Abstract
Precise gene expression is a fundamental aspect of organismal function and depends on the combinatorial interplay of transcription factors (TFs) with cis-regulatory DNA elements. While much is known about TF function in general, our understanding of their cell type-specific activities is still poor. To address how widely expressed transcriptional regulators modulate downstream gene activity with high cellular specificity, we have identified binding regions for the Hox TF Deformed (Dfd) in the Drosophila genome. Our analysis of architectural features within Hox cis-regulatory response elements (HREs) shows that HRE structure is essential for cell type-specific gene expression. We also find that Dfd and Ultrabithorax (Ubx), another Hox TF specifying different morphological traits, interact with non-overlapping regions in vivo, despite their similar DNA binding preferences. While Dfd and Ubx HREs exhibit comparable design principles, their motif compositions and motif-pair associations are distinct, explaining the highly selective interaction of these Hox proteins with the regulatory environment. Thus, our results uncover the regulatory code imprinted in Hox enhancers and elucidate the mechanisms underlying functional specificity of TFs in vivo.
Keywords: ChIP-seq, Deformed, Drosophila , Hox, transcriptional gene regulation
Introduction
Fine-tuned interactions of transcription factors (TF) and other regulatory proteins, such as transcriptional cofactors and epigenetic modifiers, with defined cis-regulatory elements in the genome orchestrate the spatiotemporal regulation of gene expression (Arnone and Davidson, 1997; Ghazi and VijayRaghavan, 2000). One highly conserved and essential class of TFs encoded by the Hox genes is critically involved in the regulation of a large number of processes and genes in diverse cell types throughout an animal’s life (McGinnis and Krumlauf, 1992). Despite their broad expression, transcriptional regulators of the Hox class activate or repress transcriptional programs with extreme spatial and temporal resolution (Lohmann et al, 2002; Lohmann and McGinnis, 2002; Merabet et al, 2005; Tour et al, 2005), making them ideal models to study the mechanisms underlying TF cell and tissue specificity. However, due to the paucity of known Hox cis-regulatory response elements (HREs), the regulatory code underlying the strict spatio-temporal control of Hox target genes is only poorly understood.
To address this question, we have quantitatively characterized hundreds of binding regions of the Drosophila Hox protein Deformed (Dfd), which we independently identified by chromatin immunoprecipitation coupled to massively parallel sequencing (ChIP-seq) and by computational methods. Our analysis revealed specific architectural features like motif-pair associations and motif distance preferences to be essential for cell type-specific expression of associated target genes. HRE features indeed determine specificity, since they alone accurately predict target gene function and expression patterns. By contrasting the in vivo binding profiles of Dfd with that of Ultrabithorax (Ubx), another Drosophila Hox protein essential for the development of distinct morphological traits, we identified common and divergent enhancer features associated with the specific functions of these TFs.
Results
Dfd binding regions function as Dfd-regulated enhancers in vivo
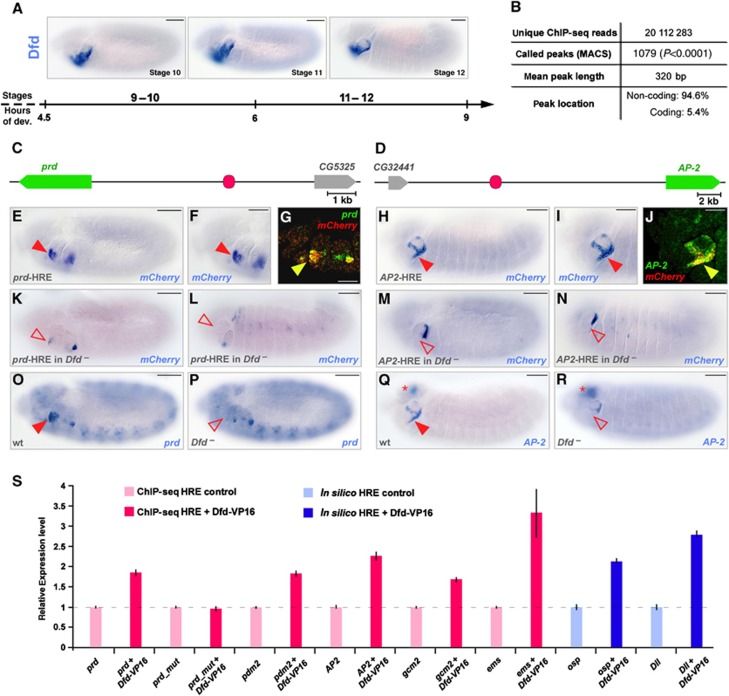
In order to quantitatively identify genomic regions bound by the Hox TF Dfd in Drosophila (Figure 1A), we employed two complementing approaches: ChIP-seq, which has been successfully applied previously to identify stage- (Zinzen et al, 2009) and tissue-specific (Visel et al, 2009) enhancer activities, and computational detection of clusters of TF binding sequences, which allows the identification of cis-regulatory modules irrespective of temporal and spatial context (Pennacchio et al, 2007). To generate genome-wide maps of Dfd binding in vivo, we performed ChIP using stage 10–12 Drosophila embryos and a Dfd-specific antibody (Figure 1A; Supplementary Figure S1). Stage-independent in silico Dfd-specific HREs were identified by searching for clusters of conserved Dfd binding motifs, as defined by a position weight matrix (PWM) (Chen et al, 2007), in the non-coding regions of the genomes of 12 distinct Drosophila species (Hueber et al, 2007). By applying both approaches, 4526 genomic regions containing clusters of Dfd binding sites and 1079 Dfd ChIP-seq enrichment peaks were identified (Figure 1B), including two out of the three well-characterized Dfd-HREs, namely rpr-4S3 (Lohmann et al, 2002) and Dfd-EAE (Kuziora and McGinnis, 1988; Supplementary Table S1). To study the regulatory capacity of novel in silico and ChIP-seq detected HREs, we first performed cell culture-based enhancer assays for 11 randomly selected HREs and found that reporter expression driven by the identified genomic regions was in all cases dependent on Dfd binding (Figure 1S). Next, we tested the in vivo activity of 21 arbitrarily selected enhancers in transgenic reporter lines (Figure 1C and D; Supplementary Figure S2), revealing that 7 out of 11 ChIP-identified (Figure 1E–J; Supplementary Figure S2A and B) and 5 out of 10 in silico-predicted (Supplementary Figure S2C, D and F) Dfd-HREs recapitulate the spatio-temporal expression of adjacent genes (Figure 1G and J). Most importantly, we were able to demonstrate Dfd-dependent regulation of both transgenic reporter expression (Figure 1K–N) and endogenous gene expression (Figure 1O–R; Supplementary Figure S2A–D and F), suggesting that they are bona fide direct Dfd target genes. Thus, the identified Dfd-HREs represent a data set of biologically relevant regulatory regions and an excellent resource to unravel sequence features within Hox responsive enhancers that might be essential for the highly selective Hox target gene regulation.
Figure 1.
Generation of a high-resolution atlas of in vivo enhancers for the Hox transcription factor Dfd. (A) Dfd mRNA expression in the maxillary and mandibular segments of stage 10–12 Drosophila embryos. Bars: 50 μm. (B) Summary of Dfd ChIP-seq results. In total, 94.6% of the 1079 MACS-called peaks are located in the Drosophila non-coding genomic regions. (C, D) Schematic representation of the genomic location of two Dfd ChIP-HREs (in pink) with the gene regulated by the ChIP-HRE indicated in green. (E–J) Reporter gene expression by the prd (E, F) and AP-2 (H, I) ChIP-HREs is directed in the maxillary segment (red arrowheads). mCherry and endogenous prd or AP-2 transcripts colocalize in the maxillary segment of the prd (G) or AP-2 (J) enhancer lines (indicated by yellow arrowheads). Bars: 50 μm (E, H), 25 μm (F, G, I, J). (K–N) Reporter gene expression by the prd (K, L) and AP-2 (M, N) ChIP-HREs in stage 11 (K, M) and stage 12 (L, N) Dfd mutant embryos. Absence of gene expression in Dfd mutant embryos is highlighted by red, open arrowheads. Bars: 50 μm. (O–R) prd (O, P) and AP-2 (Q, R) endogenous expression in wild-type (O, Q) and Dfd mutant (P, R) embryos. Absence of gene expression in Dfd mutant embryos is highlighted by red, open arrowhead in (P, R), AP-2 expression in the brain is indicated by red asterisks in (Q, R). Bars: 50 μm. (S) Relative mRNA expression of mCherry driven by different ChIP-seq-(pink) or in silico-(blue) identified Dfd-HREs, in the absence (light pink, light blue) or presence (dark pink, dark blue) of Dfd-VP16 in transiently transfected D.Mel-2 cells. Mutation of all Dfd binding sites in the prd Dfd-HRE (prd_mut) results in the inability of Dfd-VP16 to activate reporter gene expression. Data are shown as relative fold difference compared to reporter construct expression without addition of Dfd-VP16 (dashed line) (error bars, s.d.; n=3).
Architectural features of Dfd-HREs are essential for cell type-specific functions
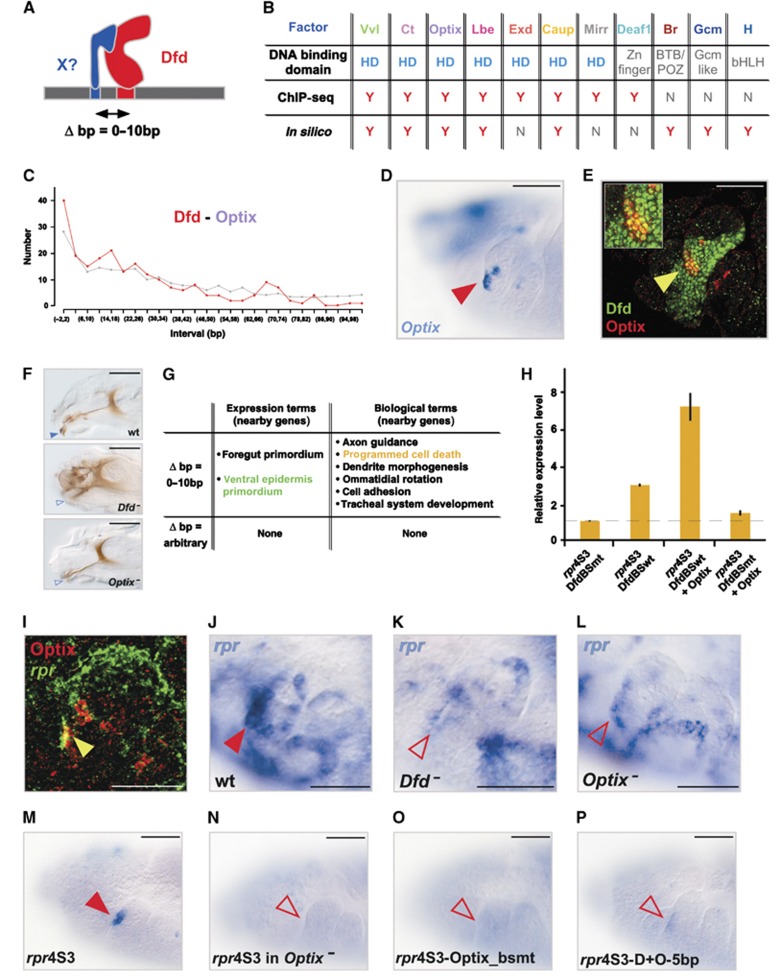
Transcriptional regulation in many cases relies on the assembly of regulatory protein complexes mediated by closely spaced TF binding sites within a cis-regulatory module (Chan et al, 1994; Makeev et al, 2003; Walsh and Carroll, 2007) and we have previously shown that Hox proteins employ this mechanism to control target gene activity in small subsets of cells (Stobe et al, 2009). Now, we systematically scanned the novel HREs for TF binding motifs appearing in close proximity to Dfd binding sites (Figure 2A). Using a statistical test for pair-wise distance distributions, we found 11 overrepresented DNA motifs for known TFs adjoining to Dfd binding sites with 5 of the motifs occurring in both the ChIP-seq and in silico-identified Dfd-HREs (Figure 2B; Supplementary Figure S3). When we analysed the expression patterns of six of these transcriptional regulators known to bind to the 11 motifs we had identified, we found colocalization with Dfd in different sub-populations of cells in all cases (Figure 2D and E; Supplementary Figure S3B and C). Colocalization was already known for two TFs, whose binding sites were coupled to Dfd motifs, including Extradenticle (Exd) (Figure 2B), which is known to cooperatively bind with Hox proteins to DNA and thereby increase Hox DNA-binding selectivity (Mann and Carroll, 2002). We next asked whether the short-distance arrangements in Dfd-HREs are of biological relevance and translated into the regulation of similar classes of target genes. To this end, we statistically tested the overrepresentation of expression and biological terms of genes associated with HREs harbouring specific combinations of Dfd and close-by motifs. This analysis revealed that only those Dfd-HREs with short distance intervals between the Dfd and adjacent motifs were coupled to similar gene classes, while random distance intervals did not show any correlation (Figure 2G; Supplementary Figure S3C). Strikingly, genes associated with specific short-distance HREs had similar expression and functional annotations as the TFs interacting with the Hox adjoining motifs (Supplementary Table S2; Supplementary Figure S3C), suggesting that time and place of Hox action is dictated by spatio-temporally restricted co-regulators. Support for this hypothesis stems from our observation that one of the close-distance partners, Optix, regulates similar processes as Dfd, since Dfd and Optix mutants displayed comparable morphological defects in the head region (Coiffier et al, 2008), such as the absence of mouth hooks (Figure 2F), a maxillary segment-derived structure known to be specified by Dfd (Regulski et al, 1987). In addition, one of the genes associated with a Dfd-Optix HRE, the known Dfd target gene reaper (rpr) (Lohmann et al, 2002), is expressed in the ventral epidermis primordium (Figure 2I) as predicted by its HRE architecture (Figure 2G), and regulated by Dfd and Optix in ventral-maxillary cells (Figure 2I–L), which also express these factors (Figure 2E). A cell-culture assay using the well-established Dfd responsive module responsible for rpr expression in a few anterior-maxillary cells, the rpr-4S3 Dfd-HRE (Figure 2M; Lohmann et al, 2002), with wild-type or mutated Dfd binding sites or reduction of Dfd levels by RNAi confirmed the requirement for simultaneous activity of Dfd and Optix on the rpr-4S3 Dfd-HRE for strong reporter gene induction (Figure 2H; Supplementary Figure S4A). Optix binding to the rpr-4S3 Dfd-HRE was additionally confirmed by electrophoretic mobility shift assay (EMSA) experiments (Supplementary Figure S4F). Furthermore, transgenic reporter expression induced by the rpr-4S3 Dfd-HRE was lost in Optix mutant embryos (Figure 2N) or when the Optix binding sites were mutated (Figure 2O). These results demonstrate that Optix, one of the newly identified factors, is a Dfd co-regulator required for proper regulation of the important Hox target gene rpr.
Figure 2.
Identification of short-distance arrangements in Hox ChIP-HREs. (A) Schematic representation of a close-distance arrangement between Dfd and a potential co-regulator on a Dfd-HRE. (B) Classification of predicted Dfd close-distance co-regulators according to their DNA binding domain (HD: homeodomain, Zn finger: Zinc finger, BTB/POZ: BR-C, ttk and bab or Pox virus and Zn finger, Gcm like: Glial cells missing like or bHLH: basic helix-loop-helix) and the presence (Y) or absence (N) of their DNA binding motifs in close vicinity to Dfd binding sequence in ChIP-seq or in in silico-predicted HREs. (C) Distribution of observed distances between Dfd and Optix binding sites in all Dfd-HREs (red line=observed distances, grey line=background). (D, E) Optix mRNA expression (red arrowhead) (D) and Optix (red) and Dfd (green) protein expression (E) in the maxillary segment of stage 12 embryos. A sub-population of Optix and Dfd expressing cells (yellow arrowhead) (E) is also shown at a higher magnification (inset). Bars: 25 μm. (F) Head morphology of wild-type, Dfd and Optix mutant embryos. The absence of the mouth hooks in the mutant embryos is highlighted by blue, open arrowheads. Bars: 50 μm. (G) Expression and biological terms of genes connected to Dfd-Optix HREs with close-by (Δbp=0–10) or arbitrary distances (Δbp=arbitrary) between Dfd and Optix binding sites. (H) Relative Firefly luciferase mRNA expression driven by the rpr4S3-DfdBSwt (wild-type Dfd binding sites) or rpr4S3-DfdBSmt (mutated Dfd binding sites) constructs, in the presence or absence of Optix, in transiently transfected D.Mel-2 cells. Data are shown as relative fold difference compared to the rpr4S3-DfdBSmt construct (dashed line) (error bars, s.d.; n=3). Due to the presence of Dfd activity in D.Mel-2 cells (Lin et al, 2011), the rpr4S3-DfdBSmt construct, which is unable to interact with Dfd protein, was used to analyse the ability of Optix to activate reporter gene expression without the assistance of Dfd. (I) Optix protein (red) and rpr RNA (green) expression in the maxillary segment of a stage 11 wild-type embryo (co-expression is highlighted by yellow arrowhead). Bar: 10 μm. (J–L) rpr RNA expression (open red arrowheads) in the anterior part of the maxillary segment of Dfd (K) or Optix (L) mutant embryos is reduced in comparison to maxillary rpr expression (red arrowhead) in wild-type embryos. Bars: 25 μm (J). (M, N) Reporter expression by the rpr4S3 HRE in wild-type (M) and Optix mutant (N) embryos. Absence of reporter expression in Optix mutant embryos is highlighted by red, open arrowhead in (N). Bars: 25 μm. (O,P) Reporter expression by the rpr4S3 HRE, with the Optix BS mutated (rpr4S3-Optix_bsmt, O) and with the Dfd and Optix BS spaced by a 5-bp insertion (rpr4S3-D+O-5 bp, P). Absence of reporter expression is highlighted by red, open arrowheads. Bars: 25 μm.
Interaction of Dfd with close-distance binding co-regulators on HREs results in spatiotemporal control of target gene expression
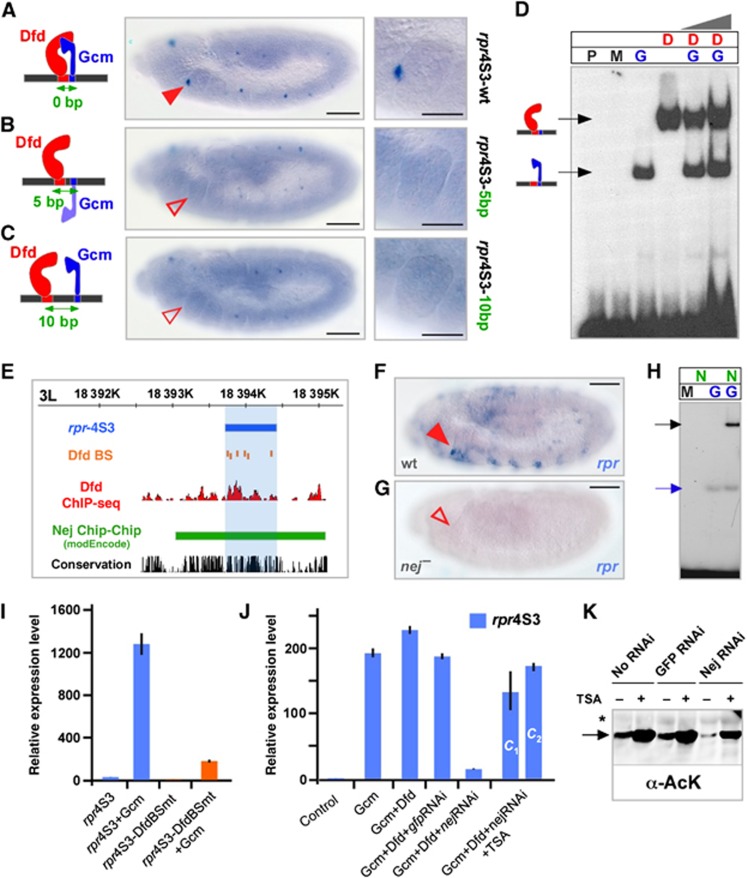
We next explored whether the precise spacing between Hox and adjacent binding sites plays a role for enhancer activity. The rpr-4S3 Dfd HRE, which induces gene expression in a few anterior-maxillary cells (Figure 3A), has previously been shown to be under the control of Dfd and Glial cells missing (Gcm) (Stobe et al, 2009), a Dfd co-regulator also identified in this study (Figure 2B; Supplementary Figure S3). Dfd and Gcm (Stobe et al, 2009) as well as Optix binding sites within the rpr-4S3 HRE are directly adjacent to each other, thus we introduced a 5- and 10-bp spacer to interfere with potential interactions of the proteins on the enhancer. In all cases, reporter gene expression was strongly reduced or completely abolished (Figures 2P, 3B and C), showing that the close-distance arrangements between Dfd and Gcm as well as Dfd and Optix are required for the in vivo activity of the rpr-4S3 enhancer.
Figure 3.
Relevance and function of short-distance arrangements in Hox ChIP-HREs. (A–C) Reporter gene expression driven by the rpr4S3-wt enhancer (A), the rpr4S3-5 bp (B) and the rpr4S3-10 bp (C) enhancers, in which the Dfd and Gcm binding sites are separated by 5 or 10 bp. Bars: 50 μm. A close-up of the maxillary segment of each genotype is shown in the right panel. Bars: 25 μm. (D) EMSA using no protein (P), control maltose-binding protein (MBP) (M), Gcm protein (G), Dfd protein (D) or combination of Dfd and Gcm (D and G). The arrows indicate specific DNA-protein complexes for Dfd (red) or Gcm (blue). (E) Schematic representation of the rpr4S3 (blue) genomic region. Dfd ChIP-seq peaks are shown in red and the Nej bound genomic region as identified by ChIP-Chip (Roy et al, 2010) in green. Dfd binding sites are shown in orange. (F, G) rpr mRNA expression in wild-type (F) and nej mutant (G) embryos. A close-up of the maxillary segment is shown on the right side of the panel. Bars: 50 μm. (H) EMSA using control MBP (M), Gcm protein (G), Nej protein (N) or combination of Gcm and Nej (N and G). The arrows indicate specific DNA–protein complexes for Gcm (blue) or Nej/Gcm (black). (I) Relative Firefly luciferase mRNA expression driven by the 4S3-DfdBSwt (wild-type Dfd binding sites) or by the 4S3-DfdBSmt (mutated Dfd binding sites) in the presence or absence of Gcm in transiently transfected D.Mel-2 cells. Data are shown as relative fold difference compared to 4S3-DfdBSmt (error bars, s.d.; n=3). Due to the presence of Dfd activity in D.Mel-2 cells (Lin et al, 2011), the rpr4S3-DfdBSmt construct, which is unable to interact with Dfd protein, was used to analyse the ability of Gcm to activate reporter gene expression without the assistance of Dfd. (J) Relative Firefly luciferase mRNA expression driven by the rpr4S3 enhancer, in the presence of the indicated expression and RNAi constructs. The HDAC inhibitor Trichostatin A was added in two different concentrations (c1=100 ng/ml and c2=200 ng/ml) to nej dsRNA-treated cells in order to compensate for the reduced acetylation levels due to Nej knockdown (error bars, s.d.; n=3). (K) Western blots using an anti-AcK antibody and total extracts from transiently transfected Drosophila cells. In Nej-RNAi-treated cells, acetylation of transiently transfected Gcm is reduced in comparison to control cells or cells treated with control RNAi against GFP. Treatment with the deacetylase inhibitor Trichostatin A in all cases resulted in an increase of Gcm acetylation. Asterisk indicates cross-reacting band confirming equal loading.
While our results regarding the close-distance arrangement of Dfd and Gcm binding sites suggested the formation of a Dfd–Gcm protein complex, like in the case of Dfd and Exd (Joshi et al, 2010), we only observed independent binding of the two proteins to the rpr-4S3 enhancer in EMSA experiments (Figure 3D; Stobe et al, 2009), supporting the idea of Hox proteins collaborating with other TFs on target HREs in the absence of physical contact (Walsh and Carroll, 2007). It has been shown before that Hox proteins together with other TFs that bind in the immediate vicinity recruit non-DNA binding cofactors to HREs (Walsh and Carroll, 2007). To test if such factors could interact with Dfd and the newly identified short distance binding TFs, we scanned the modENCODE data set (Roy et al, 2010) and found that dCBP/Nej, a member of the CBP/p300 family of transcriptional co-activators bearing acetyltransferase activity (Akimaru et al, 1997; Kalkhoven, 2004), binds to the rpr-4S3 enhancer in vivo (Figure 3E). As nej has been previously reported to genetically interact with Dfd (Florence and McGinnis, 1998), we examined its function in Dfd/Gcm-mediated transcriptional activation. Both factors, Dfd and Gcm, are required for transcriptional activation (Stobe et al, 2009), since expression of Gcm in Drosophila D.Mel-2 cells, which have basal levels of Dfd activity (Lin et al, 2011), resulted in strong induction of reporter gene expression (Figure 3I), while abolishing Dfd binding to the rpr-4S3 HRE by mutating all Dfd binding sites (rpr4S3-DfdBSmt) (Figure 3I) or by reducing Dfd protein levels in D.Mel-2 cells using RNAi (Supplementary Figure S4B), strongly reduced reporter gene expression in the presence of Gcm. Strikingly, Dfd- and Gcm-mediated reporter gene expression was strongly reduced in nej dsRNA-treated cells, whereas inhibition of protein deacetylation by Trichostatin A (TSA; Yoshida et al, 1990) restored reporter gene expression (Figure 3J). Consistently, rpr expression was abolished in nej mutant embryos (Figure 3F and G). These results demonstrate that dCBP/Nej-mediated protein acetylation/histone modification is important for the combined activity of Dfd and Gcm on the rpr-4S3 HRE. While we could not demonstrate that Nej physically interacts with Dfd protein using various assays (data not shown), our EMSA experiments show that Nej interacts with Gcm (Figure 3H). Furthermore, we detected acetylation of transiently transfected Gcm in cultured Drosophila cells (Figure 3K; Supplementary Figure S4E). Acetylation of Gcm is dependent on Nej, as it was reduced upon RNAi-mediated downregulation of Nej (Figure 3K; Supplementary Figure S4E). Our results are consistent with published work demonstrating that in human cells CBP interacts with Gcma, resulting in its acetylation and stimulation of its transcriptional activity (Chang et al, 2005). Since about 10% of all Dfd and Nej in vivo genomic binding events during embryonic stages 10–12 overlap (Supplementary Figure S4C), the functional interaction of Dfd and Nej observed at the rpr locus does not seem an exception. This finding suggests that the interaction of co-activators (and co-repressors) with Hox proteins and close distance binding TFs on enhancer modules could be a commonly used mechanism to achieve highly specific spatio-temporal control of target gene activity. In this scenario, Hox proteins would control downstream genes by direct transcriptional and/or epigenetic regulation depending on HRE composition and thus cofactor identity and recruitment.
Dfd and Ubx exclusively interact with non-overlapping genomic regions despite similar DNA binding properties
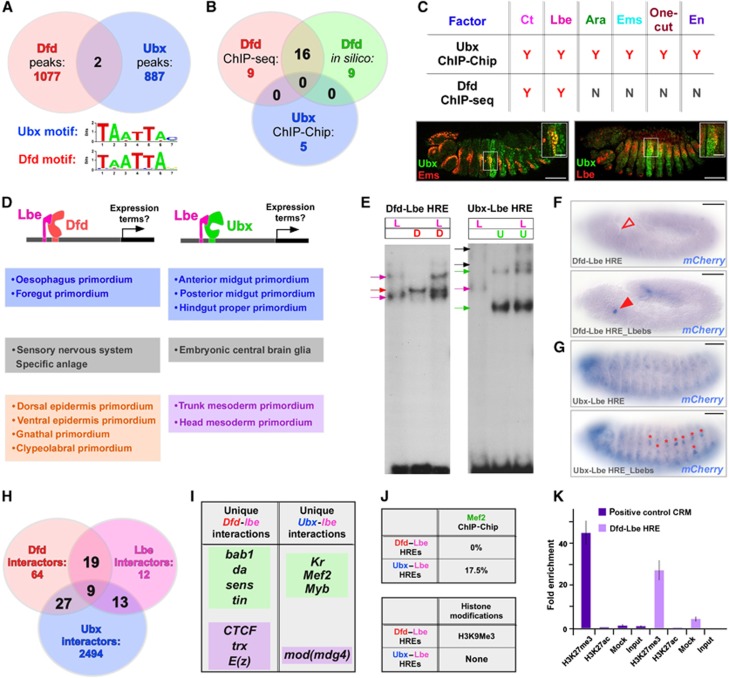
Despite very similar DNA binding behaviour in vitro (Ekker et al, 1992, 1994), Hox proteins regulate distinct morphological features along the anterior-posterior body axis in animal systems (Pearson et al, 2005). To elucidate the mechanistic basis for the differences in their regulatory properties, we compared Dfd-HREs identified in this study to genomic regions bound by the Hox TF Ultrabithorax (Ubx) at identical developmental stages, as identified by the modENCODE consortium (Roy et al, 2010). Searching for overrepresented DNA motifs in both enriched ChIP regions, we found that Dfd and Ubx bind to identical DNA sequences in vivo (Figure 4A), reminiscent to in vitro systems (Ekker et al, 1992, 1994). However, individual binding motifs seem to play only a minor role for Hox binding site selection in vivo, since our analysis revealed that Dfd and Ubx exclusively interact with non-overlapping genomic regions in embryonic stages 9–12 (Figure 4A). Consequently, Dfd- and Ubx-HREs were found to be associated with distinct classes of genes (Figure 4B), revealing that genes with roles in the epidermis are primarily under the control of Dfd at the analysed embryonic stages while genes with mesoderm-related functions are predominantly regulated by Ubx (Supplementary Table S3). Consistently, we found that the expression of tartan (trn), one of the genes associated with a Dfd-HRE, is regulated exclusively by Dfd, but not by Ubx, in epidermal cells (Supplementary Figure S7A–D), while parcas (pcs), one of the genes linked to a Ubx-HRE is under the selective control of Ubx in mesodermal cells (Supplementary Figure S7E–H). Furthermore, only Ubx-HREs were found to substantially overlap with cis-regulatory elements stage specifically bound by the mesoderm-specifying TFs Myocyte enhancer factor 2 (Mef2), Twist and Tinman (Zinzen et al, 2009; Supplementary Figure S7I). In contrast, the common ability of both Dfd and Ubx to regulate genes involved in nervous system development (Supplementary Figure S7I; Supplementary Table S3) was underlined by comparable representations of binding motifs for the neuronal-specifying TFs Asense, Deadpan and Snail in Dfd- and Ubx-HREs (Southall and Brand, 2009).
Figure 4.
Regulatory specificity of different HREs. (A) Distribution of Dfd ChIP-seq peaks (pink) identified in this study and Ubx ChIP-Chip peaks (blue) (Roy et al, 2010). Overrepresented DNA motifs identified in Ubx and Dfd ChIP-bound regions are shown at the bottom of the panel. (B) Distribution of overrepresented expression terms corresponding to genes associated with Dfd ChIP-seq peaks (pink), Dfd in silico-predicted HREs (green) and Ubx ChIP-Chip peaks (blue). (C) Top: Classification of predicted Ubx close-distance co-regulators according to the presence (Y) or absence (N) of their recognition motifs in Ubx ChIP-Chip or Dfd ChIP-seq predicted HREs. Bottom: Colocalization of Ubx (green) and Ems or Lbe (red) proteins in stage 12 embryos. Bars: 50 μm. A close-up of cells co-expressing either Ubx and Ems or Ubx and Lbe proteins is shown in the insets. Bars: 15 μm. (D) Expression terms of genes connected to Dfd-Lbe or Ubx-Lbe HREs with close-by (Δbp=0–10) distances between the respective binding sites. (E) EMSA using Lbe protein (L), Dfd protein (D) or combination of Lbe and Dfd (L and D) and a Dfd-HRE is shown on the left. The arrows indicate specific DNA-protein complexes for Lbe (pink) or Dfd (red). EMSA using Lbe protein (L), Ubx protein (U) or combination of Lbe and Ubx (L and U) and a Ubx-HRE is shown on the left. The arrows indicate specific DNA–protein complexes for Lbe (pink), Ubx (green) or Ubx/Lbe (black). (F, G) Reporter expression by a Dfd-Lbe HRE (F) and Ubx-Lbe HRE (G) with wild-type Lbe BS (upper part of each panel) or with mutated Lbe BS (Lbebs, lower part of each panel). De-repression of reporter expression in maxillary cells by the Dfd-Lbe HRE with mutated Lbe BS is highlighted by red arrowhead (F), de-repression of reporter expression in abdominal segments A1–A7 by the Ubx-Lbe HRE with mutated Lbe BS is highlighted by red asterisks. Bars: 50 μm. (H) Distribution of Dfd (orange), lbe (pink) and Ubx (blue) interactors as suggested by the DroID (Murali et al, 2011). (I) Summary of unique Dfd/lbe and Ubx/lbe interactors with function in transcriptional regulation. The green boxes highlight transcription factors, the purple boxes indicate proteins with chromatin modifying function. (J) Top: Percentage of Dfd-Lbe and Ubx-Lbe HREs overlapping with Mef2 bound genomic regions as defined by ChIP-Chip (Zinzen et al, 2009). Bottom: Percentage of Dfd-Lbe and Ubx-Lbe HREs showing and enrichment for the H3K27me3 epigenetic mark (Roy et al, 2010). (K) H3K27me3 and H3K27ac enrichment on a known Polycomb binding region (dark purple, intron 1 of AbdB) (Classen et al, 2009) and on the Dfd-Lbe HRE (light purple) in D.Mel-2 cells, as detected by ChIP. Specific enrichment of H3K27me3 and H3K27ac was assayed by quantitative real-time PCR and normalized against a negative control locus. Fold enrichment is shown in comparison to input chromatin sample (error bars, s.d.).
Motif composition and motif-pair association within Dfd- and Ubx-HREs define regulatory properties
Strikingly, we found the basic design principles of Dfd- and Ubx-HREs to be similar: like in Dfd-HREs, six binding motifs for known TFs were located adjacent to Ubx binding sites and colocalization studies showed that they are expressed in subsets of Ubx-positive cells (Figure 4C; Supplementary Figure S4B and C). Again, Ubx binding sites and motifs for potential co-regulators occurred most frequently in specific short intervals and only those Ubx-HREs with the preferred distance were associated with specific gene classes (Supplementary Figure S5). Our analysis also revealed that four of the six short-distance motifs were specific for Ubx-HREs (Figure 4C), which is consistent with our data showing that Hox proteins interact with different and spatially restricted co-regulators to control target gene expression in selected cells. Importantly, in the cases of the close-distance motifs detected in both HREs, namely the binding sites for the TFs Ladybird early (Lbe) and Cut (Ct) (Figure 4C), the associated target genes were also expressed in non-overlapping tissues (Figure 4D). This raised the question of how different Hox proteins can act on distinct target genes, even when their target HREs exhibit similar binding site compositions including short-distance arrangements. Since Lbe is active in both mesodermal and epidermal cells (Supplementary Figure S6), we exemplarily analysed one Dfd-Lbe and one Ubx-Lbe HRE and confirmed binding of Lbe protein to both HREs by EMSAs (Figure 4E), as predicted by the presence of Lbe binding sequences. Complex formation between the Hox protein and Lbe was observed in the case of Ubx and Lbe (Figure 4E, right panel) while Dfd and Lbe interact independently with the Dfd-Lbe HRE (Figure 4E, left panel), indicating that the two Hox proteins employ different mechanisms for binding to the selected HREs. Lbe interaction with the Dfd-Lbe and Ubx-Lbe HREs is essential for in vivo activity, since in both cases ectopic reporter gene expression was observed when Lbe binding sites were mutated (Figure 4F and G). Even more important, reporter gene expression was specifically changed only in segments in which either Dfd or Ubx is active, meaning in the case of the Dfd-Lbe HRE in maxillary cells and in the case of the Ubx-Lbe HRE in abdominal segments A1–A7 (Figure 4F and G). Taken together, these results demonstrate that the combined activity of Lbe and the Hox proteins Dfd or Ubx on selected HREs is critical for the precise spatiotemporal and segment-specific control of HRE activity. We next asked whether additional (DNA- and non-DNA-binding) factors contribute to the predicted cell type-specific expression of the Dfd-Lbe and Ubx-Lbe HREs (Figure 4D). Using the Drosophila Interactions Database (DroID; Murali et al, 2011) and published genome-wide DNA binding studies (Zinzen et al, 2009; Roy et al, 2010) we searched for unique Dfd-lbe and Ubx-lbe interactors and discovered that almost 20% of all Ubx-Lbe HREs but none of the Dfd-Lbe HREs were found to interact with the mesoderm-specifying factor Mef2 in vivo (Figure 4J), while H3K9me3 histone marks, which are mediated by one of the unique Dfd-lbe interactors, Enhancer of zeste E(z) (Czermin et al, 2001; Muller et al, 2002; Figure 4I), are enriched only within Dfd-Lbe HREs (Figure 4J). Interestingly, E(z) modifies chromatin also by trimethylating H3K27 residues (Tie et al, 2009), a histone mark highly enriched at the genomic region spanning the ChIP-detected Dfd-Lbe HRE (Figure 4K). Consistent with the repressive function of this histone modification (Kouzarides, 2007), loss of Lbe binding to the Dfd-Lbe HRE results in ectopic reporter gene expression (Figure 4F), suggesting that Lbe (and Dfd) recruits E(z) to the Dfd-Lbe HRE for cell type-specific target gene repression.
Taken together, these results demonstrate that Hox proteins interact with different regulatory proteins on HREs, which allows them to differentially regulate their target genes despite their similar DNA binding properties. The fact that these interactions occur only in a few cells for a short period of time is very likely one of the major reasons why the identification of factors conferring regulatory precision and specificity to Hox function has met with little success so far.
Discussion
It is well described that Hox TFs execute their regulatory function in a highly tissue-specific and context-dependent manner (Brodu et al, 2002; Lohmann and McGinnis, 2002; Lohmann et al, 2002; Merabet et al, 2005; Tour et al, 2005); however, the underlying mechanisms had been poorly understood. This is primarily due to the fact that even though regulatory Hox complexes have been identified for single HREs (Walsh and Carroll, 2007; Stobe et al, 2009), analysis of Hox enhancers at a genome-wide level has been rather limited.
In this study, we identified crucial features of HREs, which are essential for cell type-specific regulation of Hox target genes in vivo. We show that in addition to motif composition the exact spatial arrangement of TF binding elements is critical to translate Dfd function into transcriptional regulation in vivo. These architectural features of Dfd-HREs alone accurately predict target gene function and expression patterns. Furthermore, we found that epigenetic regulators bind to HREs on a genome-wide scale, suggesting that they generally collaborate with Hox proteins to achieve stable target gene regulation. This is in line with recent findings showing that chromatin modifications at enhancers strongly correlate with functional enhancer activity and tissue specificity (Heintzman et al, 2009). By comparing HREs regulated by Dfd and Ubx, two different Hox proteins with different embryonic regulatory specificities, we show that while similar design principles apply, specificity is encoded by distinct sets of co-occurring DNA motifs. Due to the highly dynamic regulatory output of Hox TFs in space and time, cell type-specific approaches are required in future to elucidate all relevant aspects of Hox-chromatin and Hox-cofactor interactions.
Materials and methods
ChIP-seq analysis
ChIP experiments were performed as described previously (Sandmann et al, 2006) using 5–9.5 h Drosophila melanogaster wild-type embryo collections and a guinea-pig anti-Dfd antibody (a gift from William McGinnis). Mock ChIP was performed using a guinea-pig anti-HRP antibody (Invitrogen) at equivalent protein concentrations as the Dfd antibody. Amplification of the rpr-4S3/3’ Dfd-HRE (positive control) and an unrelated, non-coding control locus devoid of Dfd binding sites (negative control) were analysed by quantitative real-time PCR in technical triplicates to test for enrichment of Dfd bound genomic regions. Libraries were prepared for ChIP and input samples using standard protocols for the Illumina platform, quantified using the Bioanalyzer and subjected to single-end sequencing.
Computing ChIP HREs from ChIP peaks
Immunoprecipitated DNA fragments were sequenced and aligned to the reference Drosophila genome (version 5.29; www.flybase.org) using Bowtie (Langmead, 2010) with parameter settings allowing a maximum of three mismatches and suppressing all non-unique reads. Peak calling using Model-based Analysis of ChIP-seq (MACS; Feng et al, 2011) resulted in the identification of 1079 enrichment peaks with a P-value threshold of 0.0001. Due to the fact that functional enhancers show a high degree of conservation, we defined Dfd-HREs by extending the ChIP-seq peaks taking into account the conservation and the presence of Dfd binding sites in the neighbouring genomic region. At each step, the peaks were extended 60 bp on each side and the extension was terminated when the average conservation of the 60 bp window was <0.5 and when Dfd BS were absent. In all, 389 out of the 1079 peaks (36%) were extended with the mean and median of extension being 143.8 and 120, respectively. The UCSC Genome Browser Database (Kent et al, 2002) was used to determine the location of ChIP-seq peaks in intergenic regions, introns or exons.
In silico prediction of Dfd-HREs
Stage-independent Dfd-HREs were identified by searching for clusters of Dfd binding motifs in the non-coding regions of the genomes of 12 distinct Drosophila species using the Cis-Analyst algorithm (Berman et al, 2004), as described previously (Hueber et al, 2007). A PFM based on DNaseI footprint data (Bergman et al, 2005) as well as consensus sequences from the literature was used for identifying Dfd binding sites. The P-value threshold, window length and binding sites in a cluster were identified using a training set of validated Dfd response elements (Hueber et al, 2007) and conserved clusters were identified by NCBI BLASTN.
De-novo motif search
MDScan was used to perform de-novo motif search in the identified ChIP-seq peaks (Liu et al, 2002). Genomic regions adjacent to the ChIP-seq peaks were used to define background sequence. ‘TCGTAAT’ and ‘TAATTAG’ were reported as top ranking motifs.
Pair-wise distance distribution
To investigate the pair-wise distance preferences between Dfd or Ubx and other DNA motifs, known TF binding sequences were first identified within all Dfd- or Ubx-HREs by using the method of Wasserman and Sandelin (2004) and specific Position Weight Matrices (http://jaspar.genereg.net/). A permutation test was performed to assess the significance of the frequency deviation between observed and expected distribution.
In-situ hybridization and immunohistochemistry
In-situ hybridization and immunohistochemistry experiments were performed with minor modifications as described in Tautz and Pfeifle (1989) and Patel (1994). Digoxigenin- and Biotin-labelled antisense RNA probes were made using the Roche RNA labeling system (Roche). Fluorescent mRNA/protein double labelling and fluorescent duplex in-situ hybridizations were done as described previously (Kosman et al, 2004). For probe detection, the TSA Plus TMR and Fluorescein systems from Perkin-Elmer (Waltham, MA) were used. Confocal images were taken at a Nikon A1Rsi microscope. ImageJ (NIH) and Photoshop CS (Adobe) software were used for image analysis.
Antibodies used for immunohistochemistry and immunoblotting
The following primary antibodies were used: rabbit anti-DsRed (1:200, Clontech), guinea pig anti-Dfd (1:500, W McGinnis), mouse anti-FasIII 7G10 (1:50, DSHB), mouse anti-Ct 2B10 (1:100, DSHB), mouse anti-En 4D9 (1:5, DSHB), rat anti-Ems (1:50, U Walldorf; Walldorf and Gehring, 1992), rabbit anti-Ubx (1:100, Santa Cruz), rabbit anti-Optix (1:250, F Pignoni (Kenyon et al, 2005)), mouse anti-Lbe (1:20, C Jagla; Jagla et al, 1997), rabbit anti-Nej/dCBP (A Mazo; Petruk et al, 2001), mouse anti-AcK (Abcam), anti-DIG-POD (1:200, Roche) and anti-DIG-AP (1:1000, Roche). Detection of HRP-coupled secondary antibodies was done using the Vectastain ABC Kit (Vector Laboratories).
Cell culture
D.Mel-2 cells were maintained in Express Five Serum Free Medium (Gibco, Invitrogen) at 25°C. dsRNA synthesis by in vitro transcription of a PCR generated template and dsRNA treatment of cultured Drosophila cells was performed as previously described (Worby et al, 2001). For Dfd or Nej knockdown, dsRNA was added to the cells twice in 7 days and three times in 9 days, respectively. After dsRNA treatment, the cells were transfected using Effectene (Qiagen) according to manufacturer’s instructions for suspension cells and were harvested 48 h after transfection. Trichostatin A (Sigma-Aldrich) was added 24 h after transfection, at a concentration of 100 ng/ml, 200 ng/ml (real-time PCR experiments) or 150 ng/ml (acetylation of Gcm experiment). For immunoblotting using Drosophila cell lysates, the equivalent of 105 cells was loaded on a 10% polyacrylamide (PAA) gel.
ChIP for modified histones using D.Mel-2 cells
ChIP was performed as previously described (Weinmann and Farnham, 2002) using D.Mel-2 cells and rabbit anti-H3K27me3 (Millipore), rabbit anti-H3K27ac (Abcam) and with rabbit IgG (Abcam) (mock control) at equivalent protein concentrations. Quantitative real-time PCR analysis was performed in technical triplicates to test for enrichment of H3K27me3 and H3K27ac bound genomic regions. A known Polycomb target region (intron 1 of AbdB) and an unrelated, non-Polycomb target control locus (Hsp68) (Classen et al, 2009) were used as positive and negative control, respectively. Input chromatin was used as a reference for calculating enrichment in the immunoprecipitated samples.
Quantitative real-time PCR
Total RNA was isolated from transfected D.Mel-2 cells using the RNeasy kit (Qiagen) and digested by DNase (Fermentas) to remove genomic DNA. First-strand cDNA synthesis was performed using the RevertAidTM First Strand cDNA Synthesis Kit (Fermentas). Real-time PCRs were performed using Platinum SYBR Green (Invitrogen) and an ABI StepOnePlus System (Applied Biosystems). All reactions were performed in triplicates (three technical replicates) and the results were analysed using the DDCt method (Livak and Schmittgen, 2001). Reporter values were normalized to Renilla Luciferase expression in order to account for variations in transfection efficiency. Relative expression levels are based on three biological replicates. Error bars throughout the manuscript represent standard deviation (s.d.) of three biological replicates.
Plasmid constructs
The ChIP-seq-identified and in silico-predicted HREs were amplified from genomic DNA. PCR products were cloned into the pENTR/D-TOPO vector (Invitrogen) and either swapped into the pHP-DEST-mCherry destination vector (Boy et al, 2010) or into the pBPGUwmCherry destination vector (unpublished) using the LR Clonase Enzyme Mix (Invitrogen). The Dfd binding sites of the ChIP-seq-identified prd enhancer were mutated from AATTA to CAGGA and from ATTA to AGGA, respectively, by a two-step overlap PCR. The resulting PCR product with mutated Dfd binding sites was cloned into the destination vector pBPGUwmCherry via Gateway cloning as described above, and used for cell culture experiments. The rpr-4S3 enhancer was amplified from genomic DNA of Oregon R flies. The rpr4S3-5 bp enhancer and the rpr4S3-10 bp enhancer were generated by a two-step overlap PCR to insert additional nucleotides. The resulting PCR products were cloned into the destination vector pBPGUwmCherry via Gateway cloning as described above. All oligonucleotide sequences are available upon request.
Generation of transgenic flies
The constructs osp_E/pHPdestmCherry, ia2_E/pHPdestmCherry, CG10283_E/ pHPdestmCherry, Dll_E/pHPdestmCherry and CG5756_E/pGreenH-Pelican were used to generate transgenic reporter fly lines by random P-element transformation. All other constructs were used to generate transgenic reporter lines by the PhiC31 site-specific transgenesis system using either the y1 w67c23; P{CaryP}attP2 (with M{vas-int.Dm}ZH-2A) landing site on the third chromosome (68A4) (Bloomington Stock Center) or the y1 w67c23; P{CaryP}attP40 (with M{vas-int.Dm}ZH-2A) landing site on the second chromosome (25C7) (Bloomington Stock Center). All transgenic fly lines were generated by the BestGene Drosophila Embryo Injection Service (USA).
Protein purification
N-terminal maltose-binding protein (MBP) fusions of Drosophila Dfd, Gcm, Lbe, Optix, Ubx and Nej were expressed in E. coli BL21(DE3) cells using the pMAL-c2x vector. The part of Nej protein including the bromodomain and histone acetyltransferase domain (amino acids 1705–2283) was used for protein expression. Protein expression was induced with 0.1 mM IPTG for 2–3 h at 20°C. Cells were harvested, resuspended in column buffer (20 mM Tris–HCl pH 7.4, 200 mM NaCl, 1 mM EDTA, 1 mM DTT) and lysed by direct sonication (Misonix Sonicator S-4000; 2 min process time, 5 s pulses). The soluble fraction was applied to an Amylose resin and bound proteins were eluted with column buffer supplemented with 20 mM maltose. All fractions were analysed by denaturing SDS–PAGE to monitor the efficiency of induction and purification.
Electrophoretic mobility shift assays
EMSA was performed as previously described (Lohmann et al, 2002). Complementary oligonucleotides were annealed by heating to 95°C and slowly cooling down to room temperature. Annealed oligonucleotides were radioactively end labelled with 32P-[γ]-ATP using T4 Polynucleotide Kinase (Fermentas) in the forward reaction, following manufacturer’s instructions. Subsequently, labelled oligonucleotides were purified on a native, 5% PAA gel and eluted with elution buffer at 37°C for 1 h. Oligonucleotides and purified proteins were incubated for 1 h at 4°C in binding buffer (50 mM Tris–HCl pH 8.0, 750 mM KCl, 2.5 mM EDTA, 0.5% Triton X-100, 1 mM DTT, 40% glycerol) to allow complex formation. All oligonucleotide sequences can be obtained upon request.
Supplementary Material
Acknowledgments
We thank C Jagla, J Kumar, A Mazo, H Nguyen, F Pignoni, U Walldorf, the Bloomington Stock Center and the Developmental Studies Hybridoma Bank at the University of Iowa for providing material; the Nikon Imaging Center for microscopy; and Jan U Lohmann, Alexis Maizel and Jochen Wittbrodt for critically reading the manuscript.
Author contributions: SS: design, acquisition, analysis and interpretation of data; NH: design, acquisition, analysis and interpretation of data; MP: design, acquisition, analysis and interpretation of data, drafting and writing the article; JF and DB: design, acquisition, analysis and interpretation of data; PK, MS, SO and SRH: acquisition and analysis of data; JM and JR: acquisition of data; FP: analysis of data; IL: design, analysis and interpretation of data, drafting and writing the article.
Footnotes
The authors declare that they have no conflict of interest.
References
- Akimaru H, Chen Y, Dai P, Hou DX, Nonaka M, Smolik SM, Armstrong S, Goodman RH, Ishii S (1997) Drosophila CBP is a co-activator of cubitus interruptus in hedgehog signalling. Nature 386: 735–738 [DOI] [PubMed] [Google Scholar]
- Arnone MI, Davidson EH (1997) The hardwiring of development: organization and function of genomic regulatory systems. Development 124: 1851–1864 [DOI] [PubMed] [Google Scholar]
- Bergman CM, Carlson JW, Celniker SE (2005) Drosophila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics 21: 1747–1749 [DOI] [PubMed] [Google Scholar]
- Berman BP, Pfeiffer BD, Laverty TR, Salzberg SL, Rubin GM, Eisen MB, Celniker SE (2004) Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol 5: R61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boy AL, Zhai Z, Habring-Muller A, Kussler-Schneider Y, Kaspar P, Lohmann I (2010) Vectors for efficient and high-throughput construction of fluorescent Drosophila reporters using the PhiC31 site-specific integration system. Genesis 48: 452–456 [DOI] [PubMed] [Google Scholar]
- Brodu V, Elstob PR, Gould AP (2002) Abdominal A specifies one cell type in Drosophila by regulating one principal target gene. Development 129: 2957–2963 [DOI] [PubMed] [Google Scholar]
- Chan SK, Jaffe L, Capovilla M, Botas J, Mann RS (1994) The DNA binding specificity of Ultrabithorax is modulated by cooperative interactions with extradenticle, another homeoprotein. Cell 78: 603–615 [DOI] [PubMed] [Google Scholar]
- Chang CW, Chuang HC, Yu C, Yao TP, Chen H (2005) Stimulation of GCMa transcriptional activity by cyclic AMP/protein kinase A signaling is attributed to CBP-mediated acetylation of GCMa. Mol Cell Biol 25: 8401–8414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Guo L, Fan Z, Jiang T (2007) Learning position weight matrices from sequence and expression data. Comput Syst Bioinformatics Conf 6: 249–260 [PubMed] [Google Scholar]
- Classen AK, Bunker BD, Harvey KF, Vaccari T, Bilder D (2009) A tumor suppressor activity of Drosophila Polycomb genes mediated by JAK-STAT signaling. Nat Genet 41: 1150–1155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coiffier D, Charroux B, Kerridge S (2008) Common functions of central and posterior Hox genes for the repression of head in the trunk of Drosophila. Development 135: 291–300 [DOI] [PubMed] [Google Scholar]
- Czermin B, Schotta G, Hulsmann BB, Brehm A, Becker PB, Reuter G, Imhof A (2001) Physical and functional association of SU(VAR)3-9 and HDAC1 in Drosophila. EMBO Rep 2: 915–919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker SC, Jackson DG, von Kessler DP, Sun BI, Young KE, Beachy PA (1994) The degree of variation in DNA sequence recognition among four Drosophila homeotic proteins. EMBO J 13: 3551–3560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekker SC, von Kessler DP, Beachy PA (1992) Differential DNA sequence recognition is a determinant of specificity in homeotic gene action. EMBO J 11: 4059–4072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Zhang Y (2011) Using MACS to identify peaks from ChIP-Seq data. Curr Protoc Bioinformatics Chapter 2: Unit 2, 14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florence B, McGinnis W (1998) A genetic screen of the Drosophila X chromosome for mutations that modify Deformed function. Genetics 150: 1497–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazi A, VijayRaghavan KV (2000) Developmental biology. Control by combinatorial codes. Nature 408: 419–420 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, Ching KA, Antosiewicz-Bourget JE, Liu H, Zhang X, Green RD, Lobanenkov VV, Stewart R, Thomson JA, Crawford GE, Kellis M et al. (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hueber SD, Bezdan D, Henz SR, Blank M, Wu H, Lohmann I (2007) Comparative analysis of Hox downstream genes in Drosophila. Development 134: 381–392 [DOI] [PubMed] [Google Scholar]
- Jagla K, Jagla T, Heitzler P, Dretzen G, Bellard F, Bellard M (1997) ladybird, a tandem of homeobox genes that maintain late wingless expression in terminal and dorsal epidermis of the Drosophila embryo. Development 124: 91–100 [DOI] [PubMed] [Google Scholar]
- Joshi R, Sun L, Mann R (2010) Dissecting the functional specificities of two Hox proteins. Genes Dev 24: 1533–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalkhoven E (2004) CBP and p300: HATs for different occasions. Biochem Pharmacol 68: 1145–1155 [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D (2002) The human genome browser at UCSC. Genome Res 12: 996–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon KL, Li DJ, Clouser C, Tran S, Pignoni F (2005) Fly SIX-type homeodomain proteins Sine oculis and Optix partner with different cofactors during eye development. Dev Dyn 234: 497–504 [DOI] [PubMed] [Google Scholar]
- Kosman D, Mizutani CM, Lemons D, Cox WG, McGinnis W, Bier E (2004) Multiplex detection of RNA expression in Drosophila embryos. Science 305: 846. [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Kuziora MA, McGinnis W (1988) Autoregulation of a Drosophila homeotic selector gene. Cell 55: 477–485 [DOI] [PubMed] [Google Scholar]
- Langmead B (2010) Aligning short sequencing reads with Bowtie. Curr Protoc Bioinformatics Chapter 11: Unit 11, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin GG, Kozaki T, Scott JG (2011) Hormone receptor-like in 96 and Broad-Complex modulate phenobarbital induced transcription of cytochrome P450 CYP6D1 in Drosophila S2 cells. Insect Mol Biol 20: 87–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS (2002) An algorithm for finding protein-DNA binding sites with applications to chromatin-immunoprecipitation microarray experiments. Nat Biotechnol 20: 835–839 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lohmann I, McGinnis N, Bodmer M, McGinnis W (2002) The Drosophila Hox gene deformed sculpts head morphology via direct regulation of the apoptosis activator reaper. Cell 110: 457–466 [DOI] [PubMed] [Google Scholar]
- Lohmann I, McGinnis W (2002) Hox Genes: it's all a matter of context. Curr Biol 12: R514–R516 [DOI] [PubMed] [Google Scholar]
- Makeev VJ, Lifanov AP, Nazina AG, Papatsenko DA (2003) Distance preferences in the arrangement of binding motifs and hierarchical levels in organization of transcription regulatory information. Nucleic Acids Res 31: 6016–6026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann RS, Carroll SB (2002) Molecular mechanisms of selector gene function and evolution. Curr Opin Genet Dev 12: 592–600 [DOI] [PubMed] [Google Scholar]
- McGinnis W, Krumlauf R (1992) Homeobox genes and axial patterning. Cell 68: 283–302 [DOI] [PubMed] [Google Scholar]
- Merabet S, Pradel J, Graba Y (2005) Getting a molecular grasp on Hox contextual activity. Trends Genet 21: 477–480 [DOI] [PubMed] [Google Scholar]
- Muller J, Hart CM, Francis NJ, Vargas ML, Sengupta A, Wild B, Miller EL, O'Connor MB, Kingston RE, Simon JA (2002) Histone methyltransferase activity of a Drosophila Polycomb group repressor complex. Cell 111: 197–208 [DOI] [PubMed] [Google Scholar]
- Murali T, Pacifico S, Yu J, Guest S, Roberts GG 3rd, Finley RL Jr (2011) DroID 2011: a comprehensive, integrated resource for protein, transcription factor, RNA and gene interactions for Drosophila. Nucleic Acids Res 39: D736–D743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel NH (1994) Imaging neuronal subsets and other cell types in whole-mount Drosophila embryos and larvae using antibody probes. Methods Cell Biol 44: 445–487 [DOI] [PubMed] [Google Scholar]
- Pearson JC, Lemons D, McGinnis W (2005) Modulating Hox gene functions during animal body patterning. Nat Rev Genet 6: 893–904 [DOI] [PubMed] [Google Scholar]
- Pennacchio LA, Loots GG, Nobrega MA, Ovcharenko I (2007) Predicting tissue-specific enhancers in the human genome. Genome Res 17: 201–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, Canaani E, Croce CM, Mazo A (2001) Trithorax and dCBP acting in a complex to maintain expression of a homeotic gene. Science 294: 1331–1334 [DOI] [PubMed] [Google Scholar]
- Regulski M, McGinnis N, Chadwick R, McGinnis W (1987) Developmental and molecular analysis of Deformed; a homeotic gene controlling Drosophila head development. EMBO J 6: 767–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ernst J, Kharchenko PV, Kheradpour P, Negre N, Eaton ML, Landolin JM, Bristow CA, Ma L, Lin MF, Washietl S, Arshinoff BI, Ay F, Meyer PE, Robine N, Washington NL, Di Stefano L, Berezikov E, Brown CD, Candeias R et al. (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandmann T, Jakobsen JS, Furlong EE (2006) ChIP-on-chip protocol for genome-wide analysis of transcription factor binding in Drosophila melanogaster embryos. Nat Protoc 1: 2839–2855 [DOI] [PubMed] [Google Scholar]
- Southall TD, Brand AH (2009) Neural stem cell transcriptional networks highlight genes essential for nervous system development. EMBO J 28: 3799–3807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stobe P, Stein MA, Habring-Muller A, Bezdan D, Fuchs AL, Hueber SD, Wu H, Lohmann I (2009) Multifactorial regulation of a hox target gene. PLoS Genet 5: e1000412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D, Pfeifle C (1989) A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98: 81–85 [DOI] [PubMed] [Google Scholar]
- Tie F, Banerjee R, Stratton CA, Prasad-Sinha J, Stepanik V, Zlobin A, Diaz MO, Scacheri PC, Harte PJ (2009) CBP-mediated acetylation of histone H3 lysine 27 antagonizes Drosophila Polycomb silencing. Development 136: 3131–3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tour E, Hittinger CT, McGinnis W (2005) Evolutionarily conserved domains required for activation and repression functions of the Drosophila Hox protein Ultrabithorax. Development 132: 5271–5281 [DOI] [PubMed] [Google Scholar]
- Visel A, Blow MJ, Li Z, Zhang T, Akiyama JA, Holt A, Plajzer-Frick I, Shoukry M, Wright C, Chen F, Afzal V, Ren B, Rubin EM, Pennacchio LA (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457: 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walldorf U, Gehring WJ (1992) Empty spiracles, a gap gene containing a homeobox involved in Drosophila head development. EMBO J 11: 2247–2259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CM, Carroll SB (2007) Collaboration between Smads and a Hox protein in target gene repression. Development 134: 3585–3592 [DOI] [PubMed] [Google Scholar]
- Wasserman WW, Sandelin A (2004) Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 5: 276–287 [DOI] [PubMed] [Google Scholar]
- Weinmann AS, Farnham PJ (2002) Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 26: 37–47 [DOI] [PubMed] [Google Scholar]
- Worby CA, Simonson-Leff N, Dixon JE (2001) RNA interference of gene expression (RNAi) in cultured Drosophila cells. Sci STKE 2001: pl1. [DOI] [PubMed] [Google Scholar]
- Yoshida M, Kijima M, Akita M, Beppu T (1990) Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 265: 17174–17179 [PubMed] [Google Scholar]
- Zinzen RP, Girardot C, Gagneur J, Braun M, Furlong EE (2009) Combinatorial binding predicts spatio-temporal cis-regulatory activity. Nature 462: 65–70 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.