Abstract
Here, we review the use of different biochemical approaches for biological synthesis of circular or backbone-cyclized proteins and peptides. These methods allow the production of circular polypeptides either in vitro or in vivo using standard recombinant DNA expression techniques. Protein circularization can significantly impact protein engineering and research in protein folding. Basic polymer theory predicts that circularization should lead to a net thermodynamic stabilization of a folded protein by reducing the entropy associated with the unfolded state. Protein cyclization also provides a valuable tool for exploring the effects of topology on protein folding kinetics. Furthermore, the biological production of cyclic polypeptides makes possible the production of cyclic polypeptide libraries. The generation of such libraries, which was previously restricted to the domain of synthetic chemists, now offers biologists access to highly diverse and stable molecular libraries for probing protein structure and function.
Keywords: Protein Chemical Modification, Protein Design, Protein Engineering, Protein Evolution, Protein Processing, Expressed Protein Ligation, Inteins, Native Chemical Ligation, Proteases, Protein Splicing
Introduction
Protein engineering is usually defined as the modification of the primary sequence of a protein to change in a defined way its three-dimensional structure and biological function. Site-specific mutagenesis using either recombinant DNA technology or chemical synthesis has been successfully applied to numerous proteins. Another useful approach to protein engineering involves changing backbone topology. Backbone cyclization or circularization (i.e. the covalent linkage of the N-terminal amino and C-terminal carboxylic groups) is a powerful tool for the study and manipulation of protein structure and function. The introduction of a covalent bond between the N and C termini of a protein provides a more stable topological constraint than the use of disulfide bonds. Hence, in proteins whose N and C termini are close in the native structure (a surprisingly common feature in folded proteins, particularly in single-domain proteins (1)), backbone cyclization should stabilize the protein fold by reducing the backbone entropy associated with the unfolded state of a protein. Backbone cyclization also has the added benefit of making proteins more resistant to proteolytic degradation, in particular against exoproteases (2, 3), which should increase both the in vitro stability of industrial enzymes and the in vivo stability of proteins with interesting pharmacological properties. In addition, protein cyclization provides a valuable tool for exploring fundamental questions in protein folding, in particular how topology affects the kinetics of protein folding (4).
Backbone peptide cyclization has also been widely used in bioorganic and medicinal chemistry to improve the biochemical and biophysical properties of flexible and labile peptides in the development of peptide-based drug candidates (2, 3). The cyclization of flexible linear peptides reduces their conformational freedom and creates constrained structural frameworks that often confer high receptor binding affinity, specificity, and enhanced stability (2, 3).
Backbone-cyclized polypeptides (ranging from small peptides to small proteins) are also found throughout nature from bacteria to animals serving in a variety of functional roles (5). Many of these are synthesized by multienzyme complexes such as cyclosporine from fungi and daptomycin from bacteria. However, an increasing number of circular polypeptides are being discovered that are ribosomally produced as precursors and then post-translationally modified by cyclization (5). For example, θ-defensins are broad-spectrum antimicrobial 18-residue circular peptides expressed in blood leukocytes and bone marrow of Old World monkeys and represent the only circular peptides found in mammals to date. Backbone-cyclized polypeptides are also found in plants. Sunflower trypsin inhibitor-1 (SFTI-1),2 for example, is a bicyclic 14-residue peptide found in sunflower seeds with potent trypsin inhibitory activity. Cyclotides comprise a novel family of well folded small globular backbone-cyclized microproteins (≈30 residues long) found in plants. Naturally occurring cyclotides have been characterized with insecticidal, uterotonic, anti-HIV, antimicrobial, antitumor, and antihelminthic activities (see Refs. 6 and 7 for recent reviews) and have been reported to cross cell membranes (8, 9). These properties make cyclotides highly attractive for the development of novel peptide-based therapeutics (10, 11).
Despite the fact that the chemical synthesis of backbone-cyclized peptides and small protein domains has been well explored and a number of different approaches involving solid-phase or liquid-phase chemistry exist (12), recent advances in the fields of molecular biology and protein engineering have now made possible the biosynthesis of cyclic polypeptides using standard expression systems (13). The biological synthesis (i.e. the production of polypeptides mediated by ribosomal translation) of circular polypeptides offers many advantages over chemical synthesis. Although the chemical synthesis of polypeptides is limited to sequences no longer than 100–150 residues long, biological approaches do not present such limitations. Biological approaches also allow, using standard molecular biology tools, the generation of large combinatorial libraries of circular polypeptides that can be screened inside living cells for their ability to attenuate or inhibit cellular processes analogous to the way that the yeast two-hybrid technology works (14).
Different approaches have been described for the biological generation of circular polypeptides, including the use of expressed protein ligation (15), intein- and enzyme-mediated protein trans-splicing reactions (16), and genetic code reprogramming (17). In this minireview, we examine these methods as well as their potential applications, which are summarized in Scheme 1 and Table 1.
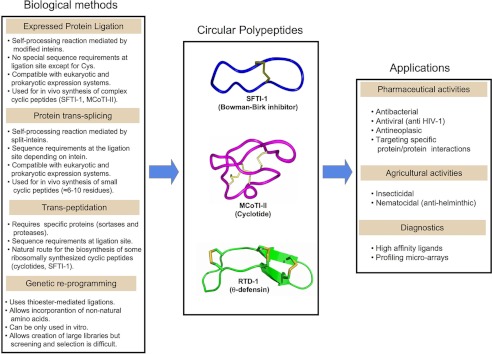
SCHEME 1.
Summary of methods employed for biological production of backbone-cyclized peptides. Left panel, these approaches rely on the ribosomal synthesis of a polypeptide precursor that undergoes cyclization mediated by inteins or proteases. Middle panel, shown are three-dimensional structures of three examples of naturally occurring circular peptides with potential biotechnological value. MCoTI-II and SFTI-1 are powerful protease inhibitors and are attractive scaffolds for the introduction of other biological activities. RTD-1 is a θ-defensin that has strong antimicrobial and anti-inflammatory properties. All have been successfully expressed in biological systems (see Table 1). Right panel, the potential biotechnological applications of circular polypeptides, which include pharmacological, agrochemical, and diagnostic applications, are summarized. Molecular structures were generated using the structures of MCoTI-II (Protein Data Bank code 1IB9), SFTI-1 (code 1JBN), and RTD-1 (code 1HVZ).
TABLE 1.
Examples of backbone-cyclized polypeptides produced by ribosomal synthesis
These were produced using EPL, PTS, SML, genetic reprogramming, and protease-mediated ligation. The sizes of the polypeptides shown range from small peptides (eight residues) to proteins (263 residues).
| Protein/peptide | No. of residues | Method | Application | Ref./source |
|---|---|---|---|---|
| c-Crk SH3 | 57 | EPL | Stability | 21 |
| Folding kinetics | 49 | |||
| TEM-1 β-lactamase | 263 | EPL | Stability | 24 |
| PTS | 34 | |||
| Kalata B1 | 29 | EPL | Natural product | 27 |
| MCoTI-I/II | 34 | EPL | Natural product | 28 |
| Library scaffold | 29 | |||
| Trypsin-mediated cyclization | Natural product | 42 | ||
| Cryptdin-4 | 32 | EPL | Stability and improved activity | 31 |
| SFTI-1 | 14 | EPL | Natural product and library scaffold | 30 |
| Trypsin-mediated cyclization | Natural product and mutants | 9,41 | ||
| Genetic reprogramming | Natural product and library scaffold | 46 | ||
| RTD-1 | 18 | EPL | Natural product | 50 |
| Genetic reprogramming | Natural product and library scaffold | 46 | ||
| Pseudostellarin F | 8 | PTS | Natural product | 16 |
| HIV protease inhibitors | 6 | PTS | Library scaffold and screening | 33 |
| GFP | 238 | EPL | Stability | 25 |
| SML | 38 | |||
| IFNα2 | 188 | SML | Thermal and serum stability | 39 |
| Histatin-1 | 38 | SML | Improve biological activity | 40 |
Backbone Cyclization Using Expressed Protein Ligation
Expressed protein ligation (EPL) (18) is an extension of native chemical ligation (NCL), a chemoselective ligation approach for the chemical synthesis of proteins (19). In this reaction, a fully unprotected peptide with an α-thioester at the C terminus reacts chemoselectively under neutral aqueous conditions with another unprotected peptide containing an N-terminal Cys residue to form a native peptide bond (19). It is also well established that when these two reactive groups are located in the same linear polypeptide precursor, the chemical ligation can proceed in an intramolecular fashion, therefore giving rise to a backbone-cyclized polypeptide (20). This reaction has been successfully used for the chemical synthesis of circular peptides and small protein domains (20).
Recent advances in protein engineering, as well as the discovery of protein splicing, have also made possible the production of recombinant proteins containing an N-terminal Cys residue and/or the C-terminal α-thioester functionality. These important developments have made it possible to perform NCL reactions between recombinant and/or synthetic polypeptides. This technology, called EPL, has provided access to a multitude of chemically engineered proteins, including backbone-cyclized polypeptides (see Ref. 20 for a recent review).
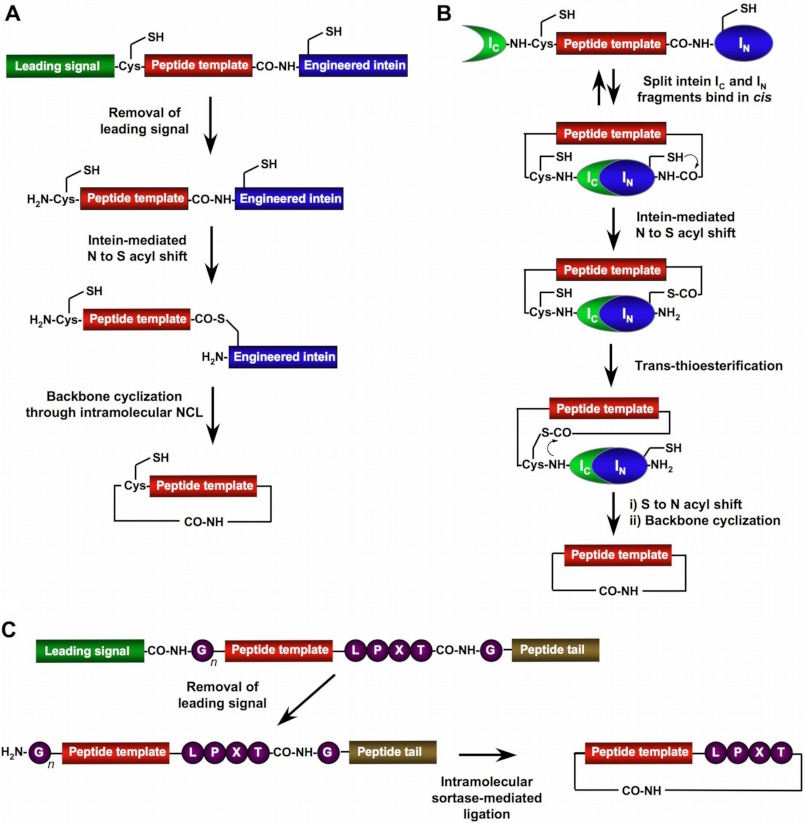
The use of EPL-mediated backbone cyclization for the biosynthesis of circular polypeptides (Fig. 1A) was first reported by Camarero and Muir in 1999 (21) using the N-terminal SH3 domain of the c-Crk protein as a model system. In this work, the SH3 domain was fused to a modified Saccharomyces cerevisiae vacuolar membrane ATPase intein at the C terminus and to the MIEGRC motif (which contains a factor Xa proteolysis site) at the N terminus. The intein fusion construct was then expressed in Escherichia coli, purified, and cleaved with factor Xa, producing an N-terminal Cys-containing SH3-intein fusion protein, which spontaneously reacted in an intramolecular fashion to yield the fully active circular SH3 domain (21). The cyclization reaction was extremely efficient and clean. It is also worth mentioning that in contrast to standard EPL protocols, which usually require denaturing conditions and the presence of thiol cofactors (18), the cyclization reaction was performed under physiological conditions at neutral pH in the absence of thiol cofactors. This was attributed to the close proximity of the α-thioester and N-terminal Cys residue in the folded state of the SH3 domain, which increased the local concentration of the reacting groups, thus facilitating the cyclization reaction. This effect has also been found in the cyclization of other small protein domains (22, 23). A similar approach has also been used by Iwai and Plückthun for the biosynthesis of circular β-lactamase (24) and GFP (25). The resulting circular proteins were biologically active and found to be more resistant against thermal denaturation (24, 25).
FIGURE 1.
Three most common approaches for biological production of backbone-cyclized polypeptides using EPL (A), PTS (B), and SML (C). IC, C-intein; IN, N-intein.
Inspired by the high efficiency of the in vitro cyclization of the SH3 domain (21), Camarero et al. (26) used a similar approach to explore the cyclization of the SH3 domain inside living cells. This work provided the first demonstration of polypeptide chemical ligation reactions performed inside the complex cytoplasmic environment of a living cell. In-cell production of circular polypeptides makes possible the generation of genetically encoded libraries that can be assayed in the intracellular milieu for rapid screening of biologically active compounds.
More recently, our laboratory has used this approach for the recombinant production of cyclotides using standard bacterial expression systems (27, 28). Cyclotides are small globular microproteins with a head-to-tail cyclized backbone, which is further stabilized by three disulfide bonds forming a cystine knot. This cystine knot motif gives cyclotides exceptional resistance to physical, chemical, and biological degradation. Our group has also pioneered the use of EPL-mediated backbone cyclization for the biosynthesis of fully folded cyclotides inside bacterial cells using standard expression systems (28, 29). We have recently used this approach for the generation of a small library based on the cyclotide MCoTI-I, in which all of the residues except Cys and those located in loop 6 were replaced by different residues (29). The resulting library was screened for biological activity and the ability to adopt a cyclotide fold, indicating that most of the residues in the cyclotide scaffold could be replaced without significantly affecting the ability of the resulting sequences to fold correctly (29). These results highlight the robustness of the cyclotide scaffold for generating large genetically encoded libraries of cyclotides that can be screened in-cell using high throughput techniques.
EPL-mediated backbone cyclization of polypeptides has also been recently used for the biosynthesis of the Bowman-Birk inhibitor SFTI-1 (30). The biosynthesis of other cyclic peptides such as backbone-cyclized α-defensins (31) and naturally occurring θ-defensins (50) has also been produced in our laboratory using bacterial expression systems, thus showing the general applicability of this method for the production of naturally occurring or engineered circular polypeptides.
Backbone Cyclization Using Intein-mediated Protein Trans-splicing
Another method to generate cyclic peptides in vivo is by protein trans-splicing (PTS) (Fig. 1B). This approach utilizes a self-processing intein that is split into two fragments, an N-intein and a C-intein. This technique was first reported by Benkovic and co-workers (32) using the naturally occurring Synechocystis sp. PCC6803 DnaE split intein. PTS is a post-translational modification similar to protein splicing with the difference that the intein self-processing domain is split into two fragments (N-intein and C-intein). The split intein fragments are not active individually; however, they can bind to each other with high specificity under appropriate conditions to form an active protein-splicing or intein domain in trans. By rearranging the order of the intein fragments through fusing the N- and C-intein fragments to the C and N termini of the polypeptide to be cyclized, the trans-splicing reaction results in the formation of a backbone-cyclized polypeptide (Fig. 1B). This methodology, often called SICLOPPS (split intein circular ligation of proteins and peptides), has been successfully used for the generation of several naturally occurring cyclic peptides as well as large genetically encoded libraries of small cyclic peptides (16). More recently, Schultz and co-workers (33) reported the use of this technology in combination with nonsense codon suppressor tRNA technology to build large libraries of cyclic hexapeptides using an expanded set of non-natural amino acid building blocks. These libraries were then used to screen for an inhibitor of HIV protease using a selection based on cellular viability (33). This was accomplished by linking cellular viability to the activity of HIV protease. In this particular case, the HIV protease cleavage site was introduced into the flexible hinge region of the tetracycline resistance gene (Tn10-HIV). The introduction of this sequence does not affect significantly the biological activity of Tn10-HIV; however, when it is coexpressed with HIV protease, resistance to tetracycline is abolished. Hence, cells coexpressing Tn10-HIV and HIV protease would be able to grow in medium containing tetracycline only if the HIV protease was inhibited. The authors used this elegant approach for screening surviving cells coexpressing HIV/Tn10-HIV and transformed with a genetically encoded library of cyclic peptides. Under these conditions, only cells expressing cyclic peptides able to inhibit HIV protease were able to grow to saturation (33). The authors were able to select different cyclic hexapeptides containing the non-natural amino acid p-benzoylphenylalanine, which contains a keto group and is able to inhibit HIV with IC50 values in the low micromolar range (33). These interesting findings suggest that the use of an expanded genetic code using novel chemical functionalities not present in nature such as a keto group could confer an evolutionary advantage in response to selective pressure.
PTS has also been used for the generation of circular proteins (32, 34). In a recent study, an artificially split intein DnaB mini-intein from the Synechocystis sp. PCC6803 strain was used for the cyclization of the TEM-1 β-lactamase in the bacterial periplasm; this was accomplished by adding TEM-1 β-lactamase periplasm export signal peptide to the split intein precursor (34). The same authors also used this approach for the production of large libraries of small circular peptides. It was estimated that ∼50% of the combinatorial peptides were able to cyclize efficiently.
Although the use of PTS is a good alternative for the biosynthesis of backbone-cyclized polypeptides and biologically encoded libraries, it should be noted, however, that it requires the presence of specific amino acid residues at both intein-extein junctions for efficient protein splicing to occur (16). To resolve this, new or engineered split inteins that are more tolerant with non-native extein-intein junction sequences are required. In contrast, the only sequence requirement for the use of EPL or intein-mediated backbone cyclization is the presence of an N-terminal Cys. Model studies have also shown that all of the 20 natural amino acids located at the C terminus of a polypeptide thioester can support ligation (35). It is worth noting, however, that the speed rate of thioester-mediated ligations depends on the nature of the amino acid at the C terminus, e.g. Gly and Ala residues react faster, whereas β-branched residues react slower and produce product in lower yields. Furthermore, the engineered inteins most commonly used for the generation of polypeptide α-thioesters have also been shown to be compatible with most amino acids upstream of the cleavage site (20).
Backbone Cyclization Using Protease-catalyzed Transpeptidation
It is well known that prokaryotes use protease-catalyzed protein splicing to attach proteins to peptidoglycan in their cell wall envelope. Sortases are transpeptidase enzymes found in most Gram-positive bacteria that are specialized in this task. Among the different bacterial sortases isolated so far, the Staphylococcus aureus sortase A (SrtA) has been by far the most widely used for protein engineering (36). SrtA recognizes peptide substrates containing the sequence LPXTG (where X can be any residue), cleaving the peptide bond between the Thr and Gly residues. The cleavage is catalyzed by an active site Cys residue that generates a transient thioacyl-enzyme intermediate. This thioester-reactive specie then reacts with the α-amino group of oligoglycine (pentaglycine in the case of SrtA) substrates to produce ligated products. In the absence of oligoglycine substrates, the thioacyl-enzyme intermediate is hydrolyzed.
Backbone-cyclized polypeptides can be obtained by using an intramolecular version of the sortase-mediated ligation (SML) (Fig. 1C). The first example of polypeptide cyclization by SML was reported by Boder and co-workers (37). In this work, a GFP construct containing the SrtA recognition sequences at the N and C termini, respectively, was backbone-cyclized through SML. Ploegh and co-workers (38) have also recently used a similar approach to obtain a circular version of GFP using SrtA. SrtA has also been used recently for the backbone cyclization of several cytokines to improve their stability and serum half-life (39). In this elegant work, SML was used in tandem for the introduction of an aminooxyacetic acid moiety at the termini of several cytokines, which were then backbone-cyclized and site-specifically PEGylated through the aminooxyacetic acid moiety for increased stability in serum (39). SrtA has also been used recently to produce a backbone-cyclized version of histatin-1, a 38-mer salivary polypeptide with antimicrobial and wound-healing activities (40). According to the authors, the SrtA-mediated cyclization of histatin-1 provided better yields than chemical synthesis. It should be highlighted, however, that in this case, the synthesis was performed entirely using a solid-phase approach, i.e. the synthesis of the linear precursor, as well as its cyclization, was carried out entirely on a solid support using standard chemical coupling reagents. This type of approach has been shown to lead to lower yields for the cyclization of long polypeptides and small protein domains (22, 23). In contrast, the use of chemoselective ligations such as NCL (which uses the same type of chemoselective ligation as EPL) (Fig. 1A) has proven to give excellent results for the backbone cyclization of polypeptides and small protein domains (22, 23).
Although the use of SML for the production of cyclic polypeptides shows great promise, it has not been shown to work in the cell yet, which limits its use for the generation of cellular libraries of circular polypeptides for genetic experiments. Moreover, the actual approach requires the use of rather large sortase recognition sequences (LPXTG5 in the case of SrtA) (Fig. 1C) that remain incorporated in the final cyclic polypeptide, which could affect the applicability of this approach for some applications.
More promising is the use of “traceless” protease-catalyzed protein splicing for cyclization of the polypeptide backbone. For example, Craik and co-workers (41) have shown that trypsin can be used for the efficient backbone cyclization of the cyclic peptide SFTI-1. A similar approach has also been used for the cyclization of the cyclotides MCoTI-I and MCoTI-II in vitro (42). MCoTI cyclotides and SFTI-1 are potent trypsin inhibitors, with Ki values in the picomolar and nanomolar range, respectively. In both cases, the proteolytic enzyme trypsin was able to resynthesize the peptide bond between the P1 and P1′ residues to form the native circular polypeptides (41, 42). Although this approach has not yet been tested in vivo, there is mounting evidence that protease-catalyzed protein splicing is used in the biosynthesis of these naturally occurring circular polypeptides in plants. Recent studies suggest that asparaginyl endopeptidases are involved in the biosynthesis of cyclotides and SFTI-1 through an asparaginyl endopeptidase-mediated transpeptidation step (43, 44).
Backbone Cyclization Using Genetic Code Reprogramming
Genetic code reprogramming is an emerging new method that allows the production in vitro of ribosomally expressed polypeptides containing non-proteinogenic amino acids (45). The key to this approach is the use of custom-made reconstituted in vitro translation systems and, more importantly, the ability to employ enzymes that are able to load specific amino acids into designed tRNAs. Among the different methods to accomplish the task is the use of an RNA-based enzyme, flexizyme, which shows great promise (45). Flexizyme is a ribozyme that catalyzes the transfer of practically any amino acid activated as a p-nitrophenyl ester into any given tRNA.
Suga and co-workers (46) have recently shown that this method can also be used for the generation of ribosomally translated backbone-cyclized polypeptides in cell-free expression systems. This was accomplished by the incorporation of the cysteinyl-prolinyl glycolic acid ester moiety at the C terminus of ribosomally produced polypeptides by reprogramming glycolic acid into a codon using the flexizyme system. Cysteinyl-prolinyl esters can rapidly rearrange at physiological pH through the formation of a diketopiperazine intermediate to produce a C-terminal thioester (46). This post-translationally formed thioester can then react in an intramolecular fashion with an N-terminal Cys residue through NCL in the same way that EPL works (Fig. 1A). The incorporation of an N-terminal Cys into polypeptides generated in vitro can be easily accomplished by adding peptide deformylase and methionine aminopeptidase to the in vitro translation system. This approach has been used for the generation of numerous backbone-cyclized peptides, including SFTI-1 and rhesus θ-defensin-1 (RTD-1) (46).
Suga and co-workers (46) have also shown that this strategy can be used for the generation of peptide libraries. However, due to the circular nature of the peptides produced, it is difficult to genetically encode (e.g. using an RNA-display approach) the different members of a particular library generated by this technique. This makes the screening of such libraries a very challenging task, which limits the complexity of the libraries that can be analyzed using this method. Another limitation of this approach is that production yields are usually low, therefore limiting the utility of this method for screening purposes.
Summary and Outlook
In this minireview, we have briefly examined the different biochemical approaches available for the biological production of backbone-cyclized polypeptides ranging from small peptides to proteins. These include the use of protein splicing catalyzed by inteins (16) or proteases (41, 42) as well as thioester-based ligation using either modified inteins (27, 29, 30) or genetic code reprogramming (46). The ability to produce circular polypeptides using the tools of molecular biology has multiple biotechnological applications (Scheme 1). For example, it makes possible the generation of large libraries of cyclic polypeptides that can be screened for biological activity. Several of the methods discussed also allow the production of libraries inside living cells (29, 30, 33). This makes possible the rapid screening and selection of novel sequences that can modulate or inhibit the biological activities of particular biomolecular targets. This approach allows the use of molecular evolution and cell-based assays for the rapid selection of novel sequences with optimal biological activity. For example, Schultz and co-workers (33) have recently used PTS in combination with nonsense codon suppressor tRNA technology to build large libraries of cyclic hexapeptides incorporating non-natural amino acids inside bacterial cells that were later screened for activity against the HIV protease. Although the biological activities for the selected circular peptides were rather modest (IC50 values in the low micromolar range), this could be attributed to the small size of the cyclic peptide scaffold used in this work (only six residues). This could be easily addressed by using a cyclic polypeptide template incorporating more than one loop for potential interactions in the same way that antibody complementarity-determining regions work. Within this context, our group has pioneered the use of EPL-mediated backbone cyclization for the biosynthesis of cyclotide-based libraries inside living cells for the high throughput selection of novel biological activities. Our group is using the cyclotide MCoTI-I as a molecular scaffold, which has up to five hypervariable loops available for the design of complex libraries that can be used for molecular evolution experiments using cell-based screening. MCoTI cyclotides are nontoxic and are powerful trypsin inhibitors, with IC50 values in the low picomolar range, which demonstrates the potential of this scaffold for selecting novel sequences that target protein-protein interactions with very high affinities. Moreover, the cyclic cystine knot motif found in cyclotides gives them enhanced thermal, chemical, and biological stabilities. All of these features make the cyclotide scaffold an ideal molecular framework for the development of novel peptide-based therapeutics.
In this context, it is worth noting that new critical information on the origin, evolution, and processing of cyclotides as well as other circular polypeptides found in plants is beginning to provide insight on how these polypeptides are post-translationally cyclized in plants. A complete elucidation of the mechanisms associated with the ribosomal biosynthesis of circular polypeptides in plants should make possible the production of pharmacologically useful circular polypeptides in genetically engineered plants in the near future.
The production of circular polypeptides using standard biological expression systems has also made possible the introduction of NMR active isotopes (15N and/or 13C) in a very inexpensive fashion (47). This should facilitate the use of NMR to study structure-activity relationships of any potential circular polypeptide and its molecular target (47). The recent development of in-cell NMR using sequential labeling approaches (48) could be easily interfaced for the in-cell screening of genetically encoded libraries of circular polypeptides.
In summary, the development of novel biochemical approaches for the biological synthesis of circular polypeptides is opening new and exciting opportunities that previously were available only to synthetic chemists. The production of complex libraries inside living cells using standard recombinant techniques offers biologists a unique opportunity to perform experiments in molecular evolution that will unravel genomic information encoding complex biochemical pathways and protein interaction networks. The use of highly stable circular polypeptide scaffolds provides an ideal molecular tool for this challenging task.
This work was supported, in whole or in part, by National Institutes of Health Research Grant R01 GM090323 and Department of Defense Congressionally Directed Medical Research Program Grant PC09305 (to J. A. C.). This is the fifth article in the Thematic Minireview Series on Circular Proteins.
- SFTI-1
- sunflower trypsin inhibitor-1
- EPL
- expressed protein ligation
- NCL
- native chemical ligation
- SH3
- Src homology 3
- PTS
- protein trans-splicing
- SML
- sortase-mediated ligation
- RTD-1
- rhesus θ-defensin-1.
REFERENCES
- 1. Thornton J. M., Sibanda B. L. (1983) Amino- and carboxyl-terminal regions in globular proteins. J. Mol. Biol. 167, 443–460 [DOI] [PubMed] [Google Scholar]
- 2. Clark R. J., Fischer H., Dempster L., Daly N. L., Rosengren K. J., Nevin S. T., Meunier F. A., Adams D. J., Craik D. J. (2005) Engineering stable peptide toxins by means of backbone cyclization: stabilization of the α-conotoxin MII. Proc. Natl. Acad. Sci. U.S.A. 102, 13767–13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Clark R. J., Akcan M., Kaas Q., Daly N. L., Craik D. J. (2012) Cyclization of conotoxins to improve their biopharmaceutical properties. Toxicon 59, 446–455 [DOI] [PubMed] [Google Scholar]
- 4. Plaxco K. W., Simons K. T., Ruczinski I., Baker D. (2000) Topology, stability, sequence, and length: defining the determinants of two-state protein folding kinetics. Biochemistry 39, 11177–11183 [DOI] [PubMed] [Google Scholar]
- 5. Craik D. J. (2006) Chemistry. Seamless proteins tie up their loose ends. Science 311, 1563–1564 [DOI] [PubMed] [Google Scholar]
- 6. Daly N. L., Rosengren K. J., Craik D. J. (2009) Discovery, structure, and biological activities of cyclotides. Adv. Drug Deliv. Rev. 61, 918–930 [DOI] [PubMed] [Google Scholar]
- 7. Garcia A. E., Camarero J. A. (2010) Biological activities of natural and engineered cyclotides, a novel molecular scaffold for peptide-based therapeutics. Curr. Mol. Pharmacol. 3, 153–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Contreras J., Elnagar A. Y., Hamm-Alvarez S. F., Camarero J. A. (2011) Cellular uptake of cyclotide MCoTI-I follows multiple endocytic pathways. J. Controlled Release 155, 134–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cascales L., Henriques S. T., Kerr M. C., Huang Y. H., Sweet M. J., Daly N. L., Craik D. J. (2011) Identification and characterization of a new family of cell-penetrating peptides. Cyclic cell-penetrating peptides. J. Biol. Chem. 286, 36932–36943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Henriques S. T., Craik D. J. (2010) Cyclotides as templates in drug design. Drug Discov. Today 15, 57–64 [DOI] [PubMed] [Google Scholar]
- 11. Jagadish K., Camarero J. A. (2010) Cyclotides, a promising molecular scaffold for peptide-based therapeutics. Biopolymers 94, 611–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. White C. J., Yudin A. K. (2011) Contemporary strategies for peptide macrocyclization. Nat. Chem. 3, 509–524 [DOI] [PubMed] [Google Scholar]
- 13. Sancheti H., Camarero J. A. (2009) “Splicing up” drug discovery. Cell-based expression and screening of genetically encoded libraries of backbone-cyclized polypeptides. Adv. Drug Deliv. Rev. 61, 908–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suter B., Kittanakom S., Stagljar I. (2008) Two-hybrid technologies in proteomics research. Curr. Opin Biotechnol. 19, 316–323 [DOI] [PubMed] [Google Scholar]
- 15. Kimura R., Camarero J. A. (2005) Expressed protein ligation: a new tool for the biosynthesis of cyclic polypeptides. Protein Pept. Lett. 12, 789–794 [DOI] [PubMed] [Google Scholar]
- 16. Tavassoli A., Benkovic S. J. (2007) Split intein-mediated circular ligation used in the synthesis of cyclic peptide libraries in E. coli. Nat. Protoc. 2, 1126–1133 [DOI] [PubMed] [Google Scholar]
- 17. Ohta A., Yamagishi Y., Suga H. (2008) Synthesis of biopolymers using genetic code reprogramming. Curr. Opin. Chem. Biol. 12, 159–167 [DOI] [PubMed] [Google Scholar]
- 18. Muir T. W. (2003) Semisynthesis of proteins by expressed protein ligation. Annu. Rev. Biochem. 72, 249–289 [DOI] [PubMed] [Google Scholar]
- 19. Dawson P. E., Kent S. B. (2000) Synthesis of native proteins by chemical ligation. Annu. Rev. Biochem. 69, 923–960 [DOI] [PubMed] [Google Scholar]
- 20. Berrade L., Camarero J. A. (2009) Expressed protein ligation: a resourceful tool to study protein structure and function. Cell. Mol. Life Sci. 66, 3909–3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Camarero J. A., Muir T. W. (1999) Biosynthesis of a head-to-tail cyclized protein with improved biological activity. J. Am. Chem. Soc. 121, 5597–5598 [Google Scholar]
- 22. Camarero J. A., Pavel J., Muir T. W. (1998) Angew. Chem. Int. Ed. Engl. 37, 347–349 [DOI] [PubMed] [Google Scholar]
- 23. Camarero J. A., Cotton G. J., Adeva A., Muir T. W. (1998) Chemical ligation of unprotected peptides directly from a solid support. J. Pept. Res. 51, 303–316 [DOI] [PubMed] [Google Scholar]
- 24. Iwai H., Plückthun A. (1999) Circular β-lactamase: stability enhancement by cyclizing the backbone. FEBS Lett. 459, 166–172 [DOI] [PubMed] [Google Scholar]
- 25. Iwai H., Lingel A., Plückthun A. (2001) Cyclic green fluorescent protein produced in vivo using an artificially split PI-PfuI intein from Pyrococcus furiosus. J. Biol. Chem. 276, 16548–16554 [DOI] [PubMed] [Google Scholar]
- 26. Camarero J. A., Fushman D., Cowburn D., Muir T. W. (2001) Peptide chemical ligation inside living cells: in vivo generation of a circular protein domain. Bioorg. Med. Chem. 9, 2479–2484 [DOI] [PubMed] [Google Scholar]
- 27. Kimura R. H., Tran A. T., Camarero J. A. (2006) Biosynthesis of the cyclotide kalata B1 by using protein splicing. Angew. Chem. Int. Ed. Engl. 45, 973–976 [DOI] [PubMed] [Google Scholar]
- 28. Camarero J. A., Kimura R. H., Woo Y. H., Shekhtman A., Cantor J. (2007) Biosynthesis of a fully functional cyclotide inside living bacterial cells. ChemBioChem 8, 1363–1366 [DOI] [PubMed] [Google Scholar]
- 29. Austin J., Wang W., Puttamadappa S., Shekhtman A., Camarero J. A. (2009) Biosynthesis and biological screening of a genetically encoded library based on the cyclotide MCoTI-I. ChemBioChem 10, 2663–2670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Austin J., Kimura R. H., Woo Y. H., Camarero J. A. (2010) In vivo biosynthesis of an Ala scan library based on the cyclic peptide SFTI-1. Amino Acids 38, 1313–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Garcia A. E., Tai K. P., Puttamadappa S. S., Shekhtman A., Ouellette A. J., Camarero J. A. (2011) Biosynthesis and antimicrobial evaluation of backbone-cyclized α-defensins. Biochemistry 50, 10508–10519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Scott C. P., Abel-Santos E., Wall M., Wahnon D. C., Benkovic S. J. (1999) Production of cyclic peptides and proteins in vivo. Proc. Natl. Acad. Sci. U.S.A. 96, 13638–13643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Young T. S., Young D. D., Ahmad I., Louis J. M., Benkovic S. J., Schultz P. G. (2011) Evolution of cyclic peptide protease inhibitors. Proc. Natl. Acad. Sci. U.S.A. 108, 11052–11056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Deschuyteneer G., Garcia S., Michiels B., Baudoux B., Degand H., Morsomme P., Soumillion P. (2010) Intein-mediated cyclization of randomized peptides in the periplasm of Escherichia coli and their extracellular secretion. ACS Chem. Biol. 5, 691–700 [DOI] [PubMed] [Google Scholar]
- 35. Hackeng T. M., Griffin J. H., Dawson P. E. (1999) Protein synthesis by native chemical ligation: expanded scope by using straightforward methodology. Proc. Natl. Acad. Sci. U.S.A. 96, 10068–10073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tsukiji S., Nagamune T. (2009) Sortase-mediated ligation: a gift from Gram-positive bacteria to protein engineering. ChemBioChem 10, 787–798 [DOI] [PubMed] [Google Scholar]
- 37. Parthasarathy R., Subramanian S., Boder E. T. (2007) Sortase A as a novel molecular “stapler” for sequence-specific protein conjugation. Bioconjug. Chem. 18, 469–476 [DOI] [PubMed] [Google Scholar]
- 38. Antos J. M., Popp M. W., Ernst R., Chew G. L., Spooner E., Ploegh H. L. (2009) A straight path to circular proteins. J. Biol. Chem. 284, 16028–16036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Popp M. W., Dougan S. K., Chuang T. Y., Spooner E., Ploegh H. L. (2011) Sortase-catalyzed transformations that improve the properties of cytokines. Proc. Natl. Acad. Sci. U.S.A. 108, 3169–3174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Bolscher J. G., Oudhoff M. J., Nazmi K., Antos J. M., Guimaraes C. P., Spooner E., Haney E. F., Garcia Vallejo J. J., Vogel H. J., van't Hof W., Ploegh H. L., Veerman E. C. (2011) Sortase A as a tool for high yield histatin cyclization. FASEB J. 25, 2650–2658 [DOI] [PubMed] [Google Scholar]
- 41. Marx U. C., Korsinczky M. L., Schirra H. J., Jones A., Condie B., Otvos L., Jr., Craik D. J. (2003) Enzymatic cyclization of a potent Bowman-Birk protease inhibitor, sunflower trypsin inhibitor-1, and solution structure of an acyclic precursor peptide. J. Biol. Chem. 278, 21782–21789 [DOI] [PubMed] [Google Scholar]
- 42. Thongyoo P., Roqué-Rosell N., Leatherbarrow R. J., Tate E. W. (2008) Chemical and biomimetic total syntheses of natural and engineered MCoTI cyclotides. Org. Biomo.l Chem. 6, 1462–1470 [DOI] [PubMed] [Google Scholar]
- 43. Gillon A. D., Saska I., Jennings C. V., Guarino R. F., Craik D. J., Anderson M. A. (2008) Biosynthesis of circular proteins in plants. Plant J. 53, 505–515 [DOI] [PubMed] [Google Scholar]
- 44. Mylne J. S., Colgrave M. L., Daly N. L., Chanson A. H., Elliott A. G., McCallum E. J., Jones A., Craik D. J. (2011) Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat. Chem. Biol. 7, 257–259 [DOI] [PubMed] [Google Scholar]
- 45. Goto Y., Katoh T., Suga H. (2011) Flexizymes for genetic code reprogramming. Nat. Protoc. 6, 779–790 [DOI] [PubMed] [Google Scholar]
- 46. Kawakami T., Ohta A., Ohuchi M., Ashigai H., Murakami H., Suga H. (2009) Diverse backbone-cyclized peptides via codon reprogramming. Nat. Chem. Biol. 5, 888–890 [DOI] [PubMed] [Google Scholar]
- 47. Puttamadappa S. S., Jagadish K., Shekhtman A., Camarero J. A. (2010) Backbone dynamics of cyclotide MCoTI-I free and complexed with trypsin. Angew. Chem. Int. Ed. Engl. 49, 7030–7034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Burz D. S., Dutta K., Cowburn D., Shekhtman A. (2006) Mapping structural interactions using in-cell NMR spectroscopy (STINT-NMR). Nat. Methods 3, 91–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Camarero J. A., Fushman D., Sato S., Giriat I., Cowburn D., Raleigh D. P., Muir T. W. (2001) Rescuing a destabilized protein fold through backbone cyclization. J. Mol. Biol. 308, 1045–1062 [DOI] [PubMed] [Google Scholar]
- 50. Gould A., Li Y., Majumder S., Garcia A. E., Carlsson P., Shekhtman A., Camarero J. A. (2012) Recombinant production of rhesus θ-defensin-1 using a bacterial expression system. Mol. Biosyst. 8, 1359–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]