Background: Cyclotides are defense-related cyclic plant peptides.
Results: Petunia cyclotides are encoded by novel cyclotide genes and occur in a discrete pattern in leaf architecture.
Conclusion: Novel cyclotides exist in the Solanaceae and are abundant in vascular tissues.
Significance: Cyclotide localization is consistent with an anti-herbivory role. Novel Solanaceae genes provide opportunities for expressing designer cyclic peptides in major crop species.
Keywords: Mass Spectrometry (MS), Peptide Biosynthesis, Peptides, Plant Defense, Proteomics, Cyclic Peptides, Cyclotides
Abstract
Cyclotides are a large family of plant peptides that are structurally defined by their cyclic backbone and a trifecta of disulfide bonds, collectively known as the cyclic cystine knot (CCK) motif. Structurally similar cyclotides have been isolated from plants within the Rubiaceae, Violaceae, and Fabaceae families and share the CCK motif with trypsin-inhibitory knottins from a plant in the Cucurbitaceae family. Cyclotides have previously been reported to be encoded by dedicated genes or as a domain within a knottin-encoding PA1-albumin-like gene. Here we report the discovery of cyclotides and related non-cyclic peptides we called “acyclotides” from petunia of the agronomically important Solanaceae plant family. Transcripts for petunia cyclotides and acyclotides encode the shortest known cyclotide precursors. Despite having a different precursor structure, their sequences suggest that petunia cyclotides mature via the same biosynthetic route as other cyclotides. We assessed the spatial distribution of cyclotides within a petunia leaf section by MALDI imaging and observed that the major cyclotide component Phyb A was non-uniformly distributed. Dissected leaf midvein extracts contained significantly higher concentrations of this cyclotide compared with the lamina and outer margins of leaves. This is the third distinct type of cyclotide precursor, and Solanaceae is the fourth phylogenetically disparate plant family to produce these structurally conserved cyclopeptides, suggesting either convergent evolution upon the CCK structure or movement of cyclotide-encoding sequences within the plant kingdom.
Introduction
Cyclotides are a family of backbone-cyclized plant peptides first discovered in Oldenlandia affinis from the Rubiaceae plant family but since found in a growing number of plants from the Violaceae, Cucurbitaceae, and Fabaceae families (1). Cyclotides are presumed to have a role in plant defense, given reports that ascribe insecticidal (2), molluscicidal (3), or anthelmintic (4) activities to isolated peptides. Since their initial discovery as the active constituents of a uterotonic traditional medicine (5), a host of other bioactivities have been attributed to cyclotides, including anti-HIV (6), cytotoxic (7), and neurotensin inhibitory activity (8).
The definitive structural feature common to cyclotides is the cyclic cystine knot (CCK)4 motif in which three disulfide bonds are entwined in a knotted conformation such that one disulfide bond is threaded through an opening bounded by two sections of the peptide backbone and the two disulfide bonds constraining them (9). The cystine knot has been demonstrated to be the feature that confers most of their stability at high temperatures, in extremes of pH, and against proteolytic enzymes (10, 11). The CCK motif is very tolerant to sequence variation of the non-Cys residues, as exemplified by the observation that it occurs in two cyclic trypsin inhibitors, MCoTI-I and MCoTI-II (12), from a Cucurbitaceae plant that differ substantially in sequence from other cyclotides and are closely related to some acyclic trypsin inhibitors from squash plants that are part of the knottin family. The stability and tolerance to sequence substitution has led to consideration of the CCK framework as a natural combinatorial template (13) with applications in drug design (14).
Several recent studies have demonstrated the suitability of the CCK framework as a stable drug design scaffold, exemplified by the synthesis of modified cyclotides to incorporate bioactive peptide epitopes that would otherwise have short in vivo half-lives. Examples include cyclotide-based vascular endothelial growth factor-A (VEGF) agonists (15) or antagonists (16) and inhibitors of tryptase β from human mast cells (17). These studies highlight the potential value cyclotides have as peptide therapeutics and provide an impetus for investigating their biosynthesis in plants, potentially opening new opportunities for the expression of “designer” cyclotides with pharmaceutical traits in plants.
In Rubiaceae and Violaceae plants, cyclotides are products of dedicated genes that comprise an endoplasmic reticulum signal sequence and a pro-region, followed by up to three cyclotide domains, each flanked by an N-terminal pro-domain and a C-terminal tail (18, 19). Recently, we reported the occurrence of cyclotides in the Fabaceae plant Clitoria ternatea (20), and subsequently it was demonstrated that the Fabaceae cyclotides are encoded within a PA1b-like albumin where the cyclotide has “replaced” the first of its usual two domains (21, 22). Typical Fabaceae albumin-1 genes encode a PA1 pro-protein that is post-translationally cleaved to liberate PA1b (a member of the knottin family) and PA1a albumins (23), whereas in the C. ternatea albumin-1 gene, a cyclotide domain has replaced the PA1b knottin domain. Despite being encoded within its unusual gene architecture, Cter M, the best characterized cyclotide from C. ternatea, shares the key structural feature of a CCK motif with cyclotides derived from dedicated cyclotide genes. Interestingly, another of the isolated peptides from C. ternatea is identical in primary sequence to a previously reported cyclotide, Psyle F from Psychotria leptothyrsa from Rubiaceae (24).
Although their gene expression does not appear to be dynamically regulated (25), cyclotides are known to be differentially expressed within a plant. In Viola hederacea, cyclotide vhr1 is specific for only root tissue (26), whereas in O. affinis, Oak4 expression and its encoded peptide kalata B2 were absent from root tissue (25). Recent work has demonstrated that GFP-tagged cyclotide precursors accumulate in plant cell vacuoles (27). Several studies have reported insecticidal activity in cyclotides (2, 21, 28) and provided the basis for further structure-activity studies (29), but little is known about the distribution of cyclotides within individual plant tissues.
Matrix-assisted laser desorption/ionization-mass spectrometric imaging (MALDI-MSI) is an analytical technique in which mass spectra are collected in a raster pattern across a tissue section to generate an average mass spectrum, which, when overlaid upon an image of the sample, can reveal the spatial distribution and relative abundances of analytes (30). MALDI-MSI (31) has been applied in the study of animal and human tissues as a research tool as well as in a medical diagnostic capacity in the study of disease pathology (32–34) and to monitor drug pharmacokinetics (35, 36). Recent examples of plant MALDI-MSI providing insights through spatial information include the peptide analytes of developing soybean cotyledons (37), secreted peptide hormones involved in plant development in Arabidopsis roots (38), and small-molecule glucosinolate derivatives involved in plant defense from Arabidopsis leaves (39).
Our discovery of cyclotides in Petunia arose from interrogating expressed sequence tag (EST) databases by tBLASTN with the Cter A peptide sequence, which yielded many matches to potential cyclotide-encoding transcripts. Here we describe the characterization of cyclotides in Petunia x hybrida following their isolation and tandem MS sequencing and report a novel architecture of their genes based upon cloning of three full-length cDNA clones and a wealth of other EST-derived sequences.
The discovery of cyclotides in the Solanaceae is significant and exciting because this plant family includes many crop species, including potato and tomato, two of the largest food crops by global yield with a combined world annual production of more than 450 million tons. Given the demonstrated potent insecticidal activity of isolated cyclotides (2, 21, 28), knowledge of the Solanaceae cyclotide gene architecture might enable their expression in important food crops to potentially provide crops protection from predation by herbivores. The combination of MALDI-MSI on cyclotide expression and localization in petunia and MS analysis of leaf region extracts provided evidence of non-uniform distribution of the major cyclotide mass consistent with location in the vasculature of the leaf. A vascular location is common for small molecule (39), physical (40), and peptidic (41–43) herbivory defense systems and would allow an additive role for cyclotides in reducing predation by herbivores. Exploitation of Solanaceae cyclotide genes might thus allow production of novel, ultrastable therapeutics, lead to the enhancement of the staple crops as “functional foods” (44–46), and/or reduce crop losses to insect attack.
Previous studies investigating the expression of the cyclotide-encoding gene Oak1 from Rubiaceae in the model plant Nicotiana benthamiana reported the production of mainly misprocessed peptides (27, 47). The discovery of a cyclotide-encoding gene from the Solanaceae has great potential to improve the value of N. benthamiana as a research tool to study cyclotide processing and also to study the effects of cyclotides in plant defense.
EXPERIMENTAL PROCEDURES
Materials
P. x hybrida seedlings were sourced from Pohlmans Nursery (Gatton, Queensland, Australia). Solid phase extraction cartridges and reverse phase HPLC columns were from Grace Vydac. All solvents and enzymes were supplied as described previously (20).
Extraction
P. x hybrida plants were rinsed extensively with distilled water to remove soil prior to separation of the various plant tissues. Fresh leaf (8.0 g) and root (5.3 g) samples were lyophilized prior to ball-milling using a Retsch MM300 homogenizer in three 30-s bursts at 25 Hz. Powdered plant samples were subsequently extracted in 60% acetonitrile (ACN), 1% formic acid with vortexing and probe sonication. Crude extracts were then centrifuged in a benchtop centrifuge at 4000 × g, and the supernatants were collected and diluted with 1% formic acid to give final solvent extract concentrations of 10% ACN, 1% formic acid, and 100 g/liter or 80 g/liter wet plant weight for leaf and root materials, respectively.
Separation
Crude extracts were separated on Grace C18-Max solid phase extraction cartridges. Briefly, cartridges were equilibrated following the manufacturer's instructions using six bed volumes of methanol followed by six bed volumes of 10% ACN, 1% formic acid. Crude extracts were applied to the cartridges and washed with six bed volumes of 10% ACN, 1% formic acid. Bound peptide components were eluted from the cartridges in a stepwise fashion, using increasing concentrations of ACN in 1% formic acid. Alternatively, crude extracts were subjected to preparative HPLC using a Grace Vydac C18 reverse phase HPLC column (250 × 20 mm, 300 Å, 15-μm particle size) with a linear 1%/min ACN gradient as supplied by a Shimadzu LC-2010 HPLC system. Eluent was monitored at 214 nm, and fractions were collected manually.
Enzyme Digestion
Enzymatic digestion of reduced and alkylated cyclotides was carried out prior to tandem MS analyses as described previously (20). Briefly, cyclotides were cleaved to produce linearized fragments following reduction and alkylation to prevent reoxidation. Lyophilized crude leaf extract (1 mg) was reconstituted in 150 μl of 100 mm ammonium bicarbonate (pH 8.0) and reduced by the addition of 15 μl of 100 mm dithiothreitol and incubated at 60 °C for 30 min under nitrogen gas. To alkylate the sample, 15 μl of 250 mm iodoacetamide was added, and the mixture was incubated for 60 min at room temperature. The alkylated sample was digested by the addition of 20 μl of 400 ng/μl endoproteinase Glu-C (P2922, Sigma) and incubated at 37 °C for 18 h. Each sample was quenched with 20 μl of 5% formic acid and stored at 4 °C until further analysis.
MALDI-MSI
Leaf and stem cryosections were applied directly to indium tin oxide-coated glass slides (Bruker) and dried in a vacuum desiccator before undergoing further washing and dehydration through submersion in cold 70% (v/v) isopropyl alcohol for 30 s, and cold 96% (v/v) isopropyl alcohol for 15 s before being returned to vacuum to dry for 20 min. Washed slides were observed under an Olympus SZX7 stereomicroscope. α-Cyano-4-hydroxycinnamic acid matrix was prepared at a concentration of 7 mg/ml in 50% ACN, 0.2% trifluoroacetic acid and misted onto the surface of the dried sample slides using a Bruker Daltonics ImagePrep matrix sprayer. Sample slides were clamped into a Bruker Daltonics MTP Slide Adapter II MALDI plate and analyzed using a Bruker Daltonics UltraFlex III MALDI-TOF instrument running flexImaging version 2.1 software. Spectra were collected in linear positive ion mode using a 100-μm raster across the leaf section over a mass range of 2500–5000 Da with signals of <1800 Da suppressed to remove matrix and polymer peaks. Following data analysis in flexImaging, positions on the leaf section corresponding to peaks of interest were respotted manually with two applications of the matrix solution prior to manual collection of MALDI-TOF spectra in reflectron positive ion mode using flexControl software. Localization of specific m/z values was determined over a window of ±5 Da centered on the peak maxima.
MALDI-TOF MS
MALDI-TOF analyses were conducted using an Applied Biosystems 4700 TOF-TOF Proteomics Analyzer. Samples were spotted 1:1 with matrix consisting of 5 mg/ml cyano-4-hydroxycinnamic acid in 50% (v/v) ACN, 1% (v/v) formic acid directly onto a stainless steel MALDI target. MALDI-TOF spectra were acquired in reflector positive operating mode with the following parameters: source voltage set at 20 kV, Grid1 voltage at 12 kV, mass range 800–5000 Da, focus mass 3000 Da, collecting 2000 shots using a random laser pattern and with a laser intensity of 5000. Spectra were externally calibrated as described previously (48) by spotting cyano-4-hydroxycinnamic acid matrix 1:1 with the ProteoMass MSCAL1 peptide and protein MALDI-MS calibration kit calibration mixture (Sigma) diluted 1:400.
Static Nanospray
Reduced and endoproteinase Glu-C-digested samples were subject to a cleanup step using C18 ZipTips (Millipore) to remove salts and elicit a solvent exchange from aqueous solution to 80% ACN, 1% formic acid. Samples (3 μl) were transferred to nanospray tips (Proxeon, ES380), and nano-electrospray ionization was induced with a voltage differential of 900 V applied to the tip on a QSTAR Pulsar i QqTOF mass spectrometer (Applied Biosystems). TOF spectra were collected over the range m/z 400–2000. Product ion spectra were collected (m/z 100–2000) using collision energy voltages ranging from 10 to 60 V. Both TOF and product ion data were acquired using Analyst QS 1.5 software, and tandem MS spectra were manually assigned.
Nano-LC-MS/MS
Reduced, carbamidomethylated, and endoproteinase Glu-C-digested (linearized) cyclotide-containing crude extracts were analyzed via LC-MS/MS with conditions as described previously (21).
Cryosectioning
Petunia leaf tissue was prepared for MALDI imaging with minor changes to a method described previously (37). Samples were cryosectioned using a Leica CM3050 cryotome with chamber temperature set at −19 °C and object temperature set at −17 °C. Frozen leaf tissue was floated on the surface of optimal cutting temperature (OCT) medium applied to the cryotome chuck and paradermal (adaxial longitudinal) cryosections sampled at 15-μm thickness.
LC/MS Analysis
The relative quantitation of Phyb A (m/z 3069) among leaf parts was performed using a method described previously (39) with some modifications. Leaves were removed from P. x hybrida plants, flash-frozen in liquid N2, and lyophilized. These leaves were subsequently dissected to yield midvein, lamina, and peripheral leaf tissue samples, which were then weighed in separate tubes, and 500 μl of water was added per mg of dried plant tissue. Sealed sample tubes were placed into a heater block set at 95 °C for 75 min, cooled to room temperature, and centrifuged at 4000 × g for 10 min. Sample supernatants were introduced to a QSTAR Pulsar i QqTOF mass spectrometer (Applied Biosystems) equipped with a Turbospray ionization source, using an Agilent 1100 binary HPLC system (Agilent). Reversed phase separation of peptide analytes was achieved using a linear gradient comprising solvent A (0.1% formic acid) and solvent B (90% ACN, 0.1% formic acid (aqueous)) at a flow rate of 200 μl/min applied to a Jupiter C18 300-Å column (Phenomenex) of dimensions 150 mm × 2.0 mm with a particle size of 5 μm. TOF spectra were collected over the range m/z 400–2000 and analyzed using Analyst QS 2.0 software.
Cloning of PETUNITIDE Genes
We used tBLASTN to search the NCBI dbEST database with the amino acid sequence of Fabaceae cyclotide Cter A (GVIPCGESCVFIPCISTVIGCSCKNKVCYRN). Several Petunia ESTs matched with Expect (E) values of ≤1e7, including FN039431, FN022914, FN016979, FN020412, FN022913, FN039432, FN017679, and FN017678. Further analysis of these ESTs indicated that they were likely to encode cyclotides. These sequences were used to retrieve similar Petunia EST sequences. An alignment of the following 32 EST sequences was made: JI334153, JI334152, JI361658, JI335590, JI335591, DC242826, FN035504, FN039432, FN022913, FN017679, JI335742, FN006075, FN000250, FN001371, JI333116, FN002680, FN020915, FN001731, FN004316, FN003549, FN001318, FN005840, FN004550, FN005530, FN035505, FN039431, FN022914, FN016979, FN020412, FN017678, FN020916, and FN021459. Based on this alignment, there appeared to be several different cyclotide-encoding genes in Petunia. Against this alignment, primers that would amplify complete cyclotide domains were designed for 5′- and 3′-RACE: JM532 (5′-CAT ATA TAT GCC CCT CTC CT-3′) for 5′-RACE bound downstream from the stop codon in 19 of 32 sequences aligned; JM533 (5′-GAC GCA CGC GTA ATG GAT-3′) for 3′-RACE bound 30 bp upstream of the region encoding the cyclotide domain in 23 of 32 sequences aligned; JM534 (5′-TCA CGT GTG TTT CTG CCA CT-3′) for 3′-RACE bound 4 bp upstream of the region encoding the cyclotide domain in 15 of 32 sequences aligned; and JM535 (5′-TGT CAC GTG TGT TTC TGC AA-3′) for 3′-RACE bound in a similar location to JM534 that is 6 bp upstream of the cyclotide domain but was specific for a different group of 6 among the 32 sequences aligned.
RNA was extracted from the leaves, flowers, and roots of P. x hybrida using phenol/chloroform extraction, and selective precipitation of RNA was performed using lithium chloride as described previously (49). Between 500 ng and 1 μg of total RNA was used to create 5′- and 3′-RACE libraries using the SMARTer RACE cDNA amplification kit (634923, Clontech) as per the manufacturer's instructions. The three 5′-RACE libraries were PCR-amplified using JM532, whereas the three 3′-RACE libraries were PCR-amplified using JM533, JM534, and JM535. The 5′- and 3′-RACE products were cloned into pGEM-T (Promega), sequenced, and aligned. These partial sequences suggested that up to five different transcripts had been amplified. Using the transcript sequences tentatively named PETUNITIDE1 to -5, we designed the following primers in the 5′- and 3′-UTRs that would be specific and amplify the full ORF: JM549 (5′-CAT ACT CAG TGA TTT CCC ACC A-3′) bound to the 3′-UTR of the transcript tentatively named PETUNITIDE1; JM552 (5′-CAT ACT CAG CTA CAC ATA GTG C-3′) bound to the 3′-UTR of PETUNITIDE3; JM550 (5′-CAT ACC CAG TGA TTT TCC ACC A-3′) bound to the 3′-UTR of PETUNITIDE4; JM551 (5′-CAT ACC CAG TAA TTT CCT ACC A-3′) bound to the 3′-UTR of PETUNITIDE5; JM554 (5′-GCC AGC TAC ACA TAG TGC T-3′) bound to the 3′-UTR of most PETUNITIDE transcripts and is upstream from the binding sites of JM550-JM552; and JM553 (5′-TGG CAA AGA TAA TAC TTT CA-3′) bound to the 5′-UTR of both PETUNITIDE2 and PETUNITIDE4.
PCR amplification of the aforementioned 5′- and 3′-RACE libraries with these primers yielded products of the expected sizes that were subsequently cloned into pGEM-T and sequenced. This sequencing revealed three different PETUNITIDE transcript products each encoding a full ORF. For each PETUNITIDE transcript, at least three independent clones were obtained.
RESULTS
Searching for Cyclotide Sequences in Petunia Transcript Databases
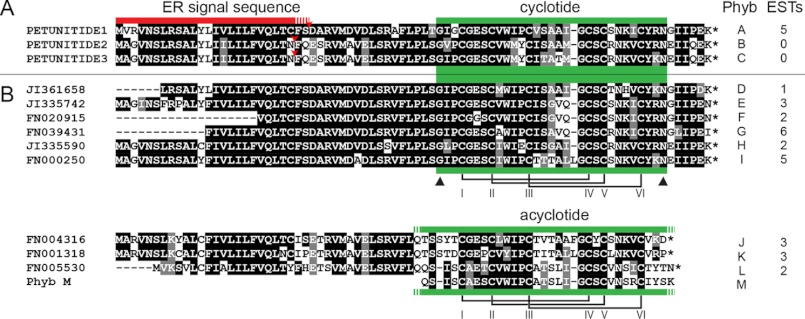
Structurally homologous cyclotides have previously been characterized from plants of the Rubiaceae, Violaceae, and Fabaceae families, with the investigated species typically having been selected on the basis of an identified bioactivity. To search for potential cyclotide-encoding genes within publicly available bioinformatic data, Fabaceae cyclotide Cter A was used as a tBLASTN search string to interrogate the EST database at NCBI. Numerous putative cyclotide-encoding ESTs from the genus Petunia matched the submitted protein sequence (summarized in Fig. 1, with their accession numbers under “Experimental Procedures”). We named these putative petunia cyclotides Phyb A through Phyb L in accordance with a previously established convention (50). PETUNITIDE1 to -3 and the related ESTs appeared to encode precursor proteins possessing an endoplasmic reticulum target signal and, toward the end, a cyclotide domain containing six cysteines of typical spacing, the highly conserved Glu in loop 1, a proto-N-terminal Gly, and the usual proto-C-terminal Asx (i.e. Asn or Asp). This arrangement differs from cyclotide precursors from the Violaceae and Rubiaceae, which have longer regions between the signal peptide region and the mature cyclotide domain(s), making the PETUNITIDE proteins essentially the same size as very recently described precursors from the Rubiaceae plant Chassalia chartacea, which are much shorter than previously described cyclotide precursors (supplemental Fig. S1).
FIGURE 1.
BOXSHADE alignment of cyclotide precursors in genus Petunia. A, PETUNITIDE sequences. B, consensus sequences derived from matched ESTs. Asterisks at the C termini of sequences indicate stop codons. Red bar denotes predicted signal motifs, and green bars denote cyclotide or acyclotide domains. Triangles denote prototerminal amino acids of encoded cyclotides. Disulfide connectivity is based upon previously characterized cyclotides. Sequences translated from EST data only are listed in supplemental Table S1 (note paired clones (F + R)).
Commonly trailing the cyclotide domains' proto-C-terminal Asx is an Ile or Leu located two residues downstream (at P2′). This residue is consistently observed among the corresponding regions of Violaceae, Rubiaceae, and Fabaceae cyclotide genes and appears to be an important residue for processing (51). In previously reported cyclotide genes, the amino acid at P1′ is typically a Gly, but the Solanaceous precursors exhibit either a Gly or Glu. We also observed that some ESTs encode PETUNITIDE proteins that are punctuated with stop codons immediately following their cyclotide domains (Fig. 1; encoding putative Phyb J, Phyb K, and Phyb L). What effect this has on peptide maturation remained to be determined by analyzing the peptide profile of petunia. A list of ESTs that would encode the putative mature cyclotides and acyclotides is given in supplemental Table S1.
Detection and Sequencing of Cyclotides and Acyclotides from P. x hybrida
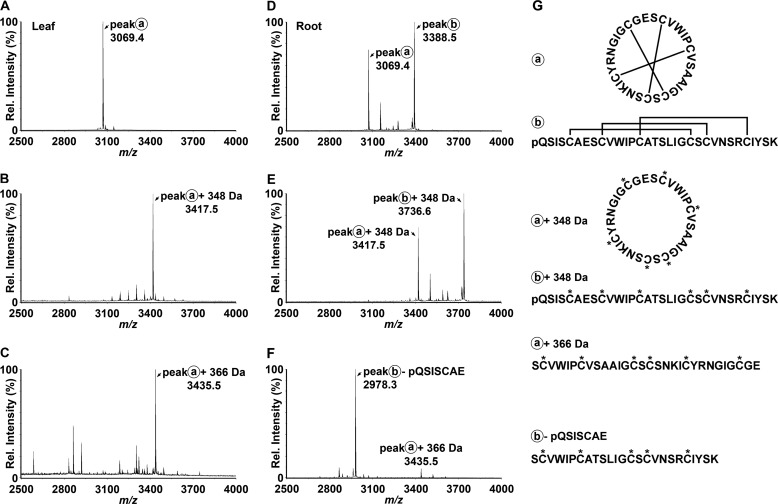
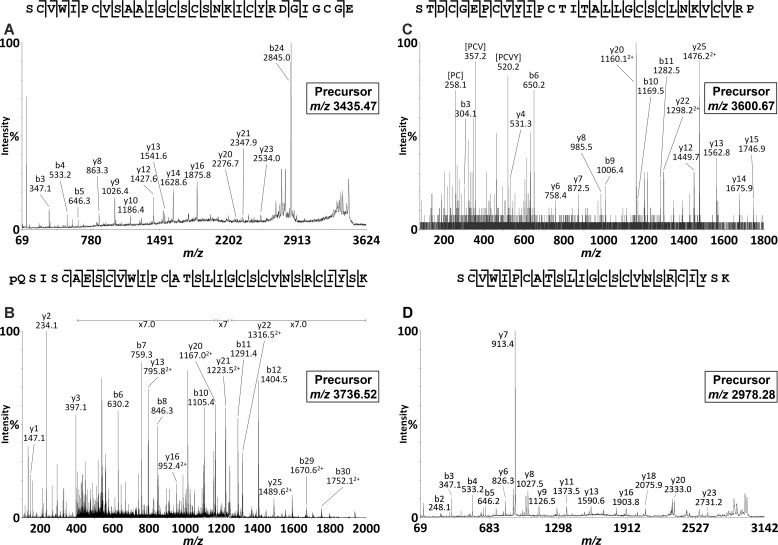
To confirm the synthesis and accumulation of the predicted cyclic and acyclic peptides, we prepared extracts of various P. x hybrida tissues and analyzed these directly using MALDI-TOF MS. As shown in Fig. 2, A and D, both leaf and root extracts exhibited signals within the mass range m/z 2800–3700 typical of cyclotides. A dominant signal observed at m/z 3069 (peak A) appeared in both leaf and root extracts and was consistent with the mass of the predicted cyclotide domain of PETUNITIDE1. After reduction and alkylation, a mass 348 Da larger than peak A was observed at m/z 3417 (Fig. 2B), consistent with the alkylation of six cysteine residues. Following digestion with endoproteinase Glu-C, an additional increase in mass of 18 Da was observed. This signal corresponded to peak A + 366 Da and appeared at m/z 3435 (Fig. 2C), consistent with a single peptide backbone cleavage, as would be expected for a cyclotide. Tandem MS characterization confirmed the identification of the peak at m/z 3435 as the bracelet cyclotide Phyb A with sequence SCVWIPCVSAAIGCSCSNKICYRNGIGCGE, in agreement with the sequence of PETUNITIDE1 (Fig. 3A).
FIGURE 2.
MALDI-TOF MS of petunia leaf and root extracts reveals putative cyclotide masses. Following reduction of disulfide bonds, free thiols were alkylated, and peptide backbones were enzymatically digested with endoproteinase Glu-C. A, native leaf extract. B, reduced and carbamidomethylated leaf extract. C, reduced, carbamidomethylated, and enzymatically digested leaf extract. D, native root extract. E, reduced and carbamidomethylated root extract. F, reduced, carbamidomethylated, and enzymatically digested root extract. G, the structures of native, alkylated, and digested Phyb A (peak a) and Phyb M (peak b) are schematically represented beside their corresponding spectra.
FIGURE 3.
MALDI-TOF/TOF analyses of cyclotide and acyclotide species isolated from petunia root. A, tandem MS analysis of the m/z 3435.47 precursor (Phyb A). B, tandem MS analysis of the m/z 3736.52 precursor (Phyb M). C, tandem MS analysis of the m/z 3600.67 precursor (Phyb K). D, tandem MS analysis of the m/z 2978.28 precursor (Phyb M fragment).
The other dominant peak observed in MALDI-TOF analyses of root extracts appeared at m/z 3388 (peak B), but no corresponding peak at +366 Da was observed in the endoproteinase Glu-C-digested sample. Apart from the peak observed at m/z 3435 (Phyb A), the dominant signal in the spectrum from endoproteinase Glu-C-digested root extract was a peak at m/z 2978 (Fig. 2F). Examination of the TOF-MS spectrum of the reduced and alkylated extract revealed a dominant peak at m/z 3736 (Fig. 2E) corresponding to the addition of 348 Da (alkylation of the six Cys residues) to the peak at m/z 3388 in the native root extract. Following endoproteinase Glu-C digestion, the peak at m/z 3736 was noticeably absent, suggesting that proteolysis had occurred. A peak at m/z 3754 would be expected following cleavage of a cyclic peptide backbone (at Glu in loop 1). However, no such peak was observed, raising two possibilities: 1) that the peptide contained more than the single Glu in loop 1 and/or 2) that the peptide was linear and was therefore cleaved into more than one fragment. Under MS/MS conditions, cyclic peptides typically do not fragment as readily as linear peptides. Extensive fragmentation of the precursor at m/z 3736 (Fig. 3B) in tandem MS indicated its peptide backbone to be non-cyclic. The assigned peptide sequence was homologous to the cyclotide-like gene product predicted by EST FN005530, with sequence pQSISCAESCVWIPCATSLIGCSCVNSRCIYSK, which we named Phyb M, and was found to incorporate a pyroglutamyl modification at its N terminus. The dominant peak at m/z 2978 observed after endoproteinase Glu-C digestion was subjected to tandem MS analysis, revealing its identity as the C-terminal portion of Phyb M.
During LC-MS/MS analysis of reduced, alkylated, and endoproteinase Glu-C-digested root extracts, another acyclic peptide was observed (Fig. 3C) with a parent ion mass and fragmentation pattern matching the cyclotide sequence encoded by EST FN001318 (STDCGEPCVYIPCTITALLGCSCLNKVCVRP) and which we named Phyb K. The masses of the characterized as well as putative cyclotides are reported in Table 1. Fig. 4 illustrates an alignment of Phyb A with the cyclotide sharing the highest sequence homology, cycloviolacin O17 from Viola odorata, along with the petunia acyclotides Phyb K and Phyb M, demonstrating the conserved cysteine spacing.
TABLE 1.
Sequence alignment of cyclotides observed in P. x hybrida extracts
| Peptide | Amino acid sequencea,b | Expt.c m/z | Expt.c mass | Theor.c mass | Errorc | Theor.d mass |
|---|---|---|---|---|---|---|
| Da | Da | ppm | Da | |||
| Phyb A | -GIGCGESCVWIPCVS-AAIGCSCSNKIC-YRN | 3434.47 | 3434.47 | 3434.55 | −23.29 | 3068.29 |
| Phyb K | -STDCGEPCVYIPCTITALLGCSCLNKVC-VRP | 1200.89 | 3599.67 | 3599.71 | −11.11 | 3251.46 |
| Phyb M | pQSISCAESCVWIPCAT-SLIGCSCVNSRCIYSK | 1246.17 | 3735.51 | 3735.73 | −58.89 | 3387.47 |
a Ile and Leu in Phyb M were assigned on the basis of homology to Phyb L.
b Phyb K and Phyb M are acyclic.
c Expt., experimental; Theor., theoretical; linearized peptide masses after reduction and carbamidomethylation of cysteines.
d Theor., theoretical; native peptide masses.
FIGURE 4.
Alignment of cylcoviolacin O17 with cyclotide Phyb A (86% homology) and acyclotides Phyb K and Phyb M from P. x hybrida. Disulfide connectivity based upon previously characterized cyclotides is denoted by solid lines between boxed cysteines, and intercysteine loops are indicated. Loop 6 is not present in Phyb K or Phyb M, which are both acyclic.
Judging from LC-MS analyses, the abundance of petunia cyclotides in source plant material was within the range previously reported for cyclotides in V. odorata and O. affinis (52), with Cter A in wet leaf material estimated to be 30.0 μg/g, whereas Cter K and Cter M were present in wet root material at 2.3 and 7.6 μg/g, respectively.
Determination of Cyclotide Distribution in Petunia Leaf Tissue
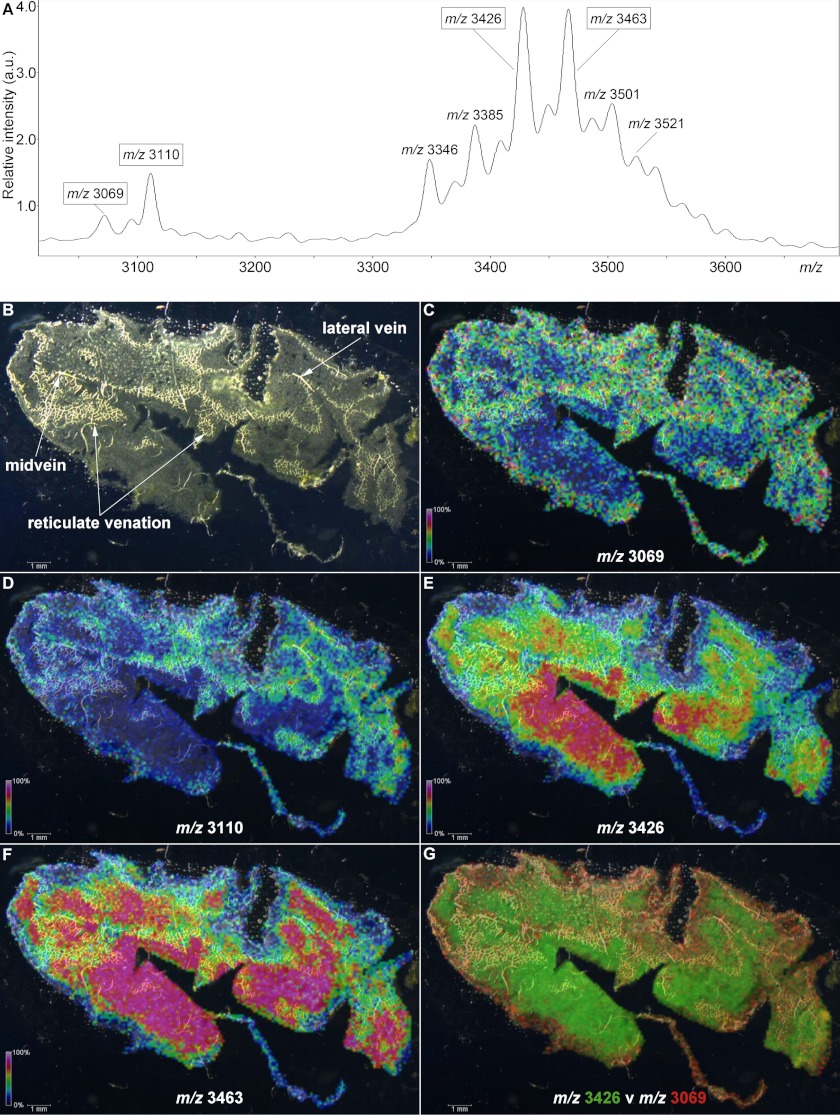
To examine the spatial distribution of petunia cyclotides within plant tissue, we used MALDI-MSI to analyze a paradermal leaf section, generating an ion intensity map. As shown in the average mass spectrum in Fig. 5A, numerous peaks were detected in the range m/z 3000–3600. Analysis of leaf extracts showed a single dominant peak at m/z 3069, which was sequenced and named Phyb A (Fig. 2A), and this peak was observed in the MALDI-MSI experiment along with peaks at m/z 3110, 3426, and 3463. Apart from m/z 3069, none of these other masses corresponded to isolated or predicted cyclotides (Figs. 1 and 2). LC-MS/MS analysis of leaf extracts was undertaken to determine if the MALDI-MSI peaks observed at m/z 3110, 3426, and 3463 were cyclotides; however, only a precursor at m/z 3424 (corresponding to m/z 3426 average mass in MALDI-MSI) was observed. Limited fragmentation of this precursor was observed, but following a reduction step, a mass increase of 2 Da was observed (supplemental Fig. S2, A and B), indicating the presence of a single intramolecular disulfide bond, and leading to extensive fragmentation in subsequent tandem MS (supplemental Fig. S3). The sequence resulting from manual de novo mass spectral interpretation (DEEPKRGTPEAKKKYSSVCVTNPTARICRY) was used in a BLAST search and found to be consistent with a translated EST (FN008610) from petunia encoding a sequence homologous to nuclear Photosystem II 5-kDa protein (PSII-T) described in other plant species. This identification was further bolstered through the observation of a 210-Da increase in mass following acetylation with acetic anhydride, consistent with modification of the four Lys side chains as well as the N-terminal primary amines in the native peptide (supplemental Fig. S2C).
FIGURE 5.
MALDI-MSI of a petunia leaf. The localizations of four distinct m/z signals are indicated with intensity scales relative to average spectrum maximum inset. A, average mass spectrum for the data acquired from the leaf section. B, dark field microscope image of a paradermal (adaxial longitudinal) cryosection of P. x hybrida leaf. C, localization of m/z 3069. D, localization of m/z 3110. E, localization of m/z 3426. F, localization of m/z 3463. G, overlay of m/z 3426 (green) and m/z 3069 (red) signals.
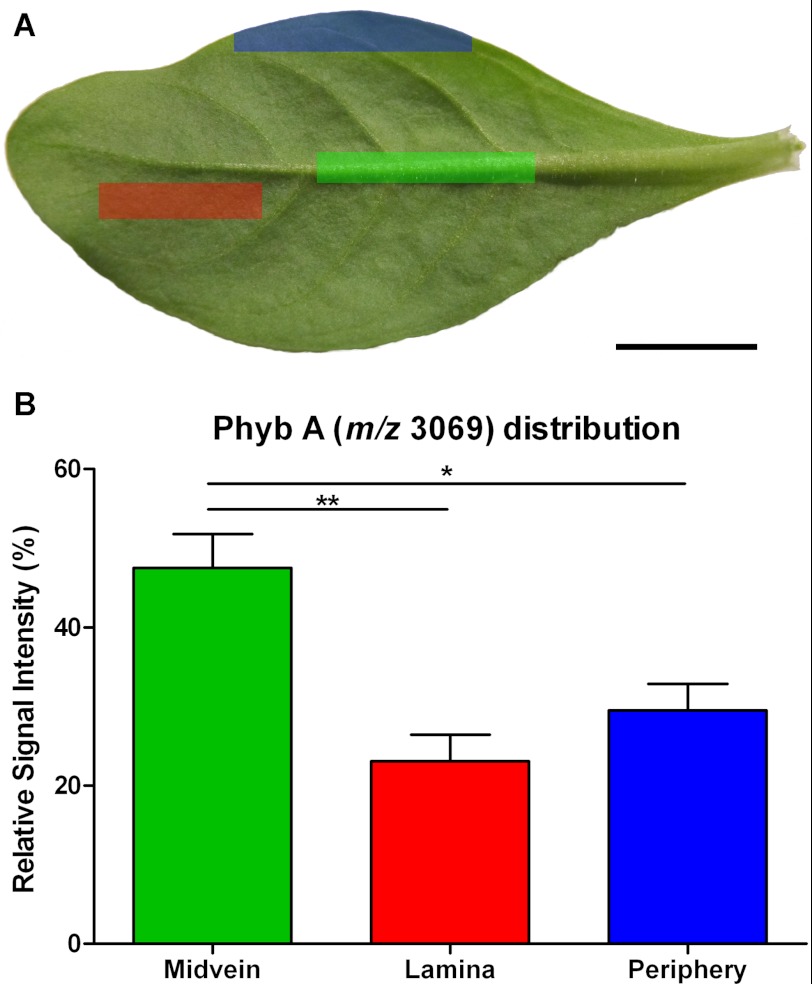
Fig. 5B highlights multiple vascular features observed in a dark field microscopy image of the leaf section analyzed in this experiment. In Fig. 5, C–F, the relative signal intensities of selected peak maxima (± 5 Da) ranging from 0% (black) to 100% (white) are superimposed upon the dark field leaf image (Fig. 5, C–F). Areas of increased signal intensity for m/z 3069 and 3110 peaks appeared to overlay with the vascular features (Fig. 5, C and D), whereas the spatial distributions and relative intensities of m/z 3426 and 3463 signals were not (Fig. 5, E and F). Signals for m/z maxima observed in the average spectrum, including the examples in Fig. 5, appeared to be differentially distributed across the sample section and localized to distinct regions, with no evidence of “hot spots” or smearing. In Fig. 5G, the relative intensities of signals for m/z 3426 and m/z 3069 are indicated over a range from transparent (0%) to bright green or red (100%), respectively, and co-localization is indicated by yellow coloration. The display of distinct green and red areas indicated heterogeneous expression patterns, with the strongest signals for m/z 3426 appearing to present within areas upon the leaf section with the least vasculature, whereas for m/z 3069, the reverse is true. To confirm the localization pattern observed for m/z 3069 in the MALDI-MSI experiment, the relative quantitation of Phyb A was determined via LC/MS in extracts of dissected petunia leaves. Approximately 2-fold higher concentrations of m/z 3069 (Phyb A) were detected in the midvein, compared with both the lamina and periphery of petunia leaves (Fig. 6B). Sixteen control signals were selected from the LC/MS data, including m/z 3424 (PSII-T), and their relative concentrations were similarly compared across leaf regions. In each case, there was either no statistically significant difference in their concentrations across the leaf or increased concentrations in the lamina or periphery (or both) compared with the midvein extracts (supplemental Fig. S4).
FIGURE 6.
Relative quantitation of Phyb A across petunia leaf regions. A, representative petunia leaf highlighting the regions sampled for LC/MS analysis. Green highlight, midvein; red highlight, lamina; blue highlight, leaf periphery. B, relative extracted ion chromatogram peak intensities for signal at 1535.02+ in LC/MS analyses (corresponds to m/z 3069 in MALDI experiments) of boiled plant extracts. Error bars, S.E. (n = 10). Asterisks denote significant differences at 0.001 < p < 0.01 (**) or 0.01 < p < 0.05 (*) following one-way analysis of variance using Bonferroni's multiple comparison test. Scale bar, 1 cm.
DISCUSSION
Here we report the discovery and characterization of cyclotides from P. x hybrida of the agronomically important Solanaceae plant family. These peptides arise from the shortest known cyclotide precursors and are distinct from previously known precursors. This is the fourth architecturally distinct precursor from which cyclotides emerge, provoking interesting questions about the evolutionary origin of their structurally identical CCK framework peptides. The new precursors present opportunities for designing synthetic peptides capable of being cyclized efficiently in planta for a range of agricultural or pharmaceutical applications. Furthermore, we have confirmed enrichment of a cyclotide in the vasculature of leaves, a finding that is consistent with a proposed general role of cyclotides in herbivory defense.
Existence of Cyclotides in the Solanaceae
The discovery of cyclotides within the Solanaceae plant family is an exciting and important development, given the significance of this plant family to human nutrition, and follows the recent landmark discovery of cyclotide genes in a member of the Fabaceae family (21, 22). The Solanaceae is host to more than 3000 species, including staple crops, such as Solanum tuberosum (potato) and Solanum lycopersicum (tomato), which constitute two of the most important vegetable crops cultivated, with combined worldwide annual production exceeding 450 million tons.
Plant species previously investigated in the search for cyclotides have typically been selected on the basis of an identified bioactivity in their extracts, such as uterotonic activity in O. affinis (53) and C. ternatea (20), anti-HIV activity in Palicourea condensata (54), hemolytic activity in Viola extracts, and trypsin inhibitory activity in Momordica cochinchinensis (12). In the current study, we examined P. x hybrida following the identification of ESTs from the genus Petunia via a database search. Further experiments confirmed petunia cyclotides to be the products of dedicated genes with a novel precursor structure.
Structural and Evolutionary Implications of the Novel Precursors from Genus Petunia
The sequences of three cyclotide-encoding genes, named PETUNITIDE1 to -3, are shown in Fig. 1 alongside the translated amino acid sequences deriving from the BLAST-matched ESTs, where they encode precursor proteins of 79 residues comprising an endoplasmic reticulum signal sequence, a pro-region of 15 residues, a single cyclotide-encoding domain, and a six-residue C-terminal tail sequence. A distinguishing feature of Solanaceae cyclotide precursors is their relatively short (15 residue) N-terminal pro-regions compared with those from Rubiaceae (22–69 residues) and Violaceae (28–45 residues) cyclotide genes. In combination with their short C-terminal tails, the Solanaceae cyclotides are encoded by relatively short cyclotide-encoding precursors, similar in size to recently reported atypical cyclotide precursors from Rubiaceae (55).
Some of the BLAST-matched Petunia ESTs appeared to terminate with stop codons directly C-terminal to the cyclotide-encoding domains, indicating that acyclic cyclotides (“acyclotides”) might be produced in planta. Accordingly, we characterized a peptide matching one of these predicted acyclic ESTs.
The first acyclotide characterized was violacin A from the Violaceae plant Viola odorata, which we referred to at the time as a “linear cyclotide” (56). Later, in O. affinis, the transcript Oak9 was found to encode kalata B20-lin, an acyclotide seemingly arising from a single nucleotide change that introduces a stop codon (25). In two recent studies of Rubiaceae plants Hedyotis biflora and Chassalia chartacea, panels of novel “linear cyclotides” were characterized and referred to as “uncyclotides” (55, 57). We prefer the term “acyclotide” for the following two reasons. 1) This is in keeping with established practices in nomenclature of organic compounds as either cyclic or acyclic (58). 2) Selectional restrictions on English language prefixes mean that the “un-“ prefix can be taken to confer two meanings (cf. “unlockable”), and when added to the word “cyclic,” the resultant “uncyclic” can be construed to convey that the item being described is “not cyclic” or alternatively that it is “no longer cyclic.” Thus, the “a-“ prefix is unambiguous and conveys only one meaning to “acyclic”: that the item is “not cyclic.” Interestingly, in some cases, the acyclotides have biological activity comparable with that of their cyclic counterparts (53), but in most cases, the linear homologues are devoid of the activity of the cyclic forms (59, 60).
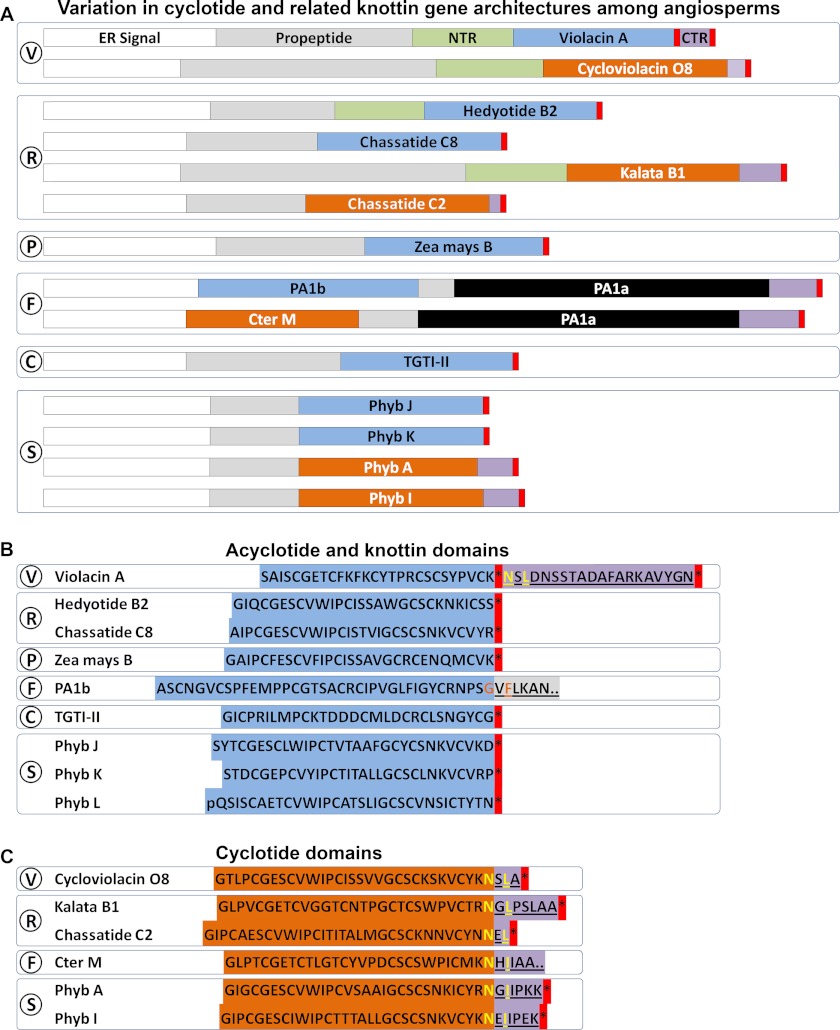
Fig. 7 illustrates a comparison of the gene structures of representative cyclotide and related knottin- or acyclotide-encoding sequences, in which PETUNITIDE genes, in terms of overall structure and size, can be seen to bear the most similarity to recently characterized CHASSATIDE genes identified within Rubiaceae plant Chassalia chartacea (55). Despite the lack of peptide evidence for cyclotide-like sequences in Poaceae, they are likely to be produced as acyclic peptides due to their truncation by a predicted stop codon, as illustrated for “Zea mays B,” and in this way bear similarity to genes encoding putative petunia acyclotides, including Phyb J, K, and L. Peptide evidence was found for hedyotide B2, an acyclotide found in Rubiaceae plant H. biflora, and the gene encoding it was found to have been truncated at the C terminus of the cyclotide domain by a stop codon (57), with the remainder of the gene exhibiting homology to other Rubiaceae cyclotide genes as indicated in Fig. 7, A and B. Other acyclotides have also been characterized from Rubiaceae plants, including kalata B20-lin from O. affinis (25) and Psyle C from P. leptothyrsa (24).
FIGURE 7.
Comparison of prototypic cyclotide and acyclotide-encoding gene structures in angiosperms. A, variation in cyclotide and related knottin gene architectures among angiosperms. B, acyclotide and knottin domains. C, cyclotide domains. Signal sequence is shown in white boxes. Knottin and acyclotide domains are shown in light blue boxes. Cyclotide domains are shown in orange boxes. N-terminal pro-domains are shown in green boxes. C-terminal repeats are shown in mauve boxes. V, Violaceae; R, Rubiaceae; P, Poaceae; F, Fabaceae; C, Cucurbitaceae; S, Solanaceae.
A single example of a linear cyclotide, violacin A, has been described in the Violaceae plant V. odorata (56). However, the gene encoding violacin A is unique compared with other acyclotide-encoding genes in that the premature stop codon does not appear after the entire peptide domain but rather appears to truncate an otherwise complete cyclotide gene. In this case, the nucleotide sequence immediately following the stop codon is replete with sequence that would encode typical proto-C-terminal and CTR amino acids, suggesting that violacin A might be the result of a single nucleotide polymorphism.
Solanaceae cyclotides are encoded by PETUNITIDE genes that incorporate sequence motifs considered integral for cyclotide biosynthesis and backbone cyclization in previously described cyclotide genes (51), including a proto-C-terminal Asx followed by a hydrophobic amino acid two residues C-terminal (e.g. –Asx-Xaa-Leu/Ile/Val–). It has been posited that an asparaginyl endopeptidase (AEP) would be the logical candidate enzyme driving cyclotide biosynthesis (47, 51), due to the demonstrated in vitro cleavage and transpeptidation (ligation) activity of jackbean AEP to produce mature concanavalin A (61) and its activity at a wide range of Asx-Xaa bonds (62). A large body of work has demonstrated that AEP can mature seed storage globulins and albumins (63–68). In sunflowers (Asteraceae), a gene encoding a napin-like preproalbumin storage PawS1 albumin gives rise to mature seed storage albumin as well as small backbone-cyclized trypsin inhibitor embedded upstream of the albumin (69). Following transformation of PawS1 into an Arabidopsis aep null mutant, it was determined that AEP was required for cleavage reactions at the proto-N terminus of SFTI, the proto-C terminus of SFTI, and the proto-N terminus of the PawS1 small albumin subunit (69), and based on this, AEP was proposed as a good candidate enzyme for mediating ligation of N and C termini of SFTI-1. This might occur through attack of the thioester acyl intermediate of AEP by the freed glycine of SFTI-1, held close to the thioester by the disulfide bond (61).
Recently, it was discovered that the butterfly pea (C. ternatea) contains pea albumin-1-like genes in which a cyclotide domain has “replaced” the first of the PA1 domains, and this cyclotide domain is trailed by residues that would enable bioprocessing via the same AEP-mediated mechanism (21, 22).
One of the peptides characterized in the current study, Phyb M, with the sequence pQSISCAESCVWIPCATSLIGCSCVNSRCIYSK is supported by EST FN005530 and incorporates a post-translational N-terminal pyro-Glu modification. The first pyroglutamyl modification of a linear cyclotide was reported recently in hedyotide B4 from H. biflora, which was reported as a degradation product of a longer linear cyclotide, hedyotide B2 (57). We did not detect peptide masses corresponding to non-pyro-Glu Phyb M, which suggests that it is not a degradation artifact from a longer mature peptide. N-terminal pyro-Glu peptides are known to be more resistant to degradation than their corresponding N-terminal Gln homologs (70), so the incorporation of a modified N-terminal residue in lieu of backbone cyclization may be an alternative strategy to provide enhanced stability toward exopeptidase activity. Thus, Phyb M bridges an evolutionary gap between Phyb K, an acyclotide with a free N terminus, and Phyb A with its “complete” CCK motif. The discovery of all three peptide forms from P. x hybrida may represent “evolution in progress.”
Despite the isolation of PETUNITIDE2 and PETUNITIDE3 transcripts, we found no mass spectrometric evidence for peptide masses for the cyclotides they would encode (Phyb B and Phyb C; Fig. 1). The sequences of these peptides as well as those encoded within the identified ESTs and the rest of the peptides characterized in this study are mostly homologous to many previously described cyclotides and incorporate permutations of previously observed amino acids within loop regions. An exception to this is the translated sequence of ESTs FN020915 and FN020916 (GIPCGGSCVWIPCISGVQGCSCSNKICYRN), in which the absolutely conserved Glu in loop 1 (Fig. 4), present in all previously characterized cyclotides, is replaced with a Gly residue.
The potentially wider prevalence of cyclotides among Solanaceae plants remains to be elucidated; however, BLAST searches using full-length PETUNITIDE sequences to query all GenBankTM nucleotide sequences, including EST databases, revealed only matches to the Petunia ESTs reported in this study. This search confirms the uniqueness of the precursor sequence, especially considering the depth of EST coverage among members of the Solanaceae (e.g. 334384 in tobacco, 297142 in tomato, 249761 in potato, and 118054 in capsicum), and suggests that cyclotides evolved independently within the Solanaceae.
Vascular Localization and Functional Significance
Trabi et al. (26) investigated the tissue-specific distribution of a panel of cyclotides from Viola hederacea by comparing LC/MS profiles of separate tissue extracts and demonstrated that cyclotides are differentially expressed among plant tissues. This phenomenon was observed in a subsequent study of cyclotide localization in O. affinis plant tissues, which, in addition to examination of extracted peptides via LC/MS, observed no cDNA encoding kalata B2 in root tissue (25). Complementary to these studies, recent work examined the subcellular location of cyclotides during their biosynthesis, the results of which indicate that they are processed and accumulate within plant cell vacuoles (27). However, despite these advances, details on the intratissue distribution of cyclotides are lacking.
To examine the localization of cyclotides within petunia leaves, we analyzed a tissue section using MALDI-MSI and observed a number of peptide masses appearing in the mass range diagnostic of cyclotides (Fig. 5A). One of the signals observed was consistent with Phyb A and appeared to correlate with the vascular structures of the prepared leaf section (Fig. 5C). Through LC/MS analysis of dissected leaf extracts, relative quantitation of Phyb A was assessed in midvein, laminar, and peripheral leaf tissues. Phyb A was found to exist at ∼2-fold higher concentrations within midvein tissue versus laminar or peripheral leaf tissue extracts (Fig. 6B). This distribution was unique compared with the trends observed for 16 control m/z signals, which were present either in equivalent abundance across the three leaf tissue areas or in higher abundance within laminar and/or peripheral leaf tissue extracts compared with midvein extracts (supplemental Fig. S4). The size and direction of the -fold change in Phyb A abundance might be of functional significance in the context of plant defense, given a previous study of Arabidopsis thaliana in which it was demonstrated that non-peptidic plant defensive glucosinolates were enriched at the midvein and the outer lamina of leaves (39). In the Arabidopsis study, it was further observed that Helicoverpa feeding preference could be influenced by as little as 1.3-fold relative changes in the concentration of indol-3-ylmethylglucosinolate, the major glucosinolate present. The observed localization pattern for Phyb A (m/z 3069) primarily in the vasculature of the leaf section mirrors the glucosinolate study and places Solanaceous cyclotides in the right location in leaves to be potential modulators of insect herbivory.
One of the limitations of MALDI-MSI is its inherent limited dynamic range, which is instrument-, matrix-, and analyte-dependent. Few studies have quantified the dynamic range of this technique, but a recent investigation demonstrated linearity of signal intensity increasing with analyte concentration from the limit of quantitation (femtomolar) over less than 2 orders of magnitude (71). Given the relative abundance of Cter A compared with other putative cyclotide signals in leaf extract (Fig. 2A), it is therefore unsurprising that low abundance putative cyclotide signals in the extract were not detected during MALDI-MSI, where the sample had not been deconvoluted through extraction, and the analyte was rather presented to the instrument in a complete, complex sample matrix.
Additional signals were observed during MALDI-MSI analysis of the leaf section that did not correspond to any of the calculated peptide masses from PETUNITIDE genes or translated EST sequences. Signals at m/z 3426 and 3463 appeared to be abundant in areas of the leaf section distinct from m/z 3069 or 3110. Tandem MS of the major peak observed at m/z 3426 (average) in the MALDI-MSI experiment (m/z 3424 monoisotopic in ESI) following reduction of a single disulfide bond permitted de novo sequencing and its further identification as nuclear PSII-T 5-kDa protein. Although a fragment of a homologous PSII-T protein has been sequenced from spinach (72), our work demonstrates the first mass spectral evidence of any nuclear PSII-T protein and describes a previously unreported disulfide bond. Given the conserved nature of the cysteines in homologous nuclear PSII-T proteins (not shown) a disulfide bond could be expected in all such proteins. The even distribution of PSII-T among all leaf areas, as shown in supplemental Fig. S4O, is consistent with the ubiquitous nature of photosynthetic proteins in leaf tissue.
Fig. 5G illustrates a difference map of signal intensities for the m/z 3426 (PSII-T) and 3069 (Phyb A) peptides and reflects the differential spatial expression of the two masses. The differential localization of the various m/z signals from the MALDI-MSI experiment indicated that the intensity of any particular signal was not significantly influenced by cell size or density and that signals for each peptide were heterogeneous across the tissue sample. This validates the sample preparation methodology and the suitability of the technique as a whole and demonstrates its ability to provide information on the spatial relative abundance of peptide analytes.
CONCLUSIONS
Here we have demonstrated the first evidence that cyclotides and acyclotides exist within the Solanaceae plant family as the products of a novel precursor structure. This work complements previous characterizations of cyclotide-encoding genes from Violaceae, Rubiaceae, and Fabaceae plant families (73). Analysis of the Solanaceae cyclotide (PETUNITIDE) genes implicates AEP in their proto-C-terminal processing, consistent with purported biosynthetic pathways of cyclotides in the literature (47) and consistent with the demonstrated requirement for AEP in processing the cyclic sunflower trypsin inhibitor SFTI-1 (69). Petunia cyclotides and their encoding genes have residues trailing the proto-C terminus consistent with those shown previously to be important for their correct biosynthesis. Similar to CHASSATIDE genes from Rubiaceae, PETUNITIDE genes are more compact than previously known cyclotide precursors. Subtle differences between the sequence motifs flanking the mature cyclotide sequences in Solanaceae and phylogenetically distinct Rubiaceae or Violaceae precursors might explain the low yields of cyclic products following expression of both natural and designed cyclotides in Solanaceae plants (47, 51). Thus, the discovery of novel cyclotide-encoding genes within the Solanaceae family might enable their application as an alternative option for circular peptide production compared with known cyclotide genes. PETUNITIDE genes might also be employed to enhance crop protection within Solanaceae species important to human nutrition, such as potato, capsicum, and tomato, through genetic incorporation of custom cyclotide and/or acyclotide-encoding domains.
Our data demonstrate that cyclotides associate with the vascular features of petunia leaf tissues, which aligns with previously characterized small molecule and peptidic mediators of plant defense. Examples include glucosinolate (39) precursors of toxic cyanocompounds in Arabidopsis, terpenes involved in squirt-gun defenses in Bursera sp. (40), pumpkin fruit trypsin inhibitor (41), cysteine proteinase inhibitors in maize (42), and defensins in capsicum (43). Hence, the localization of increased concentrations of cyclotides in these areas could modulate herbivore feeding behavior and contribute to plant defense. This work adds to the known pool of cyclotide-producing plant families and provides an impetus for the further exploration of Solanaceae species for cyclotides. Judging from the variation in cyclotide gene structures now described, it seems likely that further significant variations will be discovered in yet to be described cyclotide-containing plant families. This combined knowledge will be crucial to understanding their evolutionary origins as either the products of convergent evolution or potentially the action of transposable elements.
Acknowledgments
We thank Alun Jones and the Molecular and Cellular Proteomics Facility (University of Queensland, Institute for Molecular Bioscience) for access to mass spectrometry instrumentation and Dr. Anne Rae (CSIRO Plant Industries) for helpful discussions.
This work was supported by an Australian Research Council (ARC) grant for cyclotide analysis (Grant DP0984390), an ARC Grant for development of MALDI-MSI in plants (Grant DP0985873), and equipment purchased under ARC Linkage Grant LE0775603 housed in the ARC Centre of Excellence in Plant Energy Biology at the University of Western Australia.

This article contains supplemental Table S1 and Figs. S1–S4.
- CCK
- cyclic cystine knot
- MSI
- mass spectrometric imaging
- EST
- expressed sequence tag
- ACN
- acetonitrile
- RACE
- rapid amplification of cDNA ends
- AEP
- asparaginyl endopeptidase
- SFTI
- sunflower trypsin inhibitor.
REFERENCES
- 1. Craik D. J., Daly N. L., Bond T., Waine C. (1999) Plant cyclotides. A unique family of cyclic and knotted proteins that defines the cyclic cystine knot structural motif. J. Mol. Biol. 294, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 2. Jennings C., West J., Waine C., Craik D., Anderson M. (2001) Biosynthesis and insecticidal properties of plant cyclotides. The cyclic knotted proteins from Oldenlandia affinis. Proc. Natl. Acad. Sci. U.S.A. 98, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Plan M. R., Saska I., Cagauan A. G., Craik D. J. (2008) Backbone cyclized peptides from plants show molluscicidal activity against the rice pest Pomacea canaliculata (golden apple snail). J. Agric. Food Chem. 56, 5237–5241 [DOI] [PubMed] [Google Scholar]
- 4. Colgrave M. L., Kotze A. C., Huang Y. H., O'Grady J., Simonsen S. M., Craik D. J. (2008) Cyclotides. Natural, circular plant peptides that possess significant activity against gastrointestinal nematode parasites of sheep. Biochemistry 47, 5581–5589 [DOI] [PubMed] [Google Scholar]
- 5. Gran L. (1973) On the effect of a polypeptide isolated from “Kalata-Kalata” (Oldenlandia affinis DC) on the estrogen-dominated uterus. Acta Pharmacol. Toxicol. 33, 400–408 [DOI] [PubMed] [Google Scholar]
- 6. Gustafson K. R., Sowder R. C., Henderson L. E., Parsons I. C., Kashman Y., Cardellina J. H., McMahon J. B., Buckheit R. W., Pannell L. K., Boyd M. R. (1994) Circulins A and B: novel HIV-inhibitory macrocyclic peptides from the tropical tree Chassalia parvifolia. J. Am. Chem. Soc. 116, 9337–9338 [Google Scholar]
- 7. Lindholm P., Göransson U., Johansson S., Claeson P., Gullbo J., Larsson R., Bohlin L., Backlund A. (2002) Cyclotides. A novel type of cytotoxic agents. Mol. Cancer Ther. 1, 365–369 [PubMed] [Google Scholar]
- 8. Witherup K. M., Bogusky M. J., Anderson P. S., Ramjit H., Ransom R. W., Wood T., Sardana M. (1994) Cyclopsychotride A, a biologically active, 31-residue cyclic peptide isolated from Psychotria longipes. J. Nat. Prod. 57, 1619–1625 [DOI] [PubMed] [Google Scholar]
- 9. Saether O., Craik D. J., Campbell I. D., Sletten K., Juul J., Norman D. G. (1995) Elucidation of the primary and three-dimensional structure of the uterotonic polypeptide kalata B1. Biochemistry 34, 4147–4158 [DOI] [PubMed] [Google Scholar]
- 10. Colgrave M. L., Craik D. J. (2004) Thermal, chemical, and enzymatic stability of the cyclotide kalata B1. The importance of the cyclic cystine knot. Biochemistry 43, 5965–5975 [DOI] [PubMed] [Google Scholar]
- 11. Heitz A., Avrutina O., Le-Nguyen D., Diederichsen U., Hernandez J. F., Gracy J., Kolmar H., Chiche L. (2008) Knottin cyclization. Impact on structure and dynamics. BMC Struct. Biol. 8, 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hernandez J. F., Gagnon J., Chiche L., Nguyen T. M., Andrieu J. P., Heitz A., Trinh Hong T., Pham T. T., Le Nguyen D. (2000) Squash trypsin inhibitors from Momordica cochinchinensis exhibit an atypical macrocyclic structure. Biochemistry 39, 5722–5730 [DOI] [PubMed] [Google Scholar]
- 13. Craik D. J., Cemazar M., Wang C. K., Daly N. L. (2006) The cyclotide family of circular miniproteins. Nature's combinatorial peptide template. Biopolymers 84, 250–266 [DOI] [PubMed] [Google Scholar]
- 14. Craik D. J., Cemazar M., Daly N. L. (2006) Curr. Opin. Drug Discov. Dev. 9, 251–260 [PubMed] [Google Scholar]
- 15. Chan L. Y., Gunasekera S., Henriques S. T., Worth N. F., Le S. J., Clark R. J., Campbell J. H., Craik D. J., Daly N. L. (2011) Engineering pro-angiogenic peptides using stable, disulfide-rich cyclic scaffolds. Blood 118, 6709–6717 [DOI] [PubMed] [Google Scholar]
- 16. Gunasekera S., Foley F. M., Clark R. J., Sando L., Fabri L. J., Craik D. J., Daly N. L. (2008) Engineering stabilized vascular endothelial growth factor-A antagonists. Synthesis, structural characterization, and bioactivity of grafted analogues of cyclotides. J. Med. Chem. 51, 7697–7704 [DOI] [PubMed] [Google Scholar]
- 17. Sommerhoff C. P., Avrutina O., Schmoldt H. U., Gabrijelcic-Geiger D., Diederichsen U., Kolmar H. (2010) Engineered cystine knot miniproteins as potent inhibitors of human mast cell tryptase β. J. Mol. Biol. 395, 167–175 [DOI] [PubMed] [Google Scholar]
- 18. Dutton J. L., Renda R. F., Waine C., Clark R. J., Daly N. L., Jennings C. V., Anderson M. A., Craik D. J. (2004) Conserved structural and sequence elements implicated in the processing of gene-encoded circular proteins. J. Biol. Chem. 279, 46858–46867 [DOI] [PubMed] [Google Scholar]
- 19. Kaas Q., Craik D. J. (2010) Analysis and classification of circular proteins in CyBase. Biopolymers 94, 584–591 [DOI] [PubMed] [Google Scholar]
- 20. Poth A. G., Colgrave M. L., Philip R., Kerenga B., Daly N. L., Anderson M. A., Craik D. J. (2011) Discovery of cyclotides in the fabaceae plant family provides new insights into the cyclization, evolution, and distribution of circular proteins. ACS Chem. Biol. 6, 345–355 [DOI] [PubMed] [Google Scholar]
- 21. Poth A. G., Colgrave M. L., Lyons R. E., Daly N. L., Craik D. J. (2011) Discovery of an unusual biosynthetic origin for circular proteins in legumes. Proc. Natl. Acad. Sci. U.S.A. 108, 10127–10132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Nguyen G. K., Zhang S., Nguyen N. T., Nguyen P. Q., Chiu M. S., Hardjojo A., Tam J. P. (2011) Discovery and characterization of novel cyclotides originated from chimeric precursors consisting of albumin-1 chain a and cyclotide domains in the Fabaceae family. J. Biol. Chem. 286, 24275–24287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Higgins T. J., Chandler P. M., Randall P. J., Spencer D., Beach L. R., Blagrove R. J., Kortt A. A., Inglis A. S. (1986) Gene structure, protein structure, and regulation of the synthesis of a sulfur-rich protein in pea seeds. J. Biol. Chem. 261, 11124–11130 [PubMed] [Google Scholar]
- 24. Gerlach S. L., Burman R., Bohlin L., Mondal D., Göransson U. (2010) Isolation, characterization, and bioactivity of cyclotides from the Micronesian plant Psychotria leptothyrsa. J. Nat. Prod. 73, 1207–1213 [DOI] [PubMed] [Google Scholar]
- 25. Mylne J. S., Wang C. K., van der Weerden N. L., Craik D. J. (2010) Cyclotides are a component of the innate defense of Oldenlandia affinis. Biopolymers 94, 635–646 [DOI] [PubMed] [Google Scholar]
- 26. Trabi M., Craik D. J. (2004) Tissue-specific expression of head-to-tail cyclized miniproteins in Violaceae and structure determination of the root cyclotide Viola hederacea root cyclotide1. Plant Cell 16, 2204–2216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Conlan B. F., Gillon A. D., Barbeta B. L., Anderson M. A. (2011) Subcellular targeting and biosynthesis of cyclotides in plant cells. Am. J. Bot. 98, 2018–2026 [DOI] [PubMed] [Google Scholar]
- 28. Jennings C. V., Rosengren K. J., Daly N. L., Plan M., Stevens J., Scanlon M. J., Waine C., Norman D. G., Anderson M. A., Craik D. J. (2005) Isolation, solution structure, and insecticidal activity of kalata B2, a circular protein with a twist. Do Möbius strips exist in nature? Biochemistry 44, 851–860 [DOI] [PubMed] [Google Scholar]
- 29. Simonsen S. M., Sando L., Rosengren K. J., Wang C. K., Colgrave M. L., Daly N. L., Craik D. J. (2008) Alanine scanning mutagenesis of the prototypic cyclotide reveals a cluster of residues essential for bioactivity. J. Biol. Chem. 283, 9805–9813 [DOI] [PubMed] [Google Scholar]
- 30. Caprioli R. M., Farmer T. B., Gile J. (1997) Molecular imaging of biological samples. Localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 69, 4751–4760 [DOI] [PubMed] [Google Scholar]
- 31. Chughtai K., Heeren R. M. (2010) Mass spectrometric imaging for biomedical tissue analysis. Chem. Rev. 110, 3237–3277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Menger R. F., Stutts W. L., Anbukumar D. S., Bowden J. A., Ford D. A., Yost R. A. (2012) MALDI mass spectrometric imaging of cardiac tissue following myocardial infarction in a rat coronary artery ligation model. Anal. Chem. 84, 1117–1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ljungdahl A., Hanrieder J., Fälth M., Bergquist J., Andersson M. (2011) Imaging mass spectrometry reveals elevated nigral levels of dynorphin neuropeptides in l-DOPA-induced dyskinesia in rat model of Parkinson's disease. PLoS One 6, e25653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gemoll T., Roblick U. J., Habermann J. K. (2011) MALDI mass spectrometry imaging in oncology. Mol. Med. Rep. 4, 1045–1051 [DOI] [PubMed] [Google Scholar]
- 35. Castellino S., Groseclose M. R., Wagner D. (2011) MALDI imaging mass spectrometry. Bridging biology and chemistry in drug development. Bioanalysis 3, 2427–2441 [DOI] [PubMed] [Google Scholar]
- 36. Greer T., Sturm R., Li L. (2011) Mass spectrometry imaging for drugs and metabolites. J. Proteomics 74, 2617–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Grassl J., Taylor N. L., Millar A. H. (2011) Matrix-assisted laser desorption/ionization mass spectrometry imaging and its development for plant protein imaging. Plant Methods 7, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. (2006) A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313, 845–848 [DOI] [PubMed] [Google Scholar]
- 39. Shroff R., Vergara F., Muck A., Svatos A., Gershenzon J. (2008) Nonuniform distribution of glucosinolates in Arabidopsis thaliana leaves has important consequences for plant defense. Proc. Natl. Acad. Sci. U.S.A. 105, 6196–6201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Becerra J. X., Venable D. L. (1990) Rapid-terpene-bath and “squirt-gun” defense in Bursera schlechtendalii and the counterploy of Chrysomelid beetles. Biotropica 22, 320–323 [Google Scholar]
- 41. Dannenhoffer J. M., Suhr R. C., Thompson G. A. (2001) Phloem-specific expression of the pumpkin fruit trypsin inhibitor. Planta 212, 155–162 [DOI] [PubMed] [Google Scholar]
- 42. Lopez L., Camas A., Shivaji R., Ankala A., Williams P., Luthe D. (2007) Mir1-CP, a novel defense cysteine protease accumulates in maize vascular tissues in response to herbivory. Planta 226, 517–527 [DOI] [PubMed] [Google Scholar]
- 43. Do H. M., Lee S. C., Jung H. W., Sohn K. H., Hwang B. K. (2004) Differential expression and in situ localization of a pepper defensin (CADEF1) gene in response to pathogen infection, abiotic elicitors, and environmental stresses in Capsicum annuum. Plant Sci. 166, 1297–1305 [Google Scholar]
- 44. Thurnham D. I. (1999) Functional foods. Cholesterol-lowering benefits of plant sterols. Br. J. Nutr. 82, 255–256 [PubMed] [Google Scholar]
- 45. Ntanios F. (2001) Plant sterol-ester-enriched spreads as an example of a new functional food. Eur. J. Lipid Sci. Technol. 103, 102–106 [Google Scholar]
- 46. Ahmad A., Anjum F. M., Zahoor T., Nawaz H., Dilshad S. M. (2012) β-Glucan. A valuable functional ingredient in foods. Crit. Rev. Food Sci. Nutr. 52, 201–212 [DOI] [PubMed] [Google Scholar]
- 47. Saska I., Gillon A. D., Hatsugai N., Dietzgen R. G., Hara-Nishimura I., Anderson M. A., Craik D. J. (2007) An asparaginyl endopeptidase mediates in vivo protein backbone cyclization. J. Biol. Chem. 282, 29721–29728 [DOI] [PubMed] [Google Scholar]
- 48. Saska I., Colgrave M. L., Jones A., Anderson M. A., Craik D. J. (2008) Quantitative analysis of backbone-cyclised peptides in plants. J. Chromatogr. B 872, 107–114 [DOI] [PubMed] [Google Scholar]
- 49. Box M. S., Coustham V., Dean C., Mylne J. S. (2011) Protocol. A simple phenol-based method for 96-well extraction of high quality RNA from Arabidopsis. Plant Methods 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Broussalis A. M., Göransson U., Coussio J. D., Ferraro G., Martino V., Claeson P. (2001) First cyclotide from Hybanthus (Violaceae). Phytochemistry 58, 47–51 [DOI] [PubMed] [Google Scholar]
- 51. Gillon A. D., Saska I., Jennings C. V., Guarino R. F., Craik D. J., Anderson M. A. (2008) Biosynthesis of circular proteins in plants. Plant J. 53, 505–515 [DOI] [PubMed] [Google Scholar]
- 52. Ovesen R. G., Göransson U., Hansen S. H., Nielsen J., Hansen H. C. B. (2011) J. Chromatogr. 1218, 7964–7970 [DOI] [PubMed] [Google Scholar]
- 53. Gran L. (1970) An oxytocic principle found in Oldenlandia affinis DC. Medd. Norsk Farm. Selskap 32, 173–180 [Google Scholar]
- 54. Bokesch H. R., Pannell L. K., Cochran P. K., Sowder R. C., 2nd, McKee T. C., Boyd M. R. (2001) A novel anti-HIV macrocyclic peptide from Palicourea condensata. J. Nat. Prod. 64, 249–250 [DOI] [PubMed] [Google Scholar]
- 55. Nguyen G. K., Lim W. H., Nguyen P. Q., Tam J. P. (2012) Novel cyclotides and uncyclotides with highly shortened precursors from Chassalia chartacea and effects of methionine oxidation on bioactivities. J. Biol. Chem. 287, 17598–17607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ireland D. C., Colgrave M. L., Nguyencong P., Daly N. L., Craik D. J. (2006) Discovery and characterization of a linear cyclotide from Viola odorata. Implications for the processing of circular proteins. J. Mol. Biol. 357, 1522–1535 [DOI] [PubMed] [Google Scholar]
- 57. Nguyen G. K., Zhang S., Wang W., Wong C. T., Nguyen N. T., Tam J. P. (2011) Discovery of a linear cyclotide from the bracelet subfamily and its disulfide mapping by top-down mass spectrometry. J. Biol. Chem. 286, 44833–44844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. IUPAC (1979) Nomenclature of Organic Chemistry, Sections A, B, C, D, E, F, and H, Pergamon Press, Oxford [Google Scholar]
- 59. Barry D. G., Daly N. L., Clark R. J., Sando L., Craik D. J. (2003) Linearization of a naturally occurring circular protein maintains structure but eliminates hemolytic activity. Biochemistry 42, 6688–6695 [DOI] [PubMed] [Google Scholar]
- 60. Daly N. L., Gustafson K. R., Craik D. J. (2004) The role of the cyclic peptide backbone in the anti-HIV activity of the cyclotide kalata B1. FEBS Lett. 574, 69–72 [DOI] [PubMed] [Google Scholar]
- 61. Min W., Jones D. H. (1994) In vitro splicing of concanavalin A is catalyzed by asparaginyl endopeptidase. Nat. Struct. Biol. 1, 502–504 [DOI] [PubMed] [Google Scholar]
- 62. Abe Y., Shirane K., Yokosawa H., Matsushita H., Mitta M., Kato I., Ishii S. (1993) Asparaginyl endopeptidase of jack bean seeds. Purification, characterization, and high utility in protein sequence analysis. J. Biol. Chem. 268, 3525–3529 [PubMed] [Google Scholar]
- 63. Otegui M. S., Herder R., Schulze J., Jung R., Staehelin L. A. (2006) The proteolytic processing of seed storage proteins in Arabidopsis embryo cells starts in the multivesicular bodies. Plant Cell 18, 2567–2581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Hiraiwa N., Kondo M., Nishimura M., Hara-Nishimura I. (1997) An aspartic endopeptidase is involved in the breakdown of propeptides of storage proteins in protein-storage vacuoles of plants. Eur. J. Biochem. 246, 133–141 [DOI] [PubMed] [Google Scholar]
- 65. D'Hondt K., Bosch D., Van Damme J., Goethals M., Vandekerckhove J., Krebbers E. (1993) An aspartic proteinase present in seeds cleaves Arabidopsis 2 S albumin precursors in vitro. J. Biol. Chem. 268, 20884–20891 [PubMed] [Google Scholar]
- 66. Shimada T., Yamada K., Kataoka M., Nakaune S., Koumoto Y., Kuroyanagi M., Tabata S., Kato T., Shinozaki K., Seki M., Kobayashi M., Kondo M., Nishimura M., Hara-Nishimura I. (2003) Vacuolar processing enzymes are essential for proper processing of seed storage proteins in Arabidopsis thaliana. J. Biol. Chem. 278, 32292–32299 [DOI] [PubMed] [Google Scholar]
- 67. Gruis D., Schulze J., Jung R. (2004) Storage protein accumulation in the absence of the vacuolar processing enzyme family of cysteine proteases. Plant Cell 16, 270–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Hara-Hishimura I., Takeuchi Y., Inoue K., Nishimura M. (1993) Vesicle transport and processing of the precursor to 2S albumin in pumpkin. Plant J. 4, 793–800 [DOI] [PubMed] [Google Scholar]
- 69. Mylne J. S., Colgrave M. L., Daly N. L., Chanson A. H., Elliott A. G., McCallum E. J., Jones A., Craik D. J. (2011) Albumins and their processing machinery are hijacked for cyclic peptides in sunflower. Nat. Chem. Biol. 7, 257–259 [DOI] [PubMed] [Google Scholar]
- 70. Beck A., Bussat M. C., Klinguer-Hamour C., Goetsch L., Aubry J. P., Champion T., Julien E., Haeuw J. F., Bonnefoy J. Y., Corvaia N. (2001) Stability and CTL activity of N-terminal glutamic acid-containing peptides. J. Pept. Res. 57, 528–538 [DOI] [PubMed] [Google Scholar]
- 71. Nilsson A., Fehniger T. E., Gustavsson L., Andersson M., Kenne K., Marko-Varga G., Andrén P. E. (2010) Fine mapping the spatial distribution and concentration of unlabeled drugs within tissue microcompartments using imaging mass spectrometry. PLoS One 5, e11411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Murata N., Kajiura H., Fujimura Y., Miyao M., Murata T., Watanabe A., Shinozaki K. (1987) Progress in Photosynthesis Research, Martinus Nijhoff, Dordrecht, The Netherlands [Google Scholar]
- 73. Pinto M. F. S., Almeida R. G., Porto W. F., Fensterseifer I. C. M., Lima L. A., Dias S. C., Franco O. L. (2012) Cyclotides: From gene structure to promiscuous multifunctionality. J. Evid. Based Comp. Alt. Med. 17, 40–53 [Google Scholar]