Background: Three T6SSs are present in P. aeruginosa. H1-T6SS secretes bacteriolytic toxins.
Results: H2-T6SS is regulated by quorum sensing and Fur and modulates internalization in epithelial cells through PI3K-Akt host pathway activation.
Conclusion: H2-T6SS plays a role in virulence.
Significance: In contrast to the anti-prokaryotic H1-T6SS, H2-T6SS targets human cells. Those T6SSs can carry out different functions important in establishing infection.
Keywords: Invasion, Iron, Protein Secretion, Pseudomonas aeruginosa, Quorum Sensing, Fur, Internalization, T6SS
Abstract
The genome of Pseudomonas aeruginosa PAO1 contains three type VI secretion systems (T6SSs) called H1-, H2-, and H3-T6SS. The H1-T6SS secretes three identified toxins that target other bacteria, providing a fitness advantage for P. aeruginosa, and likely contributes to bacterial pathogenesis in chronic infections. However, no specific substrates or defined roles have been described for the two other systems. Here, we demonstrate that the expression of H2-T6SS genes of strain PAO1 is up-regulated during the transition from exponential to stationary phase growth and regulated by the Las and Rhl quorum sensing systems. In addition, we identify two putative Fur boxes in the promoter region and find that H2-T6SS transcription is negatively regulated by iron. We also show that the H2-T6SS system enhances bacterial uptake into HeLa cells (75% decrease in internalization with a H2-T6SS mutant) and into lung epithelial cells through a phosphatidylinositol 3-kinase-dependent pathway that induces Akt activation in the host cell (50% decrease in Akt phosphorylation). Finally, we show that H2-T6SS plays a role in P. aeruginosa virulence in the worm model. Thus, in contrast to H1-T6SS, H2-T6SS modulates interaction with eukaryotic host cells. Together, T6SS can carry out different functions that may be important in establishing chronic P. aeruginosa infections in the human host.
Introduction
Pseudomonas aeruginosa is one of the leading causes of nosocomial infections in immunocompromised humans and is responsible for chronic respiratory disease of patients with cystic fibrosis. A key factor in P. aeruginosa virulence is its capacity to secrete a wide range of hydrolytic enzymes and toxins to the extracellular medium as well as to inject virulence effectors directly into host cells (1, 2). Recently, a new secretion pathway, called the type VI secretion system (T6SS),4 that resembles an inverted phage tail has been described among Gram-negative bacteria, including P. aeruginosa (3–6). Substrates for this secretion system were first discovered in Vibrio cholerae by Pukatzki et al. (7), who demonstrated that secretion of proteins lacking a signal peptide, such as Hcp (hemolysin co-regulated protein) and VgrG (valine-glycine repeat), requires a functional T6SS. Gene clusters encoding T6SS machinery and putative substrates have been found in diverse animal and plant pathogens (8–10). Many bacteria encode more than one T6SS; in the case of P. aeruginosa, three T6SS, H1-T6SS to H3-T6SS, have been described. There is emerging evidence that secreted T6SS effectors can target either eukaryotic host cells or other bacteria (9, 10), and various T6SS within a single strain may serve different functions and/or be differentially regulated. V. cholerae VgrG1, which is homologous to a phage tail protein, is thought to function both as part of the secretion system and as a translocated effector (11). It encodes a C-terminal extension that can cross-link actin in vitro and in vivo and has been shown to be translocated into eukaryotic cells by a T6SS-dependent mechanism, where it is associated with cytotoxicity and virulence (12, 13). Similarly, the VgrG1 protein of Aeromonas hydrophila contains a VIP-2 (vegetative insecticidal protein domain 2) domain with an ADP-ribosyltransferase activity capable of modifying actin in vivo and is targeted to the host cytosol upon infection (14).
Interestingly, gene loci encoding T6SSs are also found in nonpathogenic bacteria and symbionts, suggesting that T6SS may not be dedicated exclusively to pathogenic interactions of bacteria with eukaryotes (9, 10). Indeed, T6SS can be potent mediators of interbacterial interactions as discovered in P. aeruginosa, where the type VI exported effectors Tse1–3 are bacteriolytic (15, 16) and may give P. aeruginosa a survival benefit in a multi-bacterial environment, in biofilms, and in polymicrobial infections, such as those encountered in cystic fibrosis patient airways (3, 17, 18).
Despite the increasing information available on the H1-T6SS, very little is known about the two other T6SSs encoded by P. aeruginosa. The H2-T6SS locus of the PA14 strain is required for virulence in the plant model Arabidopsis thaliana and in the mouse model of acute infections (19). Although the H1-T6SS loci are very similar between the P. aeruginosa strains, PA14 and PAO1, their H2-T6SS loci differ; two putative structural components that are also exported, hcp2 and vgrG2, are present in PA14 but missing in PAO1. They constitute, together with two other genes, a divergent operon to the H2-T6SS secretion machinery operon. Indeed the H2-T6SS of strain PAO1 may lack the extracytoplasmic part of the T6SS contractile phage tail-like structure that is thought to punctuate the outer membrane and host cell membrane (5). One could even ask whether the PAO1 H2-T6SS machinery can be functional. Here, we examined the regulation and function of H2-T6SS in PAO1. We find that it is subject to regulation by quorum sensing (QS) and by iron. In addition, we provide evidence that the H2 secretion machinery promotes bacterial internalization into epithelial cells, suggesting that this T6SS targets eukaryotic cells. Finally, we show that H2-T6SS plays a role in P. aeruginosa virulence in the worm model.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
The bacterial strains and plasmids used in this study are described in Table 1. LB and TSB broths and agar were used for the growth of P. aeruginosa and Escherichia coli strains at 37 °C. The cultures were inoculated at A600 of 0.1 with overnight cultures, and strains were grown at 37 °C with aeration in LB or TSB. Recombinant plasmids were introduced in P. aeruginosa using the conjugative properties of pRK2013 (see Table 1). P. aeruginosa transconjugants were selected on Pseudomonas isolation agar (Difco Laboratories) supplemented with appropriate antibiotics. The antibiotics concentrations were as follows: for E. coli, ampicillin (50 μg ml−1), kanamycin (25 μg ml−1), tetracycline (15 μg ml−1), and chloramphenicol (50 μg ml−1); and for P. aeruginosa, carbenicillin (500 μg ml−1) and tetracycline (200 μg ml−1 for plates or 50 μg ml−1 for liquid growth).
TABLE 1.
Strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Genotype, description, or sequence | Source and/or reference |
|---|---|---|
| E. coli strains | ||
| TG1 | supE, hsdΔR, thiΔ (lac-proAB), F′ (traD36, proAB+, lacIq, lacZΔM15)) | Laboratory collection |
| CC118(λpir) | (λpir) Δ (ara-leu), araD, ΔlacX74, galE, galK, phoA-20, thi-1, rpsE, rpoB, Arg(Am), recA1, Rfr (λpir) | Ref. 70 |
| TOP10F′ | F− mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 nupG recA1 araD139 Δ(ara-leu)7697 galE15 galK16 rpsL(StrR) endA1 λ− | Laboratory collection |
| MG1655 | F− λ− ilvG-, rfb-50, rph-1 | Laboratory collection |
| MG1655fur | MG1655 fur::kan | Ref. 71 |
| P. aeruginosa strains | ||
| PAO1 | Wild type, prototroph, chl-2 | B. Holloway |
| PAO1Z | Promoterless lacZ gene integrated at ctx att site in PAO1 | This work |
| PAO1TS2 | H2-T6SS promoter integrated at ctx att site in PAO1 | This work |
| PAOR | lasR mutant of PAO1, CbR | Ref. 28 |
| PAORTS2 | H2-T6SS promoter integrated at ctx att site in PAOR, CbR | This work |
| PDO100 | rhlI mutant of PAO1, HgR | Ref. 29 |
| PDO100TS2 | H2-T6SS promoter integrated at ctx att site in PDO100, HgR | This work |
| PAO1ΔclpV2 | clpV2 deletion mutant | This work |
| PAO1::gfp | miniTn7gfp integrated in PAO1 | This work |
| PAO1::gfp ΔclpV2 | miniTn7gfp integrated in PAO1ΔclpV2 | This work |
| Plasmids | ||
| pCR2.1 | TA cloning, lacZα, ColE1, f1 ori, ApR KmR | Invitrogen |
| pMini-CTX::lacZ | Ω-FRT-attP-MCS, ori, int, oriT, TcR | Ref. 21 |
| pMP220 | Broad host range lacZ transcriptional fusion, TcR | Laboratory collection |
| pTS2 | 722-bp upstream region of H2-T6SS in pMini-CTX::lacZ | This work |
| pTS28 | 722-bp upstream region of H2-T6SS in pCR2.1 | This work |
| pTS29 | 722-bp upstream region of H2-T6SS from pTS28 in pMP220 | This work |
| pRK2013 | Tra+, Mob+, ColE1, KmR | Ref. 72 |
| pRK600 | Tra+, OriT, OriV, Col E1, CmR | Ref. 73 |
| pUX-BF13 | Tra+, Mob+, R6K, providing Tn7 transposition functions in trans, ApR | Ref. 74 |
| miniTn7gfp | mob + ori; ApR GmR; miniTn7 vector containing gfpmut3 inserted into notI site | Ref. 75 |
| pKNG101 | oriR6K, mobRK2, sacBR+, SmR (suicide vector) | Ref. 76 |
| pTS24 | 500 bp upstream and 500 bp downstream clpV2 in pCR2.1 | This work |
| pTS27 | clpV2 deletion of pTS24 in pKNG101 | This work |
| pMMB67–42 | Broad host range vector adapted for the Gateway system tac promoter, ApR | This work |
| pHM49 | pMM67–42 encompassing clpV2-v5his6 | This work |
| pBLasR | lasR gene in the pBBRMCS-2, KmR | Ref. 28 |
| pBRhlRI | rhlR and rhlI genes in the pBBRMCS-2, KmR | Laboratory collection |
| Oligonucleotides | ||
| TSO1 | 5′-CCTAACCCTTCAATGCACACC-3′ | This work |
| TSO3 | 5′-GTCGACCTATTACGCTCTAAATCAGC-3′ | This work |
| TSO37 | 5′-TAATGCGAGGAGAGCTTGCTCG-3′ | This work |
| TSO38 | 5′-ATGCCGGCTGCACAGACGTTCC-3′ | This work |
| TSO49 | 5′-ACGCCCACCTCACATCCATCCTTAATGAATCTTG-3′ | This work |
| TSO50 | 5′-AAGGATGGATGTGAGGTGGGTGCGATGTTCAGC-3′ | This work |
| TSO51 | 5′-ACTGGAAACGCCTGATGCCCTACCTGG-3′ | This work |
| TSO52 | 5′-CGAACGAGCCCAGCTGGCCCAGTGGATGG-3′ | This work |
| OA14 | 5′-GGAAAGCTTTTCGCCCTCGTCGGATTG-3′ | This work |
| OA17 | 5′-AAAGAATTCGAGGCGTTGCAGCAGATG-3′ | This work |
| GeneRacer 5′ primer | 5′-CGACTGGAGCACGAGGACACTGA-3′ | Invitrogen |
| GeneRacer 5′ nested primer | 5′-GGACACTGACATGGACTGAAGGAGTA-3′ | Invitrogen |
Construction of the clpV2 Mutant
To generate clpV2 deletion, 500 bp upstream and 500 bp downstream of the clpV2 gene were amplified by overlapping PCR with High Fidelity DNA polymerase (Roche Applied Science) using TSO49, TSO50, TSO51, and TSO52 primers (see Table 1) (20). The PCR product was cloned in pCR2.1 (TA cloning kit; Invitrogen) giving pTS24, which was then sequenced (GATC) and subcloned in pKNG101 suicide vector giving the mutator pTS27. pTS27, maintained in the E. coli CC118λpir strain, was mobilized in the wild type P. aeruginosa strain PAO1. The mutants, in which the double recombination events occurred that resulted in the nonpolar deletion of clpV2 gene, were verified by PCR using external primers OA14–OA17.
lacZ Reporter Fusion and β-Galactosidase Assay
The H2-T6SS-lacZ transcriptional fusions were constructed by PCR amplification of the 722-bp upstream DNA region from the tssA2 gene by using TSO1 and TSO3 primers (see Table 1). PCR amplification products were directly cloned into the pMini-CTX::lacZ vector (21), yielding pTS2, or into pCR2.1 and pMP220, yielding pTS28 and pTS29, respectively. Nucleotide sequence was verified (GATC). The promoter fragment was integrated at the CTX phage attachment site in PAO1 and isogenic mutants using established protocols (21). The pTS29 was transferred into E. coli by conjugation.
For β-galactosidase assay, overnight bacterial culture was diluted in TSB to an A600 of 0.1. Growth and β-galactosidase activity were monitored by harvesting samples at different time intervals. β-Galactosidase activity was measured according to the Miller method, based on o-nitrophenyl-β-d-galactopyranoside hydrolysis (22), and expressed in Miller units.
RNA Extraction and Determination of the Transcription Start
PAO1 was grown in TSB at 37 °C for 5 h, and total RNA was extracted using the PureYield RNA Midiprep system (Promega). The MICROBExpress kit was used to purify mRNA (Ambion). 5′ RNA ligase-mediated rapid amplification of 5′ cDNA ends was performed using the GeneRacer kit (Invitrogen), starting at the TAP step on 1 μg of mRNA. Reverse transcription with Superscript III RT was performed with random oligonucleotides. TSO37, TSO38, GeneRacer 5′ Nested Primer, and GeneRacer 5′ Primer (see Table 1) were used for PCR amplification (high fidelity expand system; Roche Applied Science) of the first strand cDNA. The PCR product was cloned into the PCR2.1 vector using the TA cloning kit (Invitrogen) and transformed in TOP10F′ competent cells. Cloning was screened using the M13 forward primer and M13 reverse primer, and the cloned products were sequenced using the M13 forward primer (GATC).
HeLa Cell Invasion, Survival, and Adhesion Assays
For invasion assay, HeLa cells (ATCC, Manassas, VA) were cultured to 70% confluence at the day of the experiment in 24-well plates in Eagle's minimal essential medium (MEM) supplemented with 10% FBS. The cells were washed twice with sterile PBS and infected with P. aeruginosa wild type strain and the clpV2 mutant, grown as indicated in the text (log, transition, or stationary phase) at a multiplicity of infection of 10 in MEM with FBS at final volume of 1 ml. The plates were spun down at 1000 × g for 5 min to allow bacteria close contact with HeLa cells. After 3 h of infection at 37 °C in 5% CO2, the cells were washed twice with sterile PBS and incubated with MEM supplemented with 300 μg ml−1of gentamicin (Sigma) for 2 h to kill extracellular bacteria. Afterward, the cells were washed three times with room temperature PBS and osmotically shocked with ice-cold sterile water containing 0.05% Triton X-100 (Sigma) for 30 min on ice. Lysed cells were collected in Eppendorf tubes, and serial dilutions in sterile LB were plated onto Pseudomonas isolation agar plates. Colony-forming units were enumerated and reported as internalized bacteria.
For survival assay, HeLa cells were infected for 1.5 h and treated for 1 or 4 h with gentamicin 300 μg ml−1 to allow 3 h more for intracellular replication. For adhesion assays, infected cells were not incubated with gentamicin.
Calu-3 Cell Invasion Assay
Calu-3 cells, obtained from the ATCC, were maintained in MEM supplemented with 10% FBS (Invitrogen) and l-glutamate at 37 °C with 5% CO2. For experiments, the cells were grown on 12-mm Transwell filters (0.4-μm pore size; Corning) for 2 days (1 × 106 cells/well). Calu-3 cells grown on Transwell filters were washed in serum-free MEM and preincubated with LY (Sigma-Aldrich) in serum-free MEM supplemented with l-glutamine for 1 h. Transition phase grown bacteria were added to the apical chamber of the Transwell filter for 3 h at an multiplicity of infection of 10. After incubation, the cells were washed with serum-free MEM and incubated with serum-free MEM containing 250 μg ml−1 gentamicin (Fisher) for 2.5 h. The cells were washed twice in PBS to remove antibiotic and lysed in 1 ml Ca2+ Mg2+-free PBS with 0.25% Triton X-100 (Sigma-Aldrich) for 30 min. The cells were scraped off the Transwell filters, and the cell lysates were transferred to new tubes. Internalized bacteria were enumerated by plating serial dilutions of cell lysates to LB plates and counting colony-forming units. All of the assays were carried out on triplicate wells, and the results are reported as the average of three experiments ± S.D. (standard deviation).
Akt Immunoprecipitation
Calu-3 grown on Transwell filters were washed and placed in serum-free MEM supplemented with l-glutamate for 12 h. Transition phase grown bacteria were added for 1 h at an multiplicity of infection of 100. The infected monolayers were washed with cold PBS containing 1 mm sodium orthovanadate (Sigma-Aldrich). The cells were lysed in modified radioimmunoprecipitation assay buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 2 mm EDTA, 2 mm EGTA, 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS, 1 mm sodium orthovanadate, 50 mm NaF, 0.1 mm okadaic acid (Sigma-Aldrich), 1 mm phenylmethylsulfonyl fluoride (Sigma-Aldrich), and proteinase inhibitor tablets (Complete; Roche Applied Science)) for 30 min. The cells were scraped off the Transwell filters, and the cell lysates were centrifuged at 16,000 × g for 20 min. To preclear the cell lysate, the supernatant was mixed with 20 μl of protein G-Sepharose (4 Fast Flow; Amersham Biosciences), and the protein content was determined using protein assay reagent (bicinchoninic acid; Pierce). The cleared lysate (300–400 μg of protein) was incubated with polyclonal Akt antibody (Cell Signaling Technology) overnight at 4 °C and incubated for 2 h with protein G-Sepharose. The immune complexes were washed three times with modified radioimmunoprecipitation assay buffer without deoxycholate, eluted in SDS sample buffer, electrophoresed on 10% SDS-polyacrylamide gels, and transferred to polyvinylidene difluoride membranes. The membranes were blocked with PBS containing 0.05% Tween 20 (PBST) and 5% nonfat milk for 1 h at room temperature and then incubated overnight at 4 °C with a monoclonal antibody that recognizes Akt phosphorylated on serine 473 (Cell Signaling Technology). The membranes were washed with PBST, incubated with horseradish peroxidase-conjugated secondary antibody (Jackson ImmunoResearch Laboratories) for 1 h at room temperature, and developed using an enhanced chemiluminescence kit (Amersham Biosciences). The membranes were then stripped and reprobed with a monoclonal antibody that recognizes all forms of Akt (Cell Signaling Technology). Primary antibodies were diluted 1:1000 and secondary antibodies were diluted 1:3000.
Caenorhabditis elegans Killing Assay
The slow killing assay was performed as described previously with modifications based on Ref. 23. Each independent assay consisted of three replicates. E. coli OP50 was used as a control. L4 to adult stage C. elegans were removed from food and placed on unseeded NGM plates for 24 h at 25 °C. 50 worms were then picked onto plates containing overnight growth of each bacterial strain. The worms were evaluated for viability on a daily basis. The assay was performed three times. Animal survival was plotted using the PRISM 5.0 program. Survival curves were considered significantly different from the control when p values were <0.05. PRISM calculated survival fractions using the product limit (Kaplan-Meier) method and compared survival curves by two methods: the log-rank test (also called the Mantel-Cox test) and the Gehan-Breslow-Wilcoxon test.
Statistical Analysis
Paired Student's t tests were performed using Excel software. In the figures, * indicates p ≤ 0.05, ** indicates p ≤ 0.01, and *** indicates p ≤ 0.001.
RESULTS
The Expression of the H2-T6SS Genes Is Controlled by Quorum Sensing
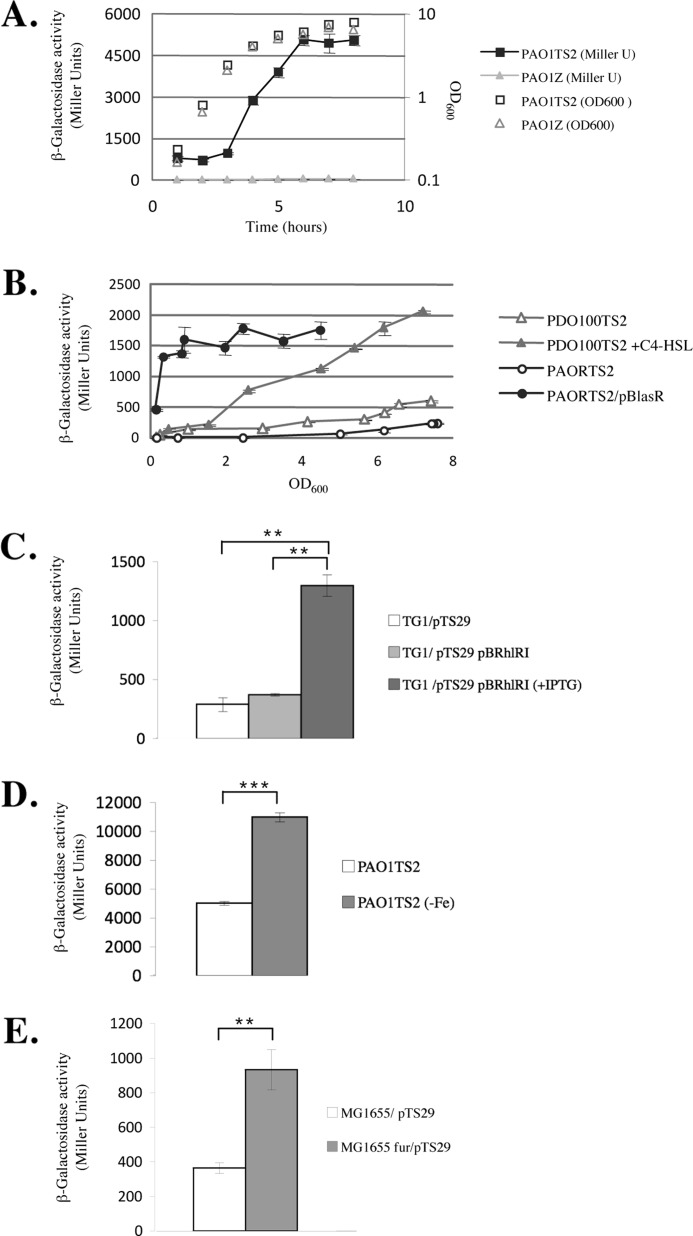
Although the H1-T6SS cluster is repressed by RetS/Gac signaling system in PAO1, the H2-T6SS and H3-T6SS are not (3, 24–26). To probe the transcriptional regulation of the H2-T6SS gene cluster, we analyzed the DNA region upstream of the first gene, hsiA2 (PA1656), of the cluster in PAO1 (Fig. 1A). Two σ70-dependent promoters were predicted in the intergenic region between the upstream gene (PA1655) and hsiA2 (PA1656) by the BProm program (Fig. 1B). We created a lacZ transcriptional fusion in which the DNA region upstream of the ATG of the first gene of the H2-T6SS cluster was fused to lacZ and integrated at the CTX phage attB site as a single copy on the chromosome, yielding strain PAO1TS2. Strain PAO1Z was similarly constructed by integrating a promoter-less lacZ gene to serve as a negative control (Table 1). The β-galactosidase activity profile associated with the transcriptional fusion indicated that the H2-T6SS gene cluster expression was low during the exponential phase but increased at 3 h of growth in TSB medium at 37 °C, during the log to stationary phase transition (Fig. 2A). The expression was at its highest (10-fold above background) in stationary phase, whereas promoter activity was undetectable in the control strain PAO1Z.
FIGURE 1.
The H2-T6SS gene cluster of P. aeruginosa PAO1. A, the genes are labeled hsiA2 to hsiJ2 and, when applicable, with the given name, i.e., clpV2 or sfa2. They are also indicated by the number of their annotation (e.g. PA1656). B, the sequence corresponding to 1173 bp upstream of the H2-T6SS gene cluster is represented. The two predicted σ70 promoters are framed in black, a Las-Rhl consensus box is indicated in bold letters, and two Fur binding sites are underlined. The experimentally demonstrated transcription initiation site and its cognate σ70 promoter are shaded in gray. The start codon of hsiA2, the first gene of the cluster, is written in gray. 722 bp upstream the start codon have been used for the transcriptional fusion encoded by pTS2 or pTS29. No promoter could be predicted in the intergenic region in between hsiF2 (PA1659) and hsiG2 (PA1660). C, alignment of the LasR-RhlR consensus sequence from Ref. 36 with the sequence framed in B. The size of each letter indicates the relative abundance at the respective position in the consensus matrix generated with MATRIX. D, alignment of the two Fur consensus sequences (37, 39) with the two regions underlined in B and overlapping with the distal promoter (left panel) or the proximal (right panel).
FIGURE 2.
The H2-T6SS gene cluster of P. aeruginosa PAO1 is induced upon iron starvation and regulated by QS. A, the expression pattern of the H2-T6SS-lacZ transcriptional fusion from the wild type PAO1 strain (PAO1TS2) (squares) and a control strain (PAO1Z) (triangles) is given in Miller units at different time points over the growth period. The A600 is also presented. B, expression of the H2-T6SS-lacZ transcriptional fusion from the PAO1 lasR strain (PAORTS2) with (filled circles) or without (open circles) the pBLasR plasmid, and from the PAO1 rhlI strain (PDO100TS2) grown in the presence (filled triangles) or absence (open triangles) of 10 μm C4-HSL. PAORTS2 pBlasR curve is significantly different from PAORTS2 (p ≤ 0.001), with the last five points of the PDO100TS2 + C4-HSL from PDO100TS2 (p ≤ 0.001). C, expression of the H2-T6SS-lacZ transcriptional fusion from the pTS29 plasmid in the E. coli TG1 with (gray bars) or without (white bar) the pBRhlRI plasmid. Expression of rhlR and rhlI gene induced in the presence of 0.5 mm IPTG (dark gray bar) or suppressed in the absence of IPTG (light gray bar). D, expression of the H2-T6SS-lacZ transcriptional fusion from the PAO1 strain (PAO1TS2) grown in high (white bar) or low iron (gray bar). E, expression of the H2-T6SS-lacZ transcriptional fusion from the pTS29 plasmid in the E. coli MG1655 (white bar) and its isogenic fur mutant (gray bar). Each experiment was done in duplicate and independently repeated three times; the error bars indicate standard deviations, and the asterisks indicate p values.
The biphasic, cell density-dependent expression profile of the reporter fusion suggested that the transcription of H2-T6SS might be regulated by QS. This could be in agreement with the regulation by LasR of the PA14 hcp2 gene, which lacks in PAO1 (19). We therefore examined H2-T6SS expression in P. aeruginosa QS mutants. Two homoserine lactone-mediated QS systems, the Las and Rhl systems, have been described in P. aeruginosa (27). The transcriptional regulators LasR and RhlR are activated by their cognate autoinducers, N-(3-oxododecanoyl)-l-homoserine lactone and N-(butanoyl)-l-HSL (C4-HSL) produced by the LasI and RhlI synthases, respectively. The pTS2 fusion was integrated on the chromosome of a lasR mutant (PAOR) (28) and that of a rhlI mutant (PDO100) (29) creating PAORTS2 and PDO100TS2 QS mutant strains. Compared with wild type PAO1, the expression of H2-T6SS was significantly delayed in the two QS mutants, becoming detectable 6 h post-inoculation, with maximal levels decreased 22- and 8.5-fold, respectively (Fig. 2B). Complementation in trans of the lasR mutation in PAORTS2 with pBLasR, a plasmid constitutively producing LasR, affected the growth of the strain (the strain stops growing at A600 = 2.5) but restored the fusion activity almost to its wild type levels at the exponential phase (filled circles in Fig. 2B). N-Acylhomoserine lactone synthase mutants, such as rhlI, can be phenotypically complemented by addition of the corresponding N-acylhomoserine lactone (30, 31). The addition of the C4-HSL (10 μm) to the rhlI mutant PDO100TS2 restored induction of the expression at the transition phase of growth, and the expression was 4-fold higher than in the mutant (Fig. 2B). One can note that the decrease in H2-T6SS expression observed with the lasR mutant is greater (2.6-fold) than with the rhlI mutant. This may imply a direct effect of LasR, as well as an indirect effect mediated through Rhl system.
Thus to confirm a direct effect of RhlR-C4-HSL on H2-T6SS expression too, the H2-T6SS promoter region was cloned into pMP220 to generate a plasmid borne lacZ transcriptional fusion whose expression was quantified in a strain of E. coli, which harbored IPTG-inducible copies of rhlR and rhlI. Upon RhlR and RhlI production, a 3.3-fold up-regulation of the H2-T6SS promoter region was observed (Fig. 2C), suggesting that the Rhl system is sufficient for the induction of H2-T6SS expression.
The promoter region of QS regulated genes in P. aeruginosa usually contains a Las-Rhl box, a 20-bp repeat located approximately between 60 and 40 bp from the start of transcription (32, 33). As shown in Fig. 1 (B and C), a putative Las-Rhl box can be identified in the upstream region of the H2-T6SS gene cluster, but far from the two predicted promoters. Using 5′ rapid amplification of 5′ cDNA ends PCR, we located a transcription initiation site 246 bp upstream of the translation initiation codon of the hsiA2 gene (the +1 position is indicated in gray in Fig. 1B). We were thus able to position −10 and −35 boxes of a σ70 promoter in proximity of the Las-Rhl box centered at −47 with respect to the transcript start site. Together, our studies support the notion that the expression of the P. aeruginosa H2-T6SS genes appears to be directly controlled by the Las and Rhl QS systems.
H2-T6SS Gene Expression Is Induced in Iron-limiting Conditions and Regulated by Fur
For many pathogens, the host environment is often iron poor, which triggers the expression of a group of genes transcriptionally regulated by iron through the ferric uptake regulator protein Fur. In addition to its role in iron homeostasis, Fur is involved in the coordinate regulation of numerous virulence genes. In the case of P. aeruginosa, Fur regulates exotoxin A and alkaline protease production (34, 35), as well as biofilm formation (36). We noted the presence of a sequence upstream of the H2-T6SS gene cluster that conformed to the E. coli and P. aeruginosa consensus Fur binding sequence (GATAATGATAATCATTATC) (37, 38) or the (NATWAT)3 sequence proposed as the E. coli Fur box (39) (Fig. 1, B and D). The two putative Fur boxes overlapped with the −10 box of the two predicted σ70 promoters, suggesting that Fur binding at this position in presence of iron prevents RNA polymerase binding to the −10 box and thus represses transcription. To test this, PAO1TS2, which harbored the H2-T6SS-lacZ transcriptional fusion, was grown in TSB in presence of the iron chelator 2,2′-bipyridyl (250 μm). The activity of the promoter was induced 2.2-fold in these conditions (Fig. 2D).
To determine whether Fur is required for H2-T6SS regulation and because Fur seems to be essential for P. aeruginosa viability (40), the expression of the plasmid borne transcriptional fusion was studied in an E. coli WT and isogenic fur mutant. After 4 h of growth, the activity of the fusion increased by 2.6-fold in the Fur mutant (Fig. 2E). Together, these data suggest that transcription of the H2-T6SS gene cluster is induced under iron-limiting conditions, an environment that the pathogen encounters in the human host.
H2-T6SS Contributes to P. aeruginosa Internalization in Non-phagocytic Cells
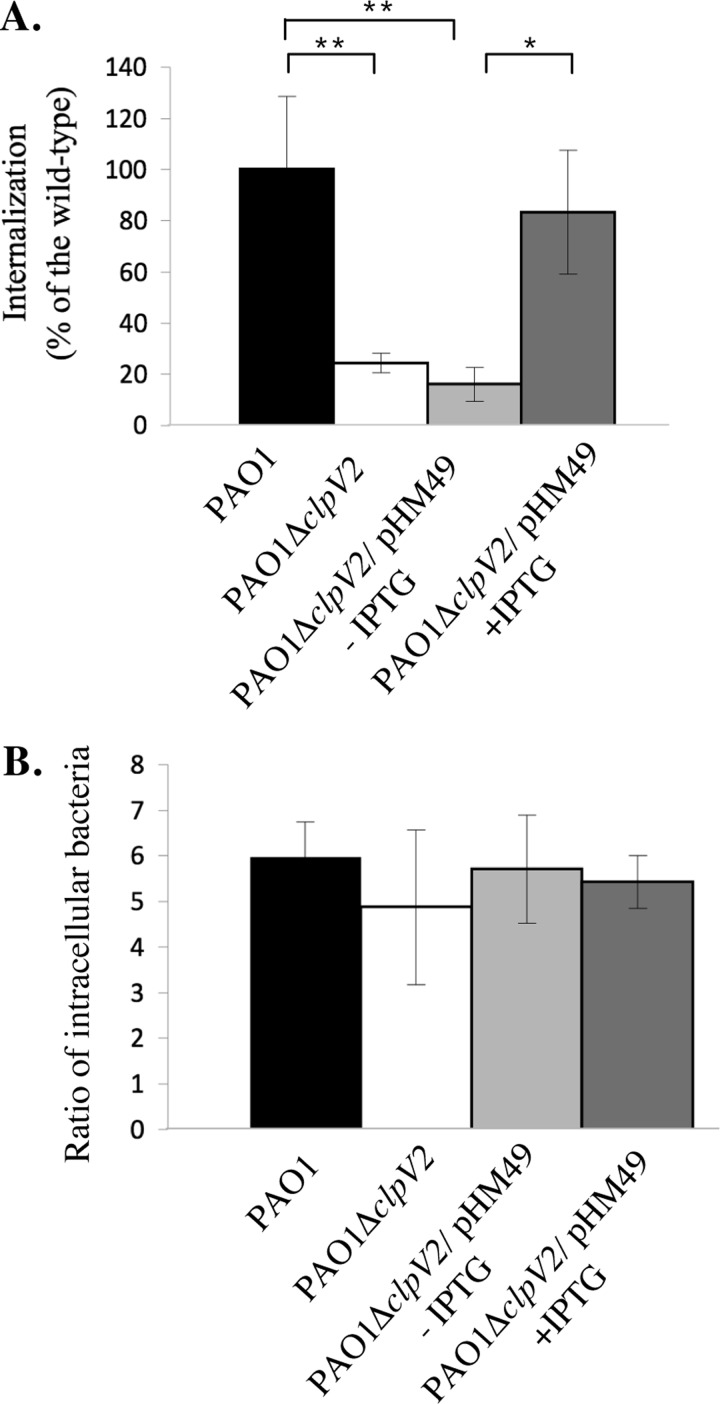
Although P. aeruginosa is considered an extracellular pathogen, it is able to measurably enter (i.e., invade) normally nonphagocytic cells ((41), for a review see Ref. 77). To address the potential role of H2-T6SS in P. aeruginosa internalization, we constructed a nonpolar mutant in the AAA+ ATPase ClpV2 whose homologue was shown to play a crucial role in the functionality of the H1-T6SS system (3, 42). We verified that the mutation does not affect the growth of the strain, nor the major adhesins of P. aeruginosa (flagellum and type IV pili), nor the secretion of ExoS, an anti-internalization factor (supplemental Fig. S1). Moreover, adhesion of the clpV2 mutant was unaffected (supplemental Fig. S2, A and B). We then compared invasion of transition phase grown wild type PAO1 with a mutant defective in H2-T6SS (PAO1ΔclpV2), using a standard antibiotic protection assay adapted from Ref. 43. As shown in Fig. 3A, PAO1 was able to invade nonphagocytic cells, with an average of 1.7% of adherent bacteria internalized (supplemental Fig. S2C), whereas PAO1ΔclpV2 entry into host cells was reduced by ∼75%. Invasion was restored to wild type levels in the complemented clpV2 mutant (Fig. 3A).
FIGURE 3.
Maximal uptake of P. aeruginosa by HeLa cells requires the H2-T6SS apparatus. Standard bacterial invasion (A) or survival (B) assays in HeLa cells upon infection with P. aeruginosa strains grown at the transition phase in TSB. A, the percent invasion of PAO1ΔclpV2 with (gray bars) or without (white bars) pHM49, normalized to PAO1 (black bars) that represents an average of 1.3 × 105 colony-forming units of internalized bacteria/well (2.5 × 105 HeLa cells). ClpV2V5/6His expression is induced with 1 mm with IPTG (dark gray bars) or remains suppressed in the absence of IPTG (light gray bars). B, the ratio between the numbers of intracellular bacteria recovered after 5.5 and 2.5 h of infection (including the gentamicin treatment). Same strain color code than in A. All of the assays were performed a minimum of three times in triplicate. The error bars represent standard deviations, and the asterisks indicate p values.
Because we previously observed a cell density expression pattern of H2-T6SS genes, we asked whether the growth phase of the bacteria impacts the internalization capacity. Although exponential grown clpV2 mutant bacteria were less internalized (∼75% reduction as with transition phase bacteria; data not shown), stationary phase bacteria were internalized at the same level as the wild type strain (supplemental Fig. S3).
Even if P. aeruginosa escapes the normal bactericidal mechanisms of epithelial cells, it only poorly replicates in A549 pneumocytes (41) or in corneal epithelial cells (43). Given the length of the infection in the invasion assay (5 h including the gentamicin treatment), to examine the possibility of an intracellular replication and/or persistence defect of the clpV2 mutant, we compared the number of intracellular bacteria after 2.5 and 5.5 h of infection for each strain. After allowing 3 h for bacterial replication, we observed the same range of data (between 4.9- and 6-fold increase) for all the strains (Fig. 3B). In conclusion, the H2-T6SS mutant displays an invasion defect.
In a complementary assay, HeLa cells were infected with GFP-labeled P. aeruginosa and visualized by laser confocal microscopy (data not shown). Few bacteria were found in HeLa cells infected with the wild type PAO1. Conversely no bacteria were found in cells infected with the clpV2 mutant. Together, these results support the assertion that the H2-T6SS system stimulates internalization of P. aeruginosa into HeLa cells.
Akt Phosphorylation upon Bacterial Invasion of Polarized Epithelial Cells Is Dependent on H2-T6SS
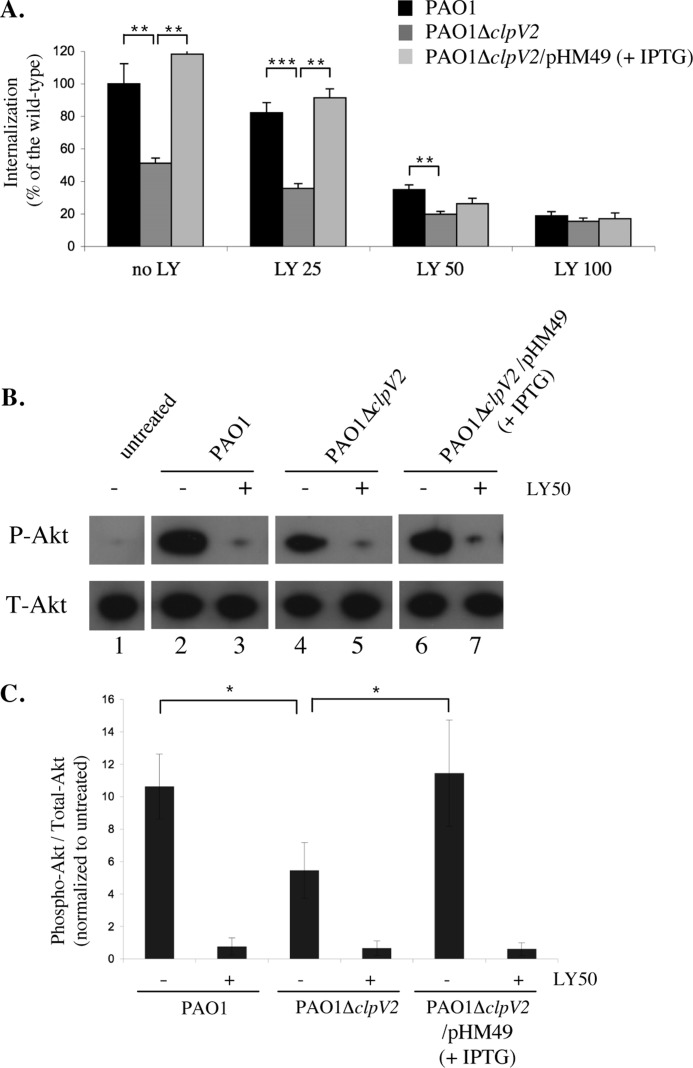
We further explored H2-T6SS-dependent bacterial entry into a more physiologically relevant cell line, Calu-3 cells, which are derived from human lung alveolar cells and which form polarized monolayers when grown on Transwell filters. P. aeruginosa binding to polarized epithelial cells activates a central host signaling molecule, PI3K, which is required for phosphatidylinositol 1,4,5-trisphosphate synthesis and for activation of a downstream effector, the protein kinase B/Akt (Akt) (44, 45). Activation of the PI3K/phosphatidylinositol 1,4,5-trisphosphate/Akt pathway was shown to be necessary and sufficient for P. aeruginosa entry from the apical surface (44). This event leads to a remarkable remodeling of the apical membrane in which protrusions enriched for phosphatidylinositol 1,4,5-trisphosphate and actin form at the apical surface at the site of bacterial entry (46). To determine whether the PI3K/Akt signaling pathway is required during the H2-T6SS-dependent uptake of P. aeruginosa by lung epithelial cells, polarized human airway epithelial Calu-3 cells were infected in the presence or absence of LY294002 (LY), a PI3K inhibitor, and adhesion, internalization, and Akt phosphorylation were determined. Although the treatment with LY did not affect bacterial adhesion, it reduced invasion of both PAO1 and the complemented PAO1ΔclpV2 mutant in a dose-dependent manner (Fig. 4A). Internalization levels of the clpV2 mutant were lower than that of PAO1 or the complemented clpV2 mutant; thus, LY treatment had little effect on the ability of the clpV2 mutant to enter host cells (Fig. 4A).
FIGURE 4.
Activation of PI3K pathway in Calu-3 cells relies on H2-T6SS-dependent PAO1 invasion. A, the percent invasion of PAO1ΔclpV2 with (gray bars) or without (white bars) pHM49, normalized to PAO1 that represents an average of 3500 colony-forming units of internalized bacteria/well (0.8. × 106 Calu-3). When indicated, the cells were incubated with a PI3K inhibitor, LY294002, at various concentrations (25, 50, and 100 μm) for 1 h prior to infection. The assays were performed a minimum of three times in triplicate. The error bars represent standard deviations, and the asterisks indicate p values. B, levels of phosphorylated Akt in Calu-3 cells infected with PAO1, PAO1ΔclpV2 with or without pHM49, or left uninfected (untreated). Akt phosphorylation was detected with anti-phospho-Akt antibodies (P-Akt), and total Akt with anti-Akt antibodies (T-Akt). When indicated (+), the cells were incubated with LY294002 at 50 μm for 1 h prior to infection. C, quantification of band density by volume analysis using ChemiDoc XRS. The graph presents phospho-Akt as a percentage of Total-Akt and normalized to untreated cells from B.
We next determined whether Akt is activated in an H2-T6SS dependent manner, as determined by phosphorylation of Akt at serine 473. Polarized Calu-3 cells were co-cultivated with PAO1, clpV2 mutant, or the complemented clpV2 mutant. At 60 min after infection, Akt was immunoprecipitated followed by immunoblotting with anti-phospho-AktSer473 antibody. The ratio of phospho-Akt to total Akt was quantified and normalized to the ratio observed in untreated cells. Infection with the clpV2 mutant resulted in ∼50% decrease in Akt phosphorylation levels compared with infection with PAO1 or with the complemented mutant (Fig. 4, B and C). The magnitude of the decrease in Akt phosphorylation was similar to the magnitude of the decrease in internalization by the clpV2 mutant (Fig. 4, B and C). LY treatment decreased Akt phosphorylation upon infection with any strain to levels observed in uninfected Calu-3 cells (Fig. 4, B and C). Together, these data suggest that H2-T6SS stimulates bacterial entry of transition phase-grown bacteria through the PI3K/Akt pathway.
H2-T6SS Contributes to P. aeruginosa Virulence in C. elegans
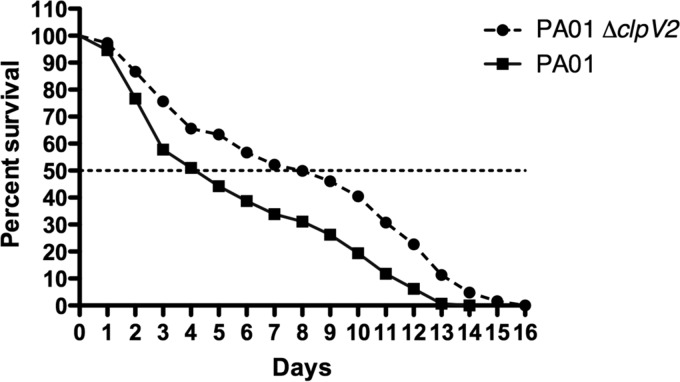
The involvement of H2-T6SS in P. aeruginosa pathogenicity led us to test its role in virulence on C. elegans. Previous studies have shown that C. elegans is a good model in which to identify P. aeruginosa virulence genes important for mammalian pathogenesis (47, 48). In a modified slow killing assay, we observed that the clpV2 mutant was significantly less virulent than the PAO1 strain (Fig. 5), indicating that H2-T6SS plays a role in P. aeruginosa virulence.
FIGURE 5.
H2-T6SS is required for virulence in C. elegans. C. elegans was infected with PAO1 and its isogenic ΔclpV2 mutant (survival curve, p value <0.0001; Prism 4 software).
DISCUSSION
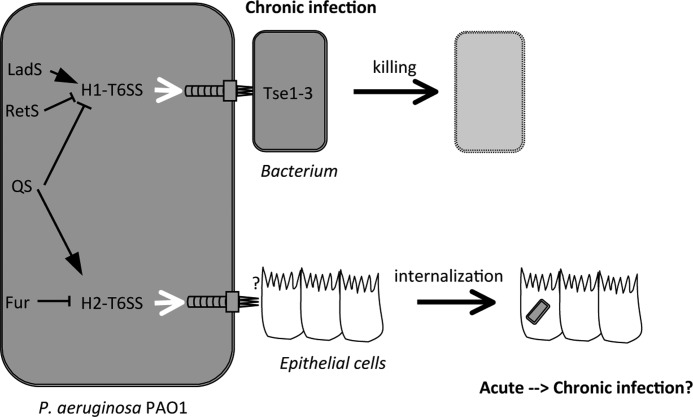
P. aeruginosa possesses three distinct T6SS genetic loci, but the function and regulation of H2-T6SS and H3-T6SS have yet to be elucidated. Here, we show that the H2-T6SS gene cluster is up-regulated in PAO1 during the transition from exponential to stationary phase growth and that its transcription is regulated by QS and induced upon iron limitation through the Fur regulator. Moreover, we demonstrate that H2-T6SS is required for PI3K/Akt-dependent internalization into epithelial cells and plays a role in P. aeruginosa virulence. Together, these data suggest that H2-T6SS is important in bacterial interactions with eukaryotic cells, which is quite distinct from the known functions of H1-T6SS. The latter is not expressed under laboratory conditions (3, 24, 25), secretes bacteriolytic effectors, and is primarily thought to give P. aeruginosa a growth advantage in mixed bacterial populations (15, 16). Thus, although the T6SS apparatus may be conserved, the diversity in secreted effectors may allow interactions with many different hosts in a broad range of habitats and conditions (Fig. 6).
FIGURE 6.
A model of T6SS functions in P. aeruginosa. H1- and H2-T6SS are differently regulated and have diverse functions. H1-T6SS targets bacteriolytic effectors in host bacteria providing a fitness advantage to P. aeruginosa presumably during chronic infection, whereas H2-T6SS mediates P. aeruginosa internalization into epithelial cells that could lead to a chronic infection.
Our results demonstrate that the LasR and RhlR QS regulators induce expression of genes coding the H2-T6SS machinery components in PAO1. In a different P. aeruginosa strain, PA14, representative gene transcripts from the three T6SS gene clusters, hcp1, hcp2, and hcp3, have also been reported to be regulated by LasR (19). As noted earlier, PAO1 and PA14 strains differ at the level of H2-T6SS loci, hcp2 is absent from PAO1 locus, and the PA14 locus contains also three other putative substrate genes, vgrG2, PA14_43090 encoding an homologue of an A. hydrophila type III secretion system (T3SS) effector and PA14_43100 encoding a putative Rhs family protein (19). From our observation, it thus stands to reason to propose that the H2-T6SS machinery of PA14 is under QS control too. Collectively, these findings tend to indicate that the H2-T6SS machinery and cognate substrates are co-regulated by QS, even if those divergences may even reflect a different function of the H2-T6SS machinery in these two P. aeruginosa strains. Indeed, QS control of the T6SS may be generally conserved, as it has also been reported for the plant pathogen Pectobacterium atrosepticum (49), V. cholerae 01 (50, 51), a clinical isolate of A. hydrophila (52), and in Yersinia pseudotuberculosis (53). This strategy may allow pathogens to coordinately regulate the T6SS with other important virulence determinants, including other secretion systems, toxins, proteases, and biofilm-promoting factors.
One challenge that many pathogens face is the low iron availability in host tissues, which sequester iron as a protective mechanism against bacterial infections. The P. aeruginosa Fur regulator responds to low iron conditions by de-repressing virulence factors, including siderophores, heme uptake system, exotoxin A, and proteases (54). We now show that H2-T6SS is also negatively regulated by the Fur system, which may facilitate uptake of the bacteria into epithelial cells. The co-regulation of iron acquisition genes and secretion machinery genes have been recently reported for other bacteria, including the T3SS genes of Bordetella pertussis (55), the T6SS of enteroaggregative E. coli (56), and the T6SS and T3SS genes of Edwarsiella tarda (57). The co-regulation of iron uptake systems and secreted virulence factors may function synergistically to improve iron availability to bacteria that are in the iron-limited environment of the host. In addition, other Fur-regulated genes may contribute to bacterial survival in the hostile environment of the host. For instance, Fur regulates the expression of stress genes involved in Helicobacter pylori acid resistance response during the gut infection (58), helping the bacteria to adapt and to adjust to the acidic conditions encountered in the stomach.
Our studies reveal that H2-T6SS mediates interactions with mammalian host cells. Interestingly, phylogenetic analysis demonstrates that the P. aeruginosa H2-T6SS is most closely related to the eukaryotic-cell targeting T6SSs of V. cholerae, A. hydrophila, or H. hepaticus (59–61). Consistent with the notion that H2-T6SS targets eukaryotic cells, we find that H2-T6SS plays a role in PI3K/Akt-dependent bacterial entry into epithelial cells; inactivation of the ATPase ClpV2 results in an ∼50% decrease in bacterial internalization and a corresponding decrease in Akt phosphorylation. This raises an interesting question about the mechanism of action of H2-T6SS in facilitating the uptake of bacteria by mammalian cells. Internalization promoting factors could act from the cellular exterior in analogy with the Listeria internalins or Yersinia invasin or from the inside as the T3SS effectors of Shigella or Salmonellae (62). Because an intracellular delivery has been demonstrated for certain VgrG proteins in other Gram-negative bacteria (11, 12, 14), we would favor that one or more H2-T6SS substrates may directly activate PI3K/Akt pathway to trigger cytoskeleton rearrangement and/or to modulate the innate immune response.
In contrast to H2-T6SS, the T3SS regulon of P. aeruginosa is negatively controlled by QS (30) and acts as an anti-internalization factor (63). Thus, H2-T6SS and T3SS are reciprocally regulated, possibly reflecting their divergent functions at different stages of the infection process (colonization versus dissemination) or the infection mode (acute virulence versus chronic persistence). One possibility could have been a direct cross-talk between the two systems, but we did not observe a type III secretion defect in the H2-T6SS mutant (supplemental Fig. S1C). We did not monitor any difference in cell rounding and toxicity (lactate dehydrogenase release) in HeLa and macrophages infected with wild type or H2-T6SS mutant strains (data not shown). This suggests that H2-T6SS is most likely important in establishing chronic infection because it does not seem to play a role in acute toxic events like T3SS.
Taken together, our observations suggest that the H2-T6SS machinery secretes proteins that target eukaryotic host cell. Despite the lack of apparent hcp or vgrG genes within the H2-T6SS locus of PAO1, two sets of protein (Hcp2a-VgrG2a (PA1512-PA1511) and Hcp2b-VgrG2b (PA0263-PA0262)) have been proposed as putative H2-T6SS substrates (3, 4, 42, 64). One can note that neither VgrG2a nor VgrG2b is an orthologue of VgrG2 from PA14, which is homologous to VgrG6 (42). Interestingly, transcriptional analysis has revealed that these putative effectors are co-regulated with the H2-T6SS machinery in PAO1 (65–69). Future studies will be aimed at identifying the secreted substrates of the H2-T6SS machinery from PAO1 and determining whether they target the host eukaryotic cell.
Acknowledgments
We thank Paul Williams for the generous gift of C4-HSL; Romé Voulhoux, Geneviève Ball, and Bérengère Ize for constant support; and Sana's thesis committee for fruitful discussions.
This work was supported, in whole or in part, by EuroPathoGenomics REX Grant LSHB-CT-2005512061-EPG), “Pathomics” ERA-net PATHO Grant ANR-08-PATH-004-01. This work was also supported by funds from the Royal Society Wellcome Trust Grant WT097939 (to A. F.), National Institutes of Health Grants P01 AI053194 and R01 AI065902 (to J. E.), a Ph.D. fellowship from the French Research Ministry (to T. G. S.), a post-doctoral fellowship from the French Research Minister and from EuroPathoGenomics network of excellence (to A. H.), and a Elizabeth Nash Memorial Fellowship from Cystic Fibrosis Research Inc. (to I. B.).

This article contains supplemental Figs. S1–S3.
- T6SS
- type VI secretion system
- T3SS
- type III secretion system
- QS
- quorum sensing
- MEM
- Eagle's minimal essential medium
- C4-HSL
- N-(butanoyl)-l-HSL
- LY
- LY294002
- TSB
- Tryptic Soy Broth.
REFERENCES
- 1. Brencic A., Lory S. (2009) Determination of the regulon and identification of novel mRNA targets of Pseudomonas aeruginosa RsmA. Mol. Microbiol. 72, 612–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bleves S., Viarre V., Salacha R., Michel G. P., Filloux A., Voulhoux R. (2010) Protein secretion systems in Pseudomonas aeruginosa. A wealth of pathogenic weapons. Int. J. Med. Microbiol. 300, 534–543 [DOI] [PubMed] [Google Scholar]
- 3. Mougous J. D., Cuff M. E., Raunser S., Shen A., Zhou M., Gifford C. A., Goodman A. L., Joachimiak G., Ordoñez C. L., Lory S., Walz T., Joachimiak A., Mekalanos J. J. (2006) A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Filloux A., Hachani A., Bleves S. (2008) The bacterial type VI secretion machine. Yet another player for protein transport across membranes. Microbiology 154, 1570–1583 [DOI] [PubMed] [Google Scholar]
- 5. Basler M., Pilhofer M., Henderson G. P., Jensen G. J., Mekalanos J. J. (2012) Type VI secretion requires a dynamic contractile phage tail-like structure. Nature 483, 182–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cascales E., Cambillau C. (2012) Structural biology of type VI secretion systems. Philos. Trans. R. Soc. Lond. 367, 1102–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pukatzki S., Ma A. T., Sturtevant D., Krastins B., Sarracino D., Nelson W. C., Heidelberg J. F., Mekalanos J. J. (2006) Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pukatzki S., McAuley S. B., Miyata S. T. (2009) The type VI secretion system. Translocation of effectors and effector-domains. Curr. Opin. Microbiol. 12, 11–17 [DOI] [PubMed] [Google Scholar]
- 9. Schwarz S., Hood R. D., Mougous J. D. (2010a) What is type VI secretion doing in all those bugs? Trends microbiol. 18, 531–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jani A. J., Cotter P. A. (2010) Type VI secretion. Not just for pathogenesis anymore. Cell Host Microbe 8, 2–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pukatzki S., Ma A. T., Revel A. T., Sturtevant D., Mekalanos J. J. (2007) Type VI secretion system translocates a phage tail spike-like protein into target cells where it cross-links actin. Proc. Natl. Acad. Sci. U.S.A. 104, 15508–15513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ma A. T., McAuley S., Pukatzki S., Mekalanos J. J. (2009) Translocation of a Vibrio cholerae type VI secretion effector requires bacterial endocytosis by host cells. Cell Host Microbe 5, 234–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ma A. T., Mekalanos J. J. (2010) In vivo actin cross-linking induced by Vibrio cholerae type VI secretion system is associated with intestinal inflammation. Proc. Natl. Acad. Sci. U.S.A. 107, 4365–4370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Suarez G., Sierra J. C., Erova T. E., Sha J., Horneman A. J., Chopra A. K. (2010) A type VI secretion system effector protein, VgrG1, from Aeromonas hydrophila that induces host cell toxicity by ADP ribosylation of actin. J. Bacteriol. 192, 155–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hood R. D., Singh P., Hsu F., Güvener T., Carl M. A., Trinidad R. R., Silverman J. M., Ohlson B. B., Hicks K. G., Plemel R. L., Li M., Schwarz S., Wang W. Y., Merz A. J., Goodlett D. R., Mougous J. D. (2010) A type VI secretion system of Pseudomonas aeruginosa targets a toxin to bacteria. Cell Host Microbe 7, 25–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Russell A. B., Hood R. D., Bui N. K., LeRoux M., Vollmer W., Mougous J. D. (2011) Type VI secretion delivers bacteriolytic effectors to target cells. Nature 475, 343–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Southey-Pillig C. J., Davies D. G., Sauer K. (2005) Characterization of temporal protein production in Pseudomonas aeruginosa biofilms. J. Bacteriol. 187, 8114–8126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Potvin E., Lehoux D. E., Kukavica-Ibrulj I., Richard K. L., Sanschagrin F., Lau G. W., Levesque R. C. (2003) In vivo functional genomics of Pseudomonas aeruginosa for high-throughput screening of new virulence factors and antibacterial targets. Environ. Microbiol. 5, 1294–1308 [DOI] [PubMed] [Google Scholar]
- 19. Lesic B., Starkey M., He J., Hazan R., Rahme L. G. (2009) Quorum sensing differentially regulates Pseudomonas aeruginosa type VI secretion locus I and homologous loci II and III, which are required for pathogenesis. Microbiology 155, 2845–2855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Vasseur P., Vallet-Gely I., Soscia C., Genin S., Filloux A. (2005) The pel genes of the Pseudomonas aeruginosa PAK strain are involved at early and late stages of biofilm formation. Microbiology 151, 985–997 [DOI] [PubMed] [Google Scholar]
- 21. Hoang T. T., Kutchma A. J., Becher A., Schweizer H. P. (2000) Integration-proficient plasmids for Pseudomonas aeruginosa. Site-specific integration and use for engineering of reporter and expression strains. Plasmid 43, 59–72 [DOI] [PubMed] [Google Scholar]
- 22. Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 23. Zaborin A., Romanowski K., Gerdes S., Holbrook C., Lepine F., Long J., Poroyko V., Diggle S. P., Wilke A., Righetti K., Morozova I., Babrowski T., Liu D. C., Zaborina O., Alverdy J. C. (2009) Red death in Caenorhabditis elegans caused by Pseudomonas aeruginosa PAO1. Proc. Natl. Acad. Sci. U.S.A. 106, 6327–6332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goodman A. L., Kulasekara B., Rietsch A., Boyd D., Smith R. S., Lory S. (2004) A signaling network reciprocally regulates genes associated with acute infection and chronic persistence in Pseudomonas aeruginosa. Dev. Cell 7, 745–754 [DOI] [PubMed] [Google Scholar]
- 25. Bordi C., Lamy M. C., Ventre I., Termine E., Hachani A., Fillet S., Roche B., Bleves S., Méjean V., Lazdunski A., Filloux A. (2010) Regulatory RNAs and the HptB/RetS signalling pathways fine-tune Pseudomonas aeruginosa pathogenesis. Mol. Microbiol. 76, 1427–1443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Moscoso J. A., Mikkelsen H., Heeb S., Williams P., Filloux A. (2011) The Pseudomonas aeruginosa sensor RetS switches type III and type VI secretion via c-di-GMP signalling. Environ. Microbiol. 13, 3128–3138 [DOI] [PubMed] [Google Scholar]
- 27. Lazdunski A. M., Ventre I., Sturgis J. N. (2004) Regulatory circuits and communication in Gram-negative bacteria. Nat. Rev. Microbiol. 2, 581–592 [DOI] [PubMed] [Google Scholar]
- 28. Latifi A., Foglino M., Tanaka K., Williams P., Lazdunski A. (1996) A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21, 1137–1146 [DOI] [PubMed] [Google Scholar]
- 29. Brint J. M., Ohman D. E. (1995) Synthesis of multiple exoproducts in Pseudomonas aeruginosa is under the control of RhlR-RhlI, another set of regulators in strain PAO1 with homology to the autoinducer-responsive LuxR-LuxI family. J. Bacteriol. 177, 7155–7163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bleves S., Soscia C., Nogueira-Orlandi P., Lazdunski A., Filloux A. (2005) Quorum sensing negatively controls type III secretion regulon expression in Pseudomonas aeruginosa PAO1. J. Bacteriol. 187, 3898–3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Winson M. K., Camara M., Latifi A., Foglino M., Chhabra S. R., Daykin M., Bally M., Chapon V., Salmond G. P., Bycroft B. W. (1995) Multiple N-acyl-l-homoserine lactone signal molecules regulate production of virulence determinants and secondary metabolites in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 92, 9427–9431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Whiteley M., Lee K. M., Greenberg E. P. (1999) Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U.S.A. 96, 13904–13909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Egland K. A., Greenberg E. P. (1999) Quorum sensing in Vibrio fischeri. Elements of the luxl promoter. Mol. Microbiol. 31, 1197–1204 [DOI] [PubMed] [Google Scholar]
- 34. Ochsner U. A., Wilderman P. J., Vasil A. I., Vasil M. L. (2002) GeneChip expression analysis of the iron starvation response in Pseudomonas aeruginosa. Identification of novel pyoverdine biosynthesis genes. Mol. Microbiol. 45, 1277–1287 [DOI] [PubMed] [Google Scholar]
- 35. Ochsner U. A., Vasil A. I., Vasil M. L. (1995) Role of the ferric uptake regulator of Pseudomonas aeruginosa in the regulation of siderophores and exotoxin A expression. Purification and activity on iron-regulated promoters. J. Bacteriol. 177, 7194–7201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Banin E., Vasil M. L., Greenberg E. P. (2005) Iron and Pseudomonas aeruginosa biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 102, 11076–11081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Lorenzo V., Wee S., Herrero M., Neilands J. B. (1987) Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169, 2624–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ochsner U. A., Johnson Z., Lamont I. L., Cunliffe H. E., Vasil M. L. (1996) Exotoxin A production in Pseudomonas aeruginosa requires the iron-regulated pvdS gene encoding an alternative sigma factor. Mol. Microbiol. 21, 1019–1028 [DOI] [PubMed] [Google Scholar]
- 39. Escolar L., Pérez-Martín J., de Lorenzo V. (1998) Binding of the fur (ferric uptake regulator) repressor of Escherichia coli to arrays of the GATAAT sequence. J. Mol. Biol. 283, 537–547 [DOI] [PubMed] [Google Scholar]
- 40. Barton H. A., Johnson Z., Cox C. D., Vasil A. I., Vasil M. L. (1996) Ferric uptake regulator mutants of Pseudomonas aeruginosa with distinct alterations in the iron-dependent repression of exotoxin A and siderophores in aerobic and microaerobic environments. Mol. Microbiol. 21, 1001–1017 [DOI] [PubMed] [Google Scholar]
- 41. Chi E., Mehl T., Nunn D., Lory S. (1991) Interaction of Pseudomonas aeruginosa with A549 pneumocyte cells. Infect. Immun. 59, 822–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hachani A., Lossi N. S., Hamilton A., Jones C., Bleves S., Albesa-Jové D., Filloux A. (2011) Type VI secretion system in Pseudomonas aeruginosa. Secretion and multimerization of VgrG proteins. J. Biol. Chem. 286, 12317–12327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Fleiszig S. M., Zaidi T. S., Pier G. B. (1995) Pseudomonas aeruginosa invasion of and multiplication within corneal epithelial cells in vitro. Infect. Immun. 63, 4072–4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kierbel A., Gassama-Diagne A., Mostov K., Engel J. N. (2005) The phosphoinositol-3-kinase-protein kinase B/Akt pathway is critical for Pseudomonas aeruginosa strain PAK internalization. Mol. Biol. Cell 16, 2577–2585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kierbel A., Gassama-Diagne A., Rocha C., Radoshevich L., Olson J., Mostov K., Engel J. (2007) Pseudomonas aeruginosa exploits a PIP3-dependent pathway to transform apical into basolateral membrane. J. Cell Biol. 177, 21–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Engel J., Eran Y. (2011) Subversion of mucosal barrier polarity by Pseudomonas aeruginosa. Front. Microbiol. 2, 114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Garvis S., Munder A., Ball G., de Bentzmann S., Wiehlmann L., Ewbank J. J., Tümmler B., Filloux A. (2009) Caenorhabditis elegans semi-automated liquid screen reveals a specialized role for the chemotaxis gene cheB2 in Pseudomonas aeruginosa virulence. PLoS Pathogens 5, e1000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Tan M. W., Ausubel F. M. (2000) Caenorhabditis elegans. A model genetic host to study Pseudomonas aeruginosa pathogenesis. Curr. Opin. Microbiol. 3, 29–34 [DOI] [PubMed] [Google Scholar]
- 49. Liu H., Coulthurst S. J., Pritchard L., Hedley P. E., Ravensdale M., Humphris S., Burr T., Takle G., Brurberg M. B., Birch P. R., Salmond G. P., Toth I. K. (2008) Quorum sensing coordinates brute force and stealth modes of infection in the plant pathogen Pectobacterium atrosepticum. PLoS Pathogens 4, e1000093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ishikawa T., Rompikuntal P. K., Lindmark B., Milton D. L., Wai S. N. (2009) Quorum sensing regulation of the two hcp alleles in Vibrio cholerae O1 strains. PLoS One 4, e6734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zheng J., Shin O. S., Cameron D. E., Mekalanos J. J. (2010) Quorum sensing and a global regulator TsrA control expression of type VI secretion and virulence in Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 107, 21128–21133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Khajanchi B. K., Sha J., Kozlova E. V., Erova T. E., Suarez G., Sierra J. C., Popov V. L., Horneman A. J., Chopra A. K. (2009) N-Acylhomoserine lactones involved in quorum sensing control the type VI secretion system, biofilm formation, protease production, and in vivo virulence in a clinical isolate of Aeromonas hydrophila. Microbiology 155, 3518–3531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zhang W., Xu S., Li J., Shen X., Wang Y., Yuan Z. (2011) Modulation of a thermoregulated type VI secretion system by AHL-dependent quorum sensing in Yersinia pseudotuberculosis. Arch. Microbiol. 193, 351–363 [DOI] [PubMed] [Google Scholar]
- 54. Cornelis P., Matthijs S., Van Oeffelen L. (2009) Iron uptake regulation in Pseudomonas aeruginosa. Biometals 22, 15–22 [DOI] [PubMed] [Google Scholar]
- 55. Brickman T. J., Cummings C. A., Liew S. Y., Relman D. A., Armstrong S. K. (2011) Transcriptional profiling of the iron starvation response in Bordetella pertussis provides new insights into siderophore utilization and virulence gene expression. J. Bacteriol. 193, 4798–4812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Brunet Y. R., Bernard C. S., Gavioli M., Lloubès R., Cascales E. (2011) An epigenetic switch involving overlapping fur and DNA methylation optimizes expression of a type VI secretion gene cluster. PLoS Genet. 7, e1002205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Chakraborty S., Sivaraman J., Leung K. Y., Mok Y. K. (2011) Two-component PhoB-PhoR regulatory system and ferric uptake regulator sense phosphate and iron to control virulence genes in type III and VI secretion systems of Edwardsiella tarda. J. Biol. Chem. 286, 39417–39430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Gancz H., Censini S., Merrell D. S. (2006) Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect. Immun. 74, 602–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Bingle L. E., Bailey C. M., Pallen M. J. (2008) Type VI secretion. A beginner's guide. Curr. Opin. Microbiol. 11, 3–8 [DOI] [PubMed] [Google Scholar]
- 60. Boyer F., Fichant G., Berthod J., Vandenbrouck Y., Attree I. (2009) Dissecting the bacterial type VI secretion system by a genome wide in silico analysis. What can be learned from available microbial genomic resources? BMC Genomics 10, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Schwarz S., West T. E., Boyer F., Chiang W. C., Carl M. A., Hood R. D., Rohmer L., Tolker-Nielsen T., Skerrett S. J., Mougous J. D. (2010b) Burkholderia type VI secretion systems have distinct roles in eukaryotic and bacterial cell interactions. PLoS Pathogens 6, e1001068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Cossart P., Sansonetti P. J. (2004) Bacterial invasion. The paradigms of enteroinvasive pathogens. Science 304, 242–248 [DOI] [PubMed] [Google Scholar]
- 63. Engel J., Balachandran P. (2009) Role of Pseudomonas aeruginosa type III effectors in disease. Curr. Opin. Microbiol. 12, 61–66 [DOI] [PubMed] [Google Scholar]
- 64. Barret M., Egan F., Fargier E., Morrissey J. P., O'Gara F. (2011) Genomic analysis of the type VI secretion systems in Pseudomonas spp. Novel clusters and putative effectors uncovered. Microbiology 157, 1726–1739 [DOI] [PubMed] [Google Scholar]
- 65. Wagner V. E., Bushnell D., Passador L., Brooks A. I., Iglewski B. H. (2003) Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons. Effects of growth phase and environment. J. Bacteriol. 185, 2080–2095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Schuster M., Lostroh C. P., Ogi T., Greenberg E. P. (2003) Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes. A transcriptome analysis. J. Bacteriol. 185, 2066–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Schuster M., Hawkins A. C., Harwood C. S., Greenberg E. P. (2004) The Pseudomonas aeruginosa RpoS regulon and its relationship to quorum sensing. Mol. Microbiol. 51, 973–985 [DOI] [PubMed] [Google Scholar]
- 68. Castang S., McManus H. R., Turner K. H., Dove S. L. (2008) H-NS family members function coordinately in an opportunistic pathogen. Proc. Natl. Acad. Sci. U.S.A. 105, 18947–18952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Déziel E., Gopalan S., Tampakaki A. P., Lépine F., Padfield K. E., Saucier M., Xiao G., Rahme L. G. (2005) The contribution of MvfR to Pseudomonas aeruginosa pathogenesis and quorum sensing circuitry regulation. Multiple quorum sensing-regulated genes are modulated without affecting lasRI, rhlRI or the production of N-acyl-l-homoserine lactones. Mol. Microbiol. 55, 998–1014 [DOI] [PubMed] [Google Scholar]
- 70. Herrero M., de Lorenzo V., Timmis K. N. (1990) Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172, 6557–6567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Touati D., Jacques M., Tardat B., Bouchard L., Despied S. (1995) Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli. Protective role of superoxide dismutase. J. Bacteriol. 177, 2305–2314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Figurski D. H., Helinski D. R. (1979) Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U.S.A. 76, 1648–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kessler B., de Lorenzo V., Timmis K. N. (1992) A general system to integrate lacZ fusions into the chromosomes of gram-negative eubacteria. Regulation of the Pm promoter of the TOL plasmid studied with all controlling elements in monocopy. Mol. Gen. Genet. 233, 293–301 [DOI] [PubMed] [Google Scholar]
- 74. Bao Y., Lies D. P., Fu H., Roberts G. P. (1991) An improved Tn7-based system for the single-copy insertion of cloned genes into chromosomes of gram-negative bacteria. Gene 109, 167–168 [DOI] [PubMed] [Google Scholar]
- 75. Koch B., Jensen L. E., Nybroe O. (2001) A panel of Tn7-based vectors for insertion of the gfp marker gene or for delivery of cloned DNA into Gram-negative bacteria at a neutral chromosomal site. J. Microbiol. Methods 45, 187–195 [DOI] [PubMed] [Google Scholar]
- 76. Kaniga K., Delor I., Cornelis G. R. (1991) A wide-host-range suicide vector for improving reverse genetics in gram-negative bacteria. Inactivation of the blaA gene of Yersinia enterocolitica. Gene 109, 137–141 [DOI] [PubMed] [Google Scholar]
- 77. Engel J. (2007) Pseudomonas: A Model System in Biology (Ramos J. L., Filloux A., eds) pp. 343–368, Springer, New York [Google Scholar]