Background: Scaffoldin structure is critical for cellulosome assembly and function.
Results: A multimodular scaffoldin fragment displays conformational flexibility and oligomerization properties and reveals a unique orientation of the type I dockerin.
Conclusion: The C terminus of the scaffoldin has unrestrained linker flexibility and may participate in higher order cellulosome organization.
Significance: Scaffoldin structure and dynamics will inform the generation of designer cellulosomes.
Keywords: Cell Surface Enzymes, Cellulase, Crystal Structure, Plant Cell Wall, Protein Complexes, Cellulosome, Cohesin, Dockerin, Modular Assembly, Scaffoldin
Abstract
Cellulosomes are multienzyme complexes responsible for efficient degradation of plant cell wall polysaccharides. The nonenzymatic scaffoldin subunit provides a platform for cellulolytic enzyme binding that enhances the overall activity of the bound enzymes. Understanding the unique quaternary structural elements responsible for the enzymatic synergy of the cellulosome is hindered by the large size and inherent flexibility of these multiprotein complexes. Herein, we have used x-ray crystallography and small angle x-ray scattering to structurally characterize a ternary protein complex from the Clostridium thermocellum cellulosome that comprises a C-terminal trimodular fragment of the CipA scaffoldin bound to the SdbA type II cohesin module and the type I dockerin module from the Cel9D glycoside hydrolase. This complex represents the largest fragment of the cellulosome solved by x-ray crystallography to date and reveals two rigid domains formed by the type I cohesin·dockerin complex and by the X module-type II cohesin·dockerin complex, which are separated by a 13-residue linker in an extended conformation. The type I dockerin modules of the four structural models found in the asymmetric unit are in an alternate orientation to that previously observed that provides further direct support for the dual mode of binding. Conserved intermolecular contacts between symmetry-related complexes were also observed and may play a role in higher order cellulosome structure. SAXS analysis of the ternary complex revealed that the 13-residue intermodular linker of the scaffoldin subunit is highly dynamic in solution. These studies provide fundamental insights into modular positioning, linker flexibility, and higher order organization of the cellulosome.
Introduction
Plant cell wall polysaccharides are the most abundant renewable carbon source on Earth. However, the composite heterogeneous structure of the plant cell wall makes it a recalcitrant substrate and an obstacle for exploiting this rich carbon source (1). Several anaerobic microorganisms have developed a specialized nanomachine, dubbed the cellulosome, capable of efficiently degrading the plant cell wall through the synergistic activity of various secreted cellulases, hemicellulases, and related hydrolytic enzymes (2–6).
The cellulosome from the thermophilic anaerobic bacterium Clostridium thermocellum was the first to be discovered, is the most thoroughly characterized, and represents the prototypical example of a cellulose-degrading multienzyme complex (2, 7–10). The central component of the cellulosome is the multimodular noncatalytic scaffoldin protein subunit, CipA, which serves as a binding platform for secreted cellulolytic enzymes while at the same time tethering the entire complex to the substrate and the bacterial cell surface (11). A family 3 cellulose-specific carbohydrate-binding module in CipA targets the multienzyme complex to its substrate (12, 13), whereas the integration of the various enzymes into the cellulosome is mediated through high affinity noncovalent interactions between the nine type I cohesin modules (CohI)4 of the scaffoldin subunit and the enzyme-borne type I dockerin modules (DocI) (14–16). An analogous interaction involving the C-terminal type II dockerin module (DocII) of CipA and the type II cohesin modules (CohII) of cell surface proteins, SdbA, Orf2p, and OlpB, fixes the entire complex to the peptidoglycan layer of the bacterium (17–21).
The concentration of cellulolytic enzymes with complementary functions into a single complex, mediated by the CipA scaffoldin subunit, promotes synergy among the enzymes that results in enhanced activity relative to enzymes free in solution (22, 23). To achieve this synergy, the structural organization of the cellulosome must offer a balance among modularity, diversity, and plasticity. Insights into these structural features have begun to emerge over the last decade. Electron microscopy imaging studies of cellulolytic bacteria revealed dynamic structures on the bacterial cell surface that house cellulosomes, which in the absence of cellulose appear as bulbous protuberances that extend and attach to substrate when it is introduced (24, 25). X-ray crystal structures of several C. thermocellum cellulosomal catalytic modules (26–28), isolated CipA scaffoldin modules (13, 29–31), and type I and type II Coh·Doc complexes have been solved (14, 15, 17). Despite these successes, a comprehensive understanding of the unique quaternary structural elements that contribute to the highly efficient cellulose-degrading properties of the cellulosome has been hindered by the large size, the heterogeneity in enzyme content, and the inherent conformational flexibility of these multiprotein complexes, all of which preclude crystallographic determination of the intact native cellulosome.
To circumvent these issues, lower resolution experimental methods and computational biology have recently been employed. Small angle x-ray scattering (SAXS) studies of catalytic subunits complexed to CipA CohI modules, either as an isolated complex or a tandem repeat and with scaffoldin linkers of varying lengths, have indicated that a conformational change occurs in the linker region connecting the catalytic domain and DocI upon binding CohI. Moreover, these studies suggest that although synergy arising from the proximity of the enzymes requires some conformational freedom in the intermodular linker regions separating the CohI modules, it is not affected by differences in linker length or sequences (32–34). Recently, cryo-electron microscopy studies of a minicellulosome comprising three consecutive cohesin modules from the C. thermocellum CipA scaffoldin bound to Cel8A enzymes revealed a mostly compact conformation with the enzymes projected away from the scaffoldin in opposite directions (35), whereas computational simulations suggested that cellulosome assembly is driven predominantly by the shape and modularity of the cellulosome components (36).
Herein, we combine x-ray crystallography and SAXS to describe the structure and dynamics of a multimodular ternary cellulosomal complex comprising the SdbA CohII, the DocI module of Cel9D (family 9 glycoside hydrolase), and the CohI9–X-DocII trimodular fragment of the CipA scaffoldin from C. thermocellum, which represents the largest fragment of the cellulosome solved by x-ray crystallography. Interscaffoldin interactions were observed between the X module of one molecule and the CohI9 module of symmetrically related scaffoldin fragment in the crystal lattice; interactions identical to those observed in the CohI9–X-DocII·CohII binary complex structure (37). However, SAXS analysis indicated that the ternary complex is monomeric in solution, which suggests a role for DocI binding in the regulation of interscaffoldin interactions. Moreover, our crystallographic and SAXS data indicate that the scaffoldin linker connecting the X module to the CohI9 is highly dynamic in solution. The DocI in all four structural models are in the same orientation and opposite to that previously reported for native DocI, lending further support for the dual mode of DocI binding to CohI. Ultimately, the work described herein has implications for understanding cellulosome assembly, higher order structure, and dynamics.
EXPERIMENTAL PROCEDURES
Crystallization and Structure Determination
Cloning, protein expression, purification, and crystallization of the Cel9D DocI·CipA CohI9–X-DocII·SdbA CohII ternary complex, along with diffraction data collection and indexing, were performed as previously described (38). The structure was solved by molecular replacement using PHASER (39). Manual fitting of the model was carried out using COOT and XFIT and refined with Phenix (40, 41). The rmsd values were calculated using DALILITE (42). The atomic coordinates for the ternary complex have been deposited in the Protein Data Bank with accession code 4FL4.
SAXS
Protein samples used for SAXS experiments were prepared as previously described (38); however, the S69A/S70A double mutation was made in the DocI module to lock the enzyme in the orientation present in the crystal structure to minimize sample heterogeneity. BSA standards were used to calibrate the I/(0) values and to assess potential aggregation of samples. Protein concentrations were determined using A280 from relative molecular mass and Abs0.1% ( = 1 g/liter) parameters calculated using ProtParam (43). A280 measurements were made using a NanoDrop ND-1000 spectrometer (NanoDrop Technologies). SAXS data were measured at the F2 station at Cornell High Energy Synchrotron Source (Ithaca, NY). All of the samples were centrifuged at 14,000 rpm for 30 min prior to data collection. The scattering intensities from each protein sample and its final dialysate were recorded for 180 s at 24 °C using an ADSC Quantum-210 CCD detector. The samples were oscillated continuously throughout measurements. Normalization for beam intensity, buffer subtraction, and merging of data were performed with Bioxtas Raw and Primus (44, 45). GNOM was used to calculate the pair distribution function and the Dmax. Theoretical SAXS data were calculated for the ternary complex using Crysol. Low resolution ab initio shapes were generated with GNOM output files and DAMMIN (46). Rigid body modeling was performed using BILBOMD (47). Molecular dynamics (MD) was used to explore the conformational space adopted by the DocI·CohI9–X-DocII·CohII complex. A minimal ensemble search (MES) was used to identify the minimal ensemble of conformations required to best fit the experimental data (47). Structural figures were prepared using PyMOL (48).
RESULTS
Crystal Structure of the Ternary Complex
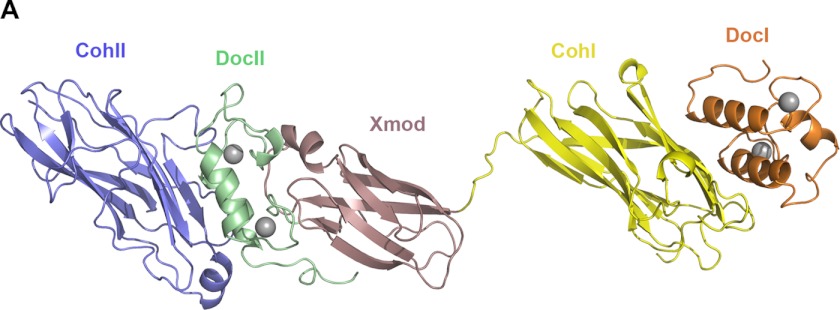
The crystal structure of the DocI·CohI9–X-DocII·CohII ternary complex was solved by molecular replacement using complex structures of X-DocII·CohII (Protein Data Bank accession code 3KCP) and S45A/S46A DocI·CohI (Protein Data Bank accession code 2CCL) as search probes (14, 17). Notably, an initial molecular replacement strategy involving the wild-type DocI·CohI complex structure (Protein Data Bank accession code 1OHZ) as a probe proved unsuccessful (15). The structure was refined to 2.8 Å resolution, and the final statistics are summarized in Table 1. Four molecules of the DocI·CohI9–X-DocII·CohII ternary complex were found in the asymmetric unit, with each displaying two well ordered regions connected by a 13-residue scaffoldin linker (Val1687–Lys1699) (Fig. 1). One region comprises the CipA X-DocII modular pair bound to SdbA CohII, and the other contains Cel9D DocI in complex with CipA CohI9. The four complex structures are also all in extended conformations spanning ∼150 Å in their longest dimension.
TABLE 1.
Data collection and refinement statistics
| Data collection | |
| Wavelength | 0.9792 |
| Resolution (Å)a | 20–2.8 (2.9–2.8) |
| Space group | P212121 |
| Cell dimensions a, b, c (Å) | 119.37, 186.31, 191.17 |
| Total reflections | 399,189 (40,808) |
| Unique reflections | 104,697 (10,403) |
| Redundancy | 3.8 (3.9) |
| Completeness (%) | 99.3 (100) |
| I/σI | 22.3 (2.1) |
| Rmerge (%)b | 4.3 (64.3) |
| Wilson B-factor (Å2) | 74.04 |
| Refinement | |
| Rwork/Rfree (%) | 19.1/23.6d |
| No. atoms | |
| Total | 16,313 |
| Protein | 15,980 |
| Average B-factors (Å2) | |
| Overall | 71.8 |
| Protein | 71.9 |
| Water | 64.7 |
| Root mean square deviations | |
| Bond lengths (Å) | 0.009 |
| Bond angles (°) | 1.189 |
| Ramachandran plot statisticsc | |
| Favored (%) | 94.2 |
| Disallowed (%) | 0.3 |
a High resolution shell is shown in parentheses.
b Rmerge = {ΣhklΣi|Ii(hkl) − i(hkl)|}/{ΣhklΣi|Ii(hkl)|}, where i(hkl) is the average value of the intensity of reflection (hkl) in the data set, and Ii(hkl) is the intensity of the ith observation of that reflection.
c Statistics calculated with MolProbity (56).
d The free R factor is calculated for a “test” set of reflections, which were not included in the refinement (5%).
FIGURE 1.
Crystal structure of the DocI·CohI9–X-DocII·CohII ternary cellulosomal complex. One representative molecule of the DocI· CohI9–X-DocII· CohII ternary complex crystal structure is shown. The backbone ribbon representation depicts SdbA CohII in blue, the CipA DocII in green, X module in rose, CohI9 in yellow, and the Cel9D DocI in orange. Calcium ions are shown as gray spheres.
The X-DocII·CohII region, defined by residues 31–195 of SdbA CohII and residues 1697–1851 of the CipA scaffoldin subunit, of the four ternary complex structures are very similar to one another (backbone rmsd of 0.23 ± 0.05 Å), to the previously reported X-DocII·CohII structure (backbone rmsd of 0.53 ± 0.14 Å) (17), and to the analogous region in the CohI9–X-DocII·CohII heterodimeric complex (backbone rmsd of 0.39 ± 0.04 Å) (37). SdbA CohII forms the typical elongated nine-stranded β-sandwich CohII fold with a crowning helix and β-flaps intervening strands 4 and 8 (17, 49–51). The CipA DocII is composed of two calcium-binding F-hand loop-helix motifs separated by a 14-residue linker. The X module adopts an Ig-like fold with two antiparallel β-sheets composed of strands 1, 4, and 7 and strands 2, 3, 5, and 6 with a short α-helix connecting strands 1 and 2 (Fig. 1).
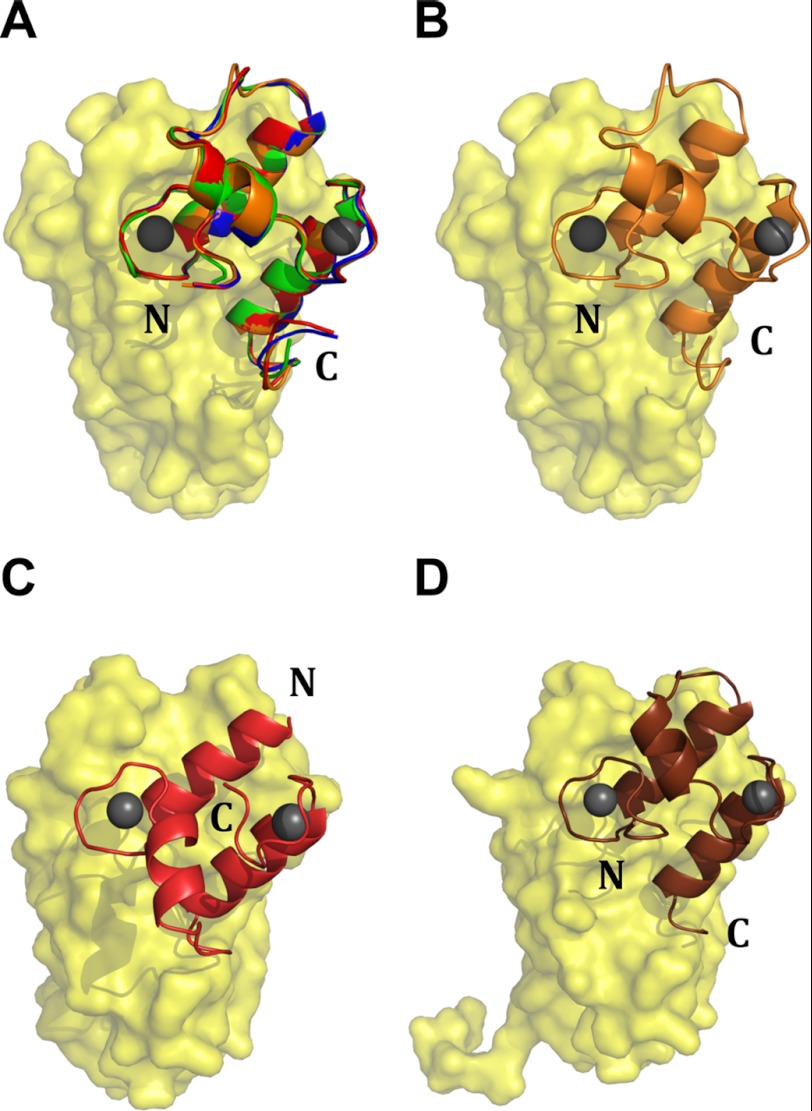
The CipA CohI9 has the typical fold seen in other CohI modules (30, 31, 52). Similar to the CipA DocII, Cel9D DocI comprises two F-hand motifs that structurally coordinate two Ca2+ ions similar to the EF-hand family of Ca2+-binding proteins (15, 53). Comparison of the DocI·CohI9 interaction in the four ternary structural models shows that each Cel9D DocI module interacts with the expected 8-3-6-5 face of CohI in an orientation (backbone rmsd of 0.22 ± 0.04 Å) that is 180° opposite to that previously reported for DocI modules on the surface of CohI modules (Fig. 2) (15, 54). Rather, the positioning is similar to those of the S45A/S46A Xyn10B and A47S/F48T Cel5A DocI mutants when in complex with their respective CohI partners (14, 54). Indeed, residues at the Cel9D DocI·CipA CohI9 interface, which participate in hydrogen bonding and van der Waals contacts (supplemental Table S1), are consistent with those residues previously identified as contributing to the S45A/S46A Xyn10B DocI·CipA CohI2 interface (14).
FIGURE 2.
The DocI modules in the DocI·CohI9–X DocII·CohII structure display a single orientation opposite to what has been seen previously. A displays an alignment of the four CohI9·DocI from the DocI·CohI9–X DocII·CohII crystal structure (Protein Data Bank code 4FL4). The CohI module is shown in yellow, and the DocI modules from molecules 1, 2, 3, and 4 are red, green, blue, and orange, respectively. B shows a representative CohI9·DocI orientation from the DocI·CohI9–X DocII·CohII crystal structure with the CohI9 and DocI modules shown in yellow and orange, respectively. C and D show the Xyn10B DocI·CohI (Protein Data Bank code 1OHZ) and the Xyn10B S54A/T46 DocI·CohI (2CCL) structures, respectively (14, 15). In both, the CohI modules are yellow. The wild-type DocI module is red, and the S54A/T46 mutant is shown in brown.
Linker Flexibility and Scaffoldin Dimerization
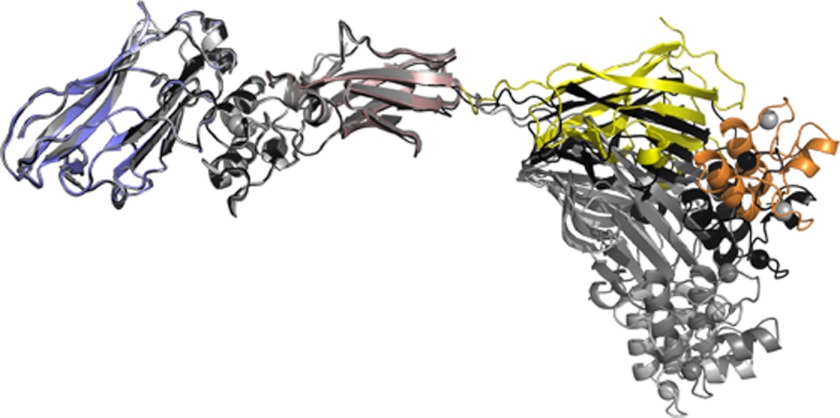
The 13-residue intermodular linker (Val1687–Lys1699) connecting CohI9 and the X module of the CipA scaffoldin, which could be fully modeled from electron density for all four molecules in the asymmetric unit, displays elevated temperature factors suggestive of a high degree of flexibility. Alignment of the X-DocII·CohII region from the four molecules of the complex, as well as the CohI9–X-DocII·CohII complex, reveals slightly different orientations of the DocI·CohI9 region, illustrating the dynamic properties of the scaffoldin linker (Fig. 3). However, considering the potential conformational variability of a dynamic 13-residue linker, the extent of flexibility observed appears to be restrained potentially by crystal packing.
FIGURE 3.
Alignment of the X-DocII·CohII region of the four DocI·CohI9—X-DocII·CohII complexes from the asymmetric unit. The DocI and CohI9 from the first molecule in the asymmetric unit are shown in orange and yellow, respectively, whereas the same modules from the other three molecules of the complex are colored gray. The X module is depicted in rose, DocII is in green, and CohII is in blue.
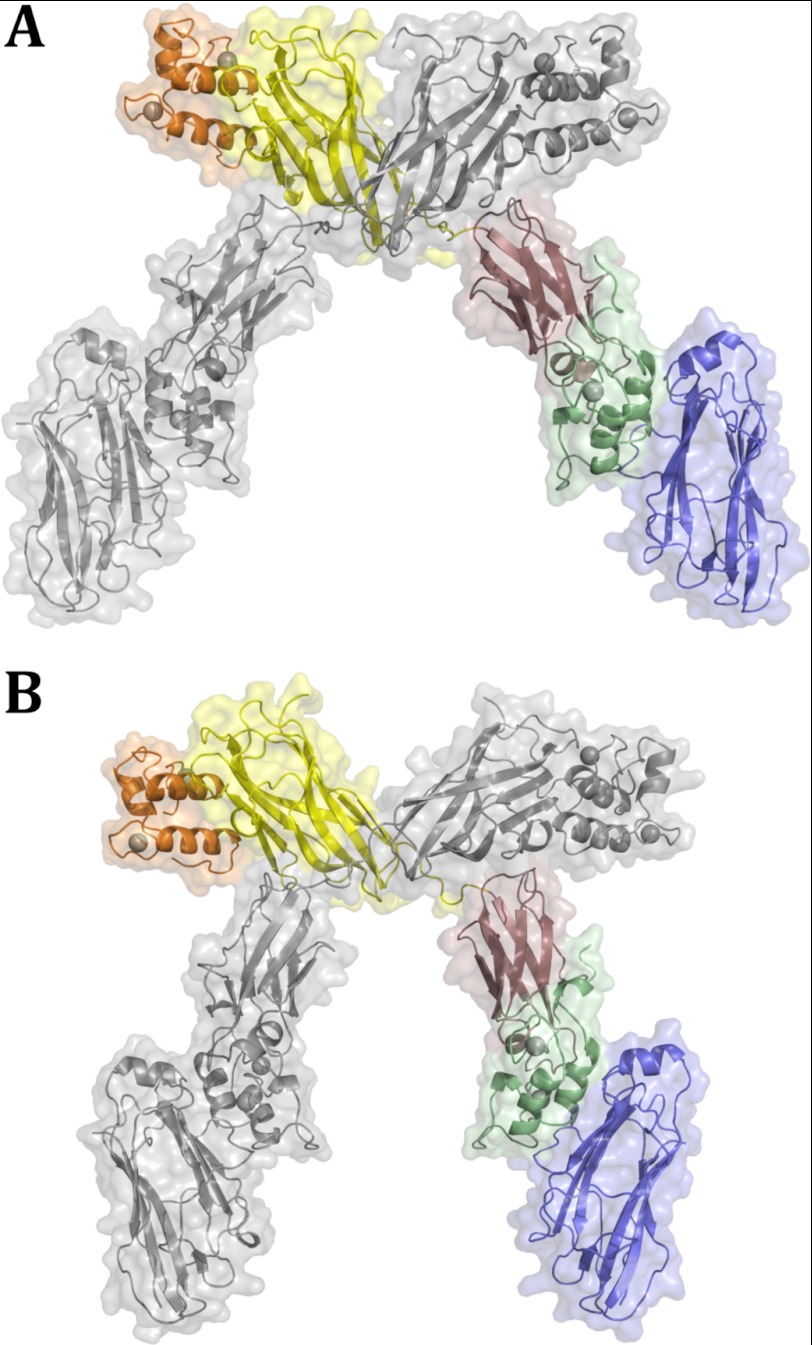
An intermolecular interface, involving residues within the linker connecting strands 4 and 5 of the X module of one ternary complex molecule and residues in strands 4 and 5 of CohI9 module from a symmetrically related complex molecule, is observed in the crystal lattice (Fig. 4 and supplemental Table S2). The resulting homodimer of the ternary complex displays symmetrical contacts between the two sets of X modules and CohI9 modules that results in an intertwining of the two complexes, similar to that observed in the CohI9–X-DocII·CohII heterodimer (37). Notably, the presence of Cel9D DocI does not disrupt the intertwined homodimer. When compared with the CohI9–X-DocII·CohII homodimer, there is a molecular rearrangement of the CohI9–X intermodular linker, which acts like a hinge to allow for positional flexibility of the X-DocII·CohII region while maintaining the intermolecular CohI–X interface (Fig. 4). However, the impact of this conformational flexibility within the linker is restrained by crystal packing. For this reason and to provide insight into the behavior of the ternary complex in solution, we complemented our crystallographic work with solution scattering studies.
FIGURE 4.
Interscaffoldin interactions observed in the DocI·CohI9–X DocII·CohII cellulosomal complex. A and B display the two homodimers of the DocI·CohI9–X DocII·CohII heterotrimeric complexes that were observed in the crystal structure. The DocI, CohI, X, DocII, and CohII are shown in orange, yellow, rose, green, and blue, respectively. Symmetry-related molecules are shown in gray.
SAXS Studies
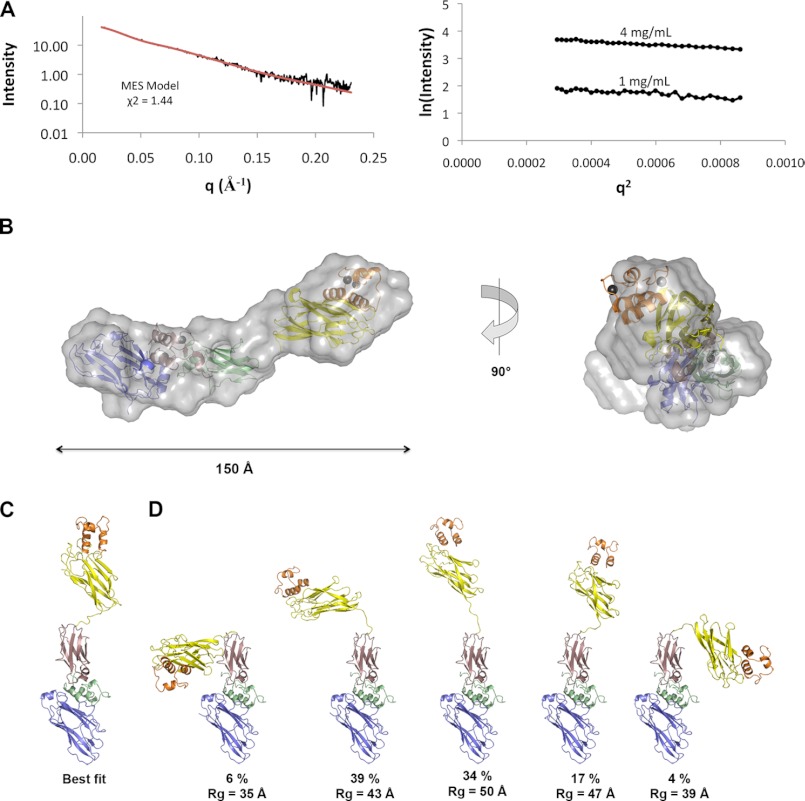
SAXS was used to investigate the structure of the DocI·CohI9–X-DocII·CohII ternary complex in solution, including the dynamic properties of the CohI9–X module intermodular linker. To decrease any potential heterogeneity in solution that could arise from the DocI dual mode of binding to CohI, mutations were made in positions 10 and 11 of the second repeat in the DocI (S69A/S70A) to preferentially select for the type I interaction in the same orientation seen in our crystal structure. The SAXS profile and linear radius of gyration (Rg = 42.6 ± 0.7 Å) of the DocI(S69A/S70A)·CohI9–X-DocII·CohII complex indicate that the complex is well behaved, monomeric, and aggregation-free in solution over a range of concentrations (1–4 mg/ml) (Fig. 5A). The maximal dimension (Dmax) of the ternary complex in solution is 146 Å. This is consistent with the extended length of the four DocI·CohI9–X-DocII·CohII ternary complexes in our crystal structure. The four ternary complexes from the asymmetric unit fit the experimental SAXS data with an average χ2 = 2.81 ± 0.19. Ten SAXS envelopes were generated by ab initio methods (supplemental Fig. S1), each revealing two domains separated by a thin connecting segment. The DocI·CohI9 and X-DocII·CohII rigid domains were manually placed within the envelope that best fit the experimental curve based on the χ2 values calculated by DAMMIN (Fig. 5B) (46). The structures fit within the two domains of the SAXS envelope with room remaining to accommodate the 13-residue linker in the thin connecting segment. This architecture suggests that the structure may be flexible in solution. Analysis of the pair distribution function and the Kratky plot are also consistent with a flexible multimodular structure (supplemental Fig. S2).
FIGURE 5.
SAXS analysis of the DocI(S69A/S70A)·CohI9–X-DocII·CohII ternary complex. A, raw SAXS data for the ternary complex (black) and theoretical SAXS curve for MES calculated minimal ensemble (red) are shown on the left, and the Guinier plots at protein concentrations of 1 and 4 mg/ml are shown on the right. B, best fit ab initio structure of DocI(S69A/S70A)·CohI9–X-DocII·CohII ternary complex and 90° rotation shown with the crystal structure colored as in Fig. 1A and manually placed within the envelope. C, best fit model of the ternary complex calculated using BILBOMD and MES. D, MES calculated minimal ensemble for the ternary complex.
To more robustly investigate the extent of flexibility of the CohI9–X module linker in solution, we utilized the BILBOMD rigid body modeling strategy, which employs MD simulations to generate thousands of different conformers, from which SAXS curves can be calculated and compared against experimental data (47). We defined regions that were resolved in our crystal structure as rigid domains, and the X-DocII·CohII positions were fixed in our analysis. The 13-residue linker connecting the X module and the CohI9 module, which displayed elevated temperature factors relative to the rest of the structure, was defined as flexible in the MD simulations along with stretches that did not show clear electron density in our crystal structure. Initial analysis was performed over Rg values between 25 and 65 Å centering around our experimentally determined Rg value of about 43 Å. However, only conformers with Rg values between 31 and 50 Å were selected based on our experimental SAXS data, which suggests that this range depicts the physical limitations of the ternary complex in solution. Consequently, subsequent MD simulations were performed using a range of 30–50 Å to better sample the conformers within this range. The best fit model (χ2 = 1.50) from the pool of approximately 10,000 calculated conformers shows an extended conformation consistent with the crystal structure and the ab initio SAXS envelope (Fig. 5C). Because flexible multimodular protein systems are not always well represented by a single model, we employed a genetic algorithm-based MES to identify conformer ensembles that optimally fit our data (47). Here, we selected for conformers that likely exist in the population and would better represent the conformational variability of the DocI(S69A/S70A)·CohI9–X-DocII·CohII ternary complex than a single model. Ensembles of two, three, four, and five conformers were generated, each showing an improvement in the fit to the experimental data with χ2 values of 1.46, 1.45, 1.44, and 1.44, respectively (Fig. 5D). Therefore, an ensemble of conformers better explains the behavior of the ternary complex in solution than a single conformer. Overall, the experimental SAXS data provide direct evidence of conformational flexibility of the CohI9–X module linker that would otherwise not be possible from the crystallographic structural studies.
DISCUSSION
The crystal structure and SAXS analysis of the DocI·CohI9–X-DocII·CohII ternary cellulosomal complex presented here illustrates the ability of a native DocI to interact with CohI using the alternative binding mode and provides insight into higher order cellulosome structure and scaffoldin linker flexibility. The signature duplicated sequence of DocI modules creates a symmetrical structure with two equivalent CohI binding surfaces, and therefore two potential binding modes that would allow for organizational plasticity during cellulosomal assembly. Carvalho et al. (14) observed that mutation of the Ser/Thr CohI recognition residues in one of the duplicated sequences resulted in preferential binding to CohI in an orientation 180° opposite to that of the native DocI. In the ternary complex presented here, all four CohI9·DocI interactions in the asymmetric unit bind in the same orientation and surprisingly in the opposite orientation to those seen in previous wild-type structures. The N-terminal region of the DocI construct, which includes a hexahistidine tag, is accommodated by a large cavity within the crystal lattice, whereas the side opposite to the termini of DocI is packed against the X-DocII region of a symmetry-related molecule. Accommodation of the DocI in the opposite orientation would therefore not be possible, because the N-terminal hexahistidine tag would very likely sterically clash with the X-DocII region of the neighboring complex molecule in the crystal. It is interesting to note that the preferential selection of this orientation during crystallization also represents the initial observation of a native DocI module that binds in an alternative orientation and thus strengthens the dual mode of binding proposed by Carvalho et al. (14).
An intermolecular interface was observed between the 4-7-2-1-9 face of CohI9 and two loops of an X module from a second complex molecule in the crystal lattice of the ternary complex. Interestingly, the same interface was observed in the heterodimeric structure, which lacks the DocI despite being crystallized under completely different crystallization conditions (37). Notably, the proximity of the X module does not inhibit DocI binding or disrupt its orientation on the 8-3-6-5 face of CohI9. Furthermore, the CohI9–X-DocII·CohII complex is in equilibrium between the monomeric and dimeric forms in solution, indicating that this phenomenon is more than a result of the local concentration effect of crystallization and may in fact occur in vivo (37).
Our solution scattering studies indicate that the DocI·CohI9–X-DocII·CohII ternary complex is monomeric in solution under buffering conditions and at concentrations at which homodimers of the CohI9–X-DocII·CohII complex form (37). However, the CohI9–X modular interface contacts still exist at much higher concentrations in the crystal lattice. This suggests that DocI binding does not prevent, but may weaken, the interscaffoldin interaction such that it is not apparent at lower concentrations. Because the DocI modules only contact the 8-3-6-5 face of the CohI9 modules in the crystal structure, it is unclear how DocI binding could affect interscaffoldin interactions. CohI9–X-mediated interscaffoldin interactions could play a role in cellulosome assembly whereby the intertwined scaffoldin structure ensures access to the 8-3-6-5 face of the enzyme-free CohI9 modules. Enzyme binding instigates rearrangement of scaffoldin interactions, which allow optimal positioning of enzymes on the substrate. The interscaffoldin interactions observed in the CohI9–X-DocII·CohII heterodimer and the DocI·CohI9–X-DocII·CohII heterotrimer crystal structures may indicate an important role for scaffoldin-scaffoldin interactions in cellulosome function. Indeed, in nature, the cellulosomes of this bacterium are housed in a highly concentrated state in cell surface protuberance-like structures (24, 25), and their native microenvironment likely emulates a condition somewhere between the crystalline and the solution states observed here. Some cell surface subunits, such as Orf2p and OlpB, comprise multiple CohII modules and thus have the ability to bind multiple scaffoldin subunits (18). In such an environment, the local concentration of the scaffoldin subunits near the bacterial cell surface may be sufficient to induce homodimerization, which may contribute to the stability of higher order cellulosome arrangements.
Cellulosomes are dynamic assemblies with structural flexibility attributed primarily to intermodular linkers of scaffoldin subunits. As a result, most structural characterization of cellulosome complexes has focused on the well structured modules. To date, only six structural studies have been published on cellulosome components that contained linkers: the CohI9–X-DocII·CohII heterodimer (37) and the type II CohB1·CohB2 dyad crystal structures (55), engineered scaffoldins studied by SAXS both bound to and free of enzymatic subunits with linkers of varying length and sequence (32–34), a MD study of cellulosome assembly (36), and a cryo-EM structure of a minicellulosome complex (35). The CohI9–X-DocII·CohII crystal structure provided a static view of the linker between the CohI and the X module because only a single conformation was trapped within the crystals. However, both the type II CohB1·CohB2 dyad crystal structure and the DocI·CohI9–X-DocII·CohII ternary crystal structure presented here reveal multiple conformations. Within the type II CohB1·CohB2 dyad crystal structure, two different conformations of the linker connecting the two CohII modules were resolved (55). Our ternary complex reveals four unique conformations of the CohI9–X linker, each one distinct from the conformation shown in the CohI9–X-DocII·CohII structure. The position of the CohI modules rotates in one axis relative to the X-DocII·CohII rigid domain. However, the linker positions are not simply shifted; each linker from each molecule takes on different conformations, which suggests that this linker region is both flexible and unstructured in solution.
Although these studies have revealed flexible regions of the cellulosome, the molecules are constrained within the crystal through crystal packing contacts and therefore do not provide an accurate view of linker flexibility. To overcome these constraints, we and others have used SAXS or EM analysis on cellulosomal complexes to investigate the cellulosome structure in a more natural environment (33, 34). SAXS studies of engineered minicellulosomes determined that the scaffoldin linker was the primary source of flexibility within the complex (32–34). Linkers between DocI and the catalytic domains contributed only small amounts of flexibility to the system (32–34). EM studies of a minicellulosome complex reveal that the inherent flexibility of the intermodular linkers of the minicellulosome allows the complex to take on a variety of different conformations where the linkers between the neighboring CohI modules are extended (35).
Here, we have established that the linker between the X module and CohI9 cohesin is the first region of flexibility of the cellulosomal scaffoldin extending from the cell surface. Other studies described above using engineered cellulosome scaffoldins have also shown that the scaffoldin linkers display unrestrained flexibility and exhibit only coincidental contacts with neighboring Coh modules. However, this is only the first study to show this using endogenous linkers and modules from a cellulosome. These studies suggest that the role of the scaffoldin linker is to provide maximal flexibility to optimally position the enzymes on the substrate. The linkers that connect the enzymatic module and its DocI module, on the other hand, have been shown to display only limited flexibility, which has been suggested to be involved in the “fine-tuning” of enzyme position.
Overall, our combined structural approach, incorporating both solution studies with a crystal structure, provides novel insight into dynamic modulation of cellulosome higher order structure assembly by DocI binding as well as a view of scaffoldin linker flexibility. The intricately regulated plasticity of cellulosome components likely reflects the heterogeneity and ever changing nature of its plant cell wall-derived cellulosic substrate during cellulosome-mediated degradation under different conditions.
Acknowledgment
We thank Dr. M. Hammel for expert assistance in the SAXS studies.
This work was supported by research grants from the Israel Science Foundation (to E. A. B.), the Natural Science and Engineering Research Council of Canada (to S. P. S.), and the Canadian Institutes of Health Research (to Z. J.).

This article contains supplemental Tables S1 and S2 and Figs. S1 and S2.
The atomic coordinates and structure factors (code 4FL4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- CohI
- type I cohesin
- CohII
- type II cohesin
- DocI
- type I dockerin
- DocII
- type II dockerin
- MD
- molecular dynamics
- SAXS
- small angle x-ray scattering
- MES
- minimal ensemble search
- rmsd
- root mean square deviation.
REFERENCES
- 1. Warren R. A. (1996) Microbial hydrolysis of polysaccharides. Annu. Rev. Microbiol. 50, 183–212 [DOI] [PubMed] [Google Scholar]
- 2. Bayer E. A., Belaich J. P., Shoham Y., Lamed R. (2004) The cellulosomes. Multienzyme machines for degradation of plant cell wall polysaccharides. Annu. Rev. Microbiol. 58, 521–554 [DOI] [PubMed] [Google Scholar]
- 3. Bayer E. A., Shimon L. J., Shoham Y., Lamed R. (1998) Cellulosomes. Structure and ultrastructure. J. Struct. Biol. 124, 221–234 [DOI] [PubMed] [Google Scholar]
- 4. Doi R. H., Kosugi A. (2004) Cellulosomes. Plant-cell-wall-degrading enzyme complexes. Nat. Rev. Microbiol. 2, 541–551 [DOI] [PubMed] [Google Scholar]
- 5. Fontes C. M., Gilbert H. J. (2010) Cellulosomes. Highly efficient nanomachines designed to deconstruct plant cell wall complex carbohydrates. Annu. Rev. Biochem. 79, 655–681 [DOI] [PubMed] [Google Scholar]
- 6. Gilbert H. J. (2007) Cellulosomes. Microbial nanomachines that display plasticity in quaternary structure. Mol. Microbiol. 63, 1568–1576 [DOI] [PubMed] [Google Scholar]
- 7. Bayer E. A., Morag E., Lamed R. (1994) The cellulosome. A treasure-trove for biotechnology. Trends Biotechnol. 12, 379–386 [DOI] [PubMed] [Google Scholar]
- 8. Béguin P., Alzari P. M. (1998) The cellulosome of Clostridium thermocellum. Biochem. Soc. Trans. 26, 178–185 [DOI] [PubMed] [Google Scholar]
- 9. Lamed R., Kenig R., Setter E., Bayer E. A. (1983) The cellulosome. A discrete cell surface organelle of Clostridium thermocellum which exhibits separate antigenic, cellulolose-binding and various cellulolytic activities. Biotechnol. Bioeng. Symp. 13, 163–181 [Google Scholar]
- 10. Lamed R., Setter E., Bayer E. A. (1983) Characterization of a cellulose-binding, cellulase-containing complex in Clostridium thermocellum. J. Bacteriol. 156, 828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gerngross U. T., Romaniec M. P., Kobayashi T., Huskisson N. S., Demain A. L. (1993) Sequencing of a Clostridium thermocellum gene (cipA) encoding the cellulosomal SL-protein reveals an unusual degree of internal homology. Mol. Microbiol. 8, 325–334 [DOI] [PubMed] [Google Scholar]
- 12. Poole D. M., Morag E., Lamed R., Bayer E. A., Hazlewood G. P., Gilbert H. J. (1992) Identification of the cellulose-binding domain of the cellulosome subunit S1 from Clostridium thermocellum YS. FEMS Microbiol. Lett. 78, 181–186 [DOI] [PubMed] [Google Scholar]
- 13. Tormo J., Lamed R., Chirino A. J., Morag E., Bayer E. A., Shoham Y., Steitz T. A. (1996) Crystal structure of a bacterial family-III cellulose-binding domain. A general mechanism for attachment to cellulose. EMBO J. 15, 5739–5751 [PMC free article] [PubMed] [Google Scholar]
- 14. Carvalho A. L., Dias F. M., Nagy T., Prates J. A., Proctor M. R., Smith N., Bayer E. A., Davies G. J., Ferreira L. M., Romão M. J., Fontes C. M., Gilbert H. J. (2007) Evidence for a dual binding mode of dockerin modules to cohesins. Proc. Natl. Acad. Sci. U.S.A. 104, 3089–3094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carvalho A. L., Dias F. M., Prates J. A., Nagy T., Gilbert H. J., Davies G. J., Ferreira L. M., Romão M. J., Fontes C. M. (2003) Cellulosome assembly revealed by the crystal structure of the cohesin-dockerin complex. Proc. Natl. Acad. Sci. U.S.A. 100, 13809–13814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schaeffer F., Matuschek M., Guglielmi G., Miras I., Alzari P. M., Béguin P. (2002) Duplicated dockerin subdomains of Clostridium thermocellum endoglucanase CelD bind to a cohesin domain of the scaffolding protein CipA with distinct thermodynamic parameters and a negative cooperativity. Biochemistry 41, 2106–2114 [DOI] [PubMed] [Google Scholar]
- 17. Adams J. J., Pal G., Jia Z., Smith S. P. (2006) Mechanism of bacterial cell-surface attachment revealed by the structure of cellulosomal type II cohesin-dockerin complex. Proc. Natl. Acad. Sci. U.S.A. 103, 305–310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Leibovitz E., Béguin P. (1996) A new type of cohesin domain that specifically binds the dockerin domain of the Clostridium thermocellum cellulosome-integrating protein CipA. J. Bacteriol. 178, 3077–3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leibovitz E., Ohayon H., Gounon P., Béguin P. (1997) Characterization and subcellular localization of the Clostridium thermocellum scaffoldin dockerin binding protein SdbA. J. Bacteriol. 179, 2519–2523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lemaire M., Miras I., Gounon P., Béguin P. (1998) Identification of a region responsible for binding to the cell wall within the S-layer protein of Clostridium thermocellum. Microbiology 144, 211–217 [DOI] [PubMed] [Google Scholar]
- 21. Lemaire M., Ohayon H., Gounon P., Fujino T., Béguin P. (1995) OlpB, a new outer layer protein of Clostridium thermocellum, and binding of its S-layer-like domains to components of the cell envelope. J. Bacteriol. 177, 2451–2459 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fierobe H. P., Bayer E. A., Tardif C., Czjzek M., Mechaly A., Bélaïch A., Lamed R., Shoham Y., Bélaïch J. P. (2002) Degradation of cellulose substrates by cellulosome chimeras. Substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277, 49621–49630 [DOI] [PubMed] [Google Scholar]
- 23. Fierobe H. P., Mingardon F., Mechaly A., Bélaïch A., Rincon M. T., Pagès S., Lamed R., Tardif C., Bélaïch J. P., Bayer E. A. (2005) Action of designer cellulosomes on homogeneous versus complex substrates. Controlled incorporation of three distinct enzymes into a defined trifunctional scaffoldin. J. Biol. Chem. 280, 16325–16334 [DOI] [PubMed] [Google Scholar]
- 24. Bayer E. A., Lamed R. (1986) Ultrastructure of the cell surface cellulosome of Clostridium thermocellum and its interaction with cellulose. J. Bacteriol. 167, 828–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mayer F., Coughlan M. P., Mori Y., Ljungdahl L. G. (1987) Macromolecular organization of the cellulolytic enzyme complex of Clostridium thermocellum as revealed by electron microscopy. Appl. Environ. Microbiol. 53, 2785–2792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Correia M. A., Prates J. A., Brás J., Fontes C. M., Newman J. A., Lewis R. J., Gilbert H. J., Flint J. E. (2008) Crystal structure of a cellulosomal family 3 carbohydrate esterase from Clostridium thermocellum provides insights into the mechanism of substrate recognition. J. Mol. Biol. 379, 64–72 [DOI] [PubMed] [Google Scholar]
- 27. Guimarães B. G., Souchon H., Lytle B. L., David Wu J. H., Alzari P. M. (2002) The crystal structure and catalytic mechanism of cellobiohydrolase CelS, the major enzymatic component of the Clostridium thermocellum cellulosome. J. Mol. Biol. 320, 587–596 [DOI] [PubMed] [Google Scholar]
- 28. Kitago Y., Karita S., Watanabe N., Kamiya M., Aizawa T., Sakka K., Tanaka I. (2007) Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum. J. Biol. Chem. 282, 35703–35711 [DOI] [PubMed] [Google Scholar]
- 29. Lytle B. L., Volkman B. F., Westler W. M., Heckman M. P., Wu J. H. (2001) Solution structure of a type I dockerin domain, a novel prokaryotic, extracellular calcium-binding domain. J. Mol. Biol. 307, 745–753 [DOI] [PubMed] [Google Scholar]
- 30. Shimon L. J., Bayer E. A., Morag E., Lamed R., Yaron S., Shoham Y., Frolow F. (1997) A cohesin domain from Clostridium thermocellum. The crystal structure provides new insights into cellulosome assembly. Structure 5, 381–390 [DOI] [PubMed] [Google Scholar]
- 31. Tavares G. A., Béguin P., Alzari P. M. (1997) The crystal structure of a type I cohesin domain at 1.7 Å resolution. J. Mol. Biol. 273, 701–713 [DOI] [PubMed] [Google Scholar]
- 32. Hammel M., Fierobe H. P., Czjzek M., Finet S., Receveur-Bréchot V. (2004) Structural insights into the mechanism of formation of cellulosomes probed by small angle xyh-ray scattering. J. Biol. Chem. 279, 55985–55994 [DOI] [PubMed] [Google Scholar]
- 33. Hammel M., Fierobe H. P., Czjzek M., Kurkal V., Smith J. C., Bayer E. A., Finet S., Receveur-Bréchot V. (2005) Structural basis of cellulosome efficiency explored by small angle x-ray scattering. J. Biol. Chem. 280, 38562–38568 [DOI] [PubMed] [Google Scholar]
- 34. Molinier A. L., Nouailler M., Valette O., Tardif C., Receveur-Bréchot V., Fierobe H. P. (2011) Synergy, structure and conformational flexibility of hybrid cellulosomes displaying various inter-cohesins linkers. J. Mol. Biol. 405, 143–157 [DOI] [PubMed] [Google Scholar]
- 35. García-Alvarez B., Melero R., Dias F. M., Prates J. A., Fontes C. M., Smith S. P., Romão M. J., Carvalho A. L., Llorca O. (2011) Molecular architecture and structural transitions of a Clostridium thermocellum mini-cellulosome. J. Mol. Biol. 407, 571–580 [DOI] [PubMed] [Google Scholar]
- 36. Bomble Y. J., Beckham G. T., Matthews J. F., Nimlos M. R., Himmel M. E., Crowley M. F. (2011) Modeling the self-assembly of the cellulosome enzyme complex. J. Biol. Chem. 286, 5614–5623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Adams J. J., Currie M. A., Ali S., Bayer E. A., Jia Z., Smith S. P. (2010) Insights into higher-order organization of the cellulosome revealed by a dissect-and-build approach. Crystal structure of interacting Clostridium thermocellum multimodular components. J. Mol. Biol. 396, 833–839 [DOI] [PubMed] [Google Scholar]
- 38. Currie M. A., Adams J. J., Ali S., Smith S. P., Jia Z. (2010) Purification and crystallization of a multimodular heterotrimeric complex containing both type-I and type-II cohesin-dockerin interactions from the cellulosome of Clostridium thermocellum. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 66, 327–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Read R. J. (2001) Pushing the boundaries of molecular replacement with maximum likelihood. Acta Crystallogr. D Biol. Crystallogr. 57, 1373–1382 [DOI] [PubMed] [Google Scholar]
- 40. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 42. Holm L., Park J. (2000) DaliLite workbench for protein structure comparison. Bioinformatics. 16, 566–567 [DOI] [PubMed] [Google Scholar]
- 43. Bairoch A., Apweiler R., Wu C. H., Barker W. C., Boeckmann B., Ferro S., Gasteiger E., Huang H., Lopez R., Magrane M., Martin M. J., Natale D. A., O'Donovan C., Redaschi N., Yeh L. S. (2005) The Universal Protein Resource (UniProt). Nucleic Acids Res. 33, D154–D159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Konarev P. V., Volkov V. V., Sokolova A. V., Koch M. H., Svergun D. I. (2003) PRIMUS. A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 36, 1277–1282 [Google Scholar]
- 45. Nielsen S. S., Toft K. N., Snakenborg D., Jeppesen M. G., Jacobsen J. K., Vestergaard B., Kutter J. P., Arleth L. (2009) BioXTAS RAW, a software program for high-throughput automated small-angle x-ray scattering data reduction and preliminary analysis. J. Appl. Crystallogr. 42, 959–964 [Google Scholar]
- 46. Svergun D. I. (1999) Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 76, 2879–2886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Pelikan M., Hura G. L., Hammel M. (2009) Structure and flexibility within proteins as identified through small angle x-ray scattering. Gen. Physiol. Biophys. 28, 174–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. DeLano W. L. (2002) The PyMOL Molecular Graphics System, DeLano Scientific LLC, San Carlos, CA [Google Scholar]
- 49. Carvalho A. L., Pires V. M., Gloster T. M., Turkenburg J. P., Prates J. A., Ferreira L. M., Romão M. J., Davies G. J., Fontes C. M., Gilbert H. J. (2005) Insights into the structural determinants of cohesin-dockerin specificity revealed by the crystal structure of the type II cohesin from Clostridium thermocellum SdbA. J. Mol. Biol. 349, 909–915 [DOI] [PubMed] [Google Scholar]
- 50. Noach I., Frolow F., Alber O., Lamed R., Shimon L. J., Bayer E. A. (2009) Intermodular linker flexibility revealed from crystal structures of adjacent cellulosomal cohesins of. Acetivibrio cellulolyticus. J. Mol. Biol. 391, 86–97 [DOI] [PubMed] [Google Scholar]
- 51. Noach I., Frolow F., Jakoby H., Rosenheck S., Shimon L. W., Lamed R., Bayer E. A. (2005) Crystal structure of a type-II cohesin module from the Bacteroides cellulosolvens cellulosome reveals novel and distinctive secondary structural elements. J. Mol. Biol. 348, 1–12 [DOI] [PubMed] [Google Scholar]
- 52. Spinelli S., Fiérobe H. P., Belaïch A., Belaïch J. P., Henrissat B., Cambillau C. (2000) Crystal structure of a cohesin module from Clostridium cellulolyticum. Implications for dockerin recognition. J. Mol. Biol. 304, 189–200 [DOI] [PubMed] [Google Scholar]
- 53. Strynadka N. C., James M. N. (1989) Crystal structures of the helix-loop-helix calcium-binding proteins. Annu. Rev. Biochem. 58, 951–998 [DOI] [PubMed] [Google Scholar]
- 54. Pinheiro B. A., Proctor M. R., Martinez-Fleites C., Prates J. A., Money V. A., Davies G. J., Bayer E. A., Fontesm C. M., Fierobe H. P., Gilbert H. J. (2008) The Clostridium cellulolyticum dockerin displays a dual binding mode for its cohesin partner. J. Biol. Chem. 283, 18422–18430 [DOI] [PubMed] [Google Scholar]
- 55. Noach I., Levy-Assaraf M., Lamed R., Shimon L. J., Frolow F., Bayer E. A. (2010) Modular arrangement of a cellulosomal scaffoldin subunit revealed from the crystal structure of a cohesin dyad. J. Mol. Biol. 399, 294–305 [DOI] [PubMed] [Google Scholar]
- 56. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity. All-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]