Background: The contribution of protein dynamics to protein evolution is poorly understood.
Results: Convergent, parallel, and divergent evolution of protein dynamics and function were found in a panel of antibodies.
Conclusion: Antibody dynamics broadens the range of targets recognized and then is tailored by evolution for specificity.
Significance: The results support emerging models for the evolution of novel protein function.
Keywords: Antibodies, Evolution, Immunology, Isothermal Titration Calorimetry, Protein Dynamics, Spectroscopy, Flexibility, Photon Echo
Abstract
The immune system is remarkable in its ability to produce antibodies (Abs) with virtually any specificity from a limited repertoire of germ line precursors. Although the contribution of sequence diversity to this molecular recognition has been studied for decades, recent models suggest that protein dynamics may also broaden the range of targets recognized. To characterize the contribution of protein dynamics to immunological molecular recognition, we report the sequence, thermodynamic, and time-resolved spectroscopic characterization of a panel of eight Abs elicited to the chromophoric antigen 8-methoxypyrene-1,3,6-trisulfonate (MPTS). Based on the sequence data, three of the Abs arose from unique germ line Abs, whereas the remaining five comprise two sets of siblings that arose by somatic mutation of a common precursor. The thermodynamic data indicate that the Abs recognize MPTS via a variety of mechanisms. Although the spectroscopic data reveal small differences in protein dynamics, the anti-MPTS Abs generally show similar levels of flexibility and conformational heterogeneity, possibly representing the convergent evolution of the dynamics necessary for function. However, one Ab is significantly more rigid and conformationally homogeneous than the others, including a sibling Ab from which it differs by only five somatic mutations. This example of divergent evolution demonstrates that point mutations are capable of fixing significant differences in protein dynamics. The results provide unique insight into how high affinity Abs may be produced that bind virtually any target and possibly, from a more general perspective, how new protein functions are evolved.
Introduction
Proteins are inherently dynamic, and there is a growing appreciation that dynamics may contribute to biological function (1–7). Molecular recognition underlies virtually all of the functions that a protein may possess, and the limiting models of molecular recognition, lock-and-key and induced fit or conformational selection, are differentiated by the underlying protein dynamics. Protein dynamics may also be critical for the evolution of new protein functions by bestowing progenitor proteins with flexibility and conformational heterogeneity, where different conformations have unique physicochemical properties (8–11). This could be the origin of the now well documented tendency of many proteins to have minor activities (variously referred to as substrate ambiguity (12), moonlighting (13–16), gene sharing (17, 18), or promiscuous activities (17, 19)), that, when beneficial, could favor the retention of gene duplicates (20–22) and eventually emerge as the major activity of the protein once the associated conformation is stabilized by mutation.
Nowhere is the importance of molecular recognition and its evolution more evident than during an immune response, where antibodies (Abs)3 specific for virtually any target may be produced within days of their first encounter. The contribution of sequence diversity to the Ab repertoire and its specificities has been studied intensively for years (23, 24). Although protein dynamics is not implicitly evoked in the widely accepted clonal selection theory of antibody generation (25, 26), its “one receptor-one target” foundation is suggestive of the lock-and-key mechanism of molecular recognition and is thus consistent with a high level of rigidity. This notion is in contrast with the older instructive hypothesis (27, 28), which evoked an induced fit- or conformational selection-like mechanism of molecular recognition and thus more flexible and conformationally heterogeneous Abs. In fact, Abs were one of the first examples where protein dynamics was speculated to contribute to molecular recognition, specifically to broaden the range of potential targets recognized (8–10, 29–36). Unfortunately, characterizing the dynamics of any protein is experimentally challenging, and thus the range of dynamics possible within Abs, and how it might contribute to Ab evolution, has remained difficult to test.
The ability to rigorously characterize the dynamics of a chromophore's environment has been revolutionized by ultrafast nonlinear optical methods, such as three-pulse photon echo peak shift (3PEPS) spectroscopy (37–39). 3PEPS spectroscopy relies on the application of a series of three short laser pulses. The first two pulses inscribe a phase grating in the sample. After a variable time delay, referred to as the population time, a third pulse triggers rephasing of the sample, but only chromophores that have an unaltered phase can contribute to rephasing and thus emit an echo signal. Fluctuations of the environment randomize the phase of the chromophore, which, with increasing population time, results in fewer chromophores contributing to rephasing and an apparent peak shift of the echo signal, commonly described by M(t), the energy gap autocorrelation function. M(t) is conveniently represented in the frequency domain with the corresponding spectral density, ρ(ω) (40), which represents the frequency and amplitude of the environment's motions. Additionally, dynamics that appear static on the time scale of the 3PEPS experiment are manifest as a non-zero asymptote of the peak shift, thus making it possible to detect and quantify static heterogeneity (39, 40).
We have developed a system that allows for the use of 3PEPS to characterize Ab dynamics (8–11, 41). To achieve this, we immunized mice with different chromophore-protein conjugates and isolated Abs that evolved to bind the chromophore. Although Abs generally evolve to bind protein targets, small molecule targets (or antigens (Ags)) have played a central role in the study of Ab structure and function, and the chromophoric Ags are expected to bind as would any other. The Ab-Ag complexes were characterized using 3PEPS and transient grating (TG) spectroscopy, and motions on several distinct sets of time scales were observed. For one anti-fluorescein (Fl) Ab, we also characterized protein dynamics as a function of somatic evolution (affinity maturation) (10, 11). We found that evolution significantly restricted the observed dynamics, consistent with the evolution of flexible germ line Abs that are polyspecific and recognize their Ags via an induced fit- or conformational selection-like mechanism, into more rigid Abs that more specifically recognize their Ags via a lock-and-key-like mechanism.
To further explore the range of dynamics inherent to an immune response and how the dynamics might be tailored by somatic evolution, we now report the detailed analysis of eight Abs that evolved in mice to bind the chromophore 8-methoxypyrene-1,3,6-trisulfonate (MPTS; Fig. 1). Sequence, thermodynamic, and dynamic characterization via 3PEPS and TG spectroscopy reveal the range in affinities and dynamics possessed by the antibodies that constitute the immune response to MPTS and also provide interesting examples of parallel, convergent, and divergent evolution.
FIGURE 1.
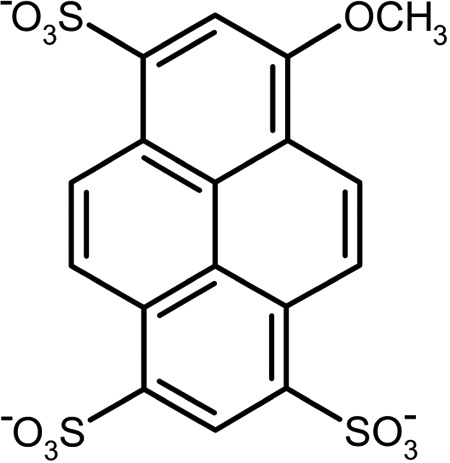
Structure of MPTS.
EXPERIMENTAL PROCEDURES
Generation and Sequencing of Anti-MPTS Ab
Production of the panel of monoclonal antibodies was performed as described previously using an MPTS-keyhole limpet hemocyanin conjugate (8). Heavy-chain variable region (VH) and light-chain variable region (VL) gene sequences were also determined as described previously (41). In brief, cDNA was synthesized using reverse transcriptase (Ambion) from mRNA isolated from fresh hybridoma cells. A primer set specifically designed for mouse variable immunoglobulin regions (42, 43) was then used to PCR-amplify the VH and VL genes from cDNA. The resulting gene fragments were sequenced using standard procedures. Nucleotide sequences of the VH and VL genes are provided in supplemental Data Set S1.
Isothermal Titration Calorimetry
Isothermal titration calorimetric experiments were performed using an MCS-ITC or a VP-ITC calorimeter (GE Healthcare) and analyzed with Origin 6.0 (Microcal) software. Titrations were performed between 20 and 35 °C. In each titration, 19 consecutive injections of MPTS every 240 s followed a single, preliminary injection into an 8 μm solution of Ab (IgG) in phosphate-buffered saline (PBS), to a final concentration of 150 μm MPTS. A titration of MPTS into buffer alone was used to correct for the heat of dilution of the titrant. The resulting ΔH0 and ΔG0 values were used to calculate TΔS0 = ΔH0 − ΔG0 at each temperature and ΔCp0 from a linear fit to ΔH0(T). The total entropy was approximated as follows,
 |
where ΔSconf0 is the entropy change from changes of internal motions of protein and ligand (41).
3PEPS and Transient Grating Experiments
Samples containing 700 μm Ab (IgG) and 700 μm MPTS in PBS were washed extensively with PBS by repeated dilution and microfiltration (10,000 molecular weight cut-off, Amicon Ultra, Millipore) to remove unbound MPTS, until the filtrate was free of MPTS as measured by UV-visible absorption. The experimental setup for 3PEPS and TG has been described previously (8). Briefly, the damped output of a Ti:sapphire regenerative amplifier system (Spectra Physics Spitfire, 5 kHz, 200-mJ pulses at 825 nm) was doubled in a 0.2-mm-thick type I BBO crystal and sent through a prism compressor, producing 416-nm pulses with ∼45-fs duration, as determined by two-photon absorption in DMSO. Typical pulse energies at the sample were ∼20 nJ/pulse. Beam splitters were then used to generate three beams of approximately equal intensity, two of which were then variably delayed using a pair of delay stages (PM6000, Kensington Laboratories).
After passing through the delay stages, the three beams were arranged in an equilateral triangle of ∼7 mm/side and focused with a 200-mm focal length plano-convex fused silica lens into a home-built spinning cell with a path length of 200 μm containing sample. The entire setup was maintained at 22 ± 1 °C. Proper masks were used to block the excitation light and allow only signal in the two phase-matching directions k1−k2+k3 and −k1+k2+k3 to pass to two silica photodiodes (DET110, Thorlabs) connected to lock-in amplifiers (SR830, Stanford). The lock-in amplifiers were referenced to a chopper (3501 Optical Chopper, New Focus, Inc.) phase-locked to the Q-switch RF frequency of the amplifier system, which was placed in the path of the fixed-delay beam. For the 3PEPS experiments, the coherence period, τ, the delay between the first and second pulses, was scanned from −150 to 150 fs for a fixed value of the population period, T, the delay between the second and third pulses. The peak shift for a given population period was then calculated from the average shift from zero of the temporal signal maximum in τ for the two phase-matching directions. As many as 40 scans at a given population time were averaged for a total of 40–50 exponentially spaced population times. To detect day-to-day instrument drift, the terminal peak shift (100 ps population time) of MPTS in a solution containing an anti-Fl antibody (700 μm) was measured on each day that experiments were performed to ensure reproducibility. At least three independent trials were performed for each 3PEPS and TG experiment. Sample degradation was eliminated as a concern by monitoring the UV-visible spectrum of the samples before and after a 3PEPS or TG experiment.
Spectroscopic Data Analysis
TG data were fit to double exponential decays with Origin 6.0 (Microcal). Parameters of the spectral densities were obtained from a simultaneous fit of the experimental 3PEPS data and the steady-state absorption spectra using Mukamel's response function formalism (40), as described in detail previously (41). In brief, the total spectral density, ρ(ω), was calculated from the sum of the vibrations of the chromophore, ρMPTS(ω), and the vibrations of the protein, ρAb(ω). Intramolecular vibrational frequencies and excitation-induced displacements of MPTS were obtained from quantum chemical calculations and validated by experimentally determined Raman frequencies, as described previously (8). ρAb(ω) was modeled as the sum of an underdamped Brownian oscillator term (which we refer to as the Brownian oscillator term, ρBO(ω)) for the fastest motions and one or two overdamped Brownian oscillator terms (which we refer to as the Kubo terms, ρKi(ω)) for slower motions. Specifically, the underdamped Brownian oscillator term,
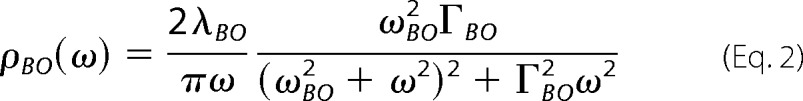 |
was used to represent the inertial subpicosecond protein dynamics, where λBO is the reorganization energy, ωBO is the frequency, and ΓBO is the damping constant of the Brownian oscillator, and the Kubo terms,
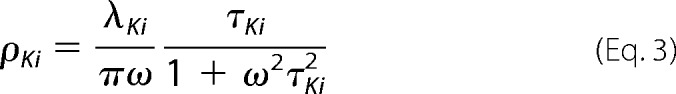 |
were used to represent the picosecond dynamics, where λK and τK are the reorganization energy and time constant, respectively. Signals for the 3PEPS experiment and the steady-state absorption spectra were calculated from the line-broadening function g(t) by using standard procedures (37, 38, 40). g(t) was calculated from ρ(ω) using the following expression.
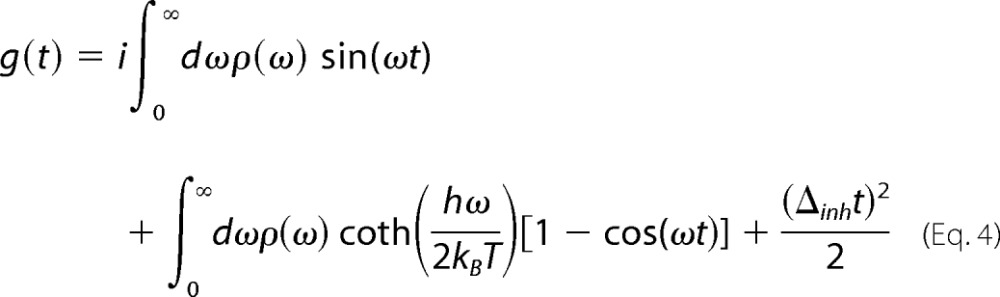 |
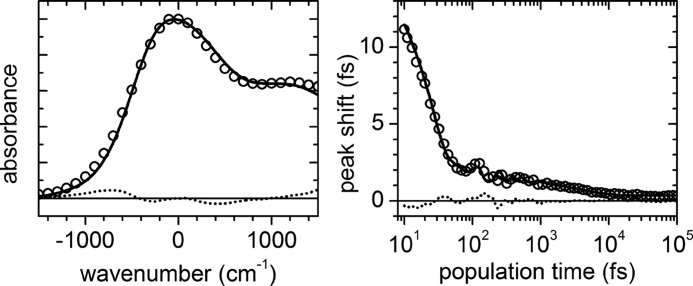
The parameters in ρAb(ω) and the amount of static inhomogeneity, Δinh, in g(t) were varied using a least-squares Simplex fit algorithm to obtain the best fit of both the experimental 3PEPS data (for population times from 10 fs to 100 ps) and the steady-state absorption spectra using a custom suite of C programs developed in our laboratory based on code kindly provided by Dr. Delmar Larsen (University of California, Davis, CA). As previously observed (11), the amplitude (λBO), frequency (ωBO), and damping constant (ΓBO) of the Brownian oscillator term could not be fit unambiguously; thus, the values of ωBO and ΓBO were fixed to average values from free fits to the 3PEPS decays. Simulation of the data with different values for these fixed terms resulted in only small variations in the remaining terms. All computations were performed on the 64-bit Linux cluster Garibaldi at the Scripps Research Institute. A representative fit is shown in Fig. 2.
FIGURE 2.
Fit to steady-state absorption spectrum and 3PEPS decay of Ab 2E8 using the nonlinear response function formalism (circles, data points; lines, best fit to data; dotted lines, residuals).
RESULTS
Production and Sequence Characterization of Anti-MPTS Abs
Anti-MPTS monoclonal IgG Abs were purified from ascites supernatants produced by standard techniques from Swiss Webster mice challenged with MPTS conjugated to keyhole limpet hemocyanin (44). Nucleotide sequences (supplemental material) were determined for each Ab from PCR-amplified VH and VL gene fragments using cDNA obtained from the corresponding hybridoma cell lines (42, 43). From the sequence data, we found that the eight mature anti-MPTS Abs arose from five different germ line precursor Abs (Figs. 3 and 4). Abs 4B2, 8H9, and 9D5 each arose from distinct germ line precursors. However, based on the nucleotide sequence of the stochastically generated V-D-J junction, it is evident that 3E6 and 7D5 are sibling Abs that diverged from a common germ line Ab and are thus differentiated only by the somatic mutations acquired after divergence. Similarly, Abs, 2E8, 3D3, and 10B8 are also siblings that diverged from a common germ line precursor.
FIGURE 3.
Anti-MPTS Ab sequences. The CDRs that form the Ab binding site are underlined. Kabat numbering is shown at the top (60).
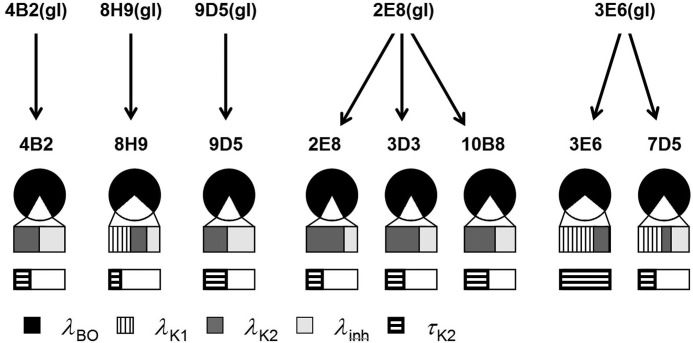
FIGURE 4.
Evolutionary relationships between germ line (top) and affinity-matured (bottom) anti-MPTS Abs. For each Ab, the fractional amplitudes of elastic (λBO) and inelastic fluctuations (λK1,K2 and λinh) are shown in the pie chart and associated expansion, respectively. The bar diagram at the bottom shows the values of the time constant associated with barrier crossing between conformational substates (τK2) for each Ab (normalized to 13 ps). See “Discussion” and Fig. 9 for definitions of the parameters.
Thermodynamics of MPTS Binding
The MPTS dissociation constant (KD) and binding enthalpy (ΔH°) were determined for each Ab by isothermal titration calorimetry and were used to calculate the binding-induced changes in free energy (ΔG0), entropy (ΔS0), and heat capacity (ΔCp0) (Table 1). Generally, KD and ΔG0 values ranged from 20 to 450 nm and from −8.7 to −10.5 kcal/mol, respectively. Large variations were also observed among the constituent ΔH0 and TΔS0 values, which varied by 4.4 and 5.6 kcal/mol, respectively. Interestingly, Ab 10B8 binds MPTS with ∼5-fold and ∼2-fold lower affinity than its two siblings 2E8 and 3D3, respectively, due to differences in both ΔH0 and ΔS0. Moreover, Ab 3E6 binds MPTS with almost 3-fold higher affinity than its sibling Ab 7D5.
TABLE 1.
Thermodynamic parameters at T = 298 K
| KD | ΔG0 | ΔH0 | TΔS0 | ΔCp0 | TΔSconf0 | |
|---|---|---|---|---|---|---|
| nm | kcal/mol | kcal/mol | kcal/mol | kcal/mol/K | kcal/mol | |
| 4B2 | 65 ± 48 | −9.9 ± 0.4 | −11.32 ± 0.20 | −1.5 ± 0.2 | −0.158 ± 0.033 | −9.9 ± 2.8 |
| 8H9 | 276 ± 65 | −9.0 ± 0.1 | −12.08 ± 0.07 | −3.1 ± 0.1 | −0.126 ± 0.012 | −10.0 ± 1.0 |
| 9D5 | 21 ± 6 | −10.5 ± 0.2 | −9.65 ± 0.07 | 0.9 ± 0.1 | −0.128 ± 0.011 | −6.1 ± 1.0 |
| 2E8 | 46 ± 10 | −10.1 ± 0.1 | −12.94 ± 0.02 | −2.9 ± 0.1 | −0.145 ± 0.003 | −10.5 ± 0.3 |
| 3D3 | 95 ± 10 | −9.6 ± 0.1 | −10.91 ± 0.04 | −1.3 ± 0.1 | −0.155 ± 0.006 | −9.9 ± 0.5 |
| 10B8 | 209 ± 15 | −9.2 ± 0.1 | −11.77 ± 0.03 | −2.6 ± 0.1 | −0.128 ± 0.005 | −11.2 ± 0.4 |
| 3E6 | 168 ± 35 | −9.3 ± 0.1 | −14.04 ± 0.04 | −4.7 ± 0.1 | −0.112 ± 0.007 | −10.0 ± 0.6 |
| 7D5 | 450 ± 110 | −8.7 ± 0.1 | −14.09 ± 0.07 | −5.4 ± 0.1 | −0.105 ± 0.012 | −10.4 ± 1.0 |
The Abs all showed negative ΔCp0 values that are somewhat smaller than those observed previously with anti-Fl Abs, suggesting that the hydrophobic surface area buried upon binding is smaller for the Ab-MPTS complexes (45), which, given the similar surface area of the chromophores, suggests that MPTS may be more solvent-exposed. The ΔCp0 values were also used to estimate the binding-induced changes in conformational entropy, ΔSconf0 (Table 1). With the exception of Ab 9D5, all of the Abs show rather similar TΔSconf0 values, varying between −9.9 and −11.2 kcal/mol. Thus, the large differences in TΔSconf0 among the different Abs appear to be due mostly to solvent release (i.e. hydrophobic contributions to binding) and not to conformational dynamics of the protein or ligand.
Spectroscopic Characterization of MPTS-Ab Complexes
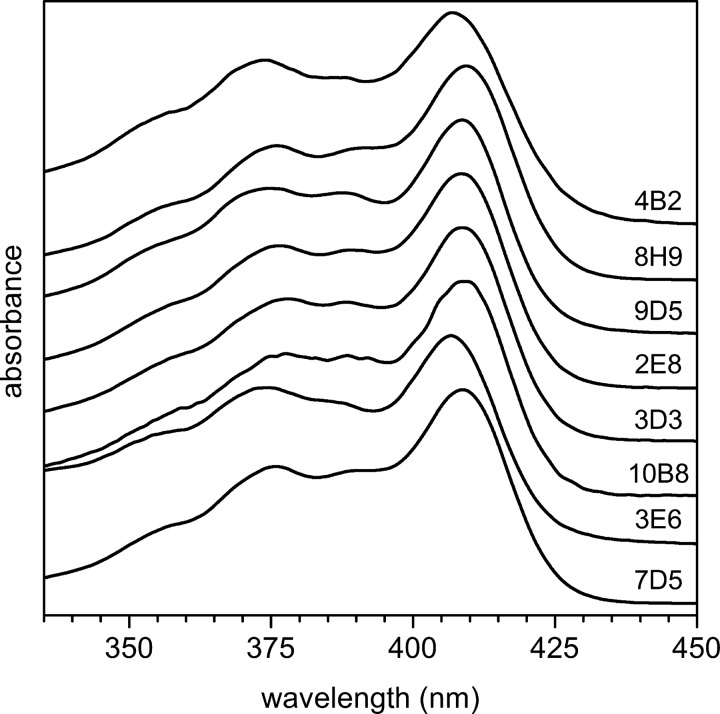
The steady-state absorption spectra show that each Ab binds MPTS in a unique environment that is distinct from free solvent (Fig. 5 and Table 2). Relative to MPTS free in solution (PBS, pH 7.4), Ab binding causes a red shift in the absorption maximum of 3–6 nm and, with the exception of Ab 4B2, a reduction of the absorption line width. Ab 9D5, which has the most positive ΔSconf0, also shows the narrowest line width. Interestingly, MPTS bound to the sibling Abs 3E6 or 7D5 exhibited distinctly different absorption maxima of 406.6 and 408.8 nm, respectively, suggesting that they evolved to bind MPTS rather differently. In contrast, despite the differences in binding thermodynamics, MPTS bound to the any of the triplet sibling Abs, 2E8, 3D3, or 10B8, exhibited nearly identical absorption maxima and line widths.
FIGURE 5.
Absorption spectra of Ab-MPTS complexes.
TABLE 2.
Absorption maxima and line widths of MPTS in water or bound to an anti-MPTS Ab
| λmax | fwhm | |
|---|---|---|
| nm | nm | |
| Water | 403.7 | 20.0 |
| 4B2 | 407.1 | 22.3 |
| 8H9 | 409.5 | 18.5 |
| 9D5 | 408.5 | 17.8 |
| 2E8 | 408.6 | 18.2 |
| 3D3 | 408.9 | 18.0 |
| 10B8 | 408.9 | 18.0 |
| 3E6 | 406.6 | 19.1 |
| 7D5 | 408.8 | 18.8 |
TG experiments were used to determine the excited state lifetime of MPTS when bound to the anti-MPTS Abs (Table 3). For all Abs, at least two exponential functions were required to fit the TG decay: a shorter time constant of 0.13–3.1 ps and a longer time constant of 84–287 ps. The decay component with the shorter time constant is attributed to coherent dynamics, except in the case of Abs 3E6, 7D5, and 8H9, where its value suggests that it reflects population dynamics. With all eight complexes, the longer time constant is attributed to the excited state lifetime of MPTS (2τTG). In addition, Abs 4B2 and 9D5 show a rise in the TG signal with a 130 fs and 220 fs time constant, respectively, which is suggestive of excited state population dynamics. Similar subpicosecond rises have previously been assigned to intersystem crossing and/or vibrational relaxation (46, 47), or to photoinduced charge or energy transfer (48).
TABLE 3.
Parameters for multiexponential fits to transient grating data
| a1 | τ1 | a2 | τ2 | |
|---|---|---|---|---|
| % | fs | % | ps | |
| 4B2 | −29 ± 12 | 130 ± 30 | 100 | 287 ± 37 |
| 8H9 | 42.0 ± 1.2 | 3150 ± 130 | 58.0 ± 1.9 | 234 ± 16 |
| 9D5 | −54.1 ± 8.8 | 224 ± 15 | 100 | 111 ± 2 |
| 2E8 | 22.2 ± 0.9 | 440 ± 60 | 77.8 ± 0.1 | 136 ± 9 |
| 3D3 | 15.1 ± 1.2 | 650 ± 250 | 84.9 ± 2.2 | 84 ± 9 |
| 10B8 | 15.5 ± 0.1 | 410 ± 80 | 84.5 ± 0.3 | 96 ± 1 |
| 3E6 | 40.3 ± 0.8 | 2910 ± 50 | 59.7 ± 1.9 | 89 ± 1 |
| 7D5 | 35.3 ± 1.1 | 2840 ± 170 | 64.7 ± 2.3 | 228 ± 6 |
The decay of the TG signals is similar for the 2E8, 3D3, and 10B8 group of siblings. Abs 3D3 and 10B8 show, within error, identical TG decays, whereas Ab 2E8 shows only small variations in the relative amplitudes and the longer time constants. The 3E6 and 7D5 siblings show indistinguishable amplitudes and time constants for the ∼3 ps decay component, and identical amplitudes for the longer decay component. However, the longer time constant varies from 89 ps for 3E6 to 228 ps for 7D5, which along with the absorption spectra discussed above, further suggests that MPTS experiences a different microenvironment in the two Abs.
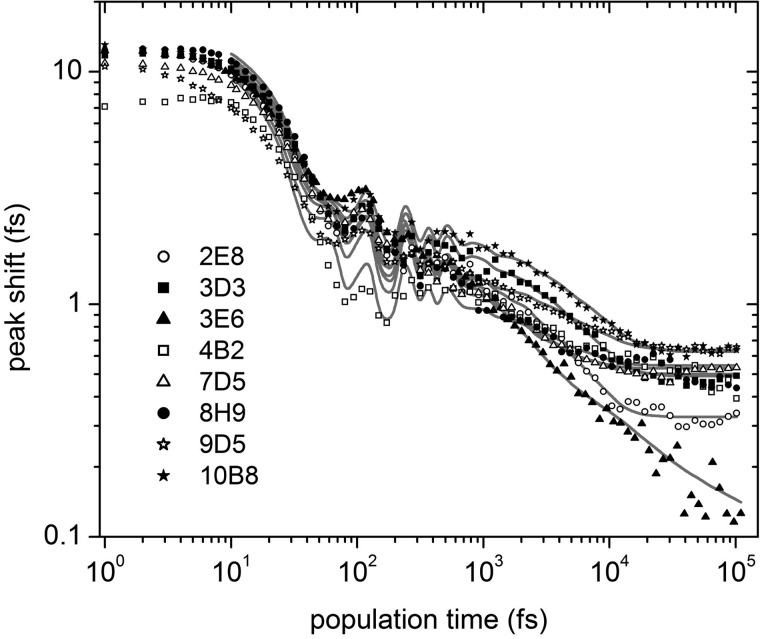
3PEPS decays were measured up to 400 ps population time, and M(t) and ρAb(ω) were calculated from the data (see “Experimental Procedures”). As previously observed for the Ab-Fl complexes (41), we found 3PEPS decays for the Ab-MPTS complexes on three time scales (Fig. 6 and Table 4): a sub-100 fs decay corresponding to inertial protein or water motions; a decay component on a 3–10 ps time scale that is assigned to subconformational dynamics (11, 41); and a longtime signal offset (i.e. corresponding to a decay on time scales longer than the experimental time window) that is assigned to conformational dynamics (11, 41). In addition, sibling Abs 3E6 and 7D5 as well as Ab 8H9 show an additional decay component with time constants of ∼1.2 and 0.5 ps, respectively. This additional decay could result from a second tier of subconformational protein dynamics or from motions of proximal water molecules whose motions are somewhat restricted relative to bulk water (a 500 fs decay component was observed previously for free MPTS in water (8)). However, as discussed above, the same three Abs also showed an atypical ∼3 ps component in the TG decay. It is thus more likely that the additional 3PEPS decay component for Abs 3E6, 7D5, and 8H9 results from spectral shifting due to excited state population dynamics. The 3PEPS decay of Ab 4B2, which shows an unusually broad absorption spectrum and an initial rise in its TG signal, also differs significantly from those of all other Abs at short population times. This further suggests that MPTS bound to Ab 4B2 undergoes a unique photoinduced process on a subpicosecond time scale.
FIGURE 6.
3PEPS decays for the Ab-MPTS complexes. Lines are fits to the data.
TABLE 4.
Fit parameters for 3PEPS data
| λBO | ωBOa | ΓBOa | λK1 | τK1 | λK2 | τK2 | Δinh | |
|---|---|---|---|---|---|---|---|---|
| cm−1 | cm−1 | cm−1 | cm−1 | fs | cm−1 | ps | cm−1 | |
| 4B2 | 699 ± 60 | 550 | 320 | 68 ± 12 | 4.1 ± 1.4 | 172 ± 34 | ||
| 8H9 | 469 ± 19 | 550 | 320 | 73 ± 19 | 448 ± 163 | 53 ± 23 | 3.2 ± 1.0 | 140 ± 3 |
| 9D5 | 560 ± 13 | 550 | 320 | 49 ± 12 | 6 ± 2 | 153 ± 7 | ||
| 2E8 | 527 ± 20 | 550 | 320 | 68 ± 9 | 4.2 ± 0.5 | 101 ± 8 | ||
| 3D3 | 489 ± 14 | 550 | 320 | 61 ± 5 | 4.8 ± 0.7 | 116 ± 5 | ||
| 10B8 | 492 ± 6 | 550 | 320 | 70 ± 6 | 6.2 ± 0.5 | 141 ± 13 | ||
| 3E6 | 490 ± 29 | 550 | 320 | 129 ± 5 | 1259 ± 383 | 61 ± 15 | 13 ± 3 | 34 ± 59 |
| 7D5 | 518 ± 7 | 550 | 320 | 59 ± 10 | 1208 ± 102 | 24 ± 6 | 4.6 ± 1.5 | 141 ± 7 |
a Parameter fixed during fit.
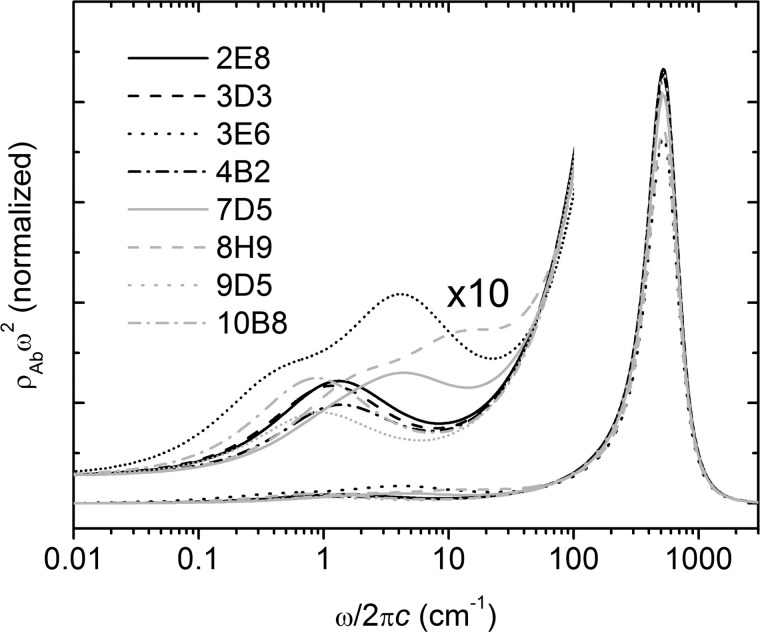
Despite these differences, the 3PEPS decays and ρAb(ω) are surprisingly uniform among the majority of the anti-MPTS Abs, both in time constants and amplitudes (Tables 4 and 5 and Figs. 6 and 7). However, Ab 3E6 was an exception, as it showed an unusually long τK2 and an unusually low λinh, both of which are not only significantly different from the other Abs with different germ line origins but also significantly different from Ab 7D5, which evolved from the same germ line precursor.
TABLE 5.
Normalized reorganization energies
| λBO | λK1 | λK2 | λinha | |
|---|---|---|---|---|
| % | % | % | % | |
| 4B2 | 83.4 ± 7.1 | 8.1 ± 1.5 | 8.5 ± 0.4 | |
| 8H9 | 73.0 ± 2.9 | 11.4 ± 3.0 | 8.2 ± 3.6 | 7.4 ± 0.1 |
| 9D5 | 84.1 ± 2.0 | 7.4 ± 1.9 | 8.5 ± 0.1 | |
| 2E8 | 85.1 ± 3.3 | 10.9 ± 1.5 | 4.0 ± 0.2 | |
| 3D3 | 83.9 ± 2.4 | 10.5 ± 0.8 | 5.6 ± 0.1 | |
| 10B8 | 80.6 ± 1.1 | 11.5 ± 0.9 | 7.9 ± 0.2 | |
| 3E6 | 71.8 ± 4.3 | 18.9 ± 0.7 | 8.9 ± 2.3 | 0.4 ± 3.5 |
| 7D5 | 79.8 ± 1.1 | 9.1 ± 1.5 | 3.7 ± 0.9 | 7.4 ± 0.1 |
a λinh = Δinh2/2kBT, T = 298 K.
FIGURE 7.
Spectral densities of Ab-MPTS complexes.
DISCUSSION
Molecular recognition is central to the biological function of any protein. Protein dynamics, by definition, plays a role in molecular recognition because it is what differentiates the limiting mechanisms of conformational selection and induced fit or lock-and-key. Moreover, it has been suggested that dynamics may broaden the range of potential activities possessed by a protein and thus facilitate the evolution of novel activities (for reviews, see Refs. 7, 49, and 50). However, the range of dynamics available to proteins are possible within a specific protein fold and how these dynamics may be tailored by evolution have remained difficult to gauge experimentally.
The immune system has long served as a paradigm for the study of molecular recognition and its evolution. In this case, protein dynamics may play an especially key role because the number of foreign molecules that must be recognized far exceeds the potential number of different Abs, which is limited by available B-cells (32). We previously characterized a panel of anti-Fl Abs (41) as well as the dynamics of one as a function of its somatic evolution, which revealed that evolution significantly rigidified the Ab and restricted its conformational heterogeneity (10, 11). To further characterize the range of dynamics possible within the immunoglobulin fold common to all Abs and how it might be tailored by somatic evolution, we raised a panel of eight Abs to the chromophoric Ag MPTS. Sequencing revealed that a wide variety of germ line Abs may serve as suitable starting points for the evolution of high affinity MPTS binders. Generally, the immunoglobulin fold of an Ab is composed of two chains, VL and VH, with the combining site composed of three hypervariable loops, or complementarity determining regions (CDRs), contributed from each chain. As is common with Abs, only VL CDR1 and VH CDR3 show variations in length among the anti-MPTS Abs, with the former being 10–16 amino acids in length and the latter being 7–13 amino acids in length. In addition to being the longest, the 4B2 VH CDR3 also contains the largest number of charged residues (one Arg, one Glu, and three Asp residues), whereas the VH CDR3 loops of 8H9, 7D5, and 3E6 are the shortest and, in the case of the 7D5 and 3E6, also the most hydrophobic. Interestingly, we also found two groups of sibling Abs, Abs that evolved via somatic mutation from a common germ line precursor (Fig. 4). Thus, this panel of Abs provides an opportunity to study the diversity of protein dynamics available within the immune response to a given Ag and potentially how it is tailored by somatic evolution for molecular recognition.
Antibody Dynamics and Molecular Recognition
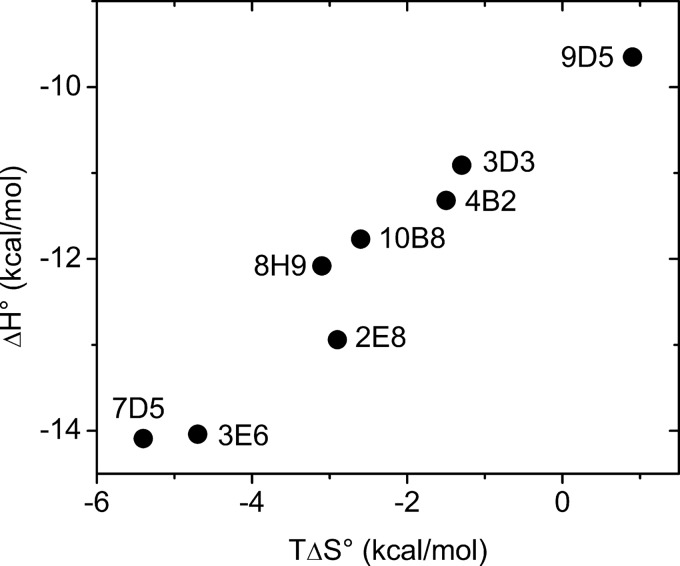
The anti-MPTS Ab evolved to bind MPTS with a relatively broad range in affinities, with KD values ranging from 20 nm (Ab 9D5) to 450 nm (Ab 7D5). These variations generally result from relatively larger variations in ΔH0 and ΔS0, which somewhat offset each other (Fig. 8). Such enthalpy-entropy compensation has been observed previously with other Abs (51), as well as with other ligand-binding proteins (52). Although the entropy differences should be interpreted cautiously because the calculated ΔSconf0 values suggest that, with the exception of Ab 9D5, the differences in binding-induced changes in conformational entropy are smaller, the data clearly demonstrate that a threshold affinity is achieved via different mechanisms of molecular recognition.
FIGURE 8.
Correlation between entropy and enthalpy of binding (T = 298 K).
As an Ab-MPTS complex fluctuates, the changing environment induces shifts in the MPTS transition frequency. In a 3PEPS experiment, this leads to dephasing and a decay in the 3PEPS signal. The time constants of the decay components observed in M(t) are related to the frequency of motions within or between discrete states of the protein, whereas the corresponding amplitudes report on the amount of phase space sampled by a particular decay component. As was observed with anti-Fl Abs (41), the distinct time scales of motion observed in the Ab-MPTS complexes are not consistent with a continuum of motions within the Ab, but rather are reminiscent of Frauenfelder's model of a hierarchical energy landscape wherein a protein exists in a limited number of conformations, each consisting of a large number of conformational substates (53, 54). Thus, we interpret the distinct time scales of motion observed in the Ab-MPTS complexes as different ranks of protein fluctuations corresponding to fluctuations between conformations as well as between and within conformational substates.
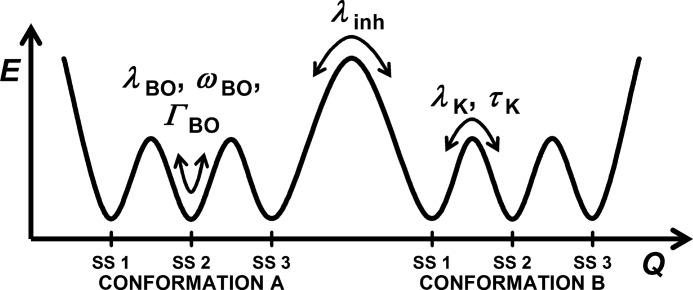
The rigidity of any material may be considered in terms of elastic and inelastic contributions. Rigid materials possess a higher level of elastic relative to inelastic motions. When described with a hierarchical energy landscape, a more rigid protein thus possesses higher levels of elastic fluctuations within conformational substates and, due to relatively higher energy barriers, lower levels of inelastic fluctuations between conformational states and substates (Fig. 9). In a 3PEPS experiment, the fast, inertial dynamics (i.e. motions within a conformational substate) are described by λBO (amplitude), ωBO (frequency), and ΓBO (damping). Little variation in elasticity, as measured with λBO, was observed among the anti-MPTS Abs, and it is probably determined largely by the motions of the charged side chains that are invariantly present within any protein.
FIGURE 9.
Schematic depiction of the protein fluctuations that give rise to λBO, ωBO, ΓBO, λK, τK, and λinh (41). SS 1–3 are different substates of conformations A and B.
The inelasticity of the Ab-MPTS complexes results from diffusive motion across barriers between different substates that are accessible on the time scale of the 3PEPS experiment, described by λK (amplitude) and τK (time scale), as well as from diffusive motion between distinct conformations that is slow on the 3PEPS time scale and thus manifest as static (conformational) inhomogeneity, λinh. The 3PEPS decays of the 3E6, 7D5, and 8H9 complexes show a component that occurs on the 0.5–1 ps time scale, which probably reflects excited state population dynamics because it also appears to be present in the TG signals. However, each Ab also shows a slower decay in its 3PEPS signals that is not present in its TG decay or with MPTS free in solution, and they are thus assigned to protein motions. The time constants for these diffusive motions (τK2) generally ranged between 3 and 6 ps. The exception is Ab 3E6, for which significantly slower diffusive motion was observed (τK2 = 13 ps). Previous studies of dyes in small molecule solvents showed diffusive motions on the 1 ps time scale (38, 55), a previous study of eosin bound to lysozyme identified diffusive motions on the 7 ps time scale (56), and five of six of the previously reported Ab-Fl complexes showed diffusive motions on the 0.6–2.9 ps time scale, with one showing a 5.2 ps time scale diffusive motion (41). Using the Eyring model (57), the observed time constants translate into barrier heights of 1.8–2.2 kcal/mol for each Ab other than 3E6 and into 2.6 kcal/mol for Ab 3E6.
The static inhomogeneity observed with all but the 3E6-MPTS complex ranged between 101 and 172 cm−1. By comparison, Δinh values for small molecule solvent are zero, whereas values of ∼500 cm−1 have been observed from the dye IR144 in acrylic glasses, which are expected to possess a large range of static environments (58, 59), and the panel of six previously characterized Ab-Fl complexes showed Δinh values that ranged from 136 to 301 cm−1. Ab 3E6 was again the exception because the Δinh value associated with this complex was within experimental error of zero. Along with the slower diffusive motions, this suggests that the 3E6-MPTS complex populates fewer conformations than the other Abs and that those conformations are separated by larger barriers.
Antibody Evolution
The majority of the anti-MPTS Abs evolved from different germ line precursors, and although differences in molecular recognition are apparent from the thermodynamic data, the flexibility and conformational heterogeneity of the Abs are generally remarkably similar. Moreover, the dynamics of the anti-MPTS Abs are similar to those of the previously characterized anti-Fl Abs (41). Although the germ lines of the anti-MPTS Abs have not yet been characterized, based on previous results (10, 11), it is likely that they are more distinct. If this is indeed the case, then the common dynamics may be the result of convergent evolution. Indeed, because germ line Abs are often broadly polyspecific and affinity-matured Abs are exquisitely specific, and because specificity is logically associated with rigidity and conformational homogeneity, it seems likely that one role of somatic mutation is to evolve rigidity. Such mutations might act, for example, by introducing hydrogen bonds or packing interactions that cross-link the CDR loops or the strands of the β-sheets that anchor the loops, as has already been suggested for an anti-Fl Ab (11). Whatever the mechanism, selecting for rigidity, as a consequence of its effect on specificity, might be a natural consequence of the competition between B cells for limiting Ag during the evolution process in an environment full of other potential binding partners (as any biological compartment is certain to be).
Abs 2E8, 3D3, and 10B8 constitute a triplet set of siblings that evolved from a common germ line Ab. By maximum parsimony, comparison of their sequences allows for the identification of the somatic mutations. The sequences reveal that Ab 3D3 retains its germ line VL sequence, whereas it acquired two VH somatic mutations, Q39L and G56D. In contrast, Ab 2E8 acquired a single VL mutation, S31N, during affinity maturation and two VH mutations, S31N and W33L. Likewise, Ab 10B8 acquired a single VL somatic mutation, M33I, and two VH mutations, G57D and A105T. Although the majority of these somatic mutations (six of eight) are in CDR loops, and some differences in affinity for MPTS are again apparent among the siblings, their flexibility and conformational heterogeneity are remarkably similar. In this case, the common dynamics are the result of parallel evolution.
Abs 3E6 and 7D5 constitute a set of twin siblings that also evolved from a common germ line Ab. Because only two sequences are available, it is not possible to identify the germ line sequence, but it was possible to identify five sites where mutations were introduced (i.e. the sites that differ). The VL chains of the Abs differ at position 17 (Lys in 7D5; Glu in 3E6), position 78, (Met in 7D5; Leu in 3E6), and position 95 (Pro in 7D5; Ser in 3E6). The VH chains differ at positions 35 (Asn in 7D5; Ser in 3E6) and 56 (Glu in 7D5; Asp in 3E6). Three of these sites are within CDR loops (Pro/Ser-95, Asn/Ser-35, and Glu/Asp-56). Abs 3E6 and 7D5 show similar values of both ΔS0 and ΔCp0, which suggests that complex formation involves less desolvation than with the other Abs. Nonetheless, Abs 3E6 and 7D5 show differences in MPTS affinity and, unlike the triplet siblings discussed above, significant differences in dynamics; Ab 3E6 binds MPTS with higher affinity than 7D5 and it is uniquely rigid and conformationally homogeneous compared with all of the other Abs characterized. It is possible that the relative flexibility of Ab 7D5 is shared with its germ line Ab, and if so, then the rigidity of Ab 3E6 would be further evidence of selection of rigidity and conformational homogeneity during somatic evolution, as already demonstrated with an anti-Fl Ab (11). Although, as mentioned above, it is not possible to rigorously identify the germ line precursor of the twin siblings, comparisons with homologous germ line genes suggest that they probably have a similar number of somatic mutations. Thus, regardless of whether the increased rigidity was itself selected for, these sibling Abs provide a clear demonstration of the power of divergent evolution to tailor protein dynamics and thus provide an opportunity for altered molecular recognition.
CONCLUSION
Eight anti-MPTS monoclonal Abs were generated and characterized, and although they clearly recognize MPTS via different mechanisms, they all show a similar level of flexibility and conformational heterogeneity that is also similar to that of a panel of anti-Fl Abs (41). It is interesting to speculate that this may reflect the levels of affinity and specificity required for immune function. The data also demonstrate that somatic evolution may have a pronounced effect on protein dynamics, which further supports a connection between rigidity, specificity, and Ab evolution. Future structural efforts will focus on elucidating the mechanisms by which the somatic mutations rigidify the Ab-combining site.
Inherently flexible germ line Abs and point mutations that rigidify them may play an important role in the evolution of molecular recognition within the immune response by maximizing the likelihood that a germ line Ab with the required activity is present and then tailoring dynamics for specificity, so that they may be produced at the high levels required to eradicate an infection without causing autoimmunity. Some level of flexibility and associated polyspecificity is inherently present in all proteins, and point mutations that favor conformations with activities and selectivities that satisfy a given selection pressure probably play an important role in the evolution of other proteins, such as enzymes, transcription factors, receptors, and components of signaling pathways.
Acknowledgments
We thank Professors Shaul Mukamel (University of California, Irvine, CA) and Peter Wolynes (University of California San Diego, La Jolla, CA) for helpful comments.
This work was supported by National Science Foundation Grant CHE-0848902 (to F. E. R.).

This article contains supplemental Data Set S1.
- Ab
- antibody
- MPTS
- 8-methoxypyrene-1,3,6-trisulfonate
- CDR
- complementarity-determining region
- VL
- light-chain variable region
- VH
- heavy-chain variable region
- 3PEPS
- three-pulse photon echo peak shift
- TG
- transient grating.
REFERENCES
- 1. Gao J., Byun K. L., Kluger R. (2004) Catalysis by enzyme conformational change. Top. Curr. Chem. 238, 113–136 [Google Scholar]
- 2. Agarwal P. K. (2006) Enzymes. An integrated view of structure, dynamics and function. Microb. Cell Fact. 5, 2–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Eisenmesser E. Z., Bosco D. A., Akke M., Kern D. (2002) Enzyme dynamics during catalysis. Science 295, 1520–1523 [DOI] [PubMed] [Google Scholar]
- 4. Daniel R. M., Dunn R. V., Finney J. L., Smith J. C. (2003) The role of dynamics in enzyme activity. Annu. Rev. Biophys. Biomol. Struct. 32, 69–92 [DOI] [PubMed] [Google Scholar]
- 5. Villa J., Warshel A. (2001) Energetics and dynamics of enzymatic reactions. J. Phys. Chem. B 105, 7887–7907 [Google Scholar]
- 6. Wright P. E., Dyson H. J. (1999) Intrinsically unstructured proteins. Reassessing the protein structure-function paradigm. J. Mol. Biol. 293, 321–331 [DOI] [PubMed] [Google Scholar]
- 7. Zimmermann J., Thielges M. C., Yu W., Romesberg F. E. (2009) Protein dynamics and the evolution of novel protein function. in Protein Engineering Handbook (Lutz S., Bornscheuer U. T., eds) pp. 147–186, Wiley-VCH, Weinheim, Germany [Google Scholar]
- 8. Jimenez R., Case D. A., Romesberg F. E. (2002) Flexibility of an antibody binding site measured with photon echo spectroscopy. J. Phys. Chem. B 106, 1090–1103 [Google Scholar]
- 9. Jimenez R., Salazar G., Baldridge K. K., Romesberg F. E. (2003) Flexibility and molecular recognition in the immune system. Proc. Natl. Acad. Sci. U.S.A. 100, 92–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jimenez R., Salazar G., Yin J., Joo T., Romesberg F. E. (2004) Protein dynamics and the immunological evolution of molecular recognition. Proc. Natl. Acad. Sci. U.S.A. 101, 3803–3808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zimmermann J., Oakman E. L., Thorpe I. F., Shi X., Abbyad P., Brooks C. L., 3rd, Boxer S. G., Romesberg F. E. (2006) Antibody evolution constrains conformational heterogeneity by tailoring protein dynamics. Proc. Natl. Acad. Sci. U.S.A. 103, 13722–13727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jensen R. A. (1976) Enzyme recruitment in evolution of new function. Annu. Rev. Microbiol. 30, 409–425 [DOI] [PubMed] [Google Scholar]
- 13. Jeffery C. J. (2003) Moonlighting proteins. Old proteins learning new tricks. Trends Genet. 19, 415–417 [DOI] [PubMed] [Google Scholar]
- 14. Copley S. D. (2003) Enzymes with extra talents. Moonlighting functions and catalytic promiscuity. Curr. Opin. Chem. Biol. 7, 265–272 [DOI] [PubMed] [Google Scholar]
- 15. Huberts D. H., van der Klei I. J. (2010) Moonlighting proteins. An intriguing mode of multitasking. Biochim. Biophys. Acta 1803, 520–525 [DOI] [PubMed] [Google Scholar]
- 16. Gancedo C., Flores C. L. (2008) Moonlighting proteins in yeasts. Microbiol. Mol. Biol. Rev. 72, 197–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Piatigorsky J., Wistow G. J. (1989) Enzyme/crystallins. Gene sharing as an evolutionary strategy. Cell 57, 197–199 [DOI] [PubMed] [Google Scholar]
- 18. Piatigorsky J. (2007) Gene Sharing and Evolution: The Diversity of Protein Functions, Harvard University Press, Cambridge, MA [Google Scholar]
- 19. O'Brien P. J., Herschlag D. (1999) Catalytic promiscuity and the evolution of new enzymatic activities. Chem. Biol. 6, R91–R105 [DOI] [PubMed] [Google Scholar]
- 20. Francino M. P. (2005) An adaptive radiation model for the origin of new gene functions. Nat. Genet. 37, 573–577 [DOI] [PubMed] [Google Scholar]
- 21. Bergthorsson U., Andersson D. I., Roth J. R. (2007) Ohno's dilemma. Evolution of new genes under continuous selection. Proc. Natl. Acad. Sci. U.S.A. 104, 17004–17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Depristo M. A. (2007) The subtle benefits of being promiscuous. Adaptive evolution potentiated by enzyme promiscuity. HFSP J. 1, 94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tonegawa S. (1983) Somatic generation of antibody diversity. Nature 302, 575–581 [DOI] [PubMed] [Google Scholar]
- 24. Steele E. J. (1990) Somatic Hypermutation in V-Regions, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 25. Jerne N. K. (1955) The natural selection theory of antibody formation. Proc. Natl. Acad. Sci. U.S.A. 41, 849–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Burnet F. M. (1957) A modification of Jerne's theory of antibody production using the concept of clonal selection. Aust. J. Sci. 20, 67–69 [DOI] [PubMed] [Google Scholar]
- 27. Breinl F., Haurowitz F. (1930) Chemical examinations on the precipitate from hemoglobin and anti-hemoglobin serum and comments on the nature of antibodies. Z. Physiol. Chem. 192, 45–57 [Google Scholar]
- 28. Pauling L. (1940) A theory of the structure and process of formation of antibodies. J. Am. Chem. Soc. 62, 2643–2657 [Google Scholar]
- 29. Patten P. A., Gray N. S., Yang P. L., Marks C. B., Wedemayer G. J., Boniface J. J., Stevens R. C., Schultz P. G. (1996) The immunological evolution of catalysis. Science 271, 1086–1091 [DOI] [PubMed] [Google Scholar]
- 30. Romesberg F. E., Spiller B., Schultz P. G., Stevens R. C. (1998) Immunological origins of binding and catalysis in a Diels-Alderase antibody. Science 279, 1929–1933 [DOI] [PubMed] [Google Scholar]
- 31. Wedemayer G. J., Patten P. A., Wang L. H., Schultz P. G., Stevens R. C. (1997) Structural insights into the evolution of an antibody-combining site. Science 276, 1665–1669 [DOI] [PubMed] [Google Scholar]
- 32. Silverstein A. M. (2003) Splitting the difference. The germ line-somatic mutation debate on generating antibody diversity. Nat. Immunol. 4, 829–833 [DOI] [PubMed] [Google Scholar]
- 33. Manivel V., Sahoo N. C., Salunke D. M., Rao K. V. S. (2000) Maturation of an antibody response is governed by modulations in flexibility of the antigen-combining site. Immnunity 13, 611–620 [DOI] [PubMed] [Google Scholar]
- 34. Foote J., Milstein C. (1994) Conformational isomerism and the diversity of antibodies. Proc. Natl. Acad. Sci. U.S.A. 91, 10370–10374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Berzofsky J. A. (1985) Intrinsic and extrinsic factors in protein antigenic structure. Science 229, 932–940 [DOI] [PubMed] [Google Scholar]
- 36. James L. C., Roversi P., Tawfik D. S. (2003) Antibody multispecificity mediated by conformational diversity. Science 299, 1362–1367 [DOI] [PubMed] [Google Scholar]
- 37. Cho M., Yu J. Y., Joo T., Nagasawa Y., Passino S. A., Fleming G. R. (1996) The integrated photon echo and solvation dynamics. J. Phys. Chem. 100, 11944–11953 [Google Scholar]
- 38. de Boeij W. P., Pshenichnikov M. S., Wiersma D. A. (1996) System bath correlation function probed by conventional and time-gated stimulated photon echo. J. Phys. Chem. 100, 11806–11823 [Google Scholar]
- 39. Fleming G. R., Cho M. H. (1996) Chromophore-solvent dynamics. Annu. Rev. Phys. Chem. 47, 109–134 [Google Scholar]
- 40. Mukamel S. (1995) Principles of Nonlinear Optical Spectroscopy, Oxford University Press, New York [Google Scholar]
- 41. Thielges M. C., Zimmermann J., Yu W., Oda M., Romesberg F. E. (2008) Exploring the energy landscape of antibody-antigen complexes. Protein dynamics, flexibility, and molecular recognition. Biochemistry 47, 7237–7247 [DOI] [PubMed] [Google Scholar]
- 42. Sastry L., Alting-Mees M., Huse W. D., Short J. M., Sorge J. A., Hay B. N., Janda K. D., Benkovic S. J., Lerner R. A. (1989) Cloning of the immunological repertoire in Escherichia coli for generation of monoclonal catalytic antibodies. Construction of a heavy chain variable region-specific cDNA library. Proc. Natl. Acad. Sci. U.S.A. 86, 5728–5732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ulrich H. D., Patten P. A., Yang P. L., Romesberg F. E., Schultz P. G. (1995) Expression studies of catalytic antibodies. Proc. Natl. Acad. Sci. U.S.A. 92, 11907–11911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Harlow E., Lane D. (1988) Antibodies: A Laboratory Manual, pp. 72–87, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 45. Spolar R. S., Livingstone J. R., Record M. T. (1992) Use of liquid-hydrocarbon and amide transfer data to estimate contributions to thermodynamic functions of protein folding from the removal of nonpolar and polar surface from water. Biochemistry 31, 3947–3955 [DOI] [PubMed] [Google Scholar]
- 46. Dietzek B., Kiefer W., Blumhoff J., Böttcher L., Rau S., Walther D., Uhlemann U., Schmitt M., Popp J. (2006) Ultrafast excited state excitation dynamics in a quasi-two-dimensional light-harvesting antenna based on ruthenium(II) and palladium(II) chromophores. Chemistry 12, 5105–5115 [DOI] [PubMed] [Google Scholar]
- 47. Wiederrecht G. P., Svec W. A., Niemczyk M. P., Wasielewski M. R. (1995) Femtosecond transient grating studies of chlorophylls and a chlorophyll-based electron donor-acceptor molecule. J. Phys. Chem. 99, 8918–8926 [Google Scholar]
- 48. Groot M. L., Yu J. Y., Agarwal R., Norris J. R., Fleming G. R. (1998) Three-pulse photon echo measurements on the accessory pigments in the reaction center of Rhodobacter sphaeroides. J. Phys. Chem. B 102, 5923–5931 [Google Scholar]
- 49. Goodman M. F., Scharff M. D., Romesberg F. E. (2007) AID-initiated purposeful mutations in immunoglobulin genes. Adv. Immunol. 94, 127–155 [DOI] [PubMed] [Google Scholar]
- 50. Tokuriki N., Tawfik D. S. (2009) Protein dynamism and evolvability. Science 324, 203–207 [DOI] [PubMed] [Google Scholar]
- 51. Kang J., Warren A. S. (2007) Enthalpy-entropy compensation in the transition of a monospecific antibody toward antigen-binding promiscuity. Mol. Immunol. 44, 3623–3624 [DOI] [PubMed] [Google Scholar]
- 52. Dunitz J. D. (1995) Win some, lose some. Enthalpy-entropy compensation in weak intermolecular interactions. Chem. Biol. 2, 709–712 [DOI] [PubMed] [Google Scholar]
- 53. Frauenfelder H., Parak F., Young R. D. (1988) Conformational substates in proteins. Annu. Rev. Biophys. Biophys Chem. 17, 451–479 [DOI] [PubMed] [Google Scholar]
- 54. Frauenfelder H., Sligar S. G., Wolynes P. G. (1991) The energy landscape and motions of proteins. Science 254, 1598–1603 [DOI] [PubMed] [Google Scholar]
- 55. Larsen D. S., Ohta K., Fleming G. R. (1999) Three-pulse photon echo studies of nondipolar solvation. Comparison with a viscoelastic model. J. Chem. Phys. 111, 8970–8979 [Google Scholar]
- 56. Jordanides X. J., Lang M. J., Song X., Fleming G. R. (1999) Solvation dynamics in protein environments studied by photon echo spectroscopy. J. Phys. Chem. B 103, 7995–8005 [Google Scholar]
- 57. Eyring H. (1935) The activated complex in chemical reactions. J. Chem. Phys. 3, 107–115 [Google Scholar]
- 58. Passino S. A., Nagasawa Y., Joo T., Fleming G. R. (1997) Three-pulse echo peak shift studies of polar solvation dynamics. J. Phys. Chem. A 101, 725–731 [Google Scholar]
- 59. Nagasawa Y., Passino S. A., Joo T., Fleming G. R. (1997) Temperature dependence of optical dephasing in an organic polymer glass (PMMA) from 300 K to 30 K. J. Chem. Phys. 106, 4840–4852 [Google Scholar]
- 60. Kabat E. A., Wu T. T. (1991) Identical V region amino acid sequences and segments of sequences in antibodies of different specificities. Relative contributions of VH and VL genes, minigenes, and complementarity-determining regions to binding of antibody-combining sites. J. Immunol. 147, 1709–1719 [PubMed] [Google Scholar]