Background: Syntaxin forms nano-sized clusters at the plasma membrane whose inner organization is unknown.
Results: In the clusters, the density of proteins gradually decreases toward the periphery.
Conclusion: Syntaxin reactivity is influenced by its location within the clusters.
Significance: dSTORM imaging combined with cluster analysis significantly contributes to understanding membranal protein distribution and cluster organization.
Keywords: Membrane, Microscopy, Neuroscience, Single Molecule Biophysics, Snare Proteins, Algorithms, Clusters, Super-resolution Microscopy
Abstract
Key synaptic proteins from the soluble SNARE (N-ethylmaleimide-sensitive factor attachment protein receptor) family, among many others, are organized at the plasma membrane of cells as clusters containing dozens to hundreds of protein copies. However, the exact membranal distribution of proteins into clusters or as single molecules, the organization of molecules inside the clusters, and the clustering mechanisms are unclear due to limitations of the imaging and analytical tools. Focusing on syntaxin 1 and SNAP-25, we implemented direct stochastic optical reconstruction microscopy together with quantitative clustering algorithms to demonstrate a novel approach to explore the distribution of clustered and nonclustered molecules at the membrane of PC12 cells with single-molecule precision. Direct stochastic optical reconstruction microscopy images reveal, for the first time, solitary syntaxin/SNAP-25 molecules and small clusters as well as larger clusters. The nonclustered syntaxin or SNAP-25 molecules are mostly concentrated in areas adjacent to their own clusters. In the clusters, the density of the molecules gradually decreases from the dense cluster core to the periphery. We further detected large clusters that contain several density gradients. This suggests that some of the clusters are formed by unification of several clusters that preserve their original organization or reorganize into a single unit. Although syntaxin and SNAP-25 share some common distributional features, their clusters differ markedly from each other. SNAP-25 clusters are significantly larger, more elliptical, and less dense. Finally, this study establishes methodological tools for the analysis of single-molecule-based super-resolution imaging data and paves the way for revealing new levels of membranal protein organization.
Introduction
Research into the membranal organization of proteins into clusters has been hampered over the years by difficulties in establishing proper methodologies (1, 2). Thus, much debate and controversy has arisen over the precise definition, size, and structure of clusters (3–5). A major limitation has been the inability to image the spatial organization and distribution of proteins at length scales smaller than the diffraction barrier of conventional light microscopy. However, in the past decade, this barrier has been mostly overcome by the development of far-field super-resolution imaging methods, including stimulated emission depletion (STED) (6), structured illumination microscopy (7), photoactivated localization microscopy (8), stochastic optical reconstruction microscopy (STORM)4 (9), and direct STORM (dSTORM) (10). Despite this tremendous progress, super-resolution imaging still requires the development of unique analytical tools to cope with the newly resolved data and to provide reliable biological interpretations. In the present study, we applied dSTORM for single-molecule-based super-resolution imaging, combined with sophisticated analytical algorithms to determine the spatial membranal distribution of syntaxin 1 and SNAP-25, essential SNARE (soluble N-ethylmaleimide-sensitive factor attachment protein receptor) proteins that mediate the fusion of intracellular membranes in synaptic transmission (11).
dSTORM employs standard fluorescent probes, such as commercially available antibodies, as immunocytochemical or chemical tags for super-resolution imaging in fixed and living cells with an optical resolution of ∼20 nm in the imaging plane (10, 12–14). The method relies on the principle that most standard fluorophores can be reversibly photo-switched between a bright fluorescent “on” state and a nonfluorescent “off” state in the presence of millimolar concentrations of thiols (12–14). Precise positional determination of the individual fluorophores when switched on (termed localization) is achieved by approximating their fluorescence emission pattern (the point spread function) with a two-dimensional Gaussian function (15). A super-resolved fluorescence image is then reconstructed by summing all localizations from a series of thousands of frames containing hundreds of thousands of “blinking” events (Fig. 1).
FIGURE 1.
dSTORM imaging of syntaxin membranal clusters versus single molecules. A, representative conventional fluorescence image of syntaxin staining in a native membrane sheet. Due to super-position of all single fluorescent fluorophores, the structure appears blurred in the conventional wide-field image. B, a dSTORM image of the membranal staining of syntaxin with superior resolution was reconstructed from thousands of single-molecule localizations. Insets in A and B are magnified sections of the same region in the membrane imaged by conventional fluorescence and by dSTORM. C, representative membranal region of 500 nm2, which includes all fluorophore localization coordinates (black circles) representing syntaxin molecules gathered during dSTORM imaging of this region. The center of mass (red points) of each putative cluster was located among the total number of localizations using ImageJ as described under “Experimental Procedures.” D, a density-based algorithm recognizes the members of each cluster (red circles) and the single-molecule localizations (blue circles) as detailed under “Experimental Procedures.”
A previous nanoscopy study based on stimulated emission depletion (16), as well as other studies (17–19), have investigated the distribution of syntaxin and other synaptic proteins on the membrane and have indicated syntaxin partitioning into nano-sized clusters (16, 17, 19–23). Sieber et al. (16) suggested the existence of two pools of syntaxin: clustered and as single freely diffusing molecules. However, direct visualization of the single-molecule pool and small clusters was not possible due to a resolution limit of 50–60 nm (16). Therefore, neither cluster size distribution nor the internal organization of the molecules in the clusters could be demonstrated. In the present study, using the improved resolution provided by dSTORM, we assemble a comprehensive picture of the distribution of syntaxin and SNAP-25 at the plasma membrane (PM) into clustered and single-molecule pools. The organization of single molecules inside the clusters is characterized for the first time, as is the distribution of single-molecule pools between these clusters. In addition, the data show that small clusters can unite at their periphery with other clusters, while retaining their internal organization, to form super-clusters.
EXPERIMENTAL PROCEDURES
Syntaxin and SNAP-25 Staining in the Inner Leaflet of the PM of PC12 Cells
To image the distribution of syntaxin and SNAP-25 in the PM, neuroendocrine PC12 cells were grown overnight on poly-l-lysine-coated glass coverslips (24, 25). Coverslips were sonicated to remove the upper parts of the cells, leaving the native PM sheets attached. These were either immediately fixed or incubated for 30 min at 37 °C in bovine serum albumin (BSA)-K-Glu (K-Glu buffer containing 3% BSA) and then fixed, washed, and immunostained as described previously (24). To detect syntaxin, we used the mouse monoclonal antibody HPC-1, and to detect SNAP-25, we used the mouse monoclonal 71.1 antibody (Synaptic Systems). Cy5-coupled goat-anti-mouse was used as secondary antibody (Dianova). Shortly before imaging, samples were incubated in a “switching buffer” consisting of PBS, pH 7.4, containing oxygen scavenger (oxygen was removed by adding 0.5 mg ml−1 glucose oxidase (Sigma), 40 μg ml−1 catalase (Roche Applied Science), 10% w/v glucose, and 100 mm β-mercaptoethylamine) (13).
dSTORM Imaging
Fluorescence imaging was performed on an Olympus IX-71 in objective-type total internal reflection fluorescence configuration using an oil immersion objective (PlanApo 60×, NA 1.45, Olympus) (10). Two continuous wave laser beams of argon-krypton laser (Innova 70C, Coherent) were selected by an acousto-optic tunable filter and used simultaneously for read-out and activation. The laser beams were coupled in the microscope objective by a polychromic beam splitter (532/647, AHF Analysentechnik). Fluorescence light was spectrally filtered with filters ET700/F75 and HQ542LP (AHF Analysentechnik) and imaged with an EMCCD camera (Andor Ixon DV897DCS-BV). Additional lenses were used to achieve a pixel size of 85 nm. The laser powers were chosen to ensure that the fraction of activated fluorophores at any given time would be sufficiently low to enable recognition of individual fluorophores. Typically, we recorded 12,000 frames at rates of 50–200 Hz. Applying a laser power of 1–15 kilowatts cm−2 at 647 nm, we detected 500–5000 photons per molecule and frame, corresponding to a localization precision of less than 10 nm (13). Optionally, irradiation at 514 nm with 0.01–0.2 kilowatt cm−2 laser power was used to increase the number of fluorescent spots in the detection area. Localizations of fluorescent molecules were extracted in three steps by the open access rapidSTORM software developed by our group for dSTORM imaging analysis (15). In the first step, candidate emission positions were found by smoothing the image data with an average mask with a width of 1 point spread function full width at half-maximum and locating the maxima in this smoothed image. In the second step, a local window with a width of 4 point spread function full width at half-maximum was cropped around each spot, and a Gaussian model of the point spread function was fitted by Levenberg-Marquardt least squares estimation to the pixels in this window. The chosen free-fit parameters were local background, number of photons, and emitter position. If the number of photons as estimated by the fitted parameters surpassed a threshold, the center of the fitted Gaussian was considered a localized fluorophore (15). In the third step, emission positions were discarded where a fit with a sum of two Gaussian functions yielded significantly better residues.
Identifying Centers of Mass of Putative Clusters
All localizations in the dSTORM images (x and y coordinates of the center of mass of each fluorophore) were systematically scanned by a clustering algorithm to identify the local maxima of clusters. The analysis proceeded as follows. First, random selected membranal regions in each of the dSTORM images were fed into ImageJ, a Java-based image-processing program (26) developed at the National Institutes of Health (rsbweb.nih.gov/ij). The images were subjected to a built-in function of the program for Fourier transform, and then a band-pass filter (set to 48–120 nm, half the spatial frequency of the actual cut-off) was operated to remove high and low spatial frequencies, to ease tracking of local intensity maxima. Then a reverse Fourier transform was performed, and a built-in algorithm of ImageJ was operated to find local intensity maxima of the potential clusters in the image (15, 26). To ensure that the located centers of mass are not artifacts, a noise level was determined to ascertain that the found local intensity maxima were higher than a certain value from the surroundings. The output obtained from the program was a list of x,y coordinates of each of the local maxima of the putative clusters.
Density-based Clustering Algorithm
The list of localizations in each dSTORM image was subjected to a second algorithm (27) that groups localizations into clusters using the local maxima coordinates found by ImageJ. Accordingly, to define localizations as part of a cluster, the neighboring area, having a predetermined radius around this localization (termed ϵ), should contain at least a minimum number of neighboring localizations, i.e. the density in the neighborhood must exceed a certain threshold of localizations (termed k). Once the threshold is met, the tested localization and the neighboring localizations within ϵ are defined as cluster members. The same procedure is performed for the rest of the localizations around the center of mass, and if the number of localizations around any one of them is lower than the threshold, the localization is defined as border localization, and grouping of localizations further away from this localization for the relevant cluster is terminated. Localizations that were not attributed to any cluster by the procedure were determined as single-molecule localizations. All localizations were confirmed as not being artifacts by exhibiting a minimum threshold of photon counts, which is calculated per movie from the background counts (typically 500 photons per fluorophore). ϵ was calculated semiautomatically by calculating the minimum distance between each localization and its 16th closest neighbor (supplemental Fig. S1). As shown in supplemental Fig. S1, ϵ was found to be 30 nm on all days of measurement by extracting the breaking point of the distribution graph of this minimum distance as described in detail in the literature (27). The parameters used for the clustering algorithm were carefully selected and optimized according to several considerations. First, the choice of parameters ensured the presence of several syntaxin molecules grouped together by determining a sufficient number of fluorophore localizations, assuming that each molecule is represented by several localizations. Second, localizations were ensured to be located in an area that is wide enough to populate several molecules considering the resolution of the method for a single molecule. Accordingly, the value of ϵ (r = 30 nm) found in supplemental Fig. S1 is three times higher than the mean localization precision of a single molecule, i.e. < 10 nm, and ensures the presence of more than a single molecule in an area of 30 nm radius. Finally, the combination of 16 localizations in a radius of 30 nm resulted in the best grouping into clusters with minimum false positives and agreement with those detected visually as compared with several other examined parameter combinations (e.g. k = 4, 8, or 64 or ϵ = 20 or 15 nm). To independently confirm that the grouping was properly performed by the algorithm, all clusters were subjected to principal component (PC) analysis (28) as detailed below. The goal was to verify that all of the localizations that were grouped together to a certain cluster by the density algorithm were indeed part of this cluster. Accordingly, the PC analysis detects outlier points by examining the distribution of the localizations in each cluster and identifying those that deviate from the expected distribution and then filters them out. Another control for the clustering algorithm was performed by determining the small angle (α) between one axis of each cluster (x′) and the original x axis of the image. A random distribution of α ensures that there was no biased preference in the detection of the clusters according to one of the original axes of the image. Supplemental Fig. S2 gives a representative example for the random distribution of α between 45° and −45° calculated for 40 clusters in one of the imaged membranes.
Analysis of Cluster Characteristics
The size of the clusters was estimated by a variation of a two-dimensional PC analysis (28). In a model that assumes elliptical shapes of clusters and a distribution of localizations in them, the area of a cluster is
 |
where M is the (2 × 2) covariance matrix, and a is the radius of the cluster in units of standard deviation. Points belong to the ellipse if, after normalization with the covariance matrix, they belong to a circle of radius a. For example, a uniform distribution implies a = 2 because in that case, the standard deviation of each coordinate is half the radius. Assuming that the density decreases toward the cluster boundaries, a larger value (a = 2.5) was chosen. All points further away were considered outliers, and the covariance matrix was recalculated with the remaining points. In the second iteration, the radius was determined to contain all of the remaining points. The localizations that were identified as outliers were removed from the clustered pool and were added to the single-molecule pool.
Nonhomogeneous Two-dimensional Stochastic Model
The in silico Monte Carlo-based dynamics simulation was based on a simulation described by Nudelman et al. (29) and was adopted to model SNARE dynamics in the presence of SNARE clusters. The model includes the individual SNARE proteins (syntaxin, SNAP-25, and synaptobrevin) visualized as freely diffusing particles in a 1-μm2 two-dimensional membrane-like surface (see Fig. 6). Each SNARE protein was assigned with its appropriate diffusion coefficient, encounter radius, and probability to form a reversible complex with the other SNARE proteins. The rates of reactions between reactants were treated as adjustable parameters or taken from Bar-On et al. (20) or Mezer et al., (30, 31) and expressed as probabilities of a certain protein to react with another once they encountered each other. By varying the probabilities of the intermediate complexes to form or dissociate, we simulated the SNARE complex assembly either in a regular membrane that includes SNARE clusters (see Fig. 6B) or in a membrane where the proteins are homogeneously randomly distributed (see Fig. 6A). In the simulation, syntaxin clusters are defined as areas in the membrane with physical boundaries where the proteins inside them can leave only with a low probability. The probability to enter/leave the clusters expresses the level of the inner density and the repulsive/attractive forces between the molecules in the clusters that define their boundaries (16). The size and density of the clusters are adjusted according to the spatial super-resolution dSTORM data obtained for the clusters and presented herein.
FIGURE 6.
Two-dimensional membrane-like grid modeling of SNARE protein distribution and kinetics. Monte Carlo-based modeling (see “Experimental Procedures”) of syntaxin (white circles) dynamics and distribution in the plasma membrane during the assembly of cis-SNARE complexes (cyan). Two simulations (100,000 steps), each representing two scenarios of SNARE protein distribution, were performed, and the amount of cis-SNARE complexes formed and their locations in the two simulations were compared. The first scenario of the simulation includes a homogeneous membrane. Syntaxin is homogeneously and randomly distributed in the membrane and can freely diffuse and interact with SNAP-25 (gray squares) to form binary complexes (green circles) and then with synaptobrevin (blue squares) to form cis-SNARE complexes (cyan circles) according to preassigned rate coefficients. A and B, two snapshots taken at different time points from the simulation showing the formation of binary and ternary SNARE complexes (C–E) In the second scenario examined using the simulation, the majority of the syntaxin population is clustered (66%), and the protein can leave the clusters and interact with the other proteins at the clusters or outside according to the same rate coefficients assigned in the first scenario (C). As can be seen, binary complexes and cis-SNARE complexes are formed first mainly between clusters (D) and then at the periphery of clusters as well (E).
RESULTS AND DISCUSSION
To fill in the crucial gaps and form a complete picture of syntaxin distribution at the PM, more than 60 regions of native membrane layers immunostained for syntaxin (20) and a secondary antibody labeled with Cy5 were randomly selected and imaged at single-molecule sensitivity. dSTORM images were then subjected to an extensive analysis characterizing the distribution of syntaxin molecules inside and outside the nano-sized clusters. Inspection of dSTORM images of syntaxin molecules showed the substantially improved resolution as compared with wide-field fluorescence (Fig. 1) and revealed localizations of single syntaxin molecules and small clusters (<60 nm) alongside large clusters that can also be easily discerned in conventional fluorescence microscopy. In previous studies, protein clusters were defined as bright spots in the image, and their size was estimated from the half-width of the spots (16). Therefore, clusters smaller than the resolution limit of the method (50–60 nm) or single molecules could not be observed. dSTORM, on the other hand, can detect the location of each individual molecule with high localization precision (10, 12–14). However, it remains challenging to group the localizations into clusters (32).
To objectively identify the distribution pattern of syntaxin molecules, i.e. individual versus clustered, we analyzed the dSTORM images by implementing a combination of two clustering algorithms (26, 27) followed by PC analysis to confirm cluster members. First, the centers of mass of high densities of localizations, i.e. putative clusters, were identified by an image-processing program (Fig. 1C, red points), and then a density-based clustering algorithm (27) that determines the density around each of the localizations was applied to scan all localizations around the identified centers of mass. This algorithm diagnoses changes in the density patterns and defines whether or not a localization is part of a cluster (Fig. 1D, red circles) (27). Finally, the distribution of all localizations in a particular cluster was examined by performing a PC analysis to detect outliers, i.e. points that deviate from the main collection of points, and attribute them to a single-molecule pool. Localizations that were not associated to clusters, exhibiting a lower density profile around the clusters than in them, were also defined as part of the single-molecule pool (Fig. 1D, blue circles). The centers of mass of the clusters were confirmed by calculating the arithmetic mean of the localizations that were attributed to each cluster by the density algorithm; these estimated centers of mass were compared with the centers of mass that had been identified by the image-processing program prior to the density-based clustering. In most cases, there were only minor differences (<20 nm, similar to the spatial resolution) between the calculated and the predetermined centers of mass (data not shown), corroborating the validity of the clustering method for proper cluster identification.
The dSTORM images, together with the density-based analysis, revealed that on average, the clustered syntaxin molecules occupy 11% of the total membranal surface area and include about 67% of the syntaxin population. A rather constant average density of ∼14 clusters/μm2 was found (Table 1), similar to a previous estimation (16) of ∼20 clusters/μm2. An average syntaxin cluster diameter of 93.4 nm (Table 1) was calculated by determining the main axes of each cluster using a variant of the two-dimensional PC analysis as detailed under “Experimental Procedures” (28). In addition, the clusters were found to be elliptical with a ratio of about 0.63 between cluster diameters.
TABLE 1.
Summary of the main characteristics of about 3000 syntaxin clusters analyzed on three independent measurement days
The table includes mean cluster diameter (MCD), percentage of the single-molecule population (SMP), ratio of diameters representing cluster shape (circular or elliptical) (RD), and the density of syntaxin clusters per μm2 (DSC). The S.E. values for each measurement day are presented in parentheses.
| 270 clusters | 1174 clusters | 1555 clusters | Average | |
|---|---|---|---|---|
| MCD (nm) | 93.3 (5.84) | 82.9 (3.14) | 104.2 (5.91) | 93.4 |
| SMP (%) | 34.2 (2.8) | 31.6 (1.7) | 33.3 (1.6) | 33 |
| RD | 0.64 (0.01) | 0.61 (0.007) | 0.64 (0.008) | 0.63 |
| DSC | 14.5 (1.15) | 17.12 (0.94) | 13.04 (0.65) | 14.89 |
In the next step, the inner organization of the molecules in the clusters was mapped in an attempt to understand how syntaxin clusters are internally organized and how they are formed. A closer look at the organization of syntaxin molecules in clusters was achieved by calculating the internal density of the molecules in the clusters and looking at their density as a function of their distance from the cluster center. Previous studies that could not discriminate between the localizations within the clusters have modeled the inner organization in the cluster as a bunch of molecules, suggesting a uniformly dense distribution of syntaxin molecules based on homomeric protein-protein interactions (16). The present analysis revealed a nonuniform distribution of localizations in the cluster with intrinsic density gradient decreasing from the dense center toward the cluster rim (Fig. 2A). The phenomenon of a density gradient in the clusters was observed in most of the clusters analyzed, but the slope of the gradient differed among the clusters.
FIGURE 2.

The inner structure of syntaxin clusters and distribution of cluster sizes. A, three-dimensional plot of a representative example of the internal density gradient of localizations in two clusters. Each of the localizations is marked with a color that represents the number of neighboring localizations in a 30-nm radius around it. The localization scale ranges from 0 to 180 (z axis). The center of the cluster exhibits a high density of localizations, and the outer parts of the cluster show a decreasing gradient of localization density. B, histogram of cluster size, represented by the mean diameter of clusters as calculated by PC analysis, reveals an exponential distribution (see “Experimental Procedures”). C, plot of a representative membrane showing the distribution and inner gradients of several syntaxin clusters. Black frame: a super-cluster composed of three smaller clusters. Blue frame: two small adjacent clusters that might be in the process of uniting. Red frame: a large cluster with a single density gradient.
The high density of syntaxin at the cluster core can be explained by homomeric interactions of syntaxin molecules forming homo-oligomers, resulting in crowding of the molecules and overcoming the repulsion forces between them (16, 22). However, the mechanism leading to a density gradient of the molecules from the cluster center to the rims and around the clusters cannot be explained by these interactions as they require direct protein-protein contact, suggesting that factors additional to syntaxin-syntaxin binding are involved in cluster formation.
A few mechanisms involving other kinds of interactions in which syntaxin is involved might explain the phenomenon of the density gradients. It is well established that lipids are distributed nonhomogeneously in the plasma membrane and can cluster into microdomains. Therefore, interaction of syntaxin with certain lipids might form the syntaxin density gradient observed in the clusters. For example, interaction of syntaxin with cholesterol might contribute to the formation of a gradient within and outside the syntaxin clusters. The effect of cholesterol removal on the arrangement of syntaxin clusters is not entirely clear. One study showed that it causes the clusters to disintegrate and changes the distribution pattern of the protein (33). However, in another study based on stimulated emission depletion microscopy, there was no change in the arrangement of syntaxin clusters following cholesterol removal (34). Because not all syntaxin clusters contain cholesterol, other lipids might be involved as well. Recently, colocalization was shown between the phosphoinositide phosphatidylinositol 4,5-bisphosphate and syntaxin clusters (35). It was suggested that syntaxin might cluster through electrostatic interactions with phosphatidylinositol 4,5-bisphosphate. However, the low colocalization percentage between syntaxin clusters and phosphatidylinositol 4,5-bisphosphate in PC12 cells, i.e. only 5–10% (35), suggests that this interaction is not the main factor in syntaxin clustering. Another explanation for the gradual decrease in the density of syntaxin molecules within and around the clusters is the presence of other proteins in the cluster periphery. Syntaxin can engage in heterodimer complex formation. These heterodimer complexes might cluster around the cores of syntaxin clusters. SNAP-25, for example, has been shown to be in close proximity to syntaxin clusters (34). Therefore, as suggested by Rickman et al. (34), it can interact with syntaxin molecules that reside in less dense areas of the clusters, or around them, to form syntaxin-SNAP-25 complexes within and around syntaxin clusters. Hence, our results, demonstrating a dense cluster core and a gradual decrease in density toward the boundaries, might support the hypothesis of Rickman and co-workers (34, 36) suggesting the formation of syntaxin-SNAP-25 complex clusters at the peripheries of syntaxin clusters, whereas the center of the syntaxin cluster acts as a reservoir for syntaxin molecules.
To evaluate whether other SNARE proteins share common distributional features with syntaxin, we analyzed the distribution pattern of SNAP-25 and compared it with that of syntaxin. Interestingly, by applying the same clustering procedure, the distribution of SNAP-25 molecules is markedly different from that of syntaxin. Based on analysis of more than 150,000 localizations and 528 SNAP-25 clusters, we found that 68% of SNAP-25 molecules fall into clusters that are much larger as compared with those of syntaxin (a mean diameter of 129.6 nm versus 93.4 nm, Table 2). The SNAP-25 clusters are significantly more elliptical, with a ratio of 0.53 between the diameters. The density of the clusters on the inner leaflet of the cell membrane is smaller than that of syntaxin, with an average of 9 clusters/1 μm2. Additionally, more SNAP-25 clusters share a border with other SNAP-25 clusters (an average of 1.94 neighboring clusters/cluster versus only 0.58 for syntaxin clusters, see Fig. 4A). Despite these differences in size, shape, and distribution between SNAP-25 and syntaxin clusters, both proteins share some common organizational features within the clusters. For example, like syntaxin, the density of clustered SNAP-25 molecules decreases gradually from the cluster core toward the periphery, forming in the clusters one or more density gradients (see Fig. 4B).
TABLE 2.
Summary of the main characteristics of 528 SNAP-25 clusters
The table includes mean cluster diameter (MCD), percentage of the single-molecule population (SMP), ratio of diameters representing cluster shape (circular or elliptical) (RD), and the density of SNAP-25 clusters per μm2 (DSC). The S.E. values are presented in parentheses.
| 528 clusters | |
|---|---|
| MCD (nm) | 129.6 (8.5) |
| SMP (%) | 32 (1.6) |
| RD | 0.52 (0.012) |
| DSC | 9.33 (0.97) |
FIGURE 4.

SNAP-25 membranal distribution. A, using the same clustering procedure, SNAP-25 clusters were identified side by side to nonclustered molecules. Each cluster is marked by the set of localizations that were attributed to it using the density-based algorithms (each cluster is presented by a different color); the nonclustered pool is marked in black circles. Please note that there are membranal areas that are completely devoid of SNAP-25. B, three-dimensional plot of a representative SNAP-25 cluster inner density gradient of localizations decreasing from the dense core to the periphery of the cluster. Each of the localizations is marked with a color that represents the number of neighboring localizations in a 30-nm radius around it. The localization scale ranges from 0 to 500 (z axis). C, histogram presenting the percentage of SNAP-25 single-molecule localizations as a function of their distance from the nearest cluster (values are presented in nm).
Next, we examined the effect of cis-SNARE complex formation on syntaxin distribution. Incubation of the membranes for 30 min (see “Experimental Procedures”), during which cis-SNARE complexes (i.e. membranal syntaxin, SNAP-25, and synaptobrevin) are accumulated (24), did not have an effect on the main characteristics of syntaxin clusters such as their size, shape, etc. (Table 3). The only change in the distribution pattern of syntaxin was a prominent increase (more than 80%) in the number of contacts between clusters following an incubation of the membrane for 30 min (0.58 at t = 0 versus 1.06 contacts/cluster at t = 30 min). It should be noted that the distribution of the minimum distance between the centers of mass of the clusters did not change (supplemental Fig. S3), suggesting a rearrangement of the syntaxin molecules only in the periphery of clusters. The cis-SNARE complexes might be formed and accumulated in the periphery of clusters and bridge between the clusters.
TABLE 3.
Summary of the main characteristics of over 7000 syntaxin clusters following incubation of the membranes for 30 min to allow cis-SNARE complex formation
The table includes mean cluster diameter (MCD), percentage of the single-molecule population (SMP), ratio of diameters representing cluster shape (circular or elliptical) (RD) and the density of Syntaxin clusters per μm2 (DSC). The S.D. values between the 3 measurements days are presented in parentheses.
| >7000 syntaxin clusters | |
|---|---|
| MCD (nm) | 94.5 (9.6) |
| SMP (%) | 31 (1.5) |
| RD | 11.4 (1.5) |
| DSC | 0.61 (0.02) |
The mechanism of cluster formation is not yet known. Nevertheless, a profile of the distribution of cluster sizes can provide some information on this mechanism (37). In several systems that investigated membranal protein clusters, it has been shown that an exponential distribution of cluster sizes can reflect their spontaneous aggregation and growth (37); based on analysis and modeling, it was suggested that protein clustering is a stochastic self-assembly process in which proteins freely diffuse in the membrane and join existing clusters or nucleate new ones (38). To characterize the size distributions of the clusters, ∼3000 clusters consisting of several hundreds of thousands of individual localizations were analyzed. We found that the majority of the clusters, i.e. 24%, are relatively small (40–60 nm) and that cluster sizes, measured according to their mean diameters, were approximately exponentially distributed (Fig. 2B). According to the stochastic self-assembly model suggested by Greenfield et al. (37), small clusters can diffuse and occasionally unite with other clusters to form larger clusters. If this is the case, using the developed analytical tools, we should be able to detect cases in our system in which small clusters with individual gradients are combined or united forming a larger super-cluster.
Indeed, a closer look at the cluster images and the inner distribution pattern revealed many cases in which a few small clusters had united at their peripheries to form a single cluster (Fig. 2C, black and blue frames). Those small clusters seemed to preserve their internal density gradients or share a joint gradient (Fig. 2C) to form a single unit with several dense cores, each surrounded by a gradually decreasing gradient (Fig. 2C). Because the dSTORM images might reflect a steady state of the cell system, our data suggest that redistribution of densities in the clusters is extremely slow and therefore the individual gradients of the smaller subclusters in the super-clusters are preserved. Large clusters containing a single density gradient with a highly dense core were also detected (Fig. 2C, red frame). The latter might be a result of cluster reorganization following the unification of several small clusters and disruption of their own inner organization to form a single gradient around a single core. These results suggest that small syntaxin clusters join together to form larger clusters that might later reorganize into a single unit, providing a glimpse of the mechanism of syntaxin cluster formation.
In the next step, we focused on characterizing the single-molecule pool of syntaxin, i.e. those molecules not grouped into any cluster, to explore how this pool relates to the clusters, i.e. whether it is completely isolated or a continuation of the cluster population and whether it is homogeneously distributed at the membrane or concentrated in a certain location. All membranes were found to contain a significant fraction of nonclustered molecules, corresponding on average to ∼33% of the total syntaxin population. Previous studies have suggested the presence of a nonclustered, presumably freely diffusing pool. However, direct visualization of that pool was not possible due to resolution limitations, and therefore, its distribution could not be defined (16, 22). Here, dSTORM images revealed singleton localizations representing the single-molecule pool of syntaxin outside the clusters. Further analysis characterized the distribution of this pool in relation to the clusters by examining the minimum distance of each of the localizations from the nearest neighbor cluster. Our findings revealed that the single syntaxin molecules are nonhomogeneously distributed in the membrane but concentrated in areas adjacent to clusters (Fig. 3A, black circles). Over 80% of syntaxin molecules are concentrated at a distance of up to 120 nm from clusters, and the percentage of molecules at longer distances decreases exponentially (Fig. 3B). Similarly, 90% of the nonclustered SNAP-25 molecules (33% of the total SNAP-25 population) are located up to 120 nm from the clusters (Fig. 4C). This finding joins previous dynamic measurements (16) in suggesting that there might be a dynamic equilibrium between clusters and single molecules at and near the cluster boundaries and that single molecules diffuse and occasionally leave or join existing clusters. The single-molecule pool is mostly located around clusters but still exhibits a different density pattern.
FIGURE 3.
Distribution of the nonclustered localizations as a function of cluster proximity. A, representative image of a membrane presenting localization of the single-molecule pool (black points) that resides close to the clusters (red points). Areas devoid of syntaxin appear in the membranal region. B, histogram presenting the percentage of single-molecule localizations as a function of their distance from the nearest cluster (values are presented in nm). The probability of finding a single molecule of syntaxin decreases exponentially as the distance to the cluster increases.
Furthermore, dSTORM images revealed that some membranal areas are completely devoid of syntaxin (Fig. 3A), implying that most of the interactions involving syntaxin molecules occur in close proximity to the clusters, where syntaxin is present. This suggests that the single-molecule pool is not freely diffusing throughout the entire membrane; rather, its diffusion is limited, perhaps by the presence of other protein clusters in the membranal environment, such as SNAP-25 clusters. Similarly to syntaxin, large areas of the membrane are devoid of SNAP-25 clusters or individual SNAP-25 molecules (Fig. 4A).
Because most of the syntaxin or the SNAP-25 molecules are concentrated within or around their own clusters and the overlap between the distribution of the populations was found to be high (34) a possible conclusion is that clusters of syntaxin reside side by side to SNAP-25 clusters and the interaction between syntaxin and SNAP-25 forming SNARE complexes might take place between or at the boundaries of the clusters.
As for syntaxin, a question still remains as to how the reactivity of syntaxin is influenced by its location in clusters or outside. Our findings clearly demonstrate three degrees of syntaxin organization: (i) densely packed in the cluster core, (ii) gradually decreasing in density toward the outer parts of the clusters, and (iii) single molecules diffusing outside the clusters (Fig. 5). The results presented herein together with the model presented in Bar-On et al. (20) for the kinetics of cis-SNARE complex formation might explain how syntaxin reactivity is affected by its position within and around clusters. The model suggests partitioning of the syntaxin molecules into three reactive states, depending on their environment. Accordingly, the densely packed molecules at the cluster center, being inaccessible to the external surroundings, are less reactive than the single syntaxin molecules that can interact freely with their partners. Moreover, the model indicates the presence of a fraction of clustered molecules that are semi-reactive and can interact with syntaxin partners, albeit with much slower kinetics (20). This fraction was suggested to represent molecules situated in the outer parts of the clusters, which are therefore more accessible to the surroundings than molecules in the center. Our results reinforce this model; we identified dense packing of syntaxin molecules in the cluster centers, which probably does not allow interaction of syntaxin with its partners. The syntaxin molecules that are part of the gradient, residing at the periphery, might represent the semi-reactive pool suggested by the kinetics model. Finally, we describe the distribution of single syntaxin molecules outside the clusters, representing the third phase of the model, i.e. molecules that are fully accessible for interaction (20). Based on the dSTORM results, we therefore interlink the organization of syntaxin molecules inside and around clusters and their ability to interact with their partners. Similarly, gradual reactivity levels can be attributed to SNAP-25 within or around clusters.
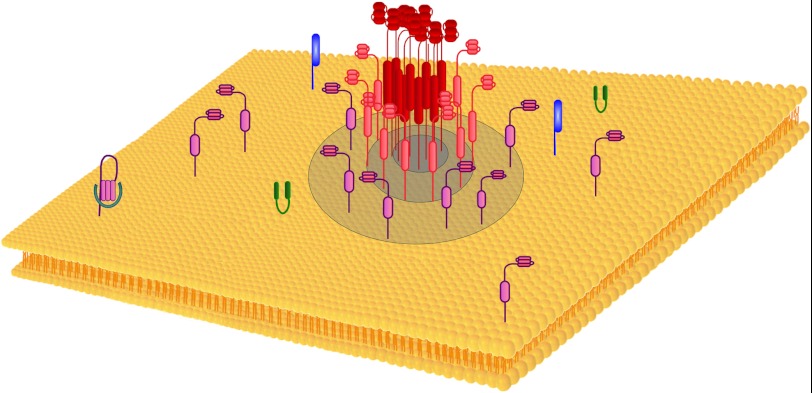
FIGURE 5.
Proposed model of the nano-scale membranal organization of syntaxin. Syntaxin distributes nonhomogeneously at the membrane, forming clusters alongside single molecules. The clusters exhibit an internal density gradient of molecules. In the example presented, a gradient of syntaxin molecules is demonstrated decreasing from the cluster center (red) to its peripheries (pink and purple). The single-molecule pool of syntaxin (purple) concentrates at and near the cluster boundaries where the molecules can react with their partners, e.g. SNAP-25 (green), synaptobrevin (blue), and Munc18 (cyan). A similar model can be applied for SNAP-25 clusters.
This hypothesis was tested by designing an in silico membrane-like grid Monte Carlo-based simulation modeling the presence of syntaxin in clusters and as single molecules around clusters (Fig. 6 and see “Experimental Procedures”). During the simulation, syntaxin can diffuse and react with membranal SNAP-25 to form a binary complex, and the latter can react with synaptobrevin to form ternary SNARE complexes according to certain probabilities to react with each other once they have encountered each other. Using the simulation, we examined specifically the effect of the presence of syntaxin in clusters on the engagement of syntaxin into membranal cis-SNARE complexes (20, 24) and the location of the newly formed cis-SNARE complexes with respect to the clusters. Accordingly, we examined cis-SNARE complex formation in two scenarios; the first is a homogeneous membrane where all proteins can diffuse freely, encounter each other, and react (Fig. 6, A and B). In the second one, we modeled the distribution of syntaxin into clusters and single molecules. Clusters were defined according to the dSTORM data presented herein (14 clusters/1 μm2 and 66% of the syntaxin molecules are clustered).
The other proteins (SNAP-25 and synaptobrevin) diffuse between the clusters (Fig. 6, C–E). For simplicity, SNAP-25 clusters were not incorporated into the model. Simulating both scenarios for the same time period (100,000 steps) using the same rate coefficients and concentrations showed that fewer cis-SNARE complexes are formed when syntaxin is mostly partitioned into clusters in comparison with when it is homogeneously distributed in the membrane, showing that cis-SNARE complex formation is slowed down due to the presence of syntaxin in clusters. Moreover, following the process of cis-SNARE complex formation in the presence of the clusters showed that the binary complexes and the cis-SNARE complexes were formed first outside the clusters (Fig. 6, C) and then in the periphery of clusters (Fig. 6, D and E), in accordance with our hypothesis that syntaxin molecules situated in the outer parts of the clusters might represent the semi-reactive pool suggested by the kinetic model.
Our results suggest, for the first time, a link between the inner organization of proteins in clusters and around them and their kinetic properties. Furthermore, the findings provide a new perspective on the role of syntaxin clusters as reaction centers releasing reactive syntaxin molecules for interactions with their partners in SNARE formation or other interactions. Syntaxin molecules have been shown to interact with many other proteins including calcium channels (39–41), Ca2+-dependent activator protein for secretion (CAPS) (42), and the cytoplasmic proteins tomosyn (43) and Munc18 (44). As syntaxin clusters might play a role in vesicle docking and fusion, a future challenge will be to characterize the relative distribution (and later also the dynamic distribution) of all of these proteins in relation to syntaxin clusters and single molecules to assemble the entire intriguing distribution hallmark of SNARE proteins and SNARE partners. The current study presents a comprehensive set of tools that can be readily applied to cope with this challenge. These tools can be used for analyzing single-molecule-based super-resolution data with emphasis on protein clustering, possibly even in living cells, with the ultimate goal of forming integral nano-scale maps of membranal protein distribution.
Acknowledgments
We thank Prof. Reinhard Jahn for constant support of the project and Dr. Oliver Keller for insightful input and assistance with the mathematics and statistics.
This work was supported by the Biophotonics Initiative of the Bundesministerium für Bildung und Forschung (to M. S.), the Israel Science Foundation Grant 1211/07 (to U. A.), National Institutes of Health Grant RO1 NS053978 (to U. A.), and German-Israeli Foundation (GIF) Grant 1125-145.1/2010 (to U. A. and M. S.).

This article contains supplemental Figs. S1–S3.
- STORM
- stochastic optical reconstruction microscopy
- dSTORM
- direct STORM
- PM
- plasma membrane
- PC
- principal component.
REFERENCES
- 1. Lingwood D., Simons K. (2010) Lipid rafts as a membrane-organizing principle. Science 327, 46–50 [DOI] [PubMed] [Google Scholar]
- 2. Hanzal-Bayer M. F., Hancock J. F. (2007) Lipid rafts and membrane traffic. FEBS Lett. 581, 2098–2104 [DOI] [PubMed] [Google Scholar]
- 3. Jacobson K., Mouritsen O. G., Anderson R. G. (2007) Lipid rafts: at a crossroad between cell biology and physics. Nat. Cell Biol. 9, 7–14 [DOI] [PubMed] [Google Scholar]
- 4. Lang T. (2007) SNARE proteins and “membrane rafts.” J. Physiol. 585, 693–698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shaw A. S. (2006) Lipid rafts: now you see them, now you don't. Nat. Immunol. 7, 1139–1142 [DOI] [PubMed] [Google Scholar]
- 6. Hell S. W. (2007) Far-field optical nanoscopy. Science 316, 1153–1158 [DOI] [PubMed] [Google Scholar]
- 7. Gustafsson M. G., Shao L., Carlton P. M., Wang C. J., Golubovskaya I. N., Cande W. Z., Agard D. A., Sedat J. W. (2008) Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957–4970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J., Hess H. F. (2006) Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642–1645 [DOI] [PubMed] [Google Scholar]
- 9. Rust M. J., Bates M., Zhuang X. (2006) Subdiffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Heilemann M., van de Linde S., Schüttpelz M., Kasper R., Seefeldt B., Mukherjee A., Tinnefeld P., Sauer M. (2008) Subdiffraction-resolution fluorescence imaging with conventional fluorescent probes. Angew. Chem. Int. Ed. Engl. 47, 6172–6176 [DOI] [PubMed] [Google Scholar]
- 11. Jahn R., Scheller R. H. (2006) SNAREs – engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7, 631–643 [DOI] [PubMed] [Google Scholar]
- 12. Klein T., Loeschberger A., Proppert S., Wolter S., van de Linde S., Sauer M. (2011) Live-cell dSTORM with SNAP tag fusion proteins. Nat. Methods 8, 7–9 [DOI] [PubMed] [Google Scholar]
- 13. van de Linde S., Löschberger A., Klein T., Heidbreder M., Wolter S., Heilemann M., Sauer M. (2011) Direct stochastic optical reconstruction microscopy with standard fluorescent probes. Nat. Protoc. 6, 991–1009 [DOI] [PubMed] [Google Scholar]
- 14. Wombacher R., Heidbreder M., van de Linde S., Sheetz M. P., Heilemann M., Cornish V. W., Sauer M. (2010) Live-cell super-resolution imaging with trimethoprim conjugates. Nat. Methods 7, 717–719 [DOI] [PubMed] [Google Scholar]
- 15. Wolter S., Schüttpelz M., Tscherepanow M., S, van de Linde S., Heilemann M., Sauer M. (2010) Real-time computation of subdiffraction-resolution fluorescence images. J. Microsc. 237, 12–22 [DOI] [PubMed] [Google Scholar]
- 16. Sieber J. J., Willig K. I., Kutzner C., Gerding-Reimers C., Harke B., Donnert G., Rammner B., Eggeling C., Hell S. W., Grubmüller H., Lang T. (2007) Anatomy and dynamics of a supramolecular membrane protein cluster. Science 317, 1072–1076 [DOI] [PubMed] [Google Scholar]
- 17. Barg S., Knowles M. K., Chen X., Midorikawa M., Almers W. (2010) Syntaxin clusters assemble reversibly at sites of secretory granules in live cells. Proc. Natl. Acad. Sci. U.S.A. 107, 20804–20809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fan J., Yang X., Lu J., Chen L., Xu P. (2007) Role of Habc domain in membrane trafficking and targeting of syntaxin 1A. Biochem. Biophys. Res. Commun. 359, 245–250 [DOI] [PubMed] [Google Scholar]
- 19. López I., Ortiz J. A., Villanueva J., Torres V., Torregrosa-Hetland C. J., del Mar Francés M., Viniegra S., Gutiérrez L. M. (2009) Vesicle motion and fusion are altered in chromaffin cells with increased SNARE cluster dynamics. Traffic 10, 172–185 [DOI] [PubMed] [Google Scholar]
- 20. Bar-On D., Gutman M., Mezer A., Ashery U., Lang T., Nachliel E. (2009) Evaluation of the heterogeneous reactivity of the syntaxin molecules on the inner leaflet of the plasma membrane. J. Neurosci. 29, 12292–12301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Murray D. H., Tamm L. K. (2009) Clustering of syntaxin-1A in model membranes is modulated by phosphatidylinositol 4,5-bisphosphate and cholesterol. Biochemistry 48, 4617–4625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sieber J. J., Willig K. I., Heintzmann R., Hell S. W., Lang T. (2006) The SNARE motif is essential for the formation of syntaxin clusters in the plasma membrane. Biophys. J. 90, 2843–2851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang X., Xu P., Xiao Y., Xiong X., Xu T. (2006) Domain requirement for the membrane trafficking and targeting of syntaxin 1A. J. Biol. Chem. 281, 15457–15463 [DOI] [PubMed] [Google Scholar]
- 24. Bar-On D., Winter U., Nachliel E., Gutman M., Fasshauer D., Lang T., Ashery U. (2008) Imaging the assembly and disassembly kinetics of cis-SNARE complexes on native plasma membranes. FEBS Lett. 582, 3563–3568 [DOI] [PubMed] [Google Scholar]
- 25. Lang T., Margittai M., Hölzler H., Jahn R. (2002) SNAREs in native plasma membranes are active and readily form core complexes with endogenous and exogenous SNAREs. J. Cell Biol. 158, 751–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abramoff M. D., Magalhães P. J., Ram S. J. (2004) Image Processing with ImageJ. Biophotonics Int. 11, 36–42 [Google Scholar]
- 27. Xu X., Ester M., Kriegel H. P., Sander J. (1998) A distribution-based clustering algorithm for mining in large spatial databases in Proceedings, 14th International Conference on Data Engineering, Orlando, FL, February 23–27, 1998, pp. 324–331, IEEE Computer Society, Washington, D. C. 20036 [Google Scholar]
- 28. Locantore N. W., Tran L. T., O'Neill R. V., McKinnis P. W., Smith E. R., O'Connell M. (2004) An overview of data integration methods for regional assessment. Environ. Monit. Assess. 94, 249–261 [DOI] [PubMed] [Google Scholar]
- 29. Nudelman G., Weigert M., Louzoun Y. (2009) In silico cell surface modeling reveals mechanism for initial steps of B-cell receptor signal transduction. Mol. Immunol. 46, 3141–3150 [DOI] [PubMed] [Google Scholar]
- 30. Mezer A., Nachliel E., Gutman M., Ashery U. (2004) A new platform to study the molecular mechanisms of exocytosis. J. Neurosci. 24, 8838–8846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mezer A., Ashery U., Gutman M., Project E., Bosis E., Fibich G., Nachliel E. (2006) Systematic search for the rate constants that control the exocytotic process from chromaffin cells by a genetic algorithm. Biochim. Biophys. Acta 1763, 345–355 [DOI] [PubMed] [Google Scholar]
- 32. Sengupta P., Jovanovic-Talisman T., Skoko D., Renz M., Veatch S. L., Lippincott-Schwartz J. (2011) Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat. Methods 8, 969–975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lang T., Bruns D., Wenzel D., Riedel D., Holroyd P., Thiele C., Jahn R. (2001) SNAREs are concentrated in cholesterol-dependent clusters that define docking and fusion sites for exocytosis. EMBO J. 20, 2202–2213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rickman C., Medine C. N., Dun A. R., Moulton D. J., Mandula O., Halemani N. D., Rizzoli S. O., Chamberlain L. H., Duncan R. R. (2010) t-SNARE protein conformations patterned by the lipid microenvironment. J. Biol. Chem. 285, 13535–13541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. van den Bogaart G., Meyenberg K., Risselada H. J., Amin H., Willig K. I., Hubrich B. E., Dier M., Hell S. W., Grubmüller H., Diederichsen U., Jahn R. (2011) Membrane protein sequestering by ionic protein-lipid interactions. Nature 479, 552–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dun A. R., Rickman C., Duncan R. R. (2010) The t-SNARE complex: a close up. Cell. Mol. Neurobiol. 30, 1321–1326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Greenfield D., McEvoy A. L., Shroff H., Crooks G. E., Wingreen N. S., Betzig E., Liphardt J. (2009) Self-organization of the Escherichia coli chemotaxis network imaged with super-resolution light microscopy. PLoS Biol. 7, e1000137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kentner D., Thiem S., Hildenbeutel M., Sourjik V. (2006) Determinants of chemoreceptor cluster formation in Escherichia coli. Mol. Microbiol. 61, 407–417 [DOI] [PubMed] [Google Scholar]
- 39. Weiss N., Hameed S., Fernández-Fernández J. M., Fablet K., Karmazinova M., Poillot C., Proft J., Chen L., Bidaud I., Monteil A., Huc-Brandt S., Lacinova L., Lory P., Zamponi G. W., De Waard M. (2012) A Cav3.2/syntaxin-1A signaling complex controls T-type channel activity and low-threshold exocytosis. J. Biol. Chem. 287, 2810–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Torregrosa-Hetland C. J., Villanueva J., López-Font I., Garcia-Martinez V., Gil A., Gonzalez-Vélez V., Segura J., Viniegra S., Gutiérrez L. M. (2010) Association of SNAREs and calcium channels with the borders of cytoskeletal cages organizes the secretory machinery in chromaffin cells. Cell. Mol. Neurobiol. 30, 1315–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Davies J. N., Jarvis S. E., Zamponi G. W. (2011) Bipartite syntaxin 1A interactions mediate CaV2.2 calcium channel regulation. Biochem. Biophys. Res. Commun. 411, 562–568 [DOI] [PubMed] [Google Scholar]
- 42. Daily N. J., Boswell K. L., James D. J., Martin T. F. (2010) Novel interactions of CAPS (Ca2+-dependent activator protein for secretion) with the three neuronal SNARE proteins required for vesicle fusion. J. Biol. Chem. 285, 35320–35329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ashery U., Bielopolski N., Barak B., Yizhar O. (2009) Friends and foes in synaptic transmission: the role of tomosyn in vesicle priming. Trends Neurosci. 32, 275–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Südhof T. C., Rothman J. E. (2009) Membrane fusion: grappling with SNARE and SM proteins. Science 323, 474–477 [DOI] [PMC free article] [PubMed] [Google Scholar]