Abstract
Tertiary interactions involving hairpin or interior loops of RNA can lead to extended quasi-continuous double helical stem regions, consisting of coaxially stacked segments of duplex RNA, bridged by single-stranded connections. This type of compact folding plays a role in various strategic regions of RNA molecules. Their role in ribosome functioning, RNA splicing and recognition of tRNA-like structures is discussed.
Full text
PDF

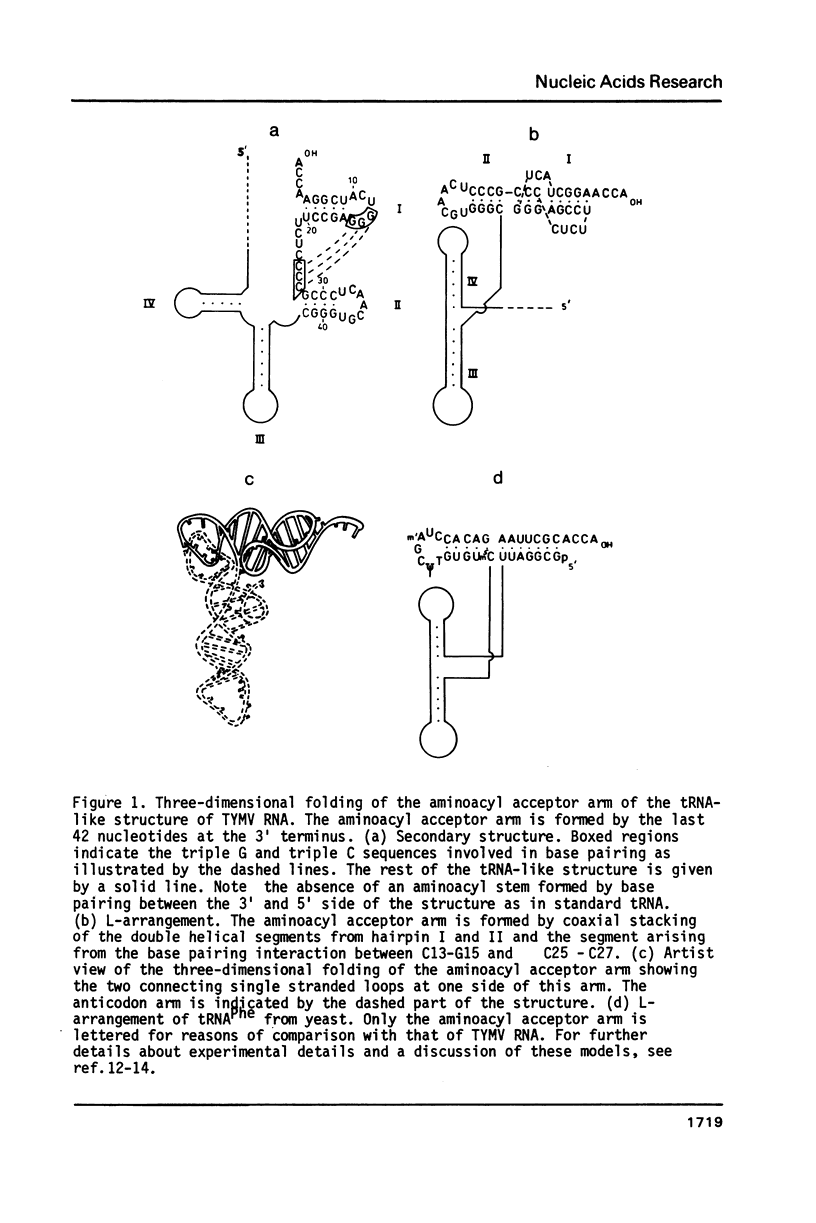


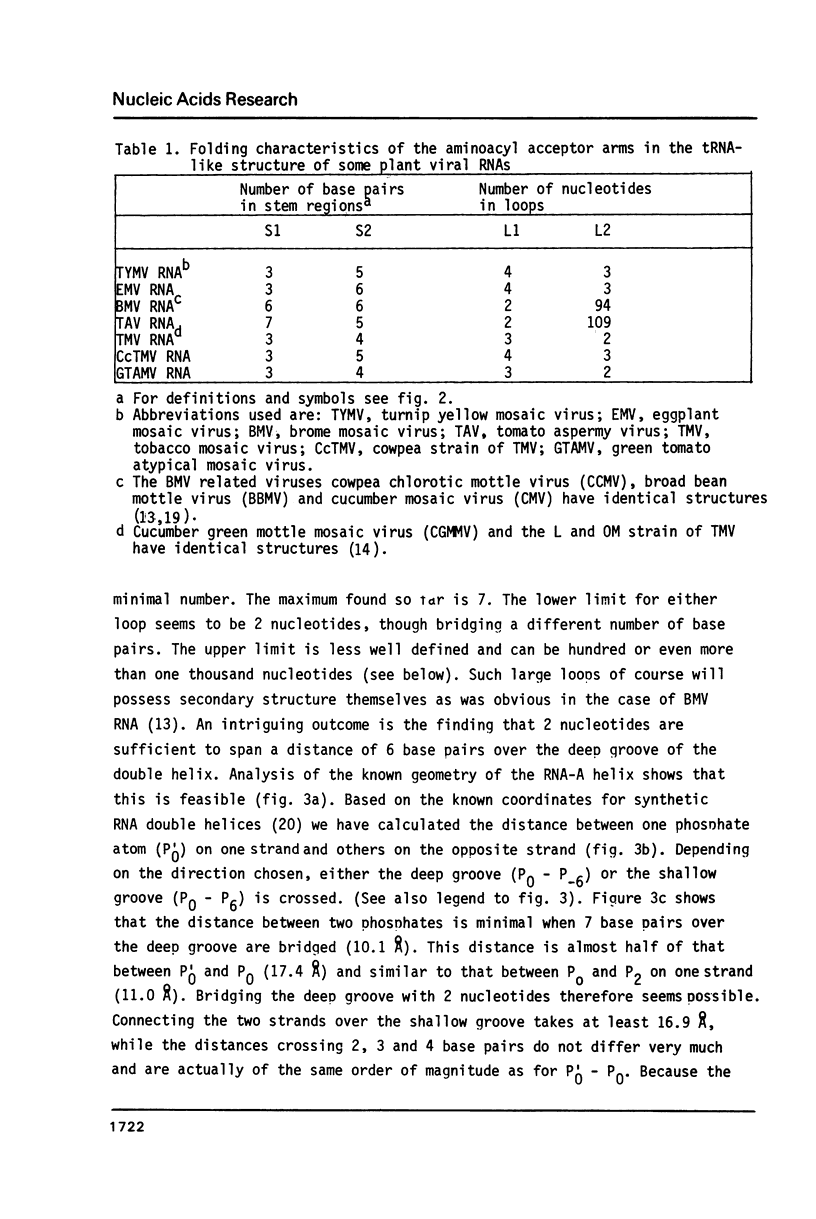
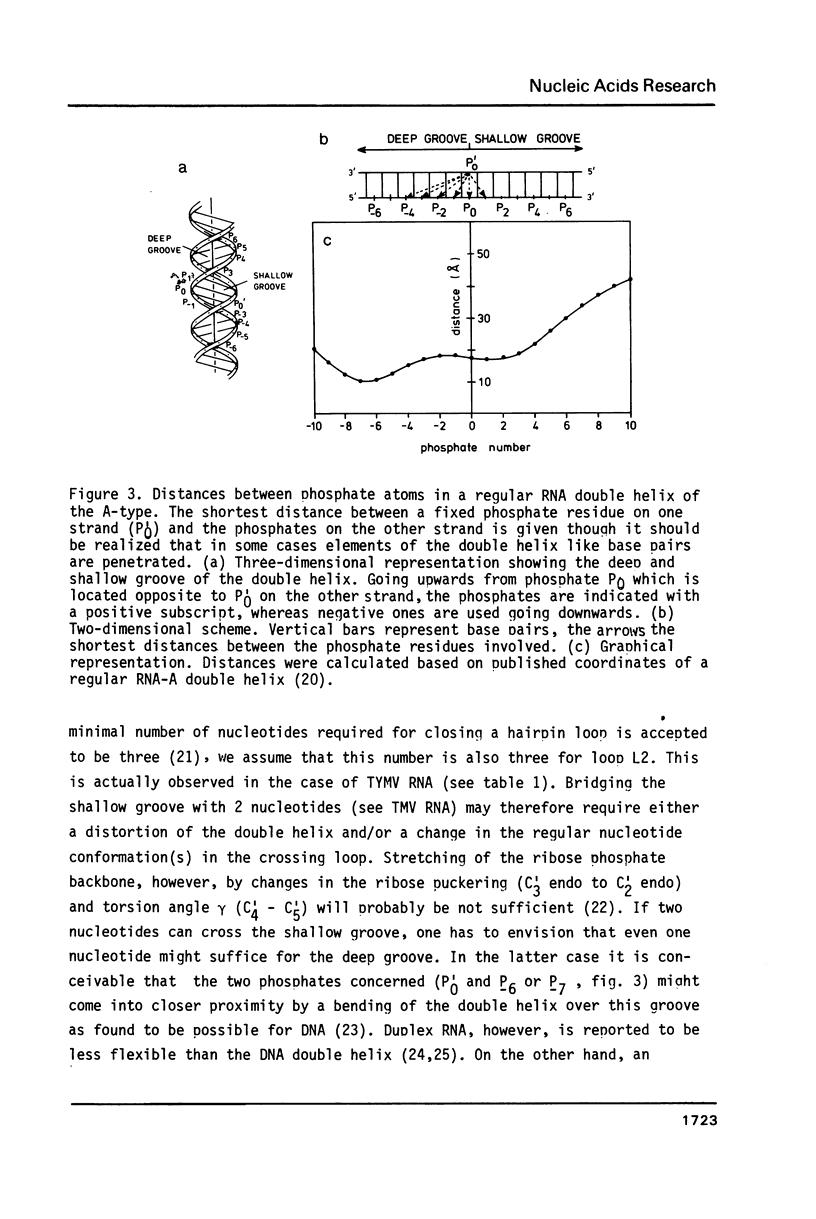


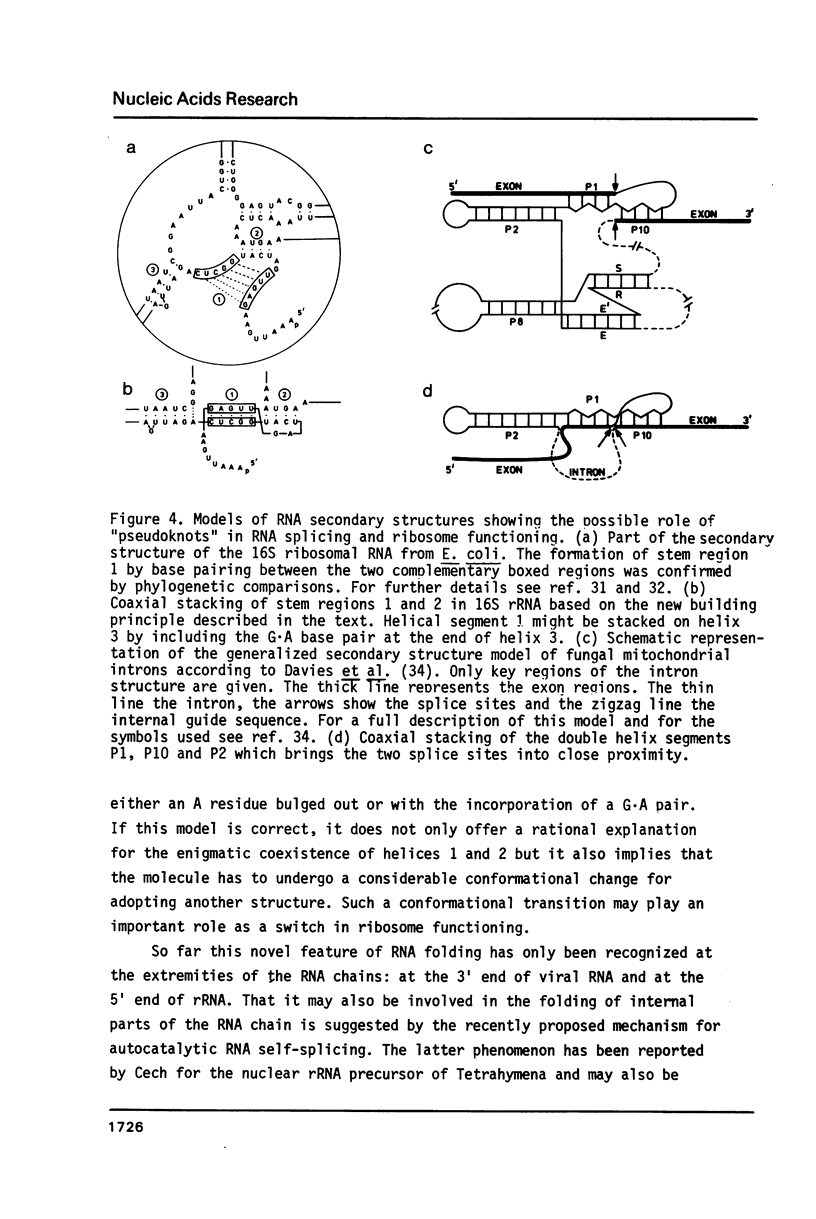




Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahlquist P., Dasgupta R., Kaesberg P. Near identity of 3- RNA secondary structure in bromoviruses and cucumber mosaic virus. Cell. 1981 Jan;23(1):183–189. doi: 10.1016/0092-8674(81)90283-x. [DOI] [PubMed] [Google Scholar]
- Altman S. Aspects of biochemical catalysis. Cell. 1984 Feb;36(2):237–239. doi: 10.1016/0092-8674(84)90216-2. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W., Dover S. D. Optimised parameters for RNA double-helices. Biochem Biophys Res Commun. 1972 Sep 26;48(6):1392–1399. doi: 10.1016/0006-291x(72)90867-4. [DOI] [PubMed] [Google Scholar]
- Auron P. E., Rindone W. P., Vary C. P., Celentano J. J., Vournakis J. N. Computer-aided prediction of RNA secondary structures. Nucleic Acids Res. 1982 Jan 11;10(1):403–419. doi: 10.1093/nar/10.1.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimacombe R., Maly P., Zwieb C. The structure of ribosomal RNA and its organization relative to ribosomal protein. Prog Nucleic Acid Res Mol Biol. 1983;28:1–48. doi: 10.1016/s0079-6603(08)60081-1. [DOI] [PubMed] [Google Scholar]
- Cech T. R. RNA splicing: three themes with variations. Cell. 1983 Oct;34(3):713–716. doi: 10.1016/0092-8674(83)90527-5. [DOI] [PubMed] [Google Scholar]
- Davies R. W., Waring R. B., Ray J. A., Brown T. A., Scazzocchio C. Making ends meet: a model for RNA splicing in fungal mitochondria. Nature. 1982 Dec 23;300(5894):719–724. doi: 10.1038/300719a0. [DOI] [PubMed] [Google Scholar]
- Florentz C., Briand J. P., Romby P., Hirth L., Ebel J. P., Glegé R. The tRNA-like structure of turnip yellow mosaic virus RNA: structural organization of the last 159 nucleotides from the 3' OH terminus. EMBO J. 1982;1(2):269–276. doi: 10.1002/j.1460-2075.1982.tb01158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gralla J., Crothers D. M. Free energy of imperfect nucleic acid helices. II. Small hairpin loops. J Mol Biol. 1973 Feb 5;73(4):497–511. doi: 10.1016/0022-2836(73)90096-x. [DOI] [PubMed] [Google Scholar]
- Hall T. C. Transfer RNA-like structures in viral genomes. Int Rev Cytol. 1979;60:1–26. doi: 10.1016/s0074-7696(08)61257-7. [DOI] [PubMed] [Google Scholar]
- Lerner M. R., Boyle J. A., Mount S. M., Wolin S. L., Steitz J. A. Are snRNPs involved in splicing? Nature. 1980 Jan 10;283(5743):220–224. doi: 10.1038/283220a0. [DOI] [PubMed] [Google Scholar]
- Levitt M. Computer simulation of DNA double-helix dynamics. Cold Spring Harb Symp Quant Biol. 1983;47(Pt 1):251–262. doi: 10.1101/sqb.1983.047.01.030. [DOI] [PubMed] [Google Scholar]
- Moras D., Comarmond M. B., Fischer J., Weiss R., Thierry J. C., Ebel J. P., Giegé R. Crystal structure of yeast tRNAAsp. Nature. 1980 Dec 25;288(5792):669–674. doi: 10.1038/288669a0. [DOI] [PubMed] [Google Scholar]
- Noller H. F., van Knippenberg P. H. Structure and function of ribosomal RNA. Horiz Biochem Biophys. 1983;7:71–99. [PubMed] [Google Scholar]
- Nussinov R., Jacobson A. B. Fast algorithm for predicting the secondary structure of single-stranded RNA. Proc Natl Acad Sci U S A. 1980 Nov;77(11):6309–6313. doi: 10.1073/pnas.77.11.6309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Gehrke L., Roth D. A., Auron P. E. Computer-aided nucleic acid secondary structure modeling incorporating enzymatic digestion data. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):347–366. doi: 10.1093/nar/12.1part1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quigley G. J., Rich A. Structural domains of transfer RNA molecules. Science. 1976 Nov 19;194(4267):796–806. doi: 10.1126/science.790568. [DOI] [PubMed] [Google Scholar]
- Quigley G. J., Teeter M. M., Rich A. Structural analysis of spermine and magnesium ion binding to yeast phenylalanine transfer RNA. Proc Natl Acad Sci U S A. 1978 Jan;75(1):64–68. doi: 10.1073/pnas.75.1.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Linschooten K., Pleij C. W., Bosch L. The three-dimensional folding of the tRNA-like structure of tobacco mosaic virus RNA. A new building principle applied twice. EMBO J. 1984 Nov;3(11):2613–2619. doi: 10.1002/j.1460-2075.1984.tb02182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Pleij C. W., Bosch L. Three-dimensional models of the tRNA-like 3' termini of some plant viral RNAs. EMBO J. 1983;2(7):1079–1085. doi: 10.1002/j.1460-2075.1983.tb01549.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietveld K., Van Poelgeest R., Pleij C. W., Van Boom J. H., Bosch L. The tRNA-like structure at the 3' terminus of turnip yellow mosaic virus RNA. Differences and similarities with canonical tRNA. Nucleic Acids Res. 1982 Mar 25;10(6):1929–1946. doi: 10.1093/nar/10.6.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertus J. D., Ladner J. E., Finch J. T., Rhodes D., Brown R. S., Clark B. F., Klug A. Structure of yeast phenylalanine tRNA at 3 A resolution. Nature. 1974 Aug 16;250(467):546–551. doi: 10.1038/250546a0. [DOI] [PubMed] [Google Scholar]
- Schevitz R. W., Podjarny A. D., Krishnamachari N., Hughes J. J., Sigler P. B., Sussman J. L. Crystal structure of a eukaryotic initiator tRNA. Nature. 1979 Mar 8;278(5700):188–190. doi: 10.1038/278188a0. [DOI] [PubMed] [Google Scholar]
- Singhal R. P., Shaw J. K. Prokaryotic and eukaryotic 5 S RNAs: primary sequences and proposed secondary structures. Prog Nucleic Acid Res Mol Biol. 1983;28:177-209, 251-2. doi: 10.1016/s0079-6603(08)60087-2. [DOI] [PubMed] [Google Scholar]
- Studnicka G. M., Rahn G. M., Cummings I. W., Salser W. A. Computer method for predicting the secondary structure of single-stranded RNA. Nucleic Acids Res. 1978 Sep;5(9):3365–3387. doi: 10.1093/nar/5.9.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Traub W., Sussman J. L. Adenine-guanine base pairing ribosomal RNA. Nucleic Acids Res. 1982 Apr 24;10(8):2701–2708. doi: 10.1093/nar/10.8.2701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo N. H., Roe B. A., Rich A. Three-dimensional structure of Escherichia coli initiator tRNAfMet. Nature. 1980 Jul 24;286(5771):346–351. doi: 10.1038/286346a0. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]



