Abstract
EMBO J advance online publication June 07 2012; doi:; DOI: 10.1016/j.molcel.2012.05.009
Heterochromatin is classically perceived to be refractory to transcription because of its compact structure. However, Keller et al (2012) now demonstrated that heterochromatic transcripts can accumulate even when heterochromatin is normally packaged. By tracking down the fate of these heterochromatic RNAs, they revealed a new post-transcriptional mechanism of silencing in heterochromatin that involves the dynamic turnover of HP1Swi6 between its free, chromatin-bound and RNA-bound forms. The latter form escorts heterochromatic RNA to degradation.
In eukaryotes, chromatin can be classified into two states: euchromatin, which is loosely packed and actively transcribed, and heterochromatin, which remains condensed during interphase. The compact structure of heterochromatin is critical for its widespread roles in chromosome integrity, stability and transposon silencing around centromeres and in other repeat-rich regions, such as subtelomeric regions.
Heterochromatin is relatively devoid of coding sequences, and reporter genes embedded are tightly repressed under most situations. The compact structure of heterochromatin was generally thought to be inert and refractory to transcription (Gasser and Cockell, 2001). However, HP1 (heterochromatin protein 1), which binds the conserved heterochromatin mark, histone H3 lysine 9 methylation (H3K9me) and serves as the structural basis for the condensed state of heterochromatin, undergoes very active turnover between the chromatin-bound and -free states (Cheutin et al, 2003; Maison and Almouzni, 2004). Furthermore, heterochromatin is not as ‘silent’ as initially thought, and undergoes substantial transcription. But the transcripts are quickly processed by RNA interference (RNAi), which utilizes 20- to 30-nt small RNA to guide cleavage or translational inhibition of target transcripts (Carmell and Hannon, 2004), and to release RNA polymerase II (Zaratiegui et al, 2011). RNA degradation also participates in this process, and its role is newly interpreted by Keller et al (2012).
Although the detailed mechanisms underlying the establishment and maintenance of heterochromatin vary in different species, the principles are conserved from yeast to human. Much work has been done in the fission yeast Schizosaccharomyces pombe to understand how the enzymes responsible for the deposition of heterochromatic marks are recruited to specific regions of the genome, and has revealed a complicated network of mechanisms both dependent and independent of RNAi (Buhler et al, 2007; Grewal and Jia, 2007). The involvement of RNA turnover in this network is known but not well understood.
Keller et al (2012) set out to understand the role of RNA degradation by tracking down the fate of heterochromatic transcripts. They used a cid14 mutant, which has defects in polyadenylation-assisted RNA turnover (Wang et al, 2008) and observed accumulation of transcripts from reporter genes embedded in heterochromatic regions. Interestingly, such derepression is not accompanied by heterochromatin decondensation. They also found a discrepancy between mRNA and protein levels, suggesting that these reporter gene transcripts are assembled into translation-incapable ribonucleoprotein particles. The authors then hypothesized that Swi6, an HP1 homologue in S. pombe, may be central to these particles by targeting and escorting heterochromatic RNA for degradation, because of its dual affinity for both H3K9me and RNA (Motamedi et al, 2008). Keller et al (2012) confirmed HP1Swi6–RNA association and further explored the structural basis of both interactions. They found that overlapping regions of HP1Swi6 were important for both interactions, and demonstrated alternation between them and induced structural change of HP1Swi6 after binding to either partner. Such alternation and structural change are important for HP1Swi6 targeting RNA from heterochromatic regions, and may prevent HP1Swi6 binding non-specifically to euchromatic mRNA. To explore the function of the newly identified HP1Swi6–RNA association, the creation of a separation-of-function mutant was necessary, as HP1Swi6 also has a structural role in heterochromatin. Guided by the structural information obtained for these interactions, the authors designed a mutant that abolishes RNA-binding, while not affecting heterochromatin structure, and indeed, observed that heterochromatic transcripts were no longer degraded, nor were they inhibited from being translated.
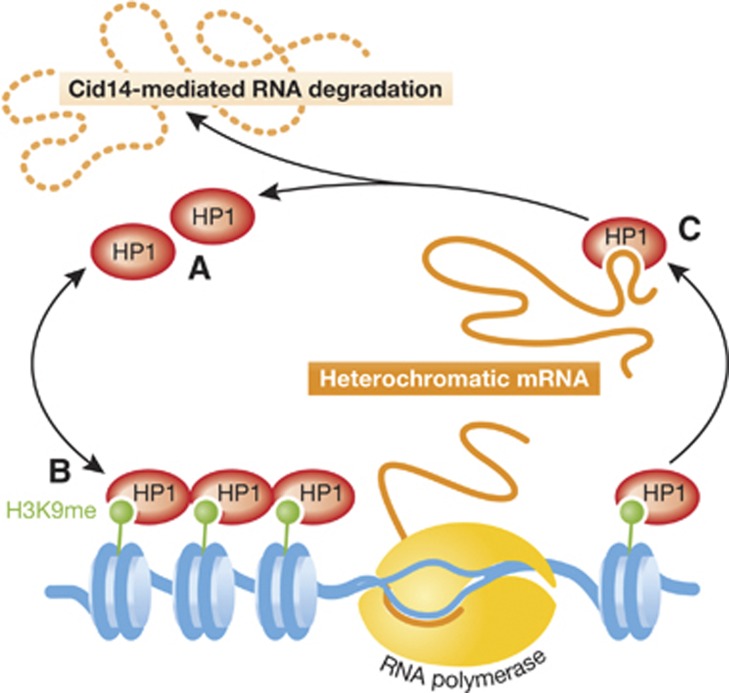
In summary, Keller et al (2012) have revealed another level of tight repression of heterochromatic genes through uncovering the dynamic turnover of HP1Swi6 between its free, H3K9me-bound and RNA-bound forms (Figure 1). The structural component of heterochromatin, HP1Swi6 serves as the unidentified link to capture heterochromatic transcripts onsite and escort them towards eventual degradation. Because of the high conservation of HP1, it is possible that a similar mechanism contributes to the tight repression of heterochromatin in higher eukaryotes.
Figure 1.
HP1Swi6 undergoes rapid turnover between its (A) free, (B) H3K9me-bound and (C) heterochromatic RNA-bound forms. The major structural component of heterochromatin, H3K9me-bound HP1Swi6 (B) exchanges dynamically with its free ensemble (A). Contrary to the classical view, RNA polymerase can get access to heterochromatin, but the transcripts are captured by HP1Swi6 (C) and escorted to Cid14-mediated RNA degradation. RNA competes with H3K9me for binding with HP1Swi6 and causes structural change to HP1Swi6. Thus, both heterochromatin and HP1Swi6–RNA association contributes to the tight repression of genes within heterochromatin.
Footnotes
The authors declare that they have no conflict of interest.
References
- Buhler M, Haas W, Gygi SP, Moazed D (2007) RNAi-dependent and -independent RNA turnover mechanisms contribute to heterochromatic gene silencing. Cell 129: 707–721 [DOI] [PubMed] [Google Scholar]
- Carmell MA, Hannon GJ (2004) RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol 11: 214–218 [DOI] [PubMed] [Google Scholar]
- Cheutin T, McNairn AJ, Jenuwein T, Gilbert DM, Singh PB, Misteli T (2003) Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science 299: 721–725 [DOI] [PubMed] [Google Scholar]
- Gasser SM, Cockell MM (2001) The molecular biology of the SIR proteins. Gene 279: 1–16 [DOI] [PubMed] [Google Scholar]
- Grewal SI, Jia S (2007) Heterochromatin revisited. Nat Rev Genet 8: 35–46 [DOI] [PubMed] [Google Scholar]
- Keller C, Adaixo R, Stunnenberg R, Woolcock KJ, Hiller S, Bühler M (2012) HP1(Swi6) mediates the recognition and destruction of heterochromatic RNA transcripts. Mol Cell (advance online publication, 7 June 2012; doi:; DOI: 10.1016/j.molcel.2012.05.009) [DOI] [PubMed] [Google Scholar]
- Maison C, Almouzni G (2004) HP1 and the dynamics of heterochromatin maintenance. Nat Rev Mol Cell Biol 5: 296–304 [DOI] [PubMed] [Google Scholar]
- Motamedi MR, Hong EJ, Li X, Gerber S, Denison C, Gygi S, Moazed D (2008) HP1 proteins form distinct complexes and mediate heterochromatic gene silencing by nonoverlapping mechanisms. Mol Cell 32: 778–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SW, Stevenson AL, Kearsey SE, Watt S, Bahler J (2008) Global role for polyadenylation-assisted nuclear RNA degradation in posttranscriptional gene silencing. Mol Cell Biol 28: 656–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaratiegui M, Castel SE, Irvine DV, Kloc A, Ren J, Li F, de Castro E, Marin L, Chang AY, Goto D, Cande WZ, Antequera F, Arcangioli B, Martienssen RA (2011) RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II. Nature 479: 135–138 [DOI] [PMC free article] [PubMed] [Google Scholar]