Abstract
Circular proteins, defined as head-to-tail cyclized polypeptides originating from ribosomal synthesis, represent a novel class of natural products attracting increasing interest. From a scientific point of view, these compounds raise questions of where and why they occur in nature and how they are formed. From a rational point of view, these proteins and their structural concept may be exploited for crop protection and novel pharmaceuticals. Here, we review the current knowledge of three protein families: cyclotides and circular sunflower trypsin inhibitors from the kingdom of plants and the Amanita toxins from fungi. A particular emphasis is placed on their biological origin, structure, and activity. In addition, the opportunity for discovery of novel circular proteins and recent insights into their mechanism of action are discussed.
Keywords: Antimicrobial Peptides, Fungi, Lipid-binding Protein, Plant Biochemistry, Protein Chemistry, Amanita Toxin, Circular Protein, Cyclotide, SFTI-1
Introduction
The destiny of proteogenic amino acid building blocks is to be joined into chains via their N- and C-terminal ends. Hence, one could argue that linear proteins with free termini are unfinished, whereas head-to-tail cyclized proteins with a full circle of amide bonds represent the only “complete” polypeptides.
Circular proteins have now been reported from all kingdoms of life. This minireview focuses on plant and fungal circular proteins of ribosomal origin, i.e. sequences that are genetically encoded. Currently, three such classes are known: the Amanita toxins from fungi and the cyclotides and sunflower trypsin inhibitors from plants. The sources, genes, and protein structures of these proteins are summarized in Fig. 1. Their diverse origin and characteristics highlight that nature has found many ways of producing circular proteins, suggesting that more classes are yet to be discovered.
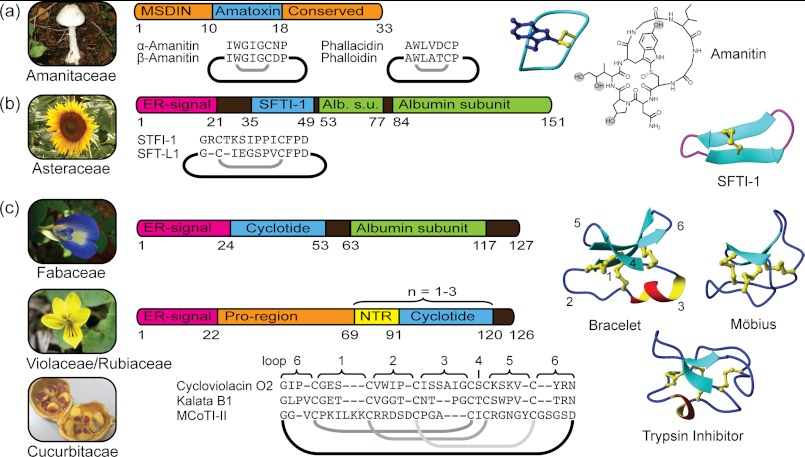
FIGURE 1.
Sources, genes, and structures of circular proteins from plants and fungi. a, amatoxins are embedded in ∼30-amino acid long precursors. Structures highlight the Cys-Trp bond and hydroxylations. b, albumin is hijacked for SFTI-1/SFT-L1 biosynthesis. The β-sheet structure is stabilized by one disulfide bond. Alb. s.u., albumin subunit. c, gene expression differs between cyclotide-expressing plant families. In Fabaceae, the gene is expressed within an albumin. Violaceae and Rubiaceae share the features of an endoplasmic reticulum (ER) signal, followed by the Pro region, the N-terminal repeat (NTR), the mature cyclotide domain, and a conserved tail region. Structures demonstrate cyclotide subfamilies and the positions of the sequence loops.
Fungal Circular Proteins
Amatoxins and phallotoxins are the major toxins of the lethal mushrooms of the family Amanitaceae (1, 2). Specifically, these toxins have been isolated from Amanita species from the section Phalloidae (including the death cap (Amanita phalloides) and the destroying angel (Amanita virosa)). The existence of “alkaloid-like” toxic compounds in Amanita has been known for almost two centuries, but their ribosomal origin was established only recently (3). Presently, the amatoxins constitute a suite of nine different 8-residue long circular peptides; the structure of α-amanitin is shown in Fig. 1a. All nine amatoxins are post-translationally modified products of the protein sequence encoding α-amanitin or β-amanitin. The post-translational process incorporates hydroxylations and an unusual cross-link between a Trp and a Cys residue via a sulfoxide. The phallotoxins are seven-membered protein circles that originate from phallacidin or phalloidin protein sequences. In analogy to amatoxins, they are stabilized by an extra cross-link; in this case, a sulfide links the Trp and Cys residues.
Amatoxins and phallotoxins share their genetic origin in that they are both products of the same gene family, MSDIN (3). The AMA1 and PHA1 genes, which encode α-amanitin and phalloidin, respectively, are expressed as 33- and 32-residue linear precursor proteins, respectively. All members of the family contain Pro residues N- and C-terminal of the mature sequence, which are likely required for the release and cyclization of the mature toxin domain by a prolyl oligopeptidase (4). Although genetics predicted 19 mature toxin sequences, only four of these have been found in modified versions at the protein level (α- and β-amanitin, phallacidin, and phalloidin). Those four sequences are the only ones containing Cys and Trp residues and thus are the only members that can contain the sulfoxide/sulfide cross-link. This suggests that the cross-link is important either for the stability and longevity or for the folding and cyclization of the peptides. However, several additional Amanita toxins lacking the link have been found at the protein level, but in contrast, the biosynthetic origin of these is unknown. For example, this is the case for antamanide (cyclo-(VPPAFFPPFF)), which contains only unmodified proteogenic amino acids and is likely also of ribosomal origin from a gene that still awaits discovery.
The amatoxins and phallotoxins are highly potent toxins. In fact, amatoxin-containing species are responsible for >90% of all fatal cases of mushroom poisoning (2). The fact that toxins survive the digestive tract is testament to the stability afforded by their cross-linked structure. The LD50 in humans for α-amanitin is ∼0.1 mg/kg, meaning that a single mushroom can contain a lethal dose. The drastic effect of these compounds is mediated through inhibition of transcription by specific interactions with RNA polymerase II (5) (Fig. 2a). This binding relies heavily on the post-translational decoration, with the constrained Trp and hydroxyl groups forming close interactions with the protein (5). Polymerases from different organisms show varying degrees of sensitivity, and the potency of the different amatoxins varies. The latter is illustrated by the LD50 values in mice: α-amanitin has an LD50 value 0.3 mg/kg, but amanullin, which lacks two hydroxyl groups, is two orders of magnitude less toxic. The phallotoxins bind to F-actin to stabilize the structure of assembled filaments (1). Phalloidin has thus become a valuable molecular tool used for cellular and molecular imaging when conjugated with fluorescent labels (6).
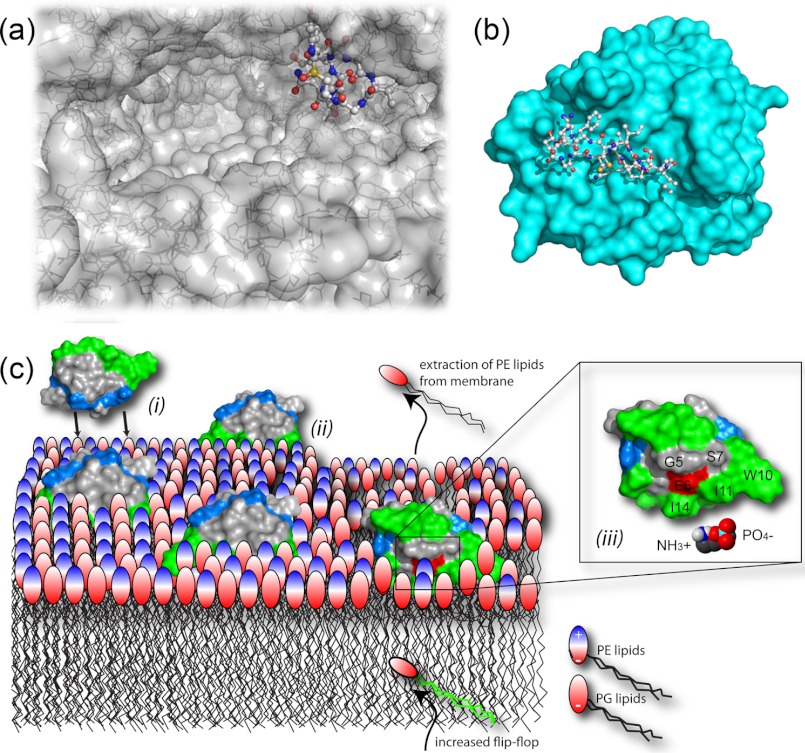
FIGURE 2.
Mechanisms of action. a, the crystal structure (Protein Data Bank code 3CQZ) shows α-amanitin binding deep in the substrate-binding channel of the large multisubunit RNA polymerase II. b, SFTI-I binds tightly to the trypsin active site (Protein Data Bank code 1SFI). A number of studies have demonstrated the ability of cyclotides to interact with and disrupt biological membranes. c, schematic model of the interaction between cycloviolacin O2 and E. coli inner membranes comprising phosphatidylglycerol (PG) and PE lipids. Accumulation of cyclotides, driven by electrostatic interaction ((i)), leads to membrane thinning ((ii)) through selective binding to and extraction of PE lipids ((iii)) and increased flip-flop of PE lipids from the inner leaflet.
Plant Circular Proteins
Sunflower Trypsin Inhibitors
Isolated from sunflower (Helianthus annuus) seeds, sunflower trypsin inhibitor-1 (SFTI-1)4 is a 14-residue cyclic peptide with subnanomolar inhibitory activity against trypsin (7). The three-dimensional structure of SFTI-1 reveals a well defined and rigid arrangement of two antiparallel β-strands that are stabilized by a single disulfide bond and an extensive internal network of hydrogen bonds (8). SFTI-1 forms a tight binding complex with bovine trypsin by an extended β-sheet across active site residues P1–P4 (trypsin/SFTI-1 Ki = 0.1 nm) (Fig. 2b) and shares a common conformation in its active loop with other serine proteases of the Bowman-Birk inhibitor family. The backbone circle in SFTI-1 is completed by a hairpin loop, which is termed the secondary loop.
The contributions made by both the circular backbone and the disulfide bond to the SFTI-I structure and activity have been examined. Synthetic disulfide-deficient but cyclic SFTI-1 is more flexible and less active than native SFTI-1, indicating a greater loss in entropy in binding to trypsin compared with native SFTI-1 (9). In contrast, the potency and hydrolysis rate of acyclic SFTI-1 opened between Asp14 and Gly1 are only slightly reduced relative to cyclic SFTI-1. The cyclic and acyclic SFTI-1 structures are essentially identical, with the exception of the loss of one hydrogen bond in the secondary loop in acyclic SFTI-1 (8). Thus, the disulfide bond, together with hydrogen bonds in the secondary loop, provides enough stability to maintain the β-sheet structure and, in turn, the rigidity of the binding loop (9). However, removal of both the disulfide and the circular backbone results in complete loss of trypsin inhibitory activity and disruption of the native-like fold (9). An alanine scan has also shown that the scaffold is tolerant to sequence modifications, with all non-cysteine residues able to be replaced without losing structural stability, with the exception of Asp14, which is involved in a hydrogen bonding network (10).
Recently, it became evident that SFTI-1 is derived from a 151-residue albumin seed storage protein and that it is also found in other Helianthus species (11). Transgenic experiments indicate that an asparaginyl endopeptidase, responsible for processing for the albumin, releases and cyclizes SFTI-1 (11). Subsequently, it was shown that another 12-residue cyclic peptide termed SFT-L1 (or SFTI-Like1) is located within the spacer region of another albumin gene. SFT-L1 currently represents the smallest plant circular protein known.
Cyclotides
The unique cyclotide structure was first described in the mid-1990s when the NMR spectroscopy analysis of kalata B1 from Oldenlandia affinis revealed the presence of both a circular peptide backbone and a so-called cystine knot, in which three conserved disulfide bonds are arranged such that one disulfide penetrates an embedded ring formed by the two other disulfides and their interconnecting backbone. Further discoveries established them as a family, and the term cyclotides (cyclo-peptides) was coined (12). Current indications point to cyclotides being one of the largest protein families known, with tens of thousands of members (13).
Structural Features
A typical cyclotide consists of ∼30 amino acids, with only 6 strictly conserved residues, the cysteines. The residues between each cysteine are defined as loops (1–6) and, in contrast, are generally highly interchangeable (Fig. 1c). The cystine knot, in combination with the additional cross-bracing afforded by the circular backbone, locks the chain into the cyclic cystine knot motif, which renders the structure as close to indestructible as a proteinaceous substance is ever likely to be. Kalata B1 in its oxidized form is fully resistant to all proteases tested, as well as thermal denaturation by boiling or unfolding by chaotropic agents (14). Numerous cyclotides have been structurally characterized, primarily by NMR spectroscopy (e.g. Ref. 15) but also by x-ray crystallographic studies (16). These studies have revealed a number of conserved features. The cyclotide backbone is tightly folded and comprises a large number of intramolecular hydrogen bonds (15). These bonds stabilize elements of secondary structure, including a β-hairpin and, in the bracelet cyclotides, a short 310 helix, which are connected by a series of well defined tight turns. The division of cyclotides into two subfamilies, Möbius and bracelets, is based on the former comprising a conserved conformation of the turn in loop 5, which includes a cis-Pro bond creating a conceptual twist of the peptide backbone (12).
A Glu residue in loop 1 is conserved throughout the family, with only a single exception among the ∼200 cyclotides known (17). This Glu has been found to coordinate a network of hydrogen bonds to amide protons in loop 3 via its carboxyl group (15, 18). This interaction is clearly a prerequisite for both structure and function of cyclotides, as replacement or modification results in both a compromised structure and significantly reduced bioactivity (19, 20).
The internal core of the cyclotide proteins is almost fully occupied by the conserved cystine knot, which gives the cyclotides a peculiar feature, namely a large number of surface-exposed hydrophobic residues. As a result, cyclotides typically have a highly amphiphilic character.
Occurrence of Cyclotides
Despite the high predictions for the number of cyclotides present in nature, to date, they have been found only in a few plant families, primarily in Violaceae and Rubiaceae. Although Rubiaceae is a large plant family, cyclotides are found only in a minority of species (13). In contrast, cyclotides have been found in all Violaceae species screened; hence, the family can be regarded as a rich source of cyclotides (21). Recently, cyclotides were also reported in one species of the Fabaceae family, Clitoria ternatea (22). Furthermore, two cyclic cystine knot peptides have been described in Momordica cochinchinensis of the Cucurbitaceae family (23), but these trypsin inhibitors are more closely related to linear squash protease inhibitors than other cyclotides. The four cyclotide-bearing plant families are phylogenetically distant, which suggests that their distribution in the plant kingdom is wider than current knowledge suggests.
Biological Activities and Mechanism of Action
Numerous biological activities have been reported for cyclotides, e.g. uterotonic, hemolytic, inhibition of neurotensin action, anti-HIV, cytotoxic, molluscicidal, anthelmintic, and antifouling effects (24). Cyclotides are also active against different bacteria (25, 26) and the insect larvae Helicoverpa punctigera and Helicoverpa armigera (27). Insecticidal and antimicrobial activities suggest that the native role of cyclotides in plants is as components of the innate defense system (28). A number of studies have shown that cyclotides interact with membranes. For example, cyclotides insert into micelles (29) and disrupt liposomes (30), bacterial membranes (31, 32), and membranes of enveloped viruses (31).
In our view, the interaction between cyclotides and the membrane has three components: (i) electrostatic interactions between charged residues and charged lipid headgroups, (ii) insertion into the membrane governed by the surface-exposed hydrophobic patch, and (iii) a specific interaction between the cyclotide and a lipid headgroup (Fig. 2c). Importantly, the contribution of each of these components to the overall affinity differs depending on the cyclotide in question and the composition of the targeted membrane, explaining differences in the bioactivity observed for different cyclotides in different assays.
(i) Electrostatic interactions between cationic peptides and anionic bacterial membranes are a key factor governing the selectivity of many antimicrobial peptides against pathogens. In contrast to the highly cationic peptides such as defensins, cyclotides carry only a few cationic residues and normally possess a net charge of +2 or less. Although this may suggest that the electrostatic effect would play little role in the mechanism of action, there is a clear correlation between overall charge and antimicrobial activity. Cyclotides such as kalata B1 and kalata B2, which carry overall charges of 0 and −1, respectively, show little activity in antibacterial assays, whereas cycloviolacin O2 (+2) and hedyotide B1 (+3) have low micromolar activity against several bacteria, including Escherichia coli (25, 26). Removal of the charges reduces the antimicrobial activity of cycloviolacin O2 (25), whereas addition of charges to kalata B1 can improve bioactivity (33).
(ii) The surface-exposed hydrophobic patch is ideally suited for insertion into membranes, and there is some correlation between the size of the hydrophobic patch and the activity in hemolytic assays (32, 34). The introduction of positive charges into the hydrophobic patch of kalata B1 has been shown to reduce bioactivity (33).
(iii) Recent studies utilizing liposome leakage assays, ellipsometry, and surface plasmon resonance techniques using model membranes with various lipid compositions demonstrated that both kalata B1 and cycloviolacin O2 have a distinct preference for membranes containing phosphatidylethanolamine (PE) lipids (31, 32). Kalata B1 can also increase the transmembrane recruitment of PE lipids from the inner leaflet (31). An alanine scan of kalata B1 has identified a region centered around the conserved Glu residue as critical for bioactivity (20). It was initially speculated that this region was critical for the self-association of kalata B1 to form membrane pores (20). However, titrations between kalata B1 and PE head groups monitored by NMR spectroscopy clearly identify this region as the site of interaction with PE. A perfect correlation between biologically inactive Ala mutants of kalata B1 and lack of binding to PE membranes demonstrates the importance of this specific interaction (31). It is possible that other cyclotides may be selective for other headgroups, allowing the targeting of different types of membranes.
The precise mechanism generating membrane leakage by cyclotides is not entirely clear. Peptide multimer formation producing a defined pore structure has been suggested based on stepwise current increase in a patch-clamp experiment (35). However, the mechanism for the cyclotide cycloviolacin O2 seems to be of a more general character. In this case, the peptide accumulation results in membrane thinning and curvature stress (32), which ultimately result in perforations of a more transient and toroidal character. These types of perforations are the predominate lytic mechanism for antimicrobial peptides, where defined multimeric pores are rare. For cycloviolacin O2, the membrane integrity was also compromised by selective PE lipid depletion from the membrane by lipid-specific micellization (32).
Reasons to Make Ends Meet
It is clear that the biosynthesis of the circular proteins described above must come with an extra cost in terms of energy for production. So what benefits do they confer to the organism that express them, and what is the role of these compounds in nature? The arguments in favor for the host defense theory are convincing. The presence of trypsin inhibitors protects against herbivores, as do the mushroom toxins. In the case of cyclotides, their insecticidal and antimicrobial effects support their role in host defense (28). In addition, phytotoxic activity and activity against soil bacteria have been demonstrated (36). The fact that cyclotides are expressed in a tissue-specific manner may be a reflection of the allelopathic role of cyclotides (37). Transgenic expression of these host defense proteins may be a potent means of crop protection (38).
The development of circular proteins coincides with the increasing interest in protein and peptides as drugs. Much of the attention attracted by circular proteins is due to one of the advantages they offer, namely their superior stability. They are thermally, chemically, and biologically stable. The circular backbone is one component underlying that stability; another key factor is side chain cross-links (14, 39). As proteins that can also accommodate a large structural diversity, these compounds have become subjects for protein design.
SFTI-1 is an appealing candidate for drug design because of its small size. The current drug design applications of SFTI-1 have significance in the field of anticancer therapeutics. For example, SFTI-1 was engineered to produce a selective inhibitor of KLK4 (Ki = 3.59 ± 0.28 nm), a protease associated with prostate cancer progression (40). Despite being larger than SFTI-1, cyclotides also are amenable to solid-phase peptide synthesis (41). Kalata B1 has been successfully grafted to contain anti-angiogenic activity (42), and MCoTI-II to display activity against foot and mouth virus (43).
In parallel to the chemical approach, an expression system of recombinant cyclotide libraries has been developed in E. coli (44) using MCoTI-II as a scaffold. Although only two relative small libraries were built in that study, proof of concept has been demonstrated that promises generation of larger libraries suitable for drug screening in a high throughput manner.
Another approach is to design stable drugs by introducing a circular backbone into naturally linear proteins. Remarkably, joining the ends of a conotoxin with a potent analgesic effect has been shown not only to increase resistance to proteolysis and to increase the protein half-life in plasma but also to allow oral administration (45).
The Never Ending Quest for Circular Proteins
Plants and fungi represent an overwhelming biological diversity. The two kingdoms together contains 2 million species (of which plants represents 400,000), each of which has its own unique setup of primary and secondary metabolites. Discovery of the proteins reviewed here is the result of observations of bioactivity, followed by targeted searches for analogous compounds. The history of the Amanita toxins dates back to early history of man and the observations of mushroom poisoning. SFTI-1 was discovered in a directed search for plant serine protease inhibitors using affinity chromatography on immobilized trypsin (7). The first cyclotide, kalata B1, was revealed to be the active compound in an herbal drug: a decoction of O. affinis used to accelerate childbirth in certain parts of central Africa (46).
But how do you target biodiscovery toward circular proteins? To determine whether a peptide or protein is cyclic at an early stage of the discovery process remains a challenge. Modern day screens for cyclotides utilize their high retention on reversed-phase HPLC, molecular weight, and cystine content (13). Recently, genetic information has been used to a larger extent: screening for cyclotide-encoding RNA has become routine. The first expressed sequence tag library of O. affinis revealed 31 cyclotide precursor expressed sequence tags (47). In comparison, only 19 cyclotides have been found at the protein level. Such discrepancies between gene and protein expression are commonly encountered; for example, cyclotide-like genes have been found in the Poaceae family, but no protein has been detected (48). In this context, it should be noted that the information from the genome sequencing was the key to uncovering the ribosomal origin of the Amanita toxins (3). Maybe genome mining for ribosomal natural products (49) and/or proteomic approaches based on mass spectrometry (50) will accelerate the discovery process in the future. It remains a problem that identification is biased toward compounds with characteristics similar to known compounds, e.g. primers are designed on conserved sequences, and only certain retention times and masses are investigated. The protein still needs to be fully sequenced before definite proof of its cyclic structure can be obtained.
Conclusions
Nature is a proven source for the discovery of new drugs and new chemical entities. In the current minireview, we have demonstrated that natural products may also play a role in the pharmaceutical and medical research of today, in which protein-based drugs become more and more important. It is clear that plant and fungi are underexplored, and many more diverse circular proteins await discovery, but the field is mushrooming.
This is the first article in the Thematic Minireview Series on Circular Proteins.
- SFTI-1
- sunflower trypsin inhibitor-1
- PE
- phosphatidylethanolamine.
REFERENCES
- 1. Vetter J. (1998) Toxins of Amanita phalloides. Toxicon 36, 13–24 [DOI] [PubMed] [Google Scholar]
- 2. Walton J. D., Hallen-Adams H. E., Luo H. (2010) Ribosomal biosynthesis of the cyclic peptide toxins of Amanita mushrooms. Biopolymers 94, 659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hallen H. E., Luo H., Scott-Craig J. S., Walton J. D. (2007) Gene family encoding the major toxins of lethal Amanita mushrooms. Proc. Natl. Acad. Sci. U.S.A. 104, 19097–19101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Luo H., Hallen-Adams H. E., Scott-Craig J. S., Walton J. D. (2010) Colocalization of amanitin and a candidate toxin-processing prolyl oligopeptidase in Amanita basidiocarps. Eukaryot. Cell 9, 1891–1900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaplan C. D., Larsson K. M., Kornberg R. D. (2008) The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by α-amanitin. Mol. Cell 30, 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Auinger S., Small J. V. (2008) Correlated light and electron microscopy of the cytoskeleton. Methods Cell Biol. 88, 257–272 [DOI] [PubMed] [Google Scholar]
- 7. Luckett S., Garcia R. S., Barker J. J., Konarev A. V., Shewry P. R., Clarke A. R., Brady R. L. (1999) High resolution structure of a potent cyclic proteinase inhibitor from sunflower seeds. J. Mol. Biol. 290, 525–533 [DOI] [PubMed] [Google Scholar]
- 8. Korsinczky M. L., Schirra H. J., Rosengren K. J., West J., Condie B. A., Otvos L., Anderson M. A., Craik D. J. (2001) Solution structures by 1H NMR of the novel cyclic trypsin inhibitor SFTI-1 from sunflower seeds and an acyclic permutant. J. Mol. Biol. 311, 579–591 [DOI] [PubMed] [Google Scholar]
- 9. Korsinczky M. L., Clark R. J., Craik D. J. (2005) Disulfide bond mutagenesis and the structure and function of the head-to-tail macrocyclic trypsin inhibitor SFTI-1. Biochemistry 44, 1145–1153 [DOI] [PubMed] [Google Scholar]
- 10. Daly N. L., Chen Y. K., Foley F. M., Bansal P. S., Bharathi R., Clark R. J., Sommerhoff C. P., Craik D. J. (2006) The absolute structural requirement for a proline in the P3′-position of Bowman-Birk protease inhibitors is surmounted in the minimized SFTI-1 scaffold. J. Biol. Chem. 281, 23668–23675 [DOI] [PubMed] [Google Scholar]
- 11. Mylne J. S., Colgrave M. L., Daly N. L., Chanson A. H., Elliott A. G., McCallum E. J., Jones A., Craik D. J. (2011) Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat. Chem. Biol. 7, 257–259 [DOI] [PubMed] [Google Scholar]
- 12. Craik D. J., Daly N. L., Bond T., Waine C. (1999) Plant cyclotides: a unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 13. Gruber C. W., Elliott A. G., Ireland D. C., Delprete P. G., Dessein S., Göransson U., Trabi M., Wang C. K., Kinghorn A. B., Robbrecht E., Craik D. J. (2008) Distribution and evolution of circular miniproteins in flowering plants. Plant Cell 20, 2471–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Colgrave M. L., Craik D. J. (2004) Thermal, chemical, and enzymatic stability of the cyclotide kalata B1: the importance of the cyclic cystine knot. Biochemistry 43, 5965–5975 [DOI] [PubMed] [Google Scholar]
- 15. Rosengren K. J., Daly N. L., Plan M. R., Waine C., Craik D. J. (2003) Twists, knots, and rings in proteins. Structural definition of the cyclotide framework. J. Biol. Chem. 278, 8606–8616 [DOI] [PubMed] [Google Scholar]
- 16. Wang C. K., Hu S. H., Martin J. L., Sjögren T., Hajdu J., Bohlin L., Claeson P., Göransson U., Rosengren K. J., Tang J., Tan N. H., Craik D. J. (2009) Combined x-ray and NMR analysis of the stability of the cyclotide cystine knot fold that underpins its insecticidal activity and potential use as a drug scaffold. J. Biol. Chem. 284, 10672–10683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaas Q., Craik D. J. (2010) Analysis and classification of circular proteins in CyBase. Biopolymers 94, 584–591 [DOI] [PubMed] [Google Scholar]
- 18. Göransson U., Herrmann A., Burman R., Haugaard-Jönsson L. M., Rosengren K. J. (2009) The conserved Glu in the cyclotide cycloviolacin O2 has a key structural role. ChemBioChem 10, 2354–2360 [DOI] [PubMed] [Google Scholar]
- 19. Herrmann A., Svangård E., Claeson P., Gullbo J., Bohlin L., Göransson U. (2006) Key role of glutamic acid for the cytotoxic activity of the cyclotide cycloviolacin O2. Cell. Mol. Life Sci. 63, 235–245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simonsen S. M., Sando L., Rosengren K. J., Wang C. K., Colgrave M. L., Daly N. L., Craik D. J. (2008) Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J. Biol. Chem. 283, 9805–9813 [DOI] [PubMed] [Google Scholar]
- 21. Burman R., Gruber C. W., Rizzardi K., Herrmann A., Craik D. J., Gupta M. P., Göransson U. (2010) Cyclotide proteins and precursors from the genus Gloeospermum: filling a blank spot in the cyclotide map of Violaceae. Phytochemistry 71, 13–20 [DOI] [PubMed] [Google Scholar]
- 22. Poth A. G., Colgrave M. L., Philip R., Kerenga B., Daly N. L., Anderson M. A., Craik D. J. (2011) Discovery of cyclotides in the Fabaceae plant family provides new insights into the cyclization, evolution, and distribution of circular proteins. ACS Chem. Biol. 6, 345–355 [DOI] [PubMed] [Google Scholar]
- 23. Hernandez J. F., Gagnon J., Chiche L., Nguyen T. M., Andrieu J. P., Heitz A., Trinh Hong T., Pham T. T., Le Nguyen D. (2000) Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 39, 5722–5730 [DOI] [PubMed] [Google Scholar]
- 24. Craik D. J. (2010) Discovery and applications of the plant cyclotides. Toxicon 56, 1092–1102 [DOI] [PubMed] [Google Scholar]
- 25. Pränting M., Lööv C., Burman R., Göransson U., Andersson D. I. (2010) The cyclotide cycloviolacin O2 from Viola odorata has potent bactericidal activity against Gram-negative bacteria. J. Antimicrob. Chemother. 65, 1964–1971 [DOI] [PubMed] [Google Scholar]
- 26. Wong C. T., Taichi M., Nishio H., Nishiuchi Y., Tam J. P. (2011) Optimal oxidative folding of the novel antimicrobial cyclotide from Hedyotis biflora requires high alcohol concentrations. Biochemistry 50, 7275–7283 [DOI] [PubMed] [Google Scholar]
- 27. Jennings C., West J., Waine C., Craik D., Anderson M. (2001) Biosynthesis and insecticidal properties of plant cyclotides: the cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. U.S.A. 98, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mylne J. S., Wang C. K., van der Weerden N. L., Craik D. J. (2010) Cyclotides are a component of the innate defense of Oldenlandia affinis. Biopolymers 94, 635–646 [DOI] [PubMed] [Google Scholar]
- 29. Shenkarev Z. O., Nadezhdin K. D., Sobol V. A., Sobol A. G., Skjeldal L., Arseniev A. S. (2006) Conformation and mode of membrane interaction in cyclotides. Spatial structure of kalata B1 bound to a dodecylphosphocholine micelle. FEBS J. 273, 2658–2672 [DOI] [PubMed] [Google Scholar]
- 30. Svangård E., Burman R., Gunasekera S., Lövborg H., Gullbo J., Göransson U. (2007) Mechanism of action of cytotoxic cyclotides: cycloviolacin O2 disrupts lipid membranes. J. Nat. Prod. 70, 643–647 [DOI] [PubMed] [Google Scholar]
- 31. Henriques S. T., Huang Y. H., Rosengren K. J., Franquelim H. G., Carvalho F. A., Johnson A., Sonza S., Tachedjian G., Castanho M. A., Daly N. L., Craik D. J. (2011) Decoding the membrane activity of the cyclotide kalata B1: the importance of phosphatidylethanolamine phospholipids and lipid organization in hemolytic and anti-HIV activities. J. Biol. Chem. 286, 24231–24241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Burman R., Strömstedt A. A., Malmsten M., Göransson U. (2011) Cyclotide-membrane interactions: defining factors of membrane binding, depletion, and disruption. Biochim. Biophys. Acta 1808, 2665–2673 [DOI] [PubMed] [Google Scholar]
- 33. Huang Y. H., Colgrave M. L., Clark R. J., Kotze A. C., Craik D. J. (2010) Lysine scanning mutagenesis reveals an amendable face of the cyclotide kalata B1 for the optimization of nematocidal activity. J. Biol. Chem. 285, 10797–10805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ireland D. C., Clark R. J., Daly N. L., Craik D. J. (2010) Isolation, sequencing, and structure-activity relationships of cyclotides. J. Nat. Prod. 73, 1610–1622 [DOI] [PubMed] [Google Scholar]
- 35. Huang Y. H., Colgrave M. L., Daly N. L., Keleshian A., Martinac B., Craik D. J. (2009) The biological activity of the prototypic cyclotide kalata b1 is modulated by the formation of multimeric pores. J. Biol. Chem. 284, 20699–20707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ovesen R. G., Brandt K. K., Göransson U., Nielsen J., Hansen H. C., Cedergreen N. (2011) Biomedicine in the environment: cyclotides constitute potent natural toxins in plants and soil bacteria. Environ. Toxicol. Chem. 30, 1190–1196 [DOI] [PubMed] [Google Scholar]
- 37. Trabi M., Svangård E., Herrmann A., Göransson U., Claeson P., Craik D. J., Bohlin L. (2004) Variations in cyclotide expression in Viola species. J. Nat. Prod. 67, 806–810 [DOI] [PubMed] [Google Scholar]
- 38. Craik D. J. (2009) Circling the enemy: cyclic proteins in plant defense. Trends Plant Sci. 14, 328–335 [DOI] [PubMed] [Google Scholar]
- 39. Colgrave M. L., Korsinczky M. J., Clark R. J., Foley F., Craik D. J. (2010) Sunflower trypsin inhibitor-1, proteolytic studies on a trypsin inhibitor peptide and its analogs. Biopolymers 94, 665–672 [DOI] [PubMed] [Google Scholar]
- 40. Swedberg J. E., Nigon L. V., Reid J. C., de Veer S. J., Walpole C. M., Stephens C. R., Walsh T. P., Takayama T. K., Hooper J. D., Clements J. A., Buckle A. M., Harris J. M. (2009) Substrate-guided design of a potent and selective kallikrein-related peptidase inhibitor for kallikrein-4. Chem. Biol. 16, 633–643 [DOI] [PubMed] [Google Scholar]
- 41. Craik D. J., Conibear A. C. (2011) The chemistry of cyclotides. J. Org. Chem. 76, 4805–4817 [DOI] [PubMed] [Google Scholar]
- 42. Gunasekera S., Foley F. M., Clark R. J., Sando L., Fabri L. J., Craik D. J., Daly N. L. (2008) Engineering stabilized vascular endothelial growth factor-A antagonists: synthesis, structural characterization, and bioactivity of grafted analogs of cyclotides. J. Med. Chem. 51, 7697–7704 [DOI] [PubMed] [Google Scholar]
- 43. Thongyoo P., Roqué-Rosell N., Leatherbarrow R. J., Tate E. W. (2008) Chemical and biomimetic total syntheses of natural and engineered MCoTI cyclotides. Org. Biomol. Chem. 6, 1462–1470 [DOI] [PubMed] [Google Scholar]
- 44. Austin J., Wang W., Puttamadappa S., Shekhtman A., Camarero J. A. (2009) Biosynthesis and biological screening of a genetically encoded library based on the cyclotide MCoTI-I. ChemBioChem 10, 2663–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Clark R. J., Jensen J., Nevin S. T., Callaghan B. P., Adams D. J., Craik D. J. (2010) The engineering of an orally active conotoxin for the treatment of neuropathic pain. Angew. Chem. Int. Ed. Engl. 49, 6545–6548 [DOI] [PubMed] [Google Scholar]
- 46. Gran L., Sandberg F., Sletten K. (2000) Oldenlandia affinis (R&S) DC. A plant containing uteroactive peptides used in African traditional medicine. J. Ethnopharmacol. 70, 197–203 [DOI] [PubMed] [Google Scholar]
- 47. Qin Q., McCallum E. J., Kaas Q., Suda J., Saska I., Craik D. J., Mylne J. S. (2010) Identification of candidates for cyclotide biosynthesis and cyclization by expressed sequence tag analysis of Oldenlandia affinis. BMC Genomics 11, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mulvenna J. P., Mylne J. S., Bharathi R., Burton R. A., Shirley N. J., Fincher G. B., Anderson M. A., Craik D. J. (2006) Discovery of cyclotide-like protein sequences in graminaceous crop plants: ancestral precursors of circular proteins? Plant Cell 18, 2134–2144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Velásquez J. E., van der Donk W. A. (2011) Genome mining for ribosomally synthesized natural products. Curr. Opin. Chem. Biol. 15, 11–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Mohimani H., Liu W. T., Mylne J. S., Poth A. G., Colgrave M. L., Tran D., Selsted M. E., Dorrestein P. C., Pevzner P. A. (2011) Cycloquest: identification of cyclopeptides via database search of their mass spectra against genome databases. J. Proteome Res. 10, 4505–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]