Background: K118(M/N) γ-actin mutations cause deafness, which indicates that they negatively affect actin structure and/or dynamics.
Results: Both mutations alter actin regulation by Arp2/3.
Conclusion: Lys-118 serves an important role in intra- and intermonomer interactions and in actin network regulation by binding proteins.
Significance: Lys-118 may play a more significant role in the internal allostery of the actin structure than previously believed.
Keywords: Actin, Cytoskeleton, Hearing, Mutagenesis Site-specific, Yeast, Arp2/3, DFNA20/26
Abstract
Hearing requires proper function of the auditory hair cell, which is critically dependent upon its actin-based cytoskeletal structure. Currently, ten point mutations in nonmuscle γ-actin have been identified as causing progressive autosomal dominant nonsyndromic hearing loss (DFNA20/26), highlighting these ten residues as functionally important to actin structure and/or regulation. Two of the mutations, K118M and K118N, are located near the putative binding site for the ubiquitously expressed Arp2/3 complex. We therefore hypothesized that these mutations may affect Arp2/3-dependent regulation of the actin cytoskeleton. Using in vitro bulk polymerization assays, we show that the Lys-118 mutations notably reduce actin + Arp2/3 polymerization rates compared with WT. Further in vitro analysis of the K118M mutant using TIRF microscopy indicates the actual number of branches formed per filament is reduced compared with WT and, surprisingly, branch location is altered such that the majority of K118M branches form near the pointed end of the filament. These results highlight a previously unknown role for the Lys-118 residue in the actin-Arp2/3 interaction and also further suggest that Lys-118 may play a more significant role in intra- and intermonomer interactions than was initially hypothesized.
Introduction
Ten point mutations in γ-actin currently have been identified as causing progressive autosomal dominant nonsyndromic hearing loss (DFNA20/26) in humans (1–3), indicating that these mutations in some way disrupt actin function. The ability of the auditory hair cell to transduce sound into nerve impulse is critically dependent upon its internal actin cytoskeleton, and thus disruption of the actin structure could lead easily to deafness. Pathogenic effects are likely caused by alteration of the actin structure itself or by changes in actin network regulation by actin binding proteins (ABPs),2 or potentially a combination of both. However, exactly how these mutations lead to deafness is currently unknown. We have thus chosen to investigate the effects of the DFNA20/26 mutations on actin function and regulation as a means to provide further understanding of actin structure and dynamics as a whole, both in deafness and in larger contexts throughout the body.
Two of the deafness-causing mutations, K118M and K118N, are located in an outward-facing and solvent-accessible residue which lies within subdomain 1 of the actin monomer (Fig. 1). This region of the protein is believed to be important for interactions between actin and its regulatory proteins, suggesting that the Lys-118 mutations may affect actin regulation as opposed to polymerization alone. Our initial results from previous studies supported this idea, indicating that the purified mutant actins polymerize similarly to WT actin in the absence of ABPs (3, 4). We thus hypothesized that characterization of the regulatory defect(s) caused by these mutations could provide detail into how the ABPs interact with actin.
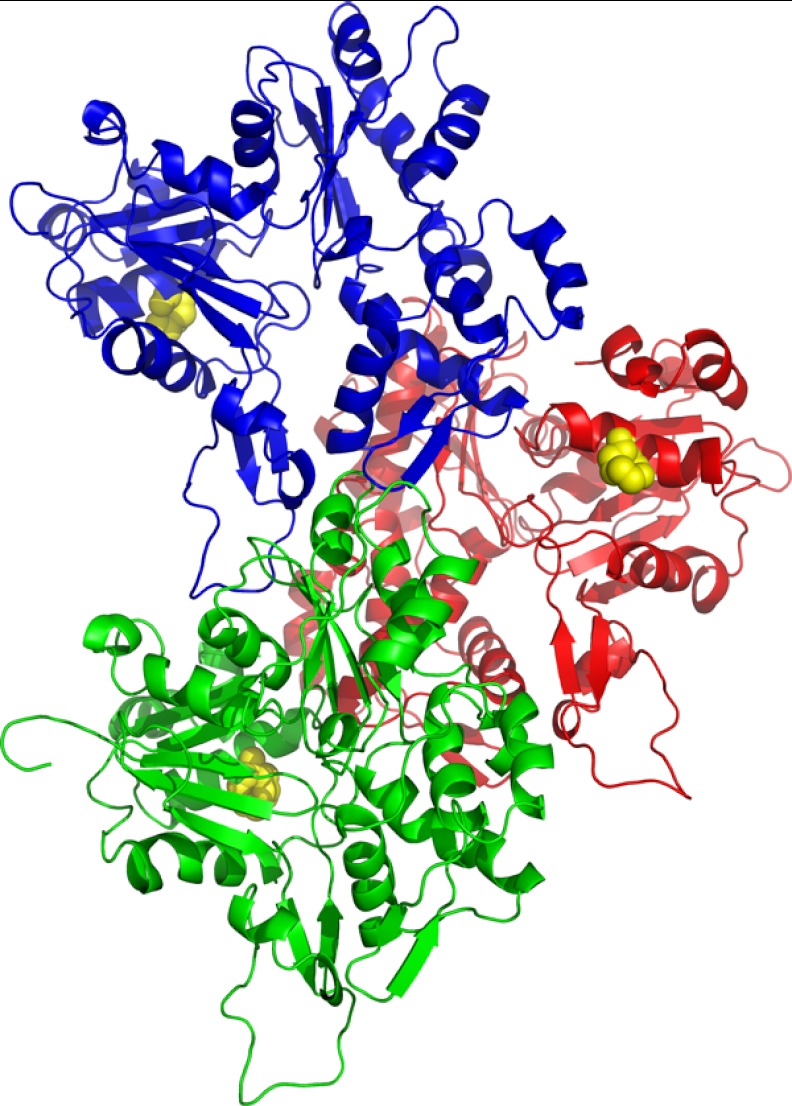
FIGURE 1.
Model of an actin trimer with monomers 1 (M1, green), 2 (M2, red), and 3 (M3, blue) based on the work of Oda and co-workers (26). Lys-118 is highlighted in yellow.
As a first step, it was necessary to narrow the pool of >100 known ABPs to those most likely to be affected by the Lys-118 mutations. The Arp2/3 branch model reported by Goley et al. (5) indicated that Lys-118 is located very close to the putative binding site for the Arp2/3 complex, and we therefore hypothesized that the residue may be involved in Arp2/3 binding to the actin filament. This interaction is depicted in Fig. 2, kindly contributed by David Sept at the University of Michigan, highlighting our residue of interest. Based upon these preliminary observations, we concluded that the Arp2/3 complex was a good first candidate for our attention. Arp2/3 is a ubiquitously expressed seven-protein complex that mediates the growth of a new “daughter” actin filament from the side of an existing “mother” filament to form a branch (6). Such Arp2/3-dependent branching plays a critical role in force generation for a wide variety of cell processes, including cell motility, endocytosis, and ciliogenesis, making it an additionally attractive subject for our study due to its broad importance within the cell.
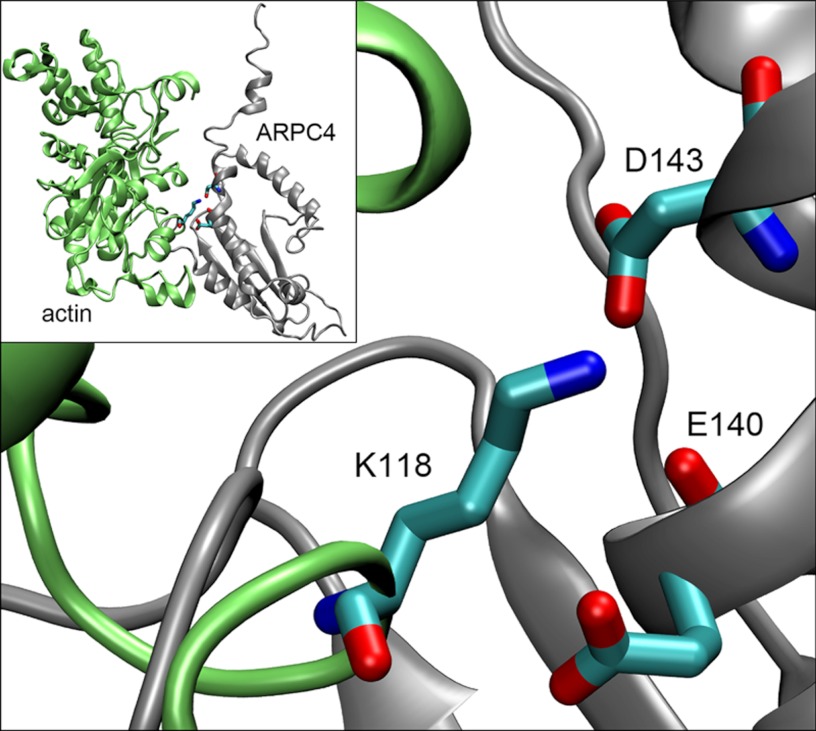
FIGURE 2.
Model of the Arp2/3 subunit ARPC4 (gray) and WT actin (green) demonstrating the location of Lys-118 in relation to the docking Arp2/3 complex. Lys-118 from actin and two neighboring residues from ARPC4, Asp-143 and Glu-140, have been modeled as sticks for detail. This figure (kindly provided by David Sept) is based upon the Arp2/3 branch model produced by Goley and co-workers (5), modified to highlight our residue of interest.
Hair cells can neither be removed from affected deafness patients nor cultured as a stable, differentiated cell line, and small animal models capable of producing biochemically significant amounts of pure mutant actin are not available. Therefore, we have introduced the K118(M/N) point mutations into actin from the budding yeast Saccharomyces cerevisiae as a model for our investigations. Although yeast actin is not identical to human γ-actin and thus may not behave identically, the two actins are 91% homologous, and all mutation sites are conserved (7). In addition, many of the same actin binding proteins that regulate actin networks in humans are also conserved in budding yeast (8), allowing for more extensive comparison of in vivo systems. One of the most significant advantages to this system, however, is that because yeast produce only one actin isoform, it is possible to obtain pure samples of mutant actin for in vitro biochemical analysis by replacing the WT actin with mutant actin. Such purification is impossible with mammalian non-muscle cells: they contain both γ- and β-actin, and the isoforms differ by only four biochemically similar amino acids at their N terminus, making it virtually impossible to separate them in active form.
Previously in our laboratory, we created two yeast strains expressing either K118M or K118N mutant actin as the sole actin in the cell. We then assessed the effects of these mutations on actin-dependent cell processes, including organelle function and growth on a variety of media (3, 4). In this paper, we have continued our investigation by observing actin cable dynamics in vivo. We then isolated the mutant actins and used both bulk solution assays and TIRF microscopy to assess the effects of the mutations on actin polymerization and Arp2/3-dependent branching.
EXPERIMENTAL PROCEDURES
Cytology
We visualized actin cable dynamics using 3XGFP-ABP140 in a manner similar to that published previously by Yang and Pon (9) with modifications. We used early log phase cells co-expressing 3XGFP-tagged ABP140 grown in YAPD medium (YPD-rich medium plus 60 mg/liter adenine hemisulfate). Movement of GFP-ABP140-marked cables was monitored using an Olympus IX81 inverted microscope with a PlanApo 60 × 1.45 oil immersion TIRF lens and Hamamatsu ORCA-R camera. Serial TIRF images of each cell were recorded for 30 s at a rate of 0.15 s/frame by Slidebook5 (3i Inc., Denver, Co.). We observed actin cables in a similar number of cells for WT, K118M, and K118N yeast. Images subsequently were enhanced for clarity using ImageJ (10); however, we were unable to track movement of mutant actins, as the ends of cables in both mutant cell types were not visible and the cables themselves appeared mostly non-motile.
Actin Purification
Yeast cakes for WT actin preparations were purchased from a local bakery. Mutant yeast strains were created as described previously (3, 4). WT and mutant actins were purified as published previously (3, 4, 11) with the exception that a 1 m NaCl wash solution was used instead of 0.6 m NaCl for the DNase I column wash. Briefly, actin was purified from clarified cell lysate via DNase I affinity chromatography (DNaseI, grade D, from Worthington was conjugated to Affi-Gel 10 from Bio-Rad), DE52 anion exchange chromatography (Whatman), and polymerization/depolymerization cycling as described previously (11). The concentration of the globular actin (G-actin) was determined by measuring A290 nm using ϵ290 nm = 25,600 m−1 cm−1, and the actin was stored in G buffer (10 mm Tris-HCl, pH 7.5, 0.2 mm CaCl2, 0.1 mm ATP, and 0.1 mm DTT) at 4 °C for no more than 4 days.
Yeast Arp2/3 and N-WASP Purification
Protein A-tagged yeast Arp2/3 was expressed and prepared from the RLY1945 yeast strain (from Dr. R. Li, Stowers Institute) as described previously (12). Briefly, cells were grown to early log phase, harvested, and lysed using a glass bead beater (Biospec). Protein A-tagged yeast Arp2/3 was affinity-purified by IgG-Sepharose affinity chromatography (Amersham Biosciences) and digested with recombinant tobacco etch virus to remove the protein A tag. Yeast Arp2/3 was further purified on a Source fast flow anionic exchange column (Amersham Biosciences). The preparations were analyzed by SDS-PAGE and visualized by Coomassie Blue staining to assess purity. Purified yeast Arp2/3 fractions were then divided into single-use aliquots, flash-frozen in liquid nitrogen, and stored at −80 °C in 10% glycerol and 0.2 mm ATP.
GST-N-WASP was overexpressed in Escherichia coli from plasmids provided by Dr. H. Higgs at Dartmouth University. Recombinant protein was purified by glutathione affinity chromatography (Amersham Biosciences) as described (13) and further purified by Source fast flow anion exchange chromatography (Amersham Biosciences). The preparation quality was checked as described above for yeast Arp2/3.
Polymerization Assays
Samples of either 5 μm or 2.5 μm unlabeled G-actin were polymerized alone or with the addition of 50 nm yeast Arp2/3. Polymerization was initiated by the addition of MgCl2 and KCl to final concentrations of 2 and 50 mm, respectively. For N-WASP studies, 1 μm actin samples were polymerized with 25 nm yeast Arp2/3 (yArp2/3) and 200 nm N-WASP with polymerization initiated as above. Actin concentration was reduced to 1 μm for assays with N-WASP to facilitate observation of differences in actin polymerization; the previous conditions polymerized too rapidly to provide meaningful data. As before, the subsequent change in light scattering was recorded as a function of time. All polymerization assays were performed in a final volume of 120 μl in a microcuvette housed in a thermostat-controlled sample compartment of a FluoroMax-3 spectrometer (Jobin Yvon-Spex) using 360 nm for both excitation and emission wavelengths. All experiments were repeated at least three times using a minimum of three different actin preparations with similar results. Elongation rates were determined from the slope of a line fit to at least four data points comprising the linear elongation phase of the polymerization curves.
TIRF Microscopy
Purified actin for TIRF microscopy was labeled at Cys-374 during the cycling stage of purification. Aliquots of purified actin were polymerized for 2 h at 25 °C in the presence of either Oregon Green® 488 maleimide (Invitrogen) or Nα-(3-maleimidylpropionyl) biocytin (Molecular Probes®) at four times the concentration of the actin. Labeling efficiency of Oregon Green was measured as a ratio of the absorbance of Oregon Green (A491) to actin (A290); biotinylation efficiency was assumed to be 100%. A 10 μm TIRF actin mix was prepared as a solution of 33% Oregon Green-labeled G-actin, 5% biotinylated G-actin, and 62% unlabeled G-actin. TIRF reactions without yArp2/3 were prepared by combining 1 μm TIRF actin with 2× TIRF buffer (1×: 10 mm imidazole, pH 7.4, 50 mm DTT, 1 mm MgCl2, 0.1 mm ATP, 20 μg/ml glucose oxidase, 0.5% methyl cellulose) (14). Reactions containing yArp2/3 were prepared similarly; however, 25 nm yArp2/3 was added to the mixture, and 0.75 μm actin was used instead of 1 μm actin to allow for clearer visualization of branching. Flow cells were prepared as described previously (15) and incubated for 10 min with 100 nm avidin prior to loading. Actin filament polymerization was visualized using an Olympus IX81 inverted microscope and Hamamatsu camera as described above. Images were captured every 10 s for ∼10 min to record the progression of filament growth. The collection of images was then adjusted for contrast and clarity using ImageJ software, and branches and filaments were counted manually every fifth frame of each time course. The number of branches was then divided by the number of filaments to obtain a branch to filament ratio, which was then plotted over time.
To support manual branch counts, overall percent area of the slide covered by filaments was calculated using ImageJ software, as the area of the slide covered by filaments appeared to correlate with the number of branches formed. To calculate percent area, we first applied a threshold to the images using the MultiThresholder plugin of ImageJ to convert all images to binary black/white. We then calculated percent area of white pixels for each frame of the image sequence and plotted these percentages over time to obtain the graphs presented herein.
To quantitate whether branching was occurring on the barbed versus pointed end of the filament, we divided 50 filaments at least 2 μm in length in half and tallied the number of branches that occurred either on the barbed half or the pointed half of the filament. This number was then divided by 50 to determine the percentage of filaments branching from the barbed end.
RESULTS
Our earlier work demonstrated that, in yeast, the K118M mutation caused a much more strikingly abnormal phenotype in vivo than the K118N mutation: K118M cells displayed abnormal mitochondria and suffered from a severe growth defect even on YPD medium, as well as on glycerol (4) and NaCl (data not shown). In sharp contrast, however, K118N cells appeared similar to WT cells both in terms of mitochondrial morphology and growth phenotype on glycerol, although K118N yeast did display a significant growth defect on hyperosmolar medium (3). As a first step to establish a basis for this difference, we examined actin cable dynamics in vivo, which often display aberrant phenotypes when actin regulation has been altered. To visualize cables, we expressed an ABP140-GFP fusion protein in wild type, K118M, and K118N cells. ABP140 is a non-essential ABP that localizes specifically to actin cables, and the binding of GFP-tagged ABP-140 to actin allows for live visualization of cable movements using fluorescence microscopy (9). Various characteristics of the cables such as overall cable organization and movement can then be assessed.
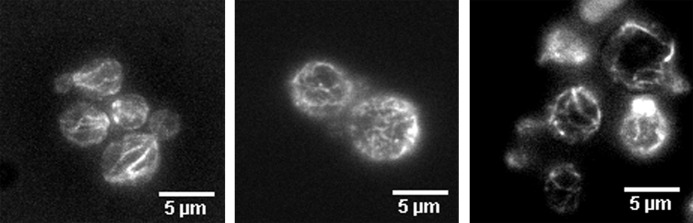
As can be observed in Fig. 3, WT yeast display polarized cables focused primarily along the mother-to-bud cell axis with an additional few cables attached to the sides of the cells in a more random orientation. In contrast, cables were difficult to detect in K118M cells, with approximately only 10% of cells displaying clearly defined cables of any sort. When cables were visible, they created a random, fragmented meshwork with no dominant polarity from mother to bud. The K118M cables also appeared more static than those of WT yeast cells, with less movement than observed in WT cables. These results suggest a defect in actin regulation in K118M mutant cells.
FIGURE 3.
ABP140-GFP labeling of actin cables in live yeast cells. WT cables appear predominantly polarized from mother to bud. K118M cables are fragmented and lack any dominant orientation, resulting in a meshwork of cables. K118N actin cables appear to maintain some mother-to-bud polarity; however, they are still notably more disorganized and random than WT cables.
The K118N cells appeared significantly more like WT cells than their K118M counterparts. Cables were clearly visible in almost all cells, and polarity was somewhat focused along the mother-to-bud cell axis. However, the cables appeared less dynamic than WT, and due to a lack of distinct markers to track, such as cable ends, we were unable to measure rates of cable movement. Virtually all cells also displayed a lack of organization, with many random cables initiating from the sides of the cells and not running along the mother-to-bud axis. These results indicate there is an effect on actin regulation in K118N cells, but it is substantially less severe than that observed in the K118M cells. The severity of the cabling phenotypes is also consistent with the observed growth phenotypes in that K118M cells display a significantly more severe phenotype than K118N cells.
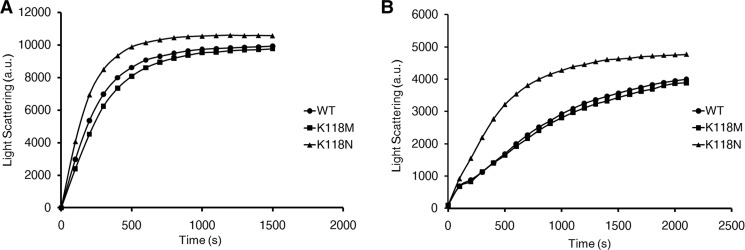
The works of Cooper and co-workers (16) and Borisy and co-workers (17, 18) suggest that Arp2/3 may be involved in actin cabling dynamics, and because Lys-118 is located near the putative Arp2/3 binding site (5), we next chose to examine the effect of the mutations on Arp2/3-dependent branching. We began our in vitro analyses by polymerizing purified WT, K118M, and K118N actins in the absence of ABPs using light scattering as a measurement. Elongation rates were then determined by the slope of the linear elongation phase of the resulting polymerization curve. As we reported previously, the K118M mutant actin polymerized identically to that of WT (4), with a rate of 2.7 a.u. per sec. for both WT and K118M, based on polymerization of 2.5 μm actin (Fig. 4). The K118N actin polymerized over twice as fast as WT actin, with a rate of 6.2 a.u./s at 2.5 μm concentration. Nevertheless, the final extent of polymerization was approximately the same for all three actins. The lack of a severe polymerization defect caused by the mutations was consistent with our hypothesis that the deafness-causing defects are due to problems in regulation and not polymerization of the actin alone.
FIGURE 4.
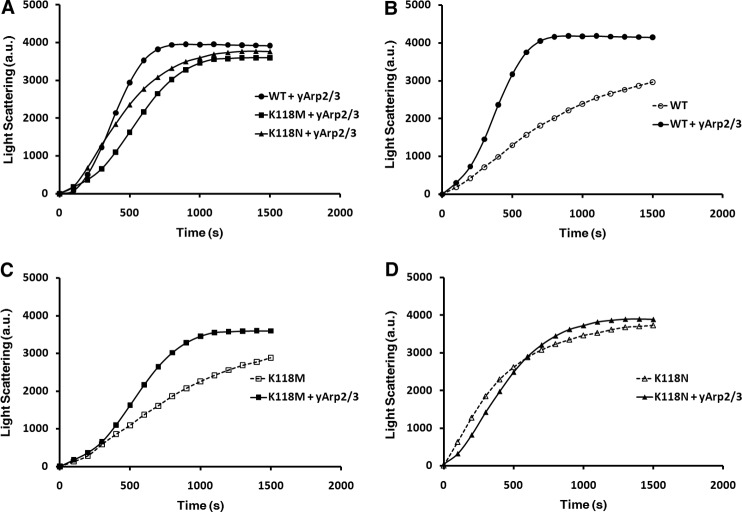
A. Polymerization of 5 μm actin (A) and 2. 5 μm actin (B) measured by light scattering. WT and K118M actins polymerize nearly identically. In contrast, K118N actin polymerizes approximately twice as fast as either WT or K118M actin and does not appear to experience the same lag during the nucleation phase.
To investigate how the Lys-118 mutations may affect regulation by Arp2/3, we polymerized 2.5 μm samples of the purified actins in the presence of 25 nm purified yArp2/3 (Fig. 5A). One significant advantage of using yArp2/3 as opposed to a mammalian variety is that the yeast complex does not require activation by another protein such as N-WASP (12), allowing us to directly assess the effects of the mutations on the yArp2/3-actin interaction. Addition of yArp2/3 to WT yeast actin results in a shortened nucleation phase followed by a rapid increase in light scattering (Fig. 5B). The rate of the elongation phase of WT + yArp2/3 was 8.2 a.u./s, 3-fold faster than in the absence of Arp2/3. In contrast, K118M actin with yArp2/3 is marked by an extended nucleation phase and a slightly slower elongation phase compared with WT + yArp2/3 (Fig. 5C). The rate of the elongation phase was 5.2 a.u./s, slightly under a 2-fold increase in elongation rate compared with polymerization of the actin alone. Because polymerization of the two actins in the absence of yArp2/3 was virtually the same, the slower elongation rate for K118M + yArp2/3 compared with WT + yArp2/3 clearly demonstrates an Arp2/3-dependent polymerization defect. These results are summarized in Table 1.
FIGURE 5.
Polymerization curves of 2. 5 μm actin samples polymerized with 25 nm yArp2/3. A, comparison of WT, K118M, and K118N actins with yArp2/3 followed by a comparison of polymerization of WT (B), K118M (C), and K118N (D) actins with and without the addition of yArp2/3.
TABLE 1.
Summary of in vitro kinetic data for purified WT and mutant actins
All actin concentrations were 2.5 μm except for assays with N-WASP, which were performed with 1 μm actin.
| Strain | Nucleation time (approx.) |
Polymerization rate |
TIRF microscopy |
|||
|---|---|---|---|---|---|---|
| −Arp2/3 | +Arp2/3 | −Arp2/3 | +Arp2/3 | +NWASP | Branch initiation | |
| s | a.u./s | |||||
| WT | 250 | 300 | 2.7 | 8.2 | 5.0 | 3.5 min |
| K118M | 250 | 350 | 2.7 | 5.2 | 3.8 | 5 min |
| K118N | NAa | 150 | 6.2 | 5.6 | 4.0 | NAb |
a No nucleation lag was apparent from the light scattering data.
b Kinetic analysis of K118N actin with TIRF was not possible due to labeling effects on the actin.
We then repeated the same experiment with K118N actin. With the addition of yArp2/3, the mutant actin continued to nucleate faster than WT actin, just as in the absence of yArp2/3 (Fig. 5A). However, the elongation rate for K118N + yArp2/3 was dramatically slower than WT + yArp2/3, with a rate of 5.6 a.u./s, compared with 8.2 a.u./s for WT. Even more notable, this elongation rate is 10% slower than for K118N actin alone (Fig. 5D). Though polymerization of K118N actin with and without Arp2/3 appeared very similar, the minor differences observed were highly repeatable. This surprising result indicates that Arp2/3 may actually inhibit K118N actin polymerization; at the very least, it fails to stimulate elongation. It can thus be argued that the K118N mutation causes a more severe Arp2/3-dependent polymerization defect than K118M. However, notably, the K118N actin still has an elongation rate slightly higher than K118M actin with yArp2/3: K118M actin displays an elongation rate 37% slower than WT + yArp2/3, and K118N actin displays an elongation rate 33% slower than WT + yArp2/3.
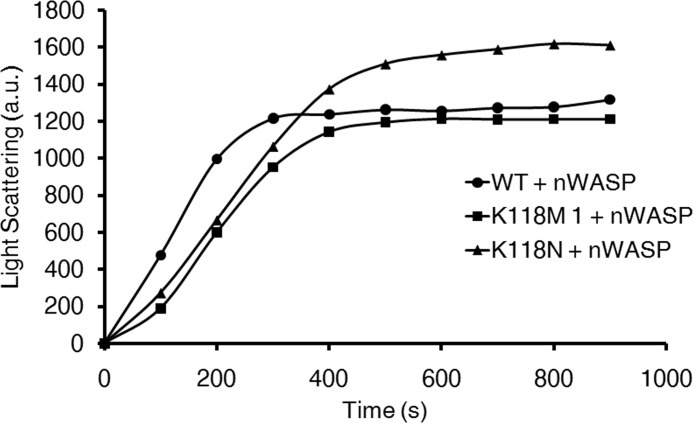
Because Arp2/3 never functions in vivo without an activator protein to facilitate its function, we next asked whether the defects we observed in polymerization rate could be rescued by the addition of the Arp2/3 activator N-WASP. We subsequently polymerized 1 μm samples of WT, K118M, and K118N actins in vitro with 25 nm yArp2/3 + 200 nm N-WASP (Fig. 6). Consistent with our assays containing only actin + yArp2/3, the K118M and K118N actins displayed a slower rate of elongation (3.8 and 4.0 a.u., respectively) compared with WT (5.0 a.u.). Notably, the final extent of light scattering for K118N actin was higher than for the other two actins. This was a consistently repeatable result over multiple N-WASP concentrations, and further experiments will be required to determine whether this is caused by bundling, altered critical concentration, increased final extent of branching, or a combination of these possibilities. Nevertheless, our results clearly demonstrate that the Arp2/3-dependent defect observed with the mutant actins is not rescued by the addition of N-WASP, and in doing so, they support our hypothesis that the defects we observe in vitro are relevant in vivo.
FIGURE 6.
Polymerization of 1 μm WT, K118M, and K118N actin with the addition of 25 nm yArp2/3 and 200 nm N-WASP.
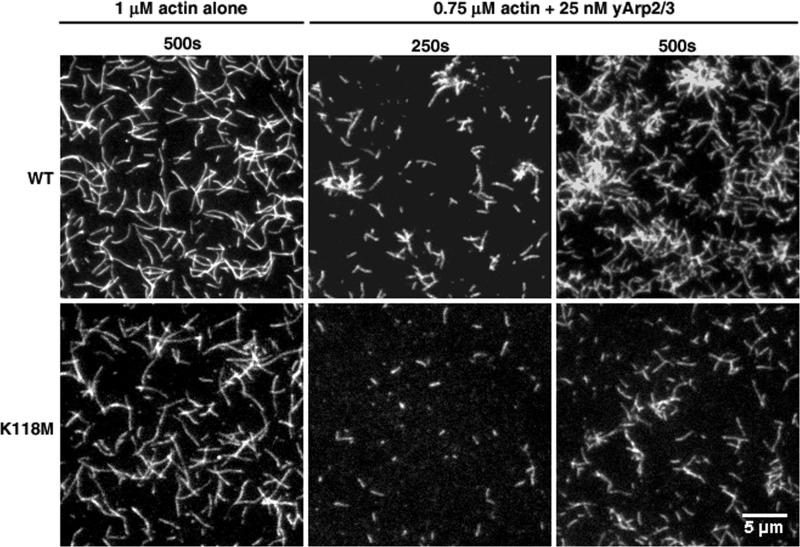
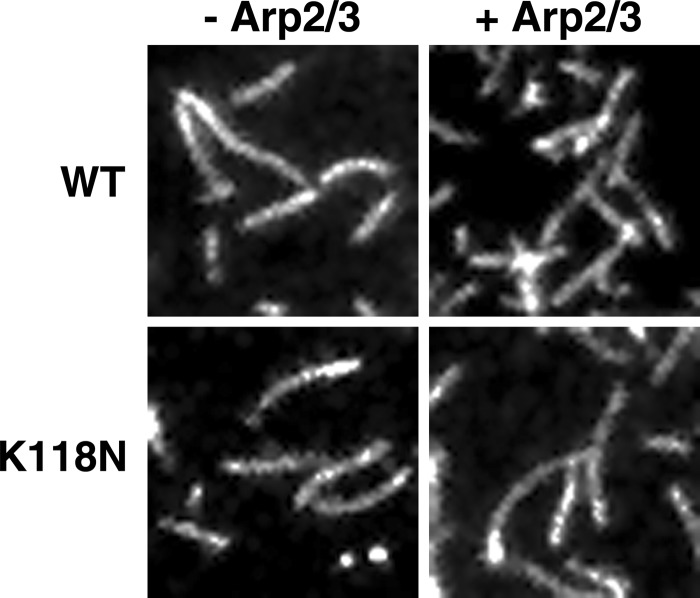
Although our light scattering results indicate that the Lys-118 mutations cause Arp2/3-dependent polymerization defects, these assays report the bulk behavior of the entire actin filament population and do not reveal whether the defect is due to reduced branching, increased fragmentation, etc. We thus re-examined the effects of the K118M and K118N mutations on Arp2/3 regulation using TIRF microscopy, which allows for visualization of single-filament behavior. When assessed in this manner, there was little difference between the polymerization of WT and K118M actins in the absence of yARP2/3, comparable with what we observed in bulk assays (Fig. 7). However, when 0.75 μm actin was polymerized in the presence of 25 nm yArp2/3, branching of the mutant actin occurred much more slowly than it did for WT actin, particularly over the first 5 min. After 250 s, branching was apparent in WT actin; however, K118M actin showed almost no branching whatsoever. After 500 s, WT actin had formed clusters of densely branched filaments, and very few filaments appeared to have no branches. In contrast, K118M actin showed almost no clustering, there were still far fewer branches than WT, and many filaments appeared to have no branches whatsoever.
FIGURE 7.
TIRF microscopy images of WT or K118M actins polymerized in the absence or presence of 25 nm yArp2/3. WT and K118M filaments appear similar without yArp2/3, but K118M actin displays a noticeable reduction in branching after the addition of yArp2/3.
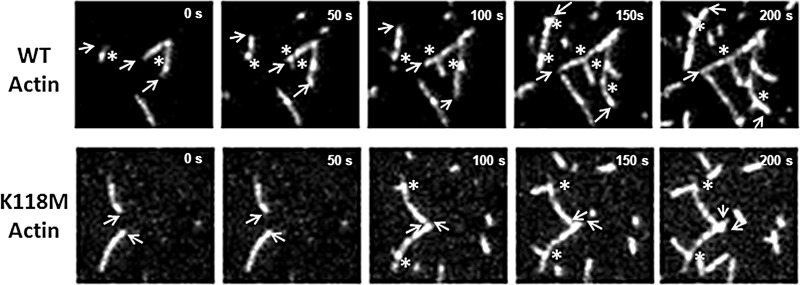
One of the most striking characteristics we observed from these TIRF images, however, was an apparent change in the predominant location of branch formation from the mother filament (Fig. 8). With WT actin, 82% of branches formed from the barbed-end half of the mother filament. In contrast, with K118M actin, only 30% of the branches occurred in the barbed-end half of the filament: 70% of branches formed from the pointed-end half of the filament.
FIGURE 8.
TIRF time course showing filament growth to allow for visualization of the barbed versus the pointed end of the filament. Growing barbed ends of filaments are marked with arrows, and newly forming branches are marked with an asterisk. As can be observed from this example, WT actin predominantly formed branches from the barbed-end half of growing filaments, whereas K118M actin predominantly formed branches toward the pointed-end half of the filament.
Somewhat surprisingly, we could not derive meaningful kinetic data from the K118N mutant using TIRF microscopy, as the biotin and Oregon Green labeling required for visualization, for an unknown reason, caused a distinct decrease in polymerization rate, both in light scattering and under the microscope. We were still able to observe branches with this mutant actin in the presence of yArp2/3 (Fig. 9), although far fewer branches formed than for WT. Because the light scattering polymerization curves of purified K118N actin with and without yArp2/3 were so similar, these TIRF results allow us to conclude that yArp2/3 is indeed able to bind to and form branches from K118N actin, supporting our conclusion that the difference in polymerization between K118N actin with and without yArp2/3 is indeed real.
FIGURE 9.
TIRF microscopy images of 1 μm WT and K118N actins with and without the addition of 25 nm yArp2/3. In the absence of yArp2/3, branches were never observed on either WT or K118N actins. With the addition of yArp2/3, branches did form from K118N actin, though much less often than compared with WT.
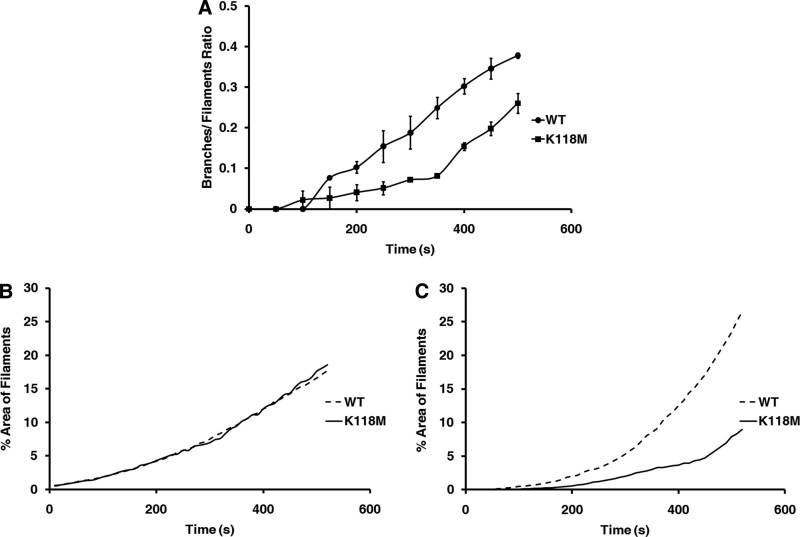
To quantitate branching in K118M and WT actins, we manually counted filaments and branches on every fifth image of the acquired image sequences. We then divided the number of branches by the number of filaments on the slide to obtain a branch per filament ratio, which we plotted over time to obtain the curve presented in Fig. 10A. WT actin + yArp2/3 began forming branches ∼3.5 min post initiation of polymerization. Consistent with previous reports (19–21), branching in WT actin showed a noticeable bias for developing at the barbed end of the filament, with newly forming branches developing from the barbed half of the filament in 82% of filaments. Branches also quickly began forming on established branches, resulting in a dense meshwork of actin. In contrast, K118M actin began forming branches notably later, ∼5 min post initiation, and there were fewer total branches formed per filament. Interestingly, K118M actin also tended to develop branches toward the pointed end of the filament: 70% of branches formed on the pointed end of the filament, compared with only 18% for WT. In addition, few new branches ever formed on already established branches for K118M actin, which, combined with the reduced total number of branches, resulted in an apparent lower overall filament density.
FIGURE 10.
A, ratio of the number of branches divided by the number of filaments based on manual branch counts from TIRF images. B, % area of the TIRF slide covered by filaments in the absence of yArp2/3. WT and K118M filament density appears the same. C, % area of the TIRF slide covered by filaments in the presence of 25 nm yArp2/3. Filament density measurement approximates the difference in branching observed in A.
Although manual branch counting was performed according to a strict protocol to minimize bias, it is both subject to human error and extremely time consuming. For this reason, we sought to develop a simple, computer-based means of estimating branching. As noted above, WT and K118M actins appeared to differ in overall density of actin filaments on the slide, and this difference appeared to correlate with the total number of branches that formed. We therefore hypothesized that if we developed a means to quantitate the difference in density, this would in turn provide an estimate of the amount of branching that is occurring. As a simple means to quantitate this difference, we determined the percent area of the slide covered by filaments and plotted this percentage over time to produce the graphs presented in Fig. 8, B and C. These results were then compared with the results obtained via hand counting. The percent areas of filaments for slides without yArp2/3 were nearly identical for both WT and K118M actins. However, with the addition of yArp2/3, percent area covered by K118M filaments was notably lower than for the WT filaments, closely mirroring the results obtained via manual counting, particularly during the early phase of polymerization. As the number of filaments on the slide increase and those filaments continue to elongate, this method loses resolution, limiting its effectiveness to approximately the first 5 min of polymerization. Nevertheless, these results support our manual counting data and affirm that the observed differences in density are due to differences in Arp2/3-dependent branching. In addition, they also validate this approach as a more time-efficient method of assessing Arp2/3-dependent action in vitro.
DISCUSSION
Of the deafness-causing actin mutations described to date, the two mutations occurring at Lys-118 present the only case in which different mutations arise at the same site, Met in one family and Asn in the other. These two substitutions, located on the exterior of the protein with their side chains facing outward, produce very different effects in vivo in yeast. The K118M cells grow poorly even on normal YPD medium and display numerous other forms of dysfunction. The K118N cells, however, behave much like WT cells. Nevertheless, both mutations lead to a significant phenotype in humans (deafness). We thus continued to characterize the effects of the K118(M/N) mutations in vivo and in vitro in an effort to better understand how they alter actin regulation and dynamics.
To extend our previously published in vivo characterizations, we began by observing the behavior of the actin in living cells expressing GFP-ABP140 as a marker of actin cables. K118M cables were rarely visible, and those that were visible were extremely fragmented and disorganized. In contrast, K118N cells displayed cables that appeared similar, although not identical, to WT cables, suggesting only a minor cabling defect. The severity of these results are consistent with the severity of the other in vivo phenotypes observed. Most importantly, however, the presence of cabling defects indicate that there is indeed a defect in actin regulation. Because Arp2/3 may be involved with actin cable initiation and Lys-118 may be near a contact site on the actin filament for the Arp2/3 complex, we proceeded to test whether Arp2/3-dependent branching might be affected by the Lys-118 mutations.
As we published previously, when K118M actin was polymerized alone, it displayed essentially the same kinetics as WT actin (4). The K118N actin, however, polymerized noticeably faster than WT, whereas previously, we reported at best a slight increase in polymerization rate (3). Subsequent studies (data not shown) revealed that this difference was most likely due to a low molecular weight contaminant in our original Asn preparations that could be removed by a more stringent 1 m salt wash while the actin was bound to the DNase I agarose column during purification.
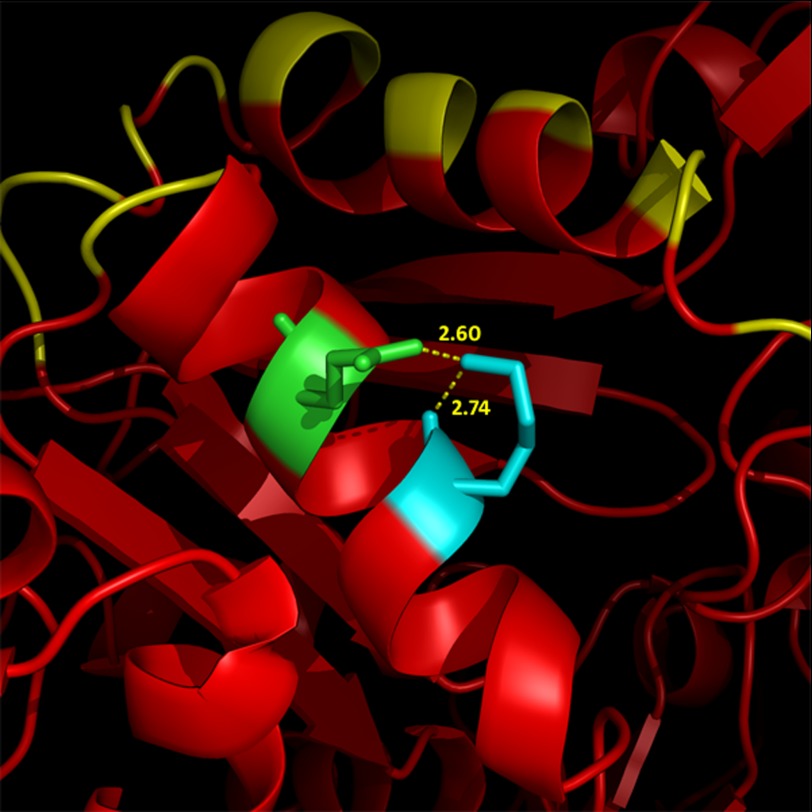
The increased rate of nucleation and polymerization observed in K118N actin likely indicates that Lys-118 plays some role either within the actin monomer or with monomer-monomer interactions. There are at least five other known disease-causing mutations in other actin isoforms in the same α-helix in which Lys-118 is located (22–25), which further emphasizes the structural importance of this region on actin function. The helix spans a region from the outside of the monomer (Fig. 1), which includes the binding site for Arp2/3, to the interface between subdomain 1 of the M2 monomer and subdomain 4 of the M3 monomer. Based on the structural model of Oda and Maéda (26), Lys-118 appears to be oriented in the helix such that it is within the standard Visual Molecular Dynamics 3.2 Å cut-off to form a salt bridge with Gln-121 (Fig. 11) (27). It is therefore possible that an interaction between Lys-118 and Gln-121 stabilizes the helix, which could have larger effects that propagate throughout the entire actin monomer or even disrupt interactions between monomers. It is also possible that Lys-118 does not normally interact, or only interacts weakly, with Gln-121, but that mutation from Lys to Asn causes a new, or stronger, interaction to form. There is some evidence that a Q-N interaction is particularly stabilizing in α-helices (28), and such a scenario could account for why the K118M actin polymerizes like WT; it would be unable to form such an interaction, whereas the K118N actin nucleates and polymerizes substantially faster than WT. Although beyond the scope of this study, elucidation of the role of this region of the actin monomer on actin function provides a promising direction for future structural investigations.
FIGURE 11.
A model of the α-helix in which Lys-118 resides. Lys-118 (cyan) and Gln-121 (green) are shown in the context of the Arp2/3 binding site as published by Goley and co-workers (yellow) (5).
Assessment of the effect of the Arp2/3 complex on bulk solution polymerization behavior yielded interesting and somewhat surprising results. Arp2/3 enhanced the rate of polymerization of the K118M actin but to a lesser extent than WT actin. Surprisingly, however, addition of Arp2/3 to the K118N actin actually slowed the rate of polymerization of the actin compared with WT, possibly due to capping of the filament barbed end by the Arp2/3 complex. This inhibition was particularly surprising considering the asparagine should have been much closer in terms of polarity to the original lysine than the methionine. Because K118N actin polymerizes notably faster than WT actin in the absence of Arp2/3, the fact that it polymerizes slower than WT with Arp2/3 indicates an overall greater decrease in polymerization rate than is observed with K118M actin. Nevertheless, the K118N actin with yArp2/3 displayed an intermediate phenotype between the K118M and WT actins, which would be consistent with what we would expect based on polarity of the mutated residues alone. It is thus possible that whatever causes the increased rate of polymerization of K118N actin without Arp2/3 is somewhat negated with the addition of Arp2/3.
Visualization of K118M actin at the single-filament level using TIRF microscopy produced additional surprises. Consistent with our bulk solution studies, the K118M mutation caused a significant lag in the onset and frequency of Arp2/3-dependent branching over the first 5 min of the reaction. At later times, this difference seemed to disappear, but the high density of the actin network at later time points made it extremely difficult to distinguish branches and the filaments from which they originated. The observed decrease in branching is likely due to either decreased binding of the Arp2/3 complex to the mother filament or decreased activation of the Arp2/3 complex once bound, or a combination of both. Pfaendtner and co-workers (29) very recently identified Lys-118 as one of several residues that appears to form a salt bridge between actin and the Arp2/3 complex, and thus mutation of Lys-118 to Met or Asn would likely weaken this interaction. However, it remains unclear whether the affected interaction is between the mother actin filament and the Arp2/3 complex or between the Arp2/3 complex and the incoming monomers that form the daughter branch.
One of the most noteworthy results of this study related to the position of branching as observed via TIRF microscopy. In our work, with WT actin, >80% of the branches occurred in the rapidly polymerizing barbed-end half of the mother filament, consistent with previous reports (19–21). In sharp contrast, with K118M actin, only 30% of the branches occurred in the barbed-end half of the filament: 70% of branches formed from the pointed-end half of the filament. There are two clear possibilities for this result: 1) it is possible that the Arp2/3 complex binds the mutant actin in the same manner as the WT, but that activation of the complex and subsequent propagation of the branch is somehow delayed. As the filament continues to elongate, by the time the branch has formed enough to be visualized, it is already localized toward the pointed end of the growing filament. 2) It is also possible that the mutation somehow alters the topology of the filament to affect the position of Arp2/3 attachment.
It is well known that following the initial polymerization of actin, conformational maturation of the filament occurs. Aebi and co-workers (30–32) first presented evidence for this based on covalent cross-linking and on an initial fuzzy filament appearance under EM that gradually converted to a more normal smooth surface. Egelman and co-workers (33, 34), using optical reconstruction of cryo-EM images, showed that the filament could exist in a number of conformations based on the twist and openness of the nucleotide cleft of monomers in the filament. They proposed that following polymerization, there were alterations both in the twist within the monomer and the size of the nucleotide cleft that led to the final filament conformational state. It is therefore possible that the chemical nature of the K118M mutant residue alters local filament surface conformation such that the preferred Arp2/3 binding site is now in the more mature area of the filament. If such a conformational change is altering the preferred binding location for Arp2/3 on the actin filament, these results will provide new and valuable insight into how filament conformation influences Arp2/3 binding. However, at the present time, resolution of the question is beyond our ability.
Our in vivo cabling data indicate that actin regulation is clearly affected by the K118M and K118N mutations. In yeast, multiple pathways exist for initiating cable formation, some of which include the involvement of Arp2/3, though the effect of the actin mutations on a non-Arp-dependent pathway may be more functionally significant. Nevertheless, our study does demonstrate experimental evidence of the predictions made from modeling studies concerning the actin-Arp2/3 interaction. It indicates both with TIRF and bulk solution studies that alterations at Lys-118 differentially affect Arp2/3-dependent nucleation of actin filament formation, and it demonstrates for the first time that an actin mutation can affect the site of branch formation on a growing filament. These findings will provide a valuable starting point for determining how filament conformation affects Arp2/3-dependent actin branching.
Acknowledgment
We thank David Sept at the University of Michigan for contribution of the model displaying Lys-118 in relation to the Arp2/3 complex.
This work was supported, in whole or in part, by National Institutes of Health Grant DC008803 from the NIDCD (to P. A. R.).
- ABP
- actin binding protein
- yArp
- yeast Arp
- a.u.
- arbitrary units
- TIRF
- total internal reflection fluorescence
- N-WASP
- neuronal Wiskott-Aldrich syndrome protein.
REFERENCES
- 1. Zhu M., Yang T., Wei S., DeWan A. T., Morell R. J., Elfenbein J. L., Fisher R. A., Leal S. M., Smith R. J., Friderici K. H. (2003) Mutations in the γ-actin gene (ACTG1) are associated with dominant progressive deafness (DFNA20/26) Am. J. Hum. Genet. 73, 1082–1091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rendtorff N. D., Zhu M., Fagerheim T., Antal T. L., Jones M., Teslovich T. M., Gillanders E. M., Barmada M., Teig E., Trent J. M., Friderici K. H., Stephan D. A., Tranebjaerg L. (2006) A novel missense mutation in ACTG1 causes dominant deafness in a Norwegian DFNA20/26 family, but ACTG1 mutations are not frequent among families with hereditary hearing impairment. Eur. J. Hum. Genet. 14, 1097–1105 [DOI] [PubMed] [Google Scholar]
- 3. Morín M., Bryan K. E., Mayo-Merino F., Goodyear R., Mencía A., Modamio-Høybjør S., del Castillo I., Cabalka J. M., Richardson G., Moreno F., Rubenstein P. A., Moreno-Pelayo M. A. (2009) In vivo and in vitro effects of two novel γ-actin (ACTG1) mutations that cause DFNA20/26 hearing impairment. Hum. Mol. Genet. 18, 3075–3089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bryan K. E., Wen K. K., Zhu M., Rendtorff N. D., Feldkamp M., Tranebjaerg L., Friderici K. H., Rubenstein P. A. (2006) Effects of human deafness γ-actin mutations (DFNA20/26) on actin function. J. Biol. Chem. 281, 20129–20139 [DOI] [PubMed] [Google Scholar]
- 5. Goley E. D., Rammohan A., Znameroski E. A., Firat-Karalar E. N., Sept D., Welch M. D. (2010) An actin-filament-binding interface on the Arp2/3 complex is critical for nucleation and branch stability. Proc. Natl. Acad. Sci. U.S.A. 107, 8159–8164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mullins R. D., Stafford W. F., Pollard T. D. (1997) Structure, subunit topology, and actin-binding activity of the Arp2/3 complex from Acanthamoeba. J. Cell Biol. 136, 331–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ng R., Abelson J. (1980) Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 77, 3912–3916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Drubin D. G. (1990) Actin and actin-binding proteins in yeast. Cell Motil. Cytoskeleton 15, 7–11 [DOI] [PubMed] [Google Scholar]
- 9. Yang H. C., Pon L. A. (2002) Actin cable dynamics in budding yeast. Proc. Natl. Acad. Sci. U.S.A. 99, 751–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Abramoff M. D., Magalhaes P. J., Ram S. J. (2004) Image Processing with Image J. Biophotonics Int. 11, 36–42 [Google Scholar]
- 11. Cook R. K., Blake W. T., Rubenstein P. A. (1992) Removal of the amino-terminal acidic residues of yeast actin. Studies in vitro and in vivo. J. Biol. Chem. 267, 9430–9436 [PubMed] [Google Scholar]
- 12. Wen K. K., Rubenstein P. A. (2005) Acceleration of yeast actin polymerization by yeast Arp2/3 complex does not require an Arp2/3-activating protein. J. Biol. Chem. 280, 24168–24174 [DOI] [PubMed] [Google Scholar]
- 13. Winter D., Lechler T., Li R. (1999) Activation of the yeast Arp2/3 complex by Bee1p, a WASP-family protein. Curr. Biol. 9, 501–504 [DOI] [PubMed] [Google Scholar]
- 14. Neidt E. M., Scott B. J., Kovar D. R. (2009) Formin differentially utilizes profilin isoforms to rapidly assemble actin filaments. J. Biol. Chem. 284, 673–684 [DOI] [PubMed] [Google Scholar]
- 15. Kuhn J. R., Pollard T. D. (2005) Real-time measurements of actin filament polymerization by total internal reflection fluorescence microscopy. Biophys. J. 88, 1387–1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karpova T. S., McNally J. G., Moltz S. L., Cooper J. A. (1998) Assembly and function of the actin cytoskeleton of yeast: Relationships between cables and patches. J. Cell Biol. 142, 1501–1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vignjevic D., Yarar D., Welch M. D., Peloquin J., Svitkina T., Borisy G. G. (2003) Formation of filopodia-like bundles in vitro from a dendritic network. J. Cell Biol. 160, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Mejillano M. R., Kojima S., Applewhite D. A., Gertler F. B., Svitkina T. M., Borisy G. G. (2004) Lamellipodial versus filopodial mode of the actin nanomachinery: Pivotal role of the filament barbed end. Cell 118, 363–373 [DOI] [PubMed] [Google Scholar]
- 19. Amann K. J., Pollard T. D. (2001) Direct real-time observation of actin filament branching mediated by Arp2/3 complex using total internal reflection fluorescence microscopy. Proc. Natl. Acad. Sci. U.S.A. 98, 15009–15013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ichetovkin I., Grant W., Condeelis J. (2002) Cofilin produces newly polymerized actin filaments that are preferred for dendritic nucleation by the Arp2/3 complex. Curr. Biol. 12, 79–84 [DOI] [PubMed] [Google Scholar]
- 21. Pollard T. D., Beltzner C. C. (2002) Structure and function of the Arp2/3 complex. Curr. Opin. Struct. Biol. 12, 768–774 [DOI] [PubMed] [Google Scholar]
- 22. Guo D. C., Pannu H., Tran-Fadulu V., Papke C. L., Yu R. K., Avidan N., Bourgeois S., Estrera A. L., Safi H. J., Sparks E., Amor D., Ades L., McConnell V., Willoughby C. E., Abuelo D., Willing M., Lewis R. A., Kim D. H., Scherer S., Tung P. P., Ahn C., Buja L. M., Raman C. S., Shete S. S., Milewicz D. M. (2007) Mutations in smooth muscle α-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat. Genet. 39, 1488–1493 [DOI] [PubMed] [Google Scholar]
- 23. Guo D. C., Papke C. L., Tran-Fadulu V., Regalado E. S., Avidan N., Johnson R. J., Kim D. H., Pannu H., Willing M. C., Sparks E., Pyeritz R. E., Singh M. N., Dalman R. L., Grotta J. C., Marian A. J., Boerwinkle E. A., Frazier L. Q., LeMaire S. A., Coselli J. S., Estrera A. L., Safi H. J., Veeraraghavan S., Muzny D. M., Wheeler D. A., Willerson J. T., Yu R. K., Shete S. S., Scherer S. E., Raman C. S., Buja L. M., Milewicz D. M. (2009) Mutations in smooth muscle α-actin (ACTA2) cause coronary artery disease, stroke, and Moyamoya disease, along with thoracic aortic disease. Am. J. Hum. Genet. 84, 617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nowak K. J., Wattanasirichaigoon D., Goebel H. H., Wilce M., Pelin K., Donner K., Jacob R. L., Hübner C., Oexle K., Anderson J. R., Verity C. M., North K. N., Iannaccone S. T., Müller C. R., Nürnberg P., Muntoni F., Sewry C., Hughes I., Sutphen R., Lacson A. G., Swoboda K. J., Vigneron J., Wallgren-Pettersson C., Beggs A. H., Laing N. G. (1999) Mutations in the skeletal muscle α-actin gene in patients with actin myopathy and nemaline myopathy. Nat. Genet. 23, 208–212 [DOI] [PubMed] [Google Scholar]
- 25. Laing N. G., Dye D. E., Wallgren-Pettersson C., Richard G., Monnier N., Lillis S., Winder T. L., Lochmüller H., Graziano C., Mitrani-Rosenbaum S., Twomey D., Sparrow J. C., Beggs A. H., Nowak K. J. (2009) Mutations and polymorphisms of the skeletal muscle α-actin gene (ACTA1) Hum. Mutat. 30, 1267–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Oda T., Maéda Y. (2010) Multiple conformations of F-actin. Structure 18, 761–767 [DOI] [PubMed] [Google Scholar]
- 27. Humphrey W., Dalke A., Schulten K. (1996) VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38 [DOI] [PubMed] [Google Scholar]
- 28. Stapley B. J., Doig A. J. (1997) Hydrogen bonding interactions between glutamine and asparagine in α-helical peptides. J. Mol. Biol. 272, 465–473 [DOI] [PubMed] [Google Scholar]
- 29. Pfaendtner J., Volkmann N., Hanein D., Dalhaimer P., Pollard T. D., Voth G. A. (2012) Key structural features of the actin filament Arp2/3 complex branch junction revealed by molecular simulation. J. Mol. Biol. 416, 148–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Bremer A., Aebi U. (1992) The structure of the F-actin filament and the actin molecule. Curr. Opin. Cell Biol. 4, 20–26 [DOI] [PubMed] [Google Scholar]
- 31. Bremer A., Millonig R. C., Sütterlin R., Engel A., Pollard T. D., Aebi U. (1991) The structural basis for the intrinsic disorder of the actin filament: the “lateral slipping” model. J. Cell Biol. 115, 689–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Millonig R., Salvo H., Aebi U. (1988) Probing actin polymerization by intermolecular cross-linking. J. Cell Biol. 106, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Galkin V. E., Orlova A., Schröder G. F., Egelman E. H. (2010) Structural polymorphism in F-actin. Nat. Struct. Mol. Biol. 17, 1318–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Orlova A., Shvetsov A., Galkin V. E., Kudryashov D. S., Rubenstein P. A., Egelman E. H., Reisler E. (2004) Actin-destabilizing factors disrupt filaments by means of a time reversal of polymerization. Proc. Natl. Acad. Sci. U.S.A. 101, 17664–17668 [DOI] [PMC free article] [PubMed] [Google Scholar]