Background: Phosphorylation of the proliferating protein PCNA through the signaling of receptor tyrosine kinases (RTKs) protects it from ubiquitylation-mediated degraded on the chromatin.
Results: The ubiquitylation enzyme CUL4A is responsible for PCNA degradation in the absence of phosphorylation.
Conclusion: Phosphorylation of PCNA stabilizes PCNA by blocking interaction between PCNA and CUL4A.
Significance: This may help sensitize cancer cells to therapeutic agents targeting RTKs.
Keywords: Breast Cancer, E3 Ubiquitin Ligase, Oncogene, Protein Kinases, Protein Stability, Receptor Tyrosine Kinase, Signal Transduction, Ubiquitylation
Abstract
Proliferating cell nuclear antigen (PCNA) is an essential component for DNA synthesis upon growth stimulation. It has been shown that phosphorylation of PCNA at Tyr-211 by the EGF receptor (EGFR) protects PCNA from polyubiquitylation and degradation, whereas blocking phosphorylation induces ubiquitylation-mediated degradation of the chromatin-bound, but not the -unbound, PCNA, and suppresses cell proliferation. However, the ubiquitin E3 ligase linking growth signaling to the proteolysis of PCNA and the underlying regulatory mechanism remain to be identified. Here we show that, in the absence of Tyr-211 phosphorylation, PCNA is subject to polyubiquitylation at Lys-164 by the CUL4A E3 ligase, resulting in the degradation of PCNA. Mutation of Lys-164 to arginine prevents PCNA ubiquitylation and rescues the degradation of the K164R/Y211F PCNA double mutant. Activation of EGFR inhibits the interaction of PCNA with CUL4A, whereas inhibition of EGFR leads to increased CUL4A-PCNA interaction and CUL4A-dependent ubiquitin-mediated degradation of PCNA. Substitution of endogenous PCNA with the Y211F mutant PCNA conveys enhanced sensitization to EGFR inhibition. Our findings identify CUL4A as the ubiquitin ligase linking the down-regulation of cell surface receptor tyrosine kinase to the nuclear DNA replication machinery in cancer cells.
Introduction
PCNA2 functions as a molecular platform at the DNA replication fork and is an indispensable protein for cell proliferation and the maintenance of genomic integrity. PCNA forms a homotrimeric ring as a sliding clamp encircling the DNA double helix. Associated with the ring are a plethora of proteins involved in DNA synthesis, such as DNA polymerases, cell cycle progression, and other cofactors required as the replication fork moves on (1). PCNA plays an essential role in coordinating the many functions and activities of the proteins involved in DNA replication and DNA damage repair.
PCNA is subject to regulation by post-translational modifications (2). Ubiquitylation of PCNA is a signal for alteration of its functional modes (2–6). In response to DNA damage stresses, PCNA is monoubiquitylated by the RAD6-RAD18 ubiquitin ligase. This modification alters the surface properties of PCNA to switch the DNA polymerase partners from those engaged in progressive DNA synthesis with those specified for error-prone translesional DNA synthesis to promote cell survival (7–10). However, polyubiquitylation of PCNA, catalyzed by RAD6-RAD18 plus the RAD5-UBC13-MMS2 E3 ligase complexes via the lysine 63 linkage of the ubiquitin, is a signal to bypass DNA damage in a error-free mode (10).
Blocking growth signals in cancer cells can also induce polyubiquitylation of PCNA that regulates its protein stability (2, 11). We previously showed that activation of the epidermal growth factor receptor (EGFR) resulted in PCNA phosphorylation at Tyr-211, which enhanced protein stability of chromatin-associated PCNA and promoted cell proliferation (11). However, inhibition of the EGFR receptor blocked Tyr-211 phosphorylation and induced proteasome-dependent degradation of PCNA (11). The role of Tyr-211 phosphorylation in the protein stability of PCNA was further demonstrated using a PCNA mutant in which the Tyr-211 residue was mutated to phenylalanine (Y211F) (11). The Y211F mutant PCNA had an elevated level of polyubiquitylation and reduced stability, which can be rescued by proteasome inhibition. We have shown that the degradation association polyubiquitylation of PCNA does not require the RAD6 or UBC13 complexes (11). However, the ubiquitin ligase involved in PCNA degradation coupled with EGFR inhibition remained to be determined.
The regulation of protein turnover through ubiquitin-mediated proteolysis is an evolutionarily conserved mechanism for controlling cell cycle progression and maintaining genome stability (12). PCNA itself has been found as an integrated component in this regulation. For example, to prevent reinitiation of DNA synthesis in S phase, the replication origin licensing factor CDT1 is directed to ubiquitin-mediated degradation after the initiation of each round of DNA replication (13–15), and the cullin 4-ring finger ligases, heretofore referred to the CUL4 ligases, are known as the E3 ligases catalyzing polyubiquitylation of CDT1 (14, 16–23). In this process, PCNA as a platform on chromatin is required to bridge the CUL4 ligase and the substrate CDT1 together for proteolysis (18, 21, 22, 24–27). PCNA is also a functional partner of CUL4 ligase in the degradation of the cyclin-dependent kinase inhibitor p21CIP1 upon low dose UV irradiation (28, 29). It has been demonstrated that binding of CUL4 ligase with the chromatin-bound PCNA creates a protein degradation complex for substrates coupled to DNA replication (30). Interestingly, it was recently shown that CUL4 monoubiquitinates PCNA to promote translesion DNA synthesis (31). In this study we show that PCNA itself is a CUL4A substrate of proteolytic polyubiquitination which is promoted by EGFR inhibition.
EXPERIMENTAL PROCEDURES
Antibodies and Chemicals
The antibodies used in this study include mouse monoclonal anti-FLAG, α-tubulin (Sigma); HA (Covance, Princeton, NJ); rabbit polyclonal anti-CUL4A, rabbit polyclonal anti-PCNA, and Tyr(P)-211 PCNA (Bethyl, Montgomery, TX); mouse monoclonal PCNA, ubiquitin (Santa Cruz Biotechnology, Santa Cruz, CA); EGFR, histone H3, Tyr(P)-1068 EGFR (Cell Signaling, Danvers, MA). The chemical used were EGF (Sigma), AG1478 and MG132 (EMD, Gibbstown, NJ), and lapatinib (LC Laboratories, Woburn, MA).
Cell Culture
The human breast cancer cell line MDA-MB-468 and the human embryonic kidney cell line HEK293T were purchased from American Type Culture Collection (ATCC, Manassas, VA). All cells were grown in DMEM/F12 (1:1) medium supplemented with 10% fetal bovine serum without antibiotics. The stable cell line expressing the FLAG-PCNA-Y211F mutant in MDA-MB-468 was maintained in the same conditions with periodic selection by supplemental neomycin/G418 (500 ng/ml).
Plasmids and DNA Transfection
Expression plasmids of wild-type CUL4A (Addgene plasmid 19907) (32) and CUL4A-DN (Addgene plasmid 15821) (33) were obtained from Addgene (Cambridge, MA). MDA-MB-468 cells were transfected using an Amaxa electroporator (Lonza, Walkersville, MD). HEK293T cells were transfected using Lipofectamine 2000 following the manufacturer's instructions (Invitrogen).
RNA Interference and Transduction of Lentiviral shRNAs
The shRNAs of CUL4A and CUL4B were cloned in the lentiviral vector pLKO.1 with the sequence CCGGGCAGAACTGATCGCAAAGCATCTCGAGATGCTTTGCGATCAGTTCTGCTTTTT and CCGGGCCATGAAAGAAGCATTTGAACTCGAGTTCAAATGCTTCTTTCATGGCTTTTT, respectively (National RNAi Core Facility, Academia Sinica, Taiwan). The control scrambled shRNA, CCTAAGGTTAAGTCGCCCTCGCTCTAGCGAGGGCGACTTAACCTTAGG, in pLKO.1 vector was obtained from Addgene (Addgene plasmid 1864) (34). Recombination with the lenti-gag/pol, envelope, and Rev plasmids in 293T cells was conducted at the Viral Vector Core at Cincinnati Children's Hospital. To generate stable cell lines, cells were infected with the corresponding lentivirus and selected briefly with puromycin. The surviving cells were then pooled for further experiments. All experiments were performed using freshly infected cells. The knock-down efficiency of CUL4A was assessed by Western blot analysis. For CUL4B, knock-down efficiency is determined by quantitative RT-PCR using the following primers: forward, GCGAAGATCACATCAAAGCACA; reverse, AGTCCCATGTCCCAAATGGAG. The siRNA of PCNA was purchased from Qiagen (SI02653287), which was transfected with Lipofectamine 2000 following the manufacturer's instructions (Invitrogen).
Immunoprecipitation and Western Blot Analysis
Cultured cells were washed twice with ice-cold PBS. The cells were then lysed in NETN buffer (150 mm NaCl, 1 mm EDTA, pH 8.0, 20 mm Tris, pH 8.0, 0.5% Nonidet P-40, 25 mm NaF, 2 mm Na3VO4, 20 μl/ml aprotinin, 0.1 m PMSF) on ice for 30 min. For immunoprecipitation, the lysate was incubated with primary antibody at 4 °C overnight. The immunocomplexes were then incubated with 40 μl of protein G-agarose (Roche Applied Science) at 4 °C for 2 h. The beads were washed four times with ice-cold NETN buffer, and the protein complexes were eluted by boiling the beads in 40 μl of 2× loading buffer. Triton X-100 extraction was performed by incubating the cell monolayer in the extraction buffer (100 mm NaCl, 300 mm sucrose, 3 mm MgCl2, 10 mm PIPES at pH 6.8, 1 mm EGTA, 0.2% Triton-X-100, 25 mm NaF, 2 mm Na3VO4, 5 mm PMSF, 2 μg/ml aprotinin) for 5 min on ice. The cells were collected by scraping and washed twice with the same buffer. The pellet was lysed in radioimmune precipitation assay buffer (150 mm NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 50 mm Tris at pH 7.5, 25 mm NaF, 2 mm Na3VO4, 5 mm PMSF, 2 μg/ml aprotinin) with sonication. Cell lysates were separated in acrylamide gels, transferred to a PVDF membrane (Bio-Rad), and probed with the indicated antibodies. Bands were visualized by a chemiluminescence-based detection method (Fisher/Pierce) that used a horseradish peroxidase-conjugated secondary antibody. Band intensities were quantitated by using the National Institutes of Health ImageJ software.
Confocal Microscopy
Cultured cells were treated with or without 10 μm AG1478 for 8 h. the cells were then fixed and permeabilized with 4% formaldehyde and 0.2% Triton X-100 for 10 min at room temperature. After four washes with PBS, the cells were blocked with 10% normal goat serum for 1 h at room temperature and then immunostained with primary antibodies (1:200 dilution in PBS with 0.2% BSA) overnight at 4 °C. After three washes with PBS, the FITC- or Texas Red-conjugated secondary antibody was applied for 45 min at room temperature. Nuclei were stained with TOPRO 3. Images were captured with a Zeiss laser scanning confocal microscope (LSM510). Colocalization of PCNA and CUL4A was quantitated by the ImageJ program (35), in which individual z-optical sections were analyzed using the colocalization plugin of the program. The results were normalized against the background colocalization using the formula: corrected colocalization = (measured colocalization − background colocalization)/(1 − background colocalization/100). Pearson's correlation was then calculated using the Correlator Plus plugin. A coefficient of +1 indicates perfect colocalization, and a coefficient of 0 indicates no colocalization. Images derived from multiple fields of at least two independent experiments were analyzed.
Generation of Adenovirus-expressing PCNA
We used the AdEasy system established by the group of Dr. Bert Vogelstein (36). Briefly, the cDNA of wild-type or Y211F FLAG-PCNA was cloned into the pAdTrack-CMV vector and cotransformed with the adenoviral backbone vector pAdEasy-1 into BJ5183 cells. The recombinant plasmid was transfected into 293 cells for viral packaging and production. The virus was then purified from the crude cell lysate with CsCl step-gradient ultracentrifugation followed by dialysis into Tris buffer (pH 7.5 with 10% glycerol).
Statistical Analysis
Data of each assay were expressed as means ± S.D. (n = 3). A difference between two groups was determined by Student's t test. A value of p < 0.05 was considered significantly significant.
RESULTS
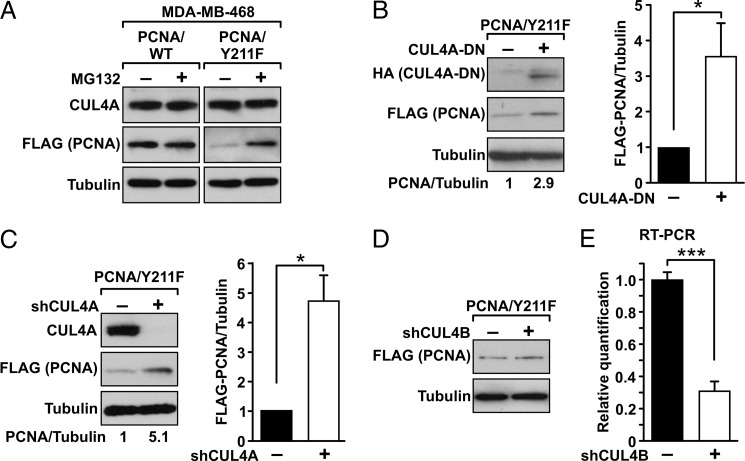
To study further how Tyr-211 phosphorylation regulates the stability of PCNA, the FLAG-tagged wild-type (FLAG-PCNA-WT) and the Y211F mutant (FLAG-PCNA-Y211F) of PCNA were stably transfected into the breast cancer cell line MDA-MB-468. The protein level of the Y211F mutant PCNA was low due to the enhanced degradation of the mutant, and treatment with the proteasome inhibitor MG132 rescued protein expression of the mutant (Fig. 1A). This result is consistent with our previous observation indicating that down-regulation of PCNA expression after inhibition of Tyr-211 phosphorylation is mediated through a proteasome-dependent mechanism (11). MDA-MB-468 cells express CUL4A, which is a known PCNA-binding E3 ligase (Fig. 1A) (21, 22). CUL4A utilizes a C-terminal domain to recruit the E2 ubiquitin-conjugating enzyme ROC-1 for substrate ubiquitylation. The mutant CUL4A lacking this ROC-1-binding motif functions as a dominant negative mutant (CUL4-DN) that specifically blocks the wild-type CUL4A (33). Expression of CUL4A-DN by transfection resulted in an increase in the protein level of the Y211F mutant PCNA (Fig. 1B). The role of CUL4A in this process was further demonstrated by silencing its expression with the specific shRNA of CUL4A (shCUL4A). Depleting of the endogenous CUL4A rescued expression level of the Y211F mutant PCNA (Fig. 1C). These results suggest that the CUL4A complex is the E3 ligase responsible for the degradation of PCNA upon inhibition of Tyr-211 phosphorylation. In contrast, silencing CUL4B, the paralog of CUL4A, did not recover the level of Y211F PCNA (Fig. 1D), indicating that the destabilization of Y211F PCNA is specifically mediated by CUL4A. The activity of the CUL4B shRNA was validated by quantitative RT-PCR (Fig. 1E).
FIGURE 1.
Y211F conveys protein instability of PCNA through a CUL4A-dependent mechanism. A, expression of the FLAG-tagged Y211F mutant PCNA in the CUL4A-expressing MDA-MB-468 cells was reduced compared with the wild-type PCNA. Treatment with the proteasome inhibitor MG132 increased the expression level of the PCNA-Y211F protein. B, transfection of CUL4A-DN into the MDA-MB-468/Y211F cells resulted in enhanced expression of the PCNA-Y211F mutant. Quantitated result of data derived from three independent experiments is shown. *, p < 0.05. C, depleting endogenous CUL4A by shRNA (shCUL4A) resulted in rescue of PCNA-Y211F expression. Levels of α-tubulin are shown as the internal loading control. Quantitated result of data derived from three independent experiments is shown. *, p < 0.05. D, depleting exogenous CUL4B by shRNA (shCUL4B) had no effect on the level of PCNA-Y211F. E, silencing efficiency of shCUL4B was determined by quantitative RT-PCR. ***, p < 0.005. Error bars, S.E.
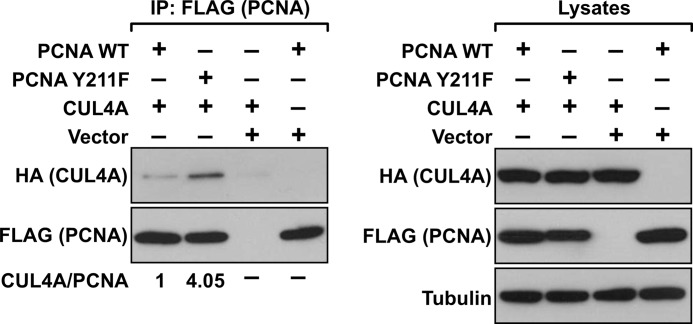
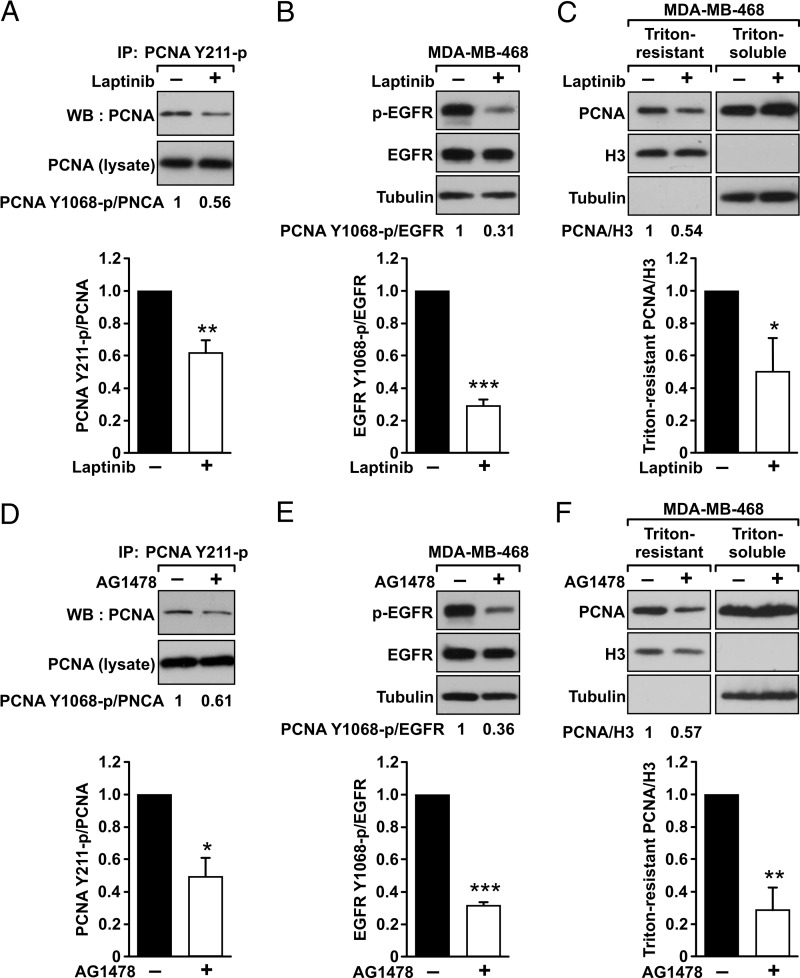
We reasoned that if CUL4A is involved in the degradation of Y211F PCNA, it should be associated preferentially with the Y211F mutant PCNA compared with wild-type PCNA. We therefore examined the association of wild-type and Y211F PCNA with CUL4A by cotransfection of different PCNA genes with CUL4A in HEK293T cells, followed by immunoprecipitation of PCNA and Western blot analysis to determine the levels of CUL4A in the immunocomplexes. Our results showed that both the wild-type and Y211F mutant PCNA interacted with CUL4A, but CUL4A associated preferentially with the Y211F mutant of PCNA (Fig. 2). These results suggest that the lack of Tyr-211 phosphorylation facilitates the interaction of PCNA with CUL4A and down-regulates the levels of PCNA. Inhibition of EGFR activity with a clinically approved anti-cancer drug lapatinib suppressed Tyr-211 phosphorylation of PCNA (Fig. 3, A and B), resulting in down-regulation of chromatin-bound PCNA (Fig. 3C). Similarly, blocking EGFR with another EGFR-specific inhibitor AG1478 led to inhibition of Tyr-211 phosphorylation and depletion of PCNA from the chromatin (Fig. 3, D–F), which is also consistent with our previous finding (11).
FIGURE 2.
CUL4A interacts preferentially with Y211F PCNA. HEK293T cells were transfected with the indicated plasmids (FLAG-PCNA-WT, FLAG-PCNA-Y211F, HA-CUL4A, and the pcDNA3 vector alone). The ectopic PCNA was immunoprecipitated by an anti-FLAG antibody, and the precipitated FLAG-PCNA and HA-CUL4A were detected by the corresponding antibodies as indicated. The input lysates were examined for the expression of the transfected PCNA and CUL4A genes. Levels of α-tubulin are shown as the internal loading control.
FIGURE 3.
Inhibition of EGFR down-regulated Tyr-211 phosphorylation and the level of chromatin-bound PCNA. MDA-MB-468 cells were treated with EGFR inhibitors lapatinib (10 μm for 24 h) (A–C) or AG1478 (10 μm for 16 h) (D–F). A and D, the endogenous PCNA was immunoprecipitated (IP) by the anti-Tyr(P)-211 PCNA. The immunoprecipitated complexes were examined by Western blot (WB) analysis using an anti-PCNA antibody. Quantitated result of data derived from three independent experiments is shown. **, p < 0.01; *, p < 0.05. B and E, after the treatment of the EGFR inhibitors, the levels of EGFR, Tyr(P)-1068 EGFR (p-EGFR), and α-tubulin are shown. Quantitated result of data derived from three independent experiments is shown. ***, p < 0.005. C and F, the treated cells were extracted with 5% of Triton X-100, and the levels of PCNA in the Triton-resistant and Triton-soluble fractions were determined by Western blot analysis. Histone H3 and α-tubulin were used as the internal control for Triton-resistant and Triton-soluble fractions, respectively. Quantitated result of data derived from three independent experiments is shown. *, p < 0.05; **, p < 0.01. Error bars, S.E.
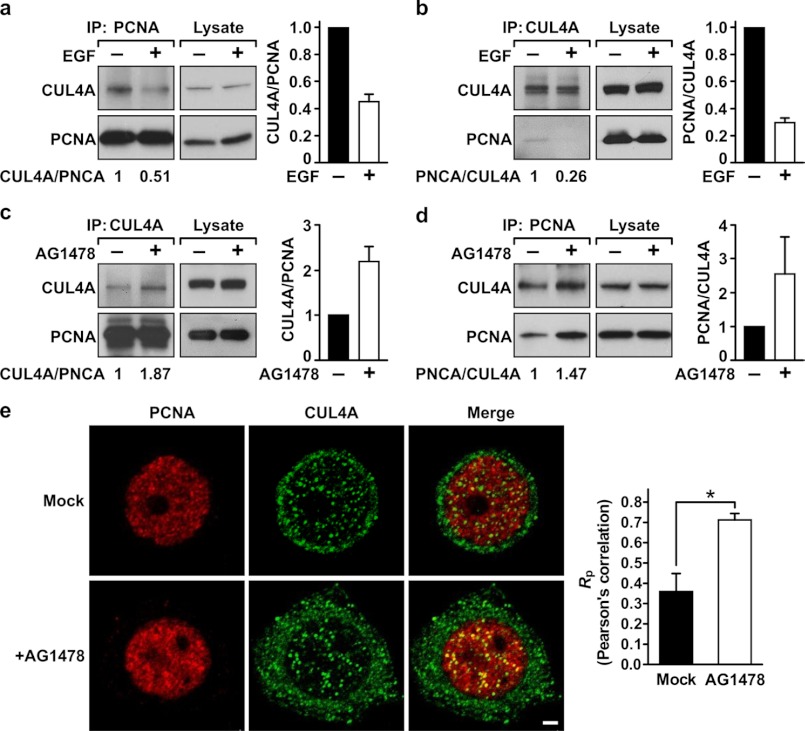
We next asked whether CUL4A is responsible for the EGFR-regulated modulation of PCNA stability. If this is the case, inhibition of EGFR should increase the interaction between PCNA and CUL4A, recapitulating the enhanced interaction between Y211F PCNA and CUL4A. However, stimulation with EGF should decrease the PCNA-CUL4A interaction. To test this hypothesis, MDA-MB-468 cells were mock-treated or stimulated with EGF. Endogenous PCNA was immunoprecipitated, and coimmunoprecipitated CUL4A was assessed by Western blot analysis. This experiment demonstrated that EGF stimulation reduced the association between CUL4A and PCNA (Fig. 4A). This result was validated by a reciprocal experiment in which CUL4A was immunoprecipitated by an anti-CUL4A antibody, followed by Western blot analysis of PCNA in the immunocomplex (Fig. 4B). However, treating the cells with AG1478 enhanced the interaction between PCNA and CUL4A as demonstrated by reciprocal immunoprecipitation experiments (Fig. 4, C and D). In cells, CUL4A is expressed in both cytoplasmic and nuclear compartments, whereas PCNA is expressed mainly in the nucleus (Fig. 4E). Immunofluorescent confocal microscopy of PCNA and CUL4A revealed a weak colocalization of these two proteins in the nuclei of mock-treated MDA-MB-468 cells, whereas pronounced colocalization was detected in cells treated with AG1478 (Fig. 4E). These results together established that the interaction between PCNA and CUL4A is negatively regulated by EGFR.
FIGURE 4.
Association between CUL4A and PCNA is negatively regulated by EGFR. A and B, MDA-MB-468 cells were serum-starved for 24 h and then stimulated by EGF (10 ng/ml) for 30 min. The cells were then lysed in NETN buffer and subjected to immunoprecipitation (IP) by the indicated antibodies. Immunoprecipitated PCNA (A) or CUL4A (B) was examined for the level of associated CUL4A or PCNA, respectively. C and D, MDA-MB-468 cells grown in normal conditions were treated with the EGFR kinase inhibitor AG1478 for 6 h. The cells were lysed, and the lysate was then subjected to immunoprecipitation for PCNA (C) or CUL4A (D). The associated CUL4A and PCNA was then assessed by Western blotting analysis. Average binding activities between these two proteins from two independent experiments are shown. E, MDA-MB-468 cells grown in normal condition were mock-treated or treated with AG1478 as described in C and then stained with primary antibodies against PCNA or CUL4A. Secondary antibodies conjugated with rhodamine and FITC were used to detect PCNA (red) and CUL4A (green), respectively. The slides were examined by fluorescence confocal microscopy, with photosections of 0.48 μm each. Note that in the merged images, colocalization of PCNA and CUL4A appears as yellow in the nucleus. Nuclear colocalization of PCNA and CUL4A was quantitated using the ImageJ program. Right, Pearson's correlation coefficients (Rp) are shown. A coefficient of +1 indicates complete colocalization. For details, see under “Experimental Procedures.” Error bars, S.E. *, p < 0.05.
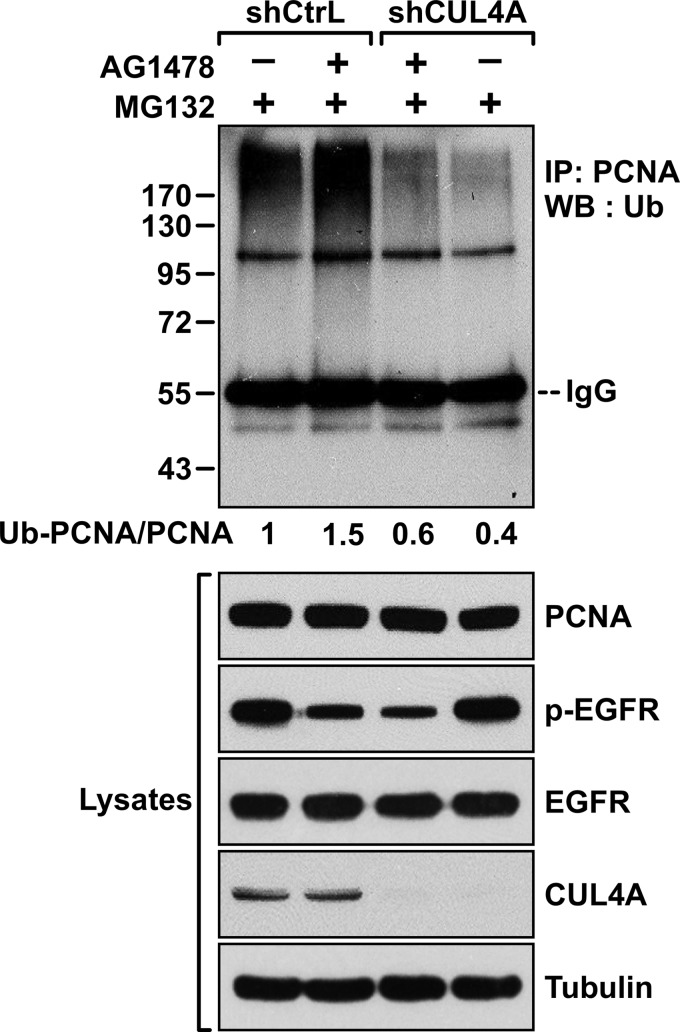
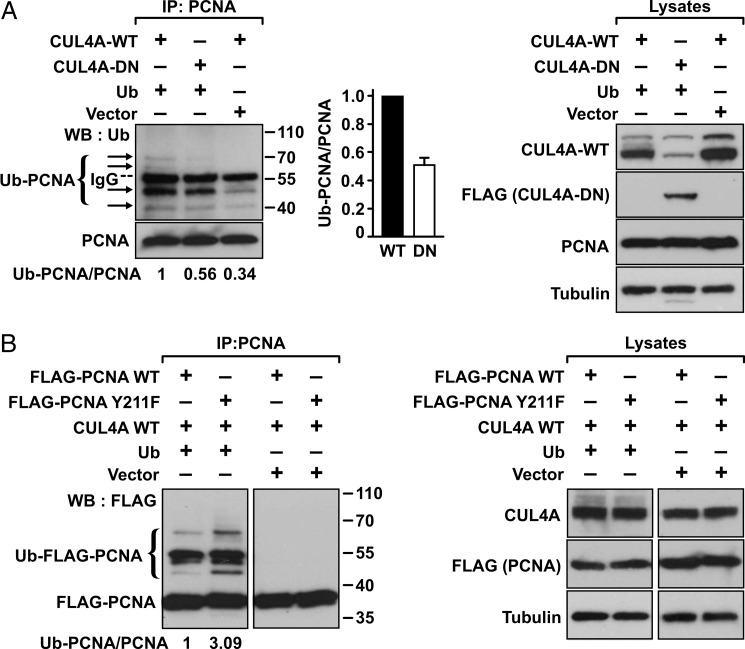
These results also suggest that, upon EGFR inhibition, CUL4A catalyzes proteolytic ubiquitylation of PCNA. To test this hypothesis, MDA-MB-468 cells were infected with a lentivirus expressing shCUL4A or the control virus, then, in the presence of MG132, treated with or without AG1478 (Fig. 5). As we reported previously, AG1478 treatment enhanced polyubiquitylation of PCNA, whereas depleting of CUL4A abrogated the ubiquitylation. To establish further that the E3 ligase activity of CUL4A was required for the AG1478-induced polyubiquitylation, the cDNA of CUL4A-WT or CUL4A-DN was transfected into HEK293T cells together with the cDNA of ubiquitin (Fig. 6A). The endogenous PCNA was then immunoprecipitated using an anti-PCNA antibody. Ubiquitylated PCNA in the immunocomplex was detected by Western blot analysis with an anti-ubiquitin antibody. The results showed that the endogenous PCNA in cells transfected with wild-type CUL4A was polyubiquitylated. However, the polyubiquitylation was suppressed by cotransfection of the CUL4A-DN mutant, indicating that polyubiquitylation of PCNA was mediated by the CUL4A E3 ligase. In light of the result that CUL4A associated preferentially with the Y211F mutant PCNA compared with the wild type (Fig. 2), it is likely that the Y211F mutant is the preferred substrate for the CUL4A-mediated ubiquitylation. To test this prediction, wild-type or Y211F mutant PCNA was cotransfected with CUL4A and ubiquitin into HEK293T cells. PCNA was then immunoprecipitated from the lysate, and the ubiquitylated ectopic wild-type or mutant PCNA was assessed (Fig. 6B). The experiment demonstrated that CUL4A mediated polyubiquitylation preferentially to the Y211F PCNA compared with the wild type.
FIGURE 5.
CUL4A is responsible for the EGFR inhibition-mediated PCNA polyubiquitylation. Upper panel, MDA-MB-468 cells were treated with and without the EGFR inhibitor AG1478 (8 μm) for 18 h in the presence of the proteasome inhibitor MG132 (5 μm). Cell lysates were then immunoprecipitated (IP) with an anti-PCNA antibody (Santa Cruz Biotechnology). The blot was then probed with an anti-ubiquitin (Ub) antibody. The numbers indicate the intensity of the ubiquitylated ladder normalized by the intensity of total PCNA. Lower panel, the input levels of PCNA, phospho-EGFR (Y1068), total EGFR, and tubulin are shown. WB, Western blotting.
FIGURE 6.
CUL4A mediates polyubiquitylation preferentially on PCNA lacking Tyr-211 phosphorylation. A, right, HEK293T cells were transfected by the indicated plasmids, and the endogenous PCNA was immunoprecipitated (IP) by an anti-PCNA antibody, followed by immunoblotting (WB) with an anti-ubiquitin (Ub) antibody. Arrows denote the ubiquitylated PCNA (Ub-PCNA). Transfection devoid of ubiquitin significantly abolished ubiquitylation. Middle, average ratio of the ubiquitylated PCNA with total PCNA was quantitated based on results derived from two independent experiments. Error bar, S.E. Left, expression levels of the transfected wild-type CUL4A (by an anti-CUL4A antibody) and the CUL4A-DN mutant (by an anti-FLAG antibody) and the endogenous PCNA (by an anti-PCNA antibody) are shown by Western blotting, with α-tubulin as the loading control. B, wild-type or Y211F PCNA (FLAG-tagged) and CUL4A were cotransfected into HEK293T cells with or without cotransfected ubiquitin cDNA. PCNA was immunoprecipitated by an anti-PCNA antibody, and the immunoblot was probed with an anti-FLAG antibody to detect the ubiquitylated PCNA molecules. Expression levels of the transfected wild-type CUL4A (by an anti-CUL4A antibody) and the transfected PCNA (by an anti-FLAG antibody) are shown by Western blotting, with α-tubulin as the loading control.
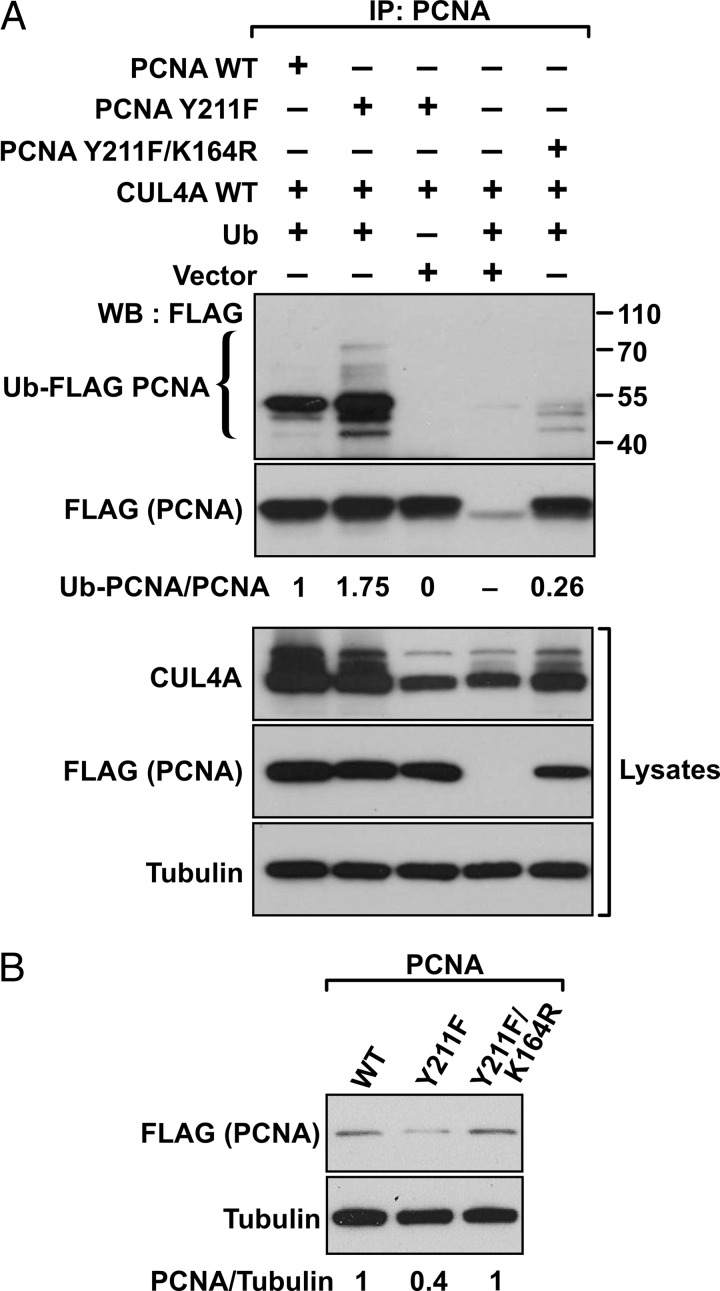
Lysine 164 is a major verified site of ubiquitylation modification for PCNA in human cells (2). We therefore asked whether the polyubiquitylation of PCNA mediated by CUL4A was catalyzed at the Lys-164 residue. To test this possibility, the wild-type, Y211F, and the double mutant of PCNA in which an additional lysine-to-arginine mutation was introduced to the Y211F mutant (Y211F/K164R) were compared for the degree of polyubiquitylation in the presence of CUL4A and ubiquitin (Fig. 7A). As shown in Fig. 6, the Y211F mutant PCNA is subject to more pronounced polyubiquitylation than the wild type, but the ubiquitylation was abolished in the Y211F/K164R double mutant, indicating that CUL4A mediates ubiquitylation of PCNA at Lys-164. This observation also predicted that abolishing the ubiquitylation site in the Y211F/K164R double mutant should overcome the intrinsic instability of Y211F PCNA. Indeed, expression of the double mutant was recovered compared with the Y211F mutant PCNA (Fig. 7B).
FIGURE 7.
CUL4A catalyzes ubiquitylation at the Lys-164 residue of PCNA. A, HEK293T cells were transfected with the indicated plasmids, and the lysates were subjected to immunoprecipitation (IP) with an anti-PCNA antibody. The Western blot (WB) was than probed with anti-FLAG antibody to detect the ubiquitylated PCNA molecules. Expression levels of the transfected genes in cell lysates are shown. B, HEK293T cells were transfected with the indicated plasmids, and the expression levels of each PCNA construct were determined by an anti-FLAG antibody with α-tubulin as the loading control.
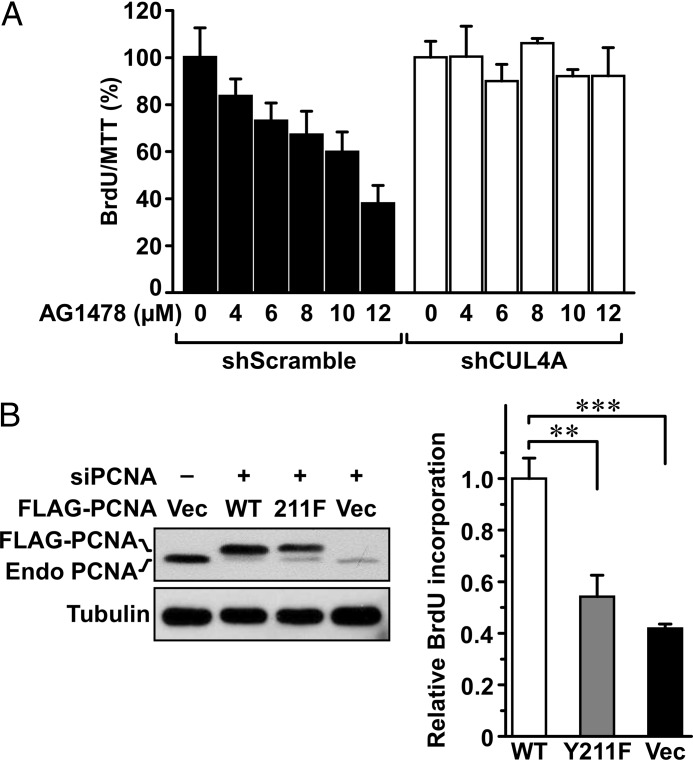
We have shown that, upon EGFR inhibition, destabilization of PCNA is confined to the chromatin-bound compartment (Fig. 3C) (11). To determine whether the CUL4A E3 ligase is responsible for the down-regulation of chromatin-bound PCNA upon EGFR inhibition, MDA-MB-468 cells harboring control shRNA (shLuc) or shCUL4A were treated with AG1478, and the chromatin-bound and -unbound fractions were isolated by Triton X-100 extraction (Fig. 8, A and B). Although inhibition of EGFR decreased the level of chromatin-bound PCNA, depleting CUL4A rescued it. No changes occurred to the chromatin-unbound PCNA. Similar observation was obtained with treatment of lapatinib (Fig. 8, C and D). These results identified CUL4A as the E3 ligase linking EGFR activity to the protein stability of PCNA on the chromatin and suggested that CUL4A can mediate the responsiveness of cancer cells to EGFR inhibition. Indeed, cells with the endogenous CUL4A depleted showed pronounced sensitivity to growth inhibition by AG1478 treatment (Fig. 9A). This result suggests that PCNA stabilization through Tyr-211 phosphorylation contributes to resistance to EGFR inhibition. This possibility was further tested by substituting the endogenous PCNA with wild-type or Y211F PCNA prior to EGFR inhibition. This was attained by infecting cells with adenovirus carrying FLAG-tagged wild-type or Y211F mutant PCNA, followed by transfection with a siRNA which targets a sequence encompassing the junction between the coding region and the 3′-untranslated region (3′-UTR) of the PCNA gene. Ectopic expression of PCNA is not affected by the siRNA because the PCNA cDNA does not contain 3′-UTR (Fig. 9B). Consistent with the decreased protein stability of PCNA in the lack of Tyr-211 phosphorylation, Y211F mutant PCNA was expressed at a lower level compared with the wild type. Upon treatment with AG1478, cells expressing wild-type PCNA showed persistent DNA synthesis activity compared with its mock treated control as measured by BrdU incorporation assay. The resistance was significantly diminished in cells infected with the Y211F mutant PCNA virus or the control virus which expresses the green fluorescence protein.
FIGURE 8.
CUL4A induced down-regulation of chromatin-bound PCNA upon EGFR inhibition. A, left, MDA-MB-468/shLuc and MDA-MB-468/shCUL4A cells grown in normal medium were treated with AG1478 (10 μm) for 16 h. The cells were then extracted with 5% of Triton X-100, and the levels of PCNA in the Triton-resistant and Triton-soluble fractions were determined by Western blot analysis. Histone H3 and α-tubulin were used as the internal control for Triton-resistant and Triton-soluble fractions, respectively. Error bar, S.E. Right, average PCNA levels were quantitated based on results derived from two independent experiments. B, expression levels of EGFR, Tyr(P)-1068 EGFR (p-EGFR), and CUL4A are shown. C and D, similar results were obtained using lapatinib (10 μm) for 24 h.
FIGURE 9.
Depleting CUL4A sensitizes MDA-MB-468 cells to EGFR inhibition. A, MDA-MB-468 cells freshly transduced with lentivirus harboring shCUL4A or the control shRNA were treated with AG1478 at the indicated concentration for 18 h, and the activities of BrdU incorporation were measured. The data were normalized by a parallel experiment of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay. B, MDA-MB-468 cells were transfected with a siRNA of PCNA to deplete the endogenous PCNA and then transduced with either a control adenovirus or an adenovirus expressing the wild-type or Y211F FLAG-tagged PCNA at 10 multiplicity of infection to reconstitute PCNA. The cells were then treated with AG1478 (5 μm) or mock-treated for 48 h. Viable cells were measured for BrdU incorporation activities, which were compared after normalization with the control treatment for each viral transduction. Ratios of the treated with the mock-treated cells based on data derived from three independent experiments were plotted. **, p < 0.01. Error bars, S.E.
DISCUSSION
PCNA level on the chromatin is subject to the regulation of growth signaling (11). Phosphorylation of PCNA at Tyr-211 by EGFR activation stabilized PCNA associated with the chromatin, and blocking the phosphorylation resulted in destabilization of the chromatin-bound PCNA through an unidentified ubiquitylation-mediated proteasome-dependent mechanism. Our current study unveils the underlying mechanism in which activation of EGFR in breast cancer cells stimulates Tyr-211 phosphorylation of PCNA and dissociates PCNA from the CUL4A E3 ligase, thus stabilizing PCNA on the chromatin (Fig. 10). Conversely, inhibition of EGFR leads to down-regulation of Tyr-211 phosphorylation of PCNA and enhanced association of PCNA with the CUL4A ligase, resulting in polyubiquitylation of PCNA at Lys-164 and the proteolysis of PCNA. Blocking this process by depleting endogenous CUL4A or overexpression of the wild-type but not the Tyr-211 mutant PCNA protected cells from EGFR inhibition.
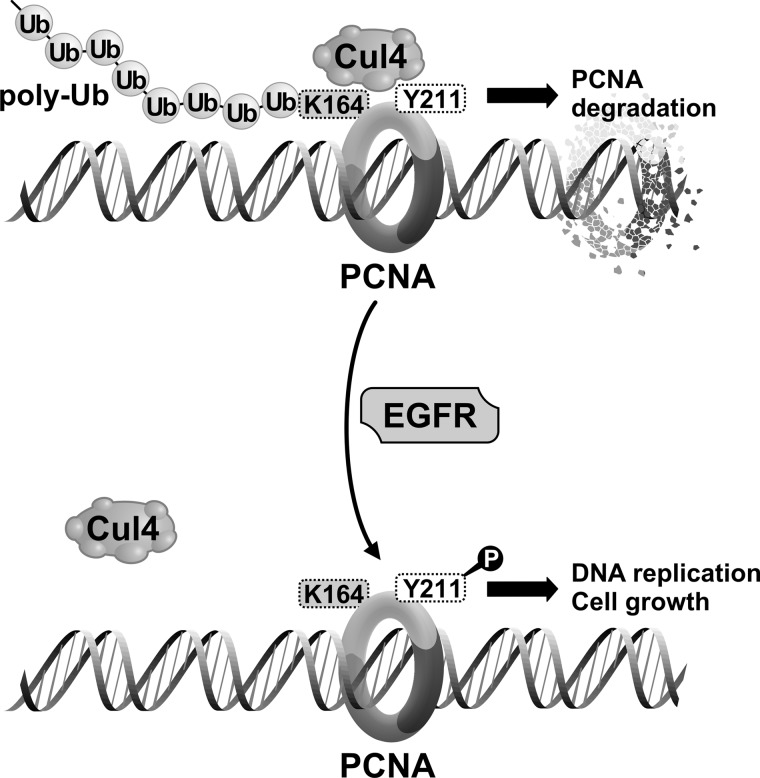
FIGURE 10.
Schematic representation of the EGFR-PCNA-CUL4A axis. We show in this study that CUL4A contributes to degradation of PCNA through Lys-164 polyubiquitylation. The ubiquitylation and PCNA degradation are enhanced when EGFR is inhibited. Tyr-211 phosphorylation of PCNA upon EGFR activation leads to dissociation of the CUL4-PCNA interaction and therefore stabilizes PCNA on the chromatin.
The concept that the protein stability of the chromatin-bound PCNA is regulated by ubiquitylation-mediated degradation is also supported by a recent report demonstrating that the E3 ligase HDM2 promoted PCNA ubiquitylation and degradation in nontransformed mammary epithelial cells (37). Interestingly, the HDM2-mediated PCNA degradation was not observed in cancer cells and seemed to be independent from the phosphorylation status of PCNA. In contrast, we have found that PCNA is tyrosine-phosphorylated in multiple cancer cell types (11, 38), which is also demonstrated in the current study in EGFR-expressing breast cancer cells. Thus, it is conceivable that degradation-associated ubiquitylation of PCNA can be mediated by different mechanisms depending on the cellular contexts and the molecular make-up. It is conceivable that, in addition to physical degradation, continuous and dynamic polyubiquitylation can also contribute to the loss of normal function of the Y211F PCNA mutant in cell proliferation.
Lys-164 monoubiquitylation of PCNA is known to be mediated by the RAD6-RAD18 and RAD5-UBC13-MMS2 (for review, see Ref. 2) or the CUL4 E3 ligases (31). Instead of proteolysis, monoubiquitylation by these enzymes resulted in functional modification of PCNA, leading to translational DNA synthesis activity of PCNA (2, 31). Thus, cells have evolved mechanisms for signaling mutually exclusive fates of PCNA by ubiquitylation. In this regard, the CUL4 complexes constitute a family of ubiquitin E3 ligases regulating fundamental biological functions including cell cycle progression and response to DNA damage (16, 39) and have been implicated in tumorigenesis through the control of genomic stability (40–42). Our study for the first time shows that CUL4A polyubiquitinates PCNA for proteolysis and that CUL4A preferentially interacts with the unphosphorylated PCNA upon suppression of EGFR activity. This result corroborates with the notion that post-translational modifications play a role in regulating substrate selection of ubiquitylation under distinct cellular environments (16).
In summary, we show here that growth signaling regulates the phosphorylation status of PCNA, which in turn modulates the stability of PCNA through differential association with its E3 ligase of the CUL4A complex. Despite extensive studies in the past decades, the current knowledge of how PCNA responds to the growth-stimulating signaling in cancer cells remains surprisingly limited. As we demonstrated recently (38), elucidating the molecular mechanisms regulating PCNA functions on the chromatin of a cancer genome will contribute to the identification of novel tumor markers with better prognostic power and the development of efficient targeted approaches for cancer therapy.
Acknowledgments
We thank Dr. I.-Fen Chen and Dr. Ou-Yang Fu for the pLKO.1/shCUL4A and PLKO.1/shCUL4B plasmids, Dr. Wade Harper for the pcDNA3-DN-hCUL4A-FLAG plasmid, Dr. David Sabatini for the pLKO.1/shScramble plasmid, and Dr. Yue Xiong for the pcDNA3-HA2-CUL4A plasmid; Dr. Jerry Lingrel for critically reading this manuscript; Birgit Ehmer for assistance in confocal microscopy; and Dr. Belinda Peace and Glenn Doerman for professional editing assistance.
This work was supported by Susan G. Komen Research Award KG080540, the Department of Defense Prostate Cancer Research Award PC073951, the Marlene Harris-Ride Cincinnati pilot grant, the University of Cincinnati and Cincinnati Children's Hospital Medical Center for Clinical and Translational Science and Training (CCTST) Junior Investigator pilot grant (to S.-C. W.), and Public Health Service Grant P30 DK078392.
- PCNA
- proliferating cell nuclear antigen
- CUL4
- cullin 4
- CUL4-DN
- dominant-negative mutant of CUL4
- EGFR
- EGF receptor.
REFERENCES
- 1. Maga G., Hubscher U. (2003) Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 116, 3051–3060 [DOI] [PubMed] [Google Scholar]
- 2. Moldovan G. L., Pfander B., Jentsch S. (2007) PCNA, the maestro of the replication fork. Cell 129, 665–679 [DOI] [PubMed] [Google Scholar]
- 3. Stoimenov I., Helleday T. (2009) PCNA on the crossroad of cancer. Biochem. Soc. Trans. 37, 605–613 [DOI] [PubMed] [Google Scholar]
- 4. Das-Bradoo S., Nguyen H. D., Wood J. L., Ricke R. M., Haworth J. C., Bielinsky A. K. (2009) Defects in DNA ligase I trigger PCNA ubiquitylation at Lys-107. Nat. Cell Biol. 12, 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Andersen P. L., Xu F., Xiao W. (2008) Eukaryotic DNA damage tolerance and translesion synthesis through covalent modifications of PCNA. Cell Res. 18, 162–173 [DOI] [PubMed] [Google Scholar]
- 6. Lehmann A. R. (2006) Translesion synthesis in mammalian cells. Exp. Cell Res. 312, 2673–2676 [DOI] [PubMed] [Google Scholar]
- 7. Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. (2004) Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23, 3886–3896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Kannouche P. L., Wing J., Lehmann A. R. (2004) Interaction of human DNA polymerase η with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, 491–500 [DOI] [PubMed] [Google Scholar]
- 9. Stelter P., Ulrich H. D. (2003) Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188–191 [DOI] [PubMed] [Google Scholar]
- 10. Hoege C., Pfander B., Moldovan G. L., Pyrowolakis G., Jentsch S. (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 [DOI] [PubMed] [Google Scholar]
- 11. Wang S. C., Nakajima Y., Yu Y. L., Xia W., Chen C. T., Yang C. C., McIntush E. W., Li L. Y., Hawke D. H., Kobayashi R., Hung M. C. (2006) Tyrosine phosphorylation controls PCNA function through protein stability. Nat. Cell Biol. 8, 1359–1368 [DOI] [PubMed] [Google Scholar]
- 12. Reed S. I. (2003) Ratchets and clocks: the cell cycle, ubiquitylation and protein turnover. Nat. Rev. Mol. Cell Biol. 4, 855–864 [DOI] [PubMed] [Google Scholar]
- 13. Higa L. A., Mihaylov I. S., Banks D. P., Zheng J., Zhang H. (2003) Radiation-mediated proteolysis of CDT1 by CUL4-ROC1 and CSN complexes constitutes a new checkpoint. Nat. Cell Biol. 5, 1008–1015 [DOI] [PubMed] [Google Scholar]
- 14. Zhong W., Feng H., Santiago F. E., Kipreos E. T. (2003) CUL-4 ubiquitin ligase maintains genome stability by restraining DNA-replication licensing. Nature 423, 885–889 [DOI] [PubMed] [Google Scholar]
- 15. Hu J., McCall C. M., Ohta T., Xiong Y. (2004) Targeted ubiquitination of CDT1 by the DDB1-CUL4A-ROC1 ligase in response to DNA damage. Nat. Cell Biol. 6, 1003–1009 [DOI] [PubMed] [Google Scholar]
- 16. Lee J., Zhou P. (2007) DCAFs, the missing link of the CUL4-DDB1 ubiquitin ligase. Mol. Cell 26, 775–780 [DOI] [PubMed] [Google Scholar]
- 17. Takeda D. Y., Parvin J. D., Dutta A. (2005) Degradation of Cdt1 during S phase is Skp2-independent and is required for efficient progression of mammalian cells through S phase. J. Biol. Chem. 280, 23416–23423 [DOI] [PubMed] [Google Scholar]
- 18. Nishitani H., Sugimoto N., Roukos V., Nakanishi Y., Saijo M., Obuse C., Tsurimoto T., Nakayama K. I., Nakayama K., Fujita M., Lygerou Z., Nishimoto T. (2006) Two E3 ubiquitin ligases, SCF-Skp2 and DDB1-Cul4, target human Cdt1 for proteolysis. EMBO J. 25, 1126–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lovejoy C. A., Lock K., Yenamandra A., Cortez D. (2006) DDB1 maintains genome integrity through regulation of Cdt1. Mol. Cell. Biol. 26, 7977–7990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li X., Zhao Q., Liao R., Sun P., Wu X. (2003) The SCF(Skp2) ubiquitin ligase complex interacts with the human replication licensing factor Cdt1 and regulates Cdt1 degradation. J. Biol. Chem. 278, 30854–30858 [DOI] [PubMed] [Google Scholar]
- 21. Hu J., Xiong Y. (2006) An evolutionarily conserved function of proliferating cell nuclear antigen for Cdt1 degradation by the Cul4-Ddb1 ubiquitin ligase in response to DNA damage. J. Biol. Chem. 281, 3753–3756 [DOI] [PubMed] [Google Scholar]
- 22. Higa L. A., Banks D., Wu M., Kobayashi R., Sun H., Zhang H. (2006) L2DTL/CDT2 interacts with the CUL4/DDB1 complex and PCNA and regulates CDT1 proteolysis in response to DNA damage. Cell Cycle 5, 1675–1680 [DOI] [PubMed] [Google Scholar]
- 23. Arias E. E., Walter J. C. (2005) Replication-dependent destruction of Cdt1 limits DNA replication to a single round per cell cycle in Xenopus egg extracts. Genes Dev. 19, 114–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Arias E. E., Walter J. C. (2006) PCNA functions as a molecular platform to trigger Cdt1 destruction and prevent re-replication. Nat. Cell Biol. 8, 84–90 [DOI] [PubMed] [Google Scholar]
- 25. Jin J., Arias E. E., Chen J., Harper J. W., Walter J. C. (2006) A family of diverse Cul4-Ddb1-interacting proteins includes Cdt2, which is required for S phase destruction of the replication factor Cdt1. Mol. Cell 23, 709–721 [DOI] [PubMed] [Google Scholar]
- 26. Senga T., Sivaprasad U., Zhu W., Park J. H., Arias E. E., Walter J. C., Dutta A. (2006) PCNA is a cofactor for Cdt1 degradation by CUL4/DDB1-mediated N-terminal ubiquitination. J. Biol. Chem. 281, 6246–6252 [DOI] [PubMed] [Google Scholar]
- 27. Raman M., Havens C. G., Walter J. C., Harper J. W. (2011) A genome-wide screen identifies p97 as an essential regulator of DNA damage-dependent CDT1 destruction. Molecular Cell 44, 72–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kim D. H., Budhavarapu V. N., Herrera C. R., Nam H. W., Kim Y. S., Yew P. R. (2010) The CRL4Cdt2 ubiquitin ligase mediates the proteolysis of cyclin-dependent kinase inhibitor Xic1 through a direct association with PCNA. Mol. Cell. Biol. 30, 4120–4133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Abbas T., Sivaprasad U., Terai K., Amador V., Pagano M., Dutta A. (2008) PCNA-dependent regulation of p21 ubiquitylation and degradation via the CRL4Cdt2 ubiquitin ligase complex. Genes Dev. 22, 2496–2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Havens C. G., Walter J. C. (2009) Docking of a specialized PIP Box onto chromatin-bound PCNA creates a degron for the ubiquitin ligase CRL4Cdt2. Mol. Cell 35, 93–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Terai K., Abbas T., Jazaeri A. A., Dutta A. (2010) CRL4(Cdt2) E3 ubiquitin ligase monoubiquitinates PCNA to promote translesion DNA synthesis. Mol. Cell 37, 143–149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hu J., Zacharek S., He Y. J., Lee H., Shumway S., Duronio R. J., Xiong Y. (2008) WD40 protein FBW5 promotes ubiquitination of tumor suppressor TSC2 by DDB1-CUL4-ROC1 ligase. Genes Dev. 22, 866–871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jin J., Ang X. L., Shirogane T., Wade Harper J. (2005) Identification of substrates for F-box proteins. Methods Enzymol. 399, 287–309 [DOI] [PubMed] [Google Scholar]
- 34. Sarbassov D. D., Guertin D. A., Ali S. M., Sabatini D. M. (2005) Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 307, 1098–1101 [DOI] [PubMed] [Google Scholar]
- 35. Martineau M., Galli T., Baux G., Mothet J. P. (2008) Confocal imaging and tracking of the exocytotic routes for d-serine-mediated gliotransmission. Glia 56, 1271–1284 [DOI] [PubMed] [Google Scholar]
- 36. He T. C., Zhou S., da Costa L. T., Yu J., Kinzler K. W., Vogelstein B. (1998) A simplified system for generating recombinant adenoviruses. Proc. Natl. Acad. Sci. U.S.A. 95, 2509–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Groehler A. L., Lannigan D. A. (2010) A chromatin-bound kinase, ERK8, protects genomic integrity by inhibiting HDM2-mediated degradation of the DNA clamp PCNA. J. Cell Biol. 190, 575–586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Zhao H., Lo Y. H., Ma L., Waltz S. E., Gray J. K., Hung M. C., Wang S. C. (2011) Targeting tyrosine phosphorylation of PCNA inhibits prostate cancer growth. Mol. Cancer Ther. 10, 29–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Dai Q., Wang H. (2006) Cullin 4 makes its mark on chromatin. Cell Div. 1, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kopanja D., Stoyanova T., Okur M. N., Huang E., Bagchi S., Raychaudhuri P. (2009) Proliferation defects and genome instability in cells lacking Cul4A. Oncogene 28, 2456–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu L., Lee S., Zhang J., Peters S. B., Hannah J., Zhang Y., Yin Y., Koff A., Ma L., Zhou P. (2009) CUL4A abrogation augments DNA damage response and protection against skin carcinogenesis. Mol. Cell 34, 451–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sugasawa K. (2009) The CUL4 enigma: culling DNA repair factors. Mol. Cell 34, 403–404 [DOI] [PubMed] [Google Scholar]