its side-chain conformation. Furthermore, Tsi1 Phe-106 interacts with Tse1 Val-117 through side-chain hydrophobic interactions, further stabilizing the Tsi1-Tse1 interaction (Fig. 5B) (2). The Tse1 β3-β4 loop and α5-β6 loop protrude into the low lying area of Tsi1, contacting the Tsi1 βe-βf loop and βg-βh loop (Fig. 5, A and C). Tsi1 Glu-65 forms three hydrogen bonds with the side chains of Tse1, Tyr-101, and Arg-135, respectively, playing an important role in the Tsi1-Tse1 interaction in this region. In addition, Tsi1 Glu-53 forms a salt bridge with Tse1 Arg-102. As a supplement, Tsi1 Ile-86 interacts with the main chain of Tse1 Val-129 through van der Waals interaction and constrains Tsi1 Phe-106 in the central region through hydrophobic interactions (Fig. 5C) (3). The extrusion of the Tse1 β4-β5 loop extends into the cleft of Tsi1 and is embraced by the Tsi1 βd-βe turn and βm-βn loop (Fig. 5, A and D). On one side, Tsi1 Arg-133 forms a salt bridge with Tse1 Asp-28. On the other side, Tsi1 Asp-61 interacts with Tse1 Lys-124 through a side-chain hydrogen bond. In the middle, Tsi1 Glu-150 forms a hydrogen bond with the main chain of Tse1 Ala-116 (Fig. 5D).
Structural Determinants of Tsi1-Tse1 Recognition
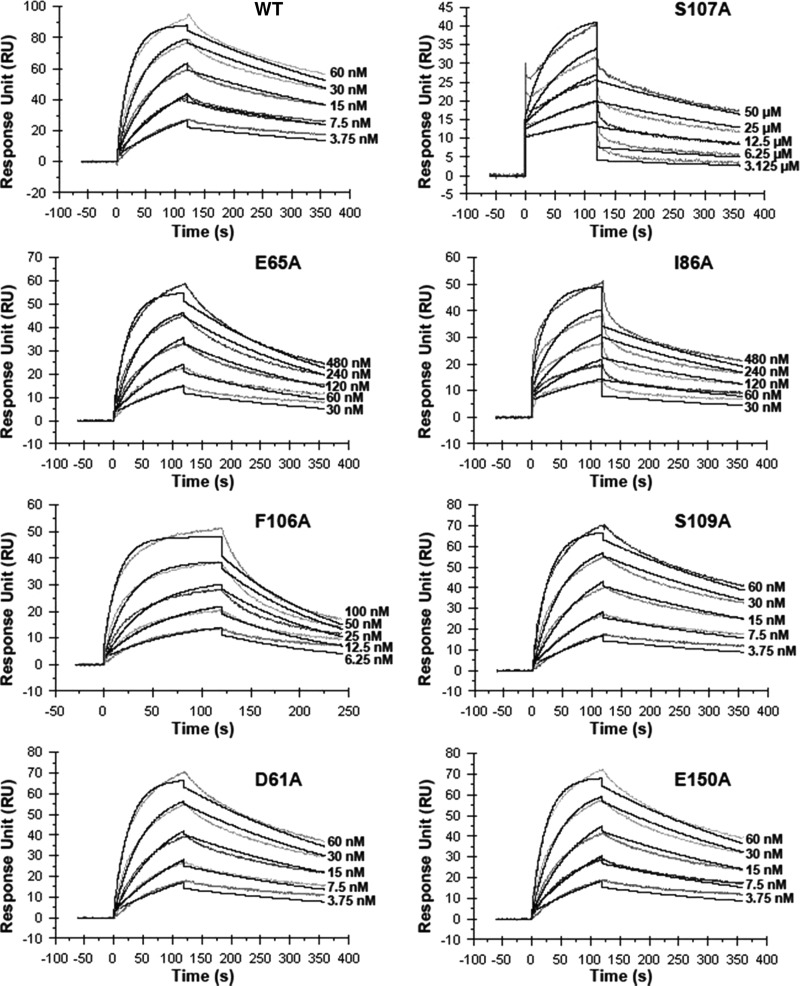
The specific recognition mode and extensive interactions undoubtedly endow Tsi1 and Tse1 with tight binding. This speculation from the structural analyses was further confirmed by surface plasmon resonance experiments with a Biacore T100. WT-Tse1 was covalently coupled to the chip, and WT or mutant Tsi1 proteins were injected in the microfluidic channel. The WT-Tsi1 and WT-Tse1 yielded a dissociation constant (Kd) of 2.77 nm (Table 2 and Fig. 6), indicative of a strong binding between Tsi1 and Tse1, even comparable with the binding affinity between antigen and antibody. However, upon mutating Tsi1 Ser-107 to an alanine, the Tsi1-S107A mutant to WT-Tse1 Kd value markedly decreased to 4.00 μm, an ∼1,500-fold reduction. In accordance with the change of binding affinity, the Tsi1-S107A mutant to WT-Tse1 association rate exhibited the same degree of reduction (Table 2 and Fig. 6). These observations indicate that Tsi1 Ser-107 in the βi-βj turn plays an indispensable role in Tsi1-Tse1-specific recognition. This finding is consistent with the broad interactions between this residue and neighboring Tse1 residues in the complex structure (Fig. 5B). Moreover, substituting an alanine for Tsi1 Glu-65 or Ile-86 in the second Tse1-contacting region led to a 15- or 11-fold reduction, respectively, of the binding affinity, accompanied by a similar 9-fold decrease of the association rate (Table 2 and Fig. 6). These results suggest that both residues also play important roles in Tsi1 recognizing Tse1. From the complex structure perspective, Tsi1 Glu-65 and Ile-86 protrude from the low lying area with their side chains responsible for specifically interacting with corresponding structural elements of Tse1 (Fig. 5C). In addition, the Tsi1-F106A mutant also exhibited a 5-fold reduction in binding affinity to WT-Tse1. However, the Phe-106 residue mainly contributes to the stabilization of the Tsi1-Tse1 interaction, as suggested by the notable 4-fold increase of the dissociation rate compared with WT and other mutant Tsi1 proteins (Table 2 and Fig. 6). Other Tsi1 residues involved in the Tsi1-Tse1 interaction (Ser-109, Asp-61, and Glu-150) were further confirmed to play subsidiary roles in the Tsi1-Tse1 interaction because mutations of these residues did not have any noticeable effects on any kinetic value compared with WT-Tsi1.
TABLE 2.
Kinetics and affinity constants for wild-type and mutant Tsi1 proteins binding to Tse1
| Protein | Association rate (ka) | Dissociation rate (kd) | Binding affinity (Ka) |
|---|---|---|---|
| m−1 s−1 | s−1 | m | |
| WT | 7.32 × 105 | 2.03 × 10−3 | 2.77 × 10−9 |
| S107A | 4.57 × 102 | 1.83 × 10−3 | 4.00 × 10−6 |
| E65A | 8.14 × 104 | 3.42 × 10−3 | 4.20 × 10−8 |
| I86A | 8.00 × 104 | 2.44 × 10−3 | 3.04 × 10−8 |
| F106A | 6.12 × 105 | 8.04 × 10−3 | 1.31 × 10−8 |
| S109A | 6.14 × 105 | 2.04 × 10−3 | 3.33 × 10−9 |
| D61A | 6.08 × 105 | 2.54 × 10−3 | 4.17 × 10−9 |
| E150A | 6.53 × 105 | 2.35 × 10−3 | 3.61 × 10−9 |
FIGURE 6.
Measure of the interactions of WT-Tsi1 and mutant Tsi1 with Tse1 by surface plasmon resonance experiments. Sensorgram indicating the titrations of Tse1 by WT-Tsi1 and mutant Tsi1 proteins at different concentrations.
DISCUSSION
Tse1 Adopts a Papain-like Structure to Hydrolyze Cross-linked Murein Peptide
Tse1 is a novel T6SS virulence effector employed by P. aeruginosa to compete with other bacteria. In this study, we found that Tse1 adopts a typical papain-like structure and shares a high structural homology with known NlpC/P60 dl-endopeptidases (Fig. 1C). However, unlike other NlpC/P60 dl-endopeptidases, Tse1 comprises a sole NlpC/P60 domain without any N-terminal signal peptide or fusion with other auxiliary domains, such as the bacterial Src homology 3-like domain (Fig. 1C).
Tse1 retains the conserved catalytic Cys-His pair of papain-like cysteine peptidases. Although harboring a novel variation of the third residue, the catalytic site of Tse1 features a spatial configuration closely resembling that of other papain-like cysteine peptidases (Fig. 2A). Therefore, the catalytic mechanism for γ-d-glutamyl-DAP bond cleavage by Tse1 is expected to be analogous to that of papain-like cysteine peptidases (26, 31). His-91 serves as a general base and deprotonates the sulfhydryl group of Cys-30 to form a thiolate anion. The thiolate anion of Cys-30 acts as a catalytic nucleophile and attacks the carbonyl carbon of the scissile γ-d-glutamyl-DAP bond in the murein peptide to turn the carbonyl oxygen into an oxyanion. The oxyanion is held by a side chain of Ser-112 that forms an oxyanion hole. The reaction is completed by activating a water molecule to hydrolyze the acyl-thioester bond to release the produced peptide and regenerate Tse1.
Prior to study of the P. aeruginosa T6SS effector Tse1, the bacterial NlpC/P60 dl-endopeptidases were characterized as endogenous hydrolases involved in the peptidoglycan recycling process during bacterial cell division and growth (32) and the peptidoglycan degradation process during bacterial cell autolysis (33). Several of the endopeptidases act on the simple murein tetrapeptide comprising the specific γ-d-glutamyl-DAP bond. The fused auxiliary domains confer substrate specificity on these peptidoglycan hydrolases (24, 26). However, as an exogenous peptidoglycan hydrolase delivered by T6SS, Tse1 directly faces the intact cell wall peptidoglycan and targets its specific γ-d-glutamyl-DAP linkage occurring in the cross-linked murein peptide. The Tse1 structure presents an extensive substrate-binding site specific for cross-linked murein peptide recognition (Fig. 3D). In fact, our structural analyses of the specific recognition for cross-linked murein peptide by Tse1 are consistent with previous biochemical studies. In cell wall peptidoglycan degradation assays, Tse1 exhibits a notable preference toward the breaking of the cross-linked murein peptide (16), supporting the specific recognition of Tse1 of the amide bond in the cross-linked murein peptide as its best substrate. It is well known that the glycan chain linkage and the murein peptide cross-linkage are the main linkages maintaining the mesh-shaped architecture of the murein of Gram-negative bacteria. Together with another T6SS effector Tse3 that breaks the glycan chain linkage (16), Tse1 enables P. aeruginosa to destroy the structure of the murein of target cells and achieve bacteriolysis.
Tsi1 Inactivates Tse1 by Blocking the Substrate Binding and Cleavage of Tse1
Tsi1 adopts a novel three β-sheet structure to specifically interact with Tse1 via a unique interactive recognition mode (Fig. 5A). Notably, the Tsi1-binding sites in Tse1 overlap with a large area of the substrate-binding sites of Tse1, including the cavity accommodating the cross-linkage in the cross-linked murein peptide substrate and E2 groove where the nonhydrolyzed murein tetrapeptide is located (Fig. 3A). The high affinity and specificity of the Tsi1-Tse1 interaction indicate that the occupancy of the substrate-binding sites of Tse1 by Tsi1 probably prevent Tse1 from binding to its cross-linked murein peptide substrate. Interestingly, in the structure of apo-Tse1, the flexible C-terminal loop of the symmetry-related molecule inserts into the substrate-binding sites of Tse1 and disappears in the structure of the Tse1-Tsi1 complex, indicating possible occupancy by Tsi1. Furthermore, the reactive Cys-30 of Tse1 responsible for attacking the scissile γ-d-glutamyl-DAP bond in the substrate lies away from the Tsi1-binding sites on Tse1 and, in turn, might be incapable of breaking Tsi1 even with Tse1 as a cysteine peptidase. Therefore, we conclude that the possible structural mechanism for protection of P. aeruginosa against mutual bacteriolysis by Tsi1 is inactivation of Tse1. This inactivation is accomplished by Tsi1 functioning as an inhibitor occupying the substrate-binding sites of exogenous Tse1 and blocking the cross-linked murein peptide binding and cleavage of Tse1.
It is noteworthy that recent studies have found that Tse1 and its immune protein Tsi1 are not confined to P. aeruginosa. They also have homologues in other Gram-negative bacteria, suggestive of the generality of the competitive strategy adopted by P. aeruginosa (34). Additionally, Tse1 comprises only a minimal functional unit of the papain-like cysteine peptidase, but it serves as an efficient factor for the killing of other bacteria by P. aeruginosa. Its specific toxicity to an essential bacterial cell component makes it a good antibacterial agent fighting against a number of Gram-negative pathogenic bacteria. The finding of an effective way to export this protein instead of T6SS is the next goal of application research. However, the important roles of Tse1 and Tsi1 for P. aeruginosa in killing rival cells and obtaining a growth advantage against other bacteria prompts the speculation that the proteins could also be potential anti-P. aeruginosa targets.
Acknowledgments
We thank Yuanyuan Chen (Institute of Biophysics, Chinese Academy of Sciences) for assistance in Biacore analysis and the staff at the beamline 3W1A of the Beijing Synchrotron Radiation Facility (Beijing, China) and at the beamline 17A of the Photon Factory (KEK, Tsukuba, Japan) for technical assistance during data collection.
This work was supported by Chinese Ministry of Science and Technology 973 Program Grants 2011CB910304 and 2011CB911103, National Natural Science Foundation of China Grant 31100535, and Chinese Academy of Sciences Grant KSCX2-EW-J-3.

This article contains supplemental Figs. 1–3.
The atomic coordinates and structure factors (codes 3VPI and 3VPJ) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- T6SS
- type VI secretion system
- Tse1
- type VI secretion exported 1
- Tsi1
- Tse1-specific immunity protein
- γ-d-glutamyl-DAP
- γ-d-glutamyl-l-meso-diaminopimelic acid
- IPTG
- isopropyl β-d-1-thiogalactopyranoside
- SeMet
- selenomethionine-substituted
- SPR
- surface plasmon resonance
- PDB
- Protein Data Bank
- r.m.s.d
- root mean square deviation.
REFERENCES
- 1. Reid G., Howard J., Gan B. S. (2001) Can bacterial interference prevent infection? Trends Microbiol. 9, 424–428 [DOI] [PubMed] [Google Scholar]
- 2. Brogden K. A., Guthmiller J. M., Taylor C. E. (2005) Human polymicrobial infections. Lancet 365, 253–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gjødsbøl K., Christensen J. J., Karlsmark T., Jørgensen B., Klein B. M., Krogfelt K. A. (2006) Multiple bacterial species reside in chronic wounds. A longitudinal study. Int. Wound J. 3, 225–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hibbing M. E., Fuqua C., Parsek M. R., Peterson S. B. (2010) Bacterial competition. Surviving and thriving in the microbial jungle. Nat. Rev. Microbiol. 8, 15–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bingle L. E., Bailey C. M., Pallen M. J. (2008) Type VI secretion. A beginner's guide. Curr. Opin. Microbiol. 11, 3–8 [DOI] [PubMed] [Google Scholar]
- 6. Cascales E. (2008) The type VI secretion toolkit. EMBO Rep. 9, 735–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Jani A. J., Cotter P. A. (2010) Type VI secretion. Not just for pathogenesis anymore. Cell Host Microbe 8, 2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schwarz S., Hood R. D., Mougous J. D. (2010) What is type VI secretion doing in all those bugs? Trends Microbiol. 18, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwarz S., West T. E., Boyer F., Chiang W. C., Carl M. A., Hood R. D., Rohmer L., Tolker-Nielsen T., Skerrett S. J., Mougous J. D. (2010) Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathog. 6, e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., Goodman A. L., Joachimiak G., Ordoñez C. L., Lory S., Walz T., Joachimiak A., Mekalanos J. J. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Leiman P. G., Basler M., Ramagopal U. A., Bonanno J. B., Sauder J. M., Pukatzki S., Burley S. K., Almo S. C., Mekalanos J. J. (2009) Type VI secretion apparatus and phage tail-associated protein complexes share a common evolutionary origin. Proc. Natl. Acad. Sci. U.S.A. 106, 4154–4159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pell L. G., Kanelis V., Donaldson L. W., Howell P. L., Davidson A. R. (2009) The phage λ major tail protein structure reveals a common evolution for long-tailed phages and the type VI bacterial secretion system. Proc. Natl. Acad. Sci. U.S.A. 106, 4160–4165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kanamaru S., Leiman P. G., Kostyuchenko V. A., Chipman P. R., Mesyanzhinov V. V., Arisaka F., Rossmann M. G. (2002) Structure of the cell-puncturing device of bacteriophage T4. Nature 415, 553–557 [DOI] [PubMed] [Google Scholar]
- 14. Kanamaru S. (2009) Structural similarity of tailed phages and pathogenic bacterial secretion systems. Proc. Natl. Acad. Sci. U.S.A. 106, 4067–4068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., Silverman J. M., Ohlson B. B., Hicks K. G., Plemel R. L., Li M., Schwarz S., Wang W. Y., Merz A. J., Goodlett D. R., Mougous J. D. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Russell A. B., Hood R. D., Bui N. K., LeRoux M., Vollmer W., Mougous J. D. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Collaborative Computational Project No. 4 (1994) The CCP4 suite. Programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 18. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX. Building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 19. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 20. Davis I. W., Leaver-Fay A., Chen V. B., Block J. N., Kapral G. J., Wang X., Murray L. W., Arendall W. B., 3rd, Snoeyink J., Richardson J. S., Richardson D. C. (2007) MolProbity. All-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 35, W375–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chenna R., Sugawara H., Koike T., Lopez R., Gibson T. J., Higgins D. G., Thompson J. D. (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res. 31, 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gouet P., Courcelle E., Stuart D. I., Métoz F. (1999) ESPript. Analysis of multiple sequence alignments in PostScript. Bioinformatics 15, 305–308 [DOI] [PubMed] [Google Scholar]
- 23. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu Q., Abdubek P., Astakhova T., Axelrod H. L., Bakolitsa C., Cai X., Carlton D., Chen C., Chiu H. J., Chiu M., Clayton T., Das D., Deller M. C., Duan L., Ellrott K., Farr C. L., Feuerhelm J., Grant J. C., Grzechnik A., Han G. W., Jaroszewski L., Jin K. K., Klock H. E., Knuth M. W., Kozbial P., Krishna S. S., Kumar A., Lam W. W., Marciano D., Miller M. D., Morse A. T., Nigoghossian E., Nopakun A., Okach L., Puckett C., Reyes R., Tien H. J., Trame C. B., van den Bedem H., Weekes D., Wooten T., Yeh A., Hodgson K. O., Wooley J., Elsliger M. A., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2010) Structure of the γ-d-glutamyl-l-diamino acid endopeptidase YkfC from Bacillus cereus in complex with l-Ala-γ-d-Glu. Insights into substrate recognition by NlpC/P60 cysteine peptidases. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 1354–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Aramini J. M., Rossi P., Huang Y. J., Zhao L., Jiang M., Maglaqui M., Xiao R., Locke J., Nair R., Rost B., Acton T. B., Inouye M., Montelione G. T. (2008) Solution NMR structure of the NlpC/P60 domain of lipoprotein Spr from Escherichia coli. Structural evidence for a novel cysteine peptidase catalytic triad. Biochemistry 47, 9715–9717 [DOI] [PubMed] [Google Scholar]
- 26. Xu Q., Sudek S., McMullan D., Miller M. D., Geierstanger B., Jones D. H., Krishna S. S., Spraggon G., Bursalay B., Abdubek P., Acosta C., Ambing E., Astakhova T., Axelrod H. L., Carlton D., Caruthers J., Chiu H. J., Clayton T., Deller M. C., Duan L., Elias Y., Elsliger M. A., Feuerhelm J., Grzechnik S. K., Hale J., Han G. W., Haugen J., Jaroszewski L., Jin K. K., Klock H. E., Knuth M. W., Kozbial P., Kumar A., Marciano D., Morse A. T., Nigoghossian E., Okach L., Oommachen S., Paulsen J., Reyes R., Rife C. L., Trout C. V., van den Bedem H., Weekes D., White A., Wolf G., Zubieta C., Hodgson K. O., Wooley J., Deacon A. M., Godzik A., Lesley S. A., Wilson I. A. (2009) Structural basis of murein peptide specificity of a γ-d-glutamyl-l-diamino acid endopeptidase. Structure 17, 303–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ruggiero A., Marasco D., Squeglia F., Soldini S., Pedone E., Pedone C., Berisio R. (2010) Structure and functional regulation of RipA, a mycobacterial enzyme essential for daughter cell separation. Structure 18, 1184–1190 [DOI] [PubMed] [Google Scholar]
- 28. Böth D., Schneider G., Schnell R. (2011) Peptidoglycan remodeling in Mycobacterium tuberculosis. Comparison of structures and catalytic activities of RipA and RipB. J. Mol. Biol. 413, 247–260 [DOI] [PubMed] [Google Scholar]
- 29. Anantharaman V., Aravind L. (2003) Evolutionary history, structural features, and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 4, R11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nakajima Y., Ito K., Toshima T., Egawa T., Zheng H., Oyama H., Wu Y. F., Takahashi E., Kyono K., Yoshimoto T. (2008) Dipeptidyl aminopeptidase IV from Stenotrophomonas maltophilia exhibits activity against a substrate containing a 4-hydroxyproline residue. J. Bacteriol. 190, 7819–7829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Storer A. C., Ménard R. (1994) Catalytic mechanism in papain family of cysteine peptidases. Methods Enzymol. 244, 486–500 [DOI] [PubMed] [Google Scholar]
- 32. Park J. T., Uehara T. (2008) How bacteria consume their own exoskeletons (turnover and recycling of cell wall peptidoglycan). Microbiol. Mol. Biol. Rev. 72, 211–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Smith T. J., Blackman S. A., Foster S. J. (2000) Autolysins of Bacillus subtilis. Multiple enzymes with multiple functions. Microbiology 146, 249–262 [DOI] [PubMed] [Google Scholar]
- 34. Russell A. B., Singh P., Brittnacher M., Bui N. K., Hood R. D., Carl M. A., Agnello D. M., Schwarz S., Goodlett D. R., Vollmer W., Mougous J. D. (2012) A widespread bacterial type VI secretion effector superfamily identified using a heuristic approach. Cell Host Microbe 11, 538–549 [DOI] [PMC free article] [PubMed] [Google Scholar]