Abstract
In the eukaryotic transcriptome, both the numbers of genes and different RNA species produced by each gene contribute to the overall complexity. These RNA species are generated by the utilization of different transcriptional initiation or termination sites, or more commonly, from different messenger RNA (mRNA) splicing events. Among the 30 000+ genes in human genome, it is estimated that more than 95% of them can generate more than one gene product via alternative RNA splicing. The protein products generated from different RNA splicing variants can have different intracellular localization, activity, or tissue-distribution. Therefore, alternative RNA splicing is an important molecular process that contributes to the overall complexity of the genome and the functional specificity and diversity among different cell types. In this review, we will discuss current efforts to unravel the full complexity of the cardiac transcriptome using a deep-sequencing approach, and highlight the potential of this technology to uncover the global impact of RNA splicing on the transcriptome during development and diseases of the heart.
Keywords: Alternative RNA splicing, Transcriptome, Gene regulation, Heart, RNA-seq
1. Alternative RNA splicing
The complexity of the eukaryotic transcriptome was first fully revealed at the genome scale with single-base resolution by a powerful deep RNA-sequencing (RNA-seq) technology, using next generation sequencers and newly developed bioinformatic tools (Bland et al., 2010; Hallegger et al., 2010). It is estimated that transcripts from ~95% of multi-exon genes undergo alternative splicing and that there are ~100 000 intermediate to high abundance alternative splicing events in major human tissues (Pan et al., 2008). RNA splicing is a ubiquitous post-transcriptional process in all eukaryotes. It involves removing intronic sequences from pre-messenger RNA (pre-mRNA) and linking exons to generate mature mRNA for translation (Chen and Manley, 2009). RNA splicing for constitutively spliced exons is carried out by a defined molecular machinery involving cis-acting regulatory sequences (splice sites) located at exon-intron boundaries, as well as trans-acting factors as part of the spliceosome (de la Grange et al., 2010). However, in many cases, the splice sites are altered, leading to different exon sizes in the final transcripts. Alternatively, certain exons can be differentially included or excluded in the final transcripts due to exon skipping. These non-constitutive RNA splicing activities are collectively called alternative RNA splicing. Through these different kinds of alternative pre-mRNA processing, individual eukaryotic genes can produce multiple mRNA and protein isoforms that may have related, distinct or even opposing functions (Wang et al., 2008; Buljan et al., 2012). Therefore, alternative RNA splicing is an important molecular step that contributes to the total complexity of the transcriptome and proteome.
2. Regulation of alternative RNA splicin
Alternative RNA splicing is a highly regulated process mediated by cis-regulatory enhancers and silencers in pre-mRNA and trans-acting splicing factors, including heterogeneous nuclear ribonucleoprotein (hnRNP) and serine-arginine rich proteins (SR proteins). The molecular nature of these regulatory elements is yet to be fully uncovered and understood. Tissue specific alternative splicing is usually regulated by a combination of tissue-specific and ubiquitously expressed splicing factors (Pan et al., 2008; Sultan et al., 2008; Chen and Manley, 2009; Bland et al., 2010) and has been demonstrated to play an important role in regulation of tissue-specific protein interaction networks (Buljan et al., 2012). In addition, mis-regulated alternative RNA splicing events have a significant role in human diseases, cell cycle, and cell death (Hallegger et al., 2010; Gang et al., 2011; Honda et al., 2012; Raghavachari, 2012; Yae et al., 2012). For example, a specific isoform of pyruvate kinase resulting from hnRNP-mediated mRNA alternative splicing is required for tumor cell proliferation (David et al., 2010).
3. Alternative RNA splicing in cardiac diseases
It is well established that alternative splicing of mRNA is tightly associated with the development of heart failure. Structural proteins, such as cardiac troponin T, or important signaling molecules, such as Ca2+/calmodulin-dependent protein kinase (CaM kinase), are subjected to alternative splicing in heart diseases (Ramchatesingh et al., 1995; Ding et al., 2004; Xu et al., 2005). Moreover, depletion of critical splicing regulators, including SC35 and RBM20, has been found to cause dilated cardiomyopathy in mouse and rat (Ding et al., 2004; Guo et al., 2012; Linke and Bucker, 2012; Refaat et al., 2012). In addition to classic SR and hnRNP proteins, CUG-BP1 and ETR-like factors (CELF)/Bruno-like family of RNA binding proteins and muscleblind-like proteins (MBNL proteins) have also been found to regulate both cardiac development and function (Warf and Berglund, 2007; Kalsotra et al., 2010; Koshelev et al., 2010; Dasgupta and Ladd, 2012). Therefore, alternative RNA splicing is essential for normal cardiac function and mis-regulated RNA splicing may have an important role in the pathogenesis of heart failure. Yet, little knowledge is available about the scope of alternative splicing at the whole genome level in normal and diseased hearts and even less about the mechanisms underlying the regulation of mRNA splicing in response to pathological injury in the heart. Recent studies have begun to fill this critical gap of information by establishing the total transcriptome, including RNA splicing variants, in normal and diseased hearts using RNA-seq and extensive bioinformatic, molecular, cellular, and functional analyses (Fig. 1).
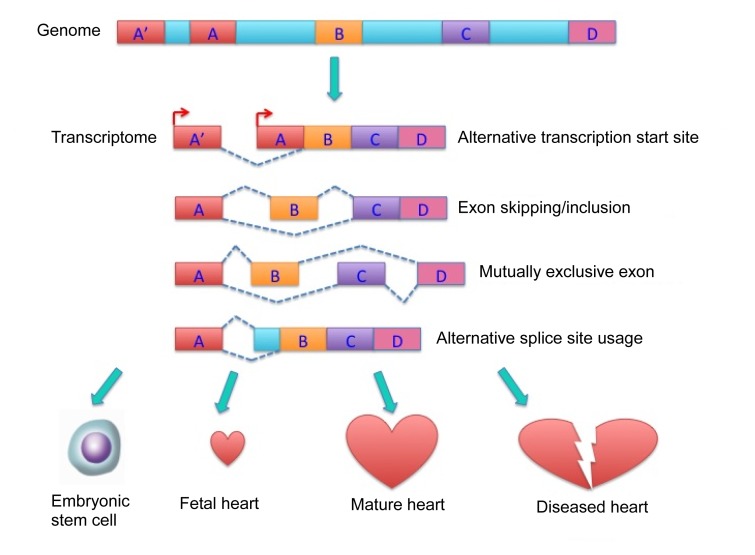
Fig. 1.
Alternative RNA splicing contributing to the overall complexity of the cardiac transcriptome during cardiomyocyte differentiation, cardiac development, and response to pathological stress
4. Experimental approaches for total cardiac transcriptome analysis
The main tools necessary for total transcriptome studies include mRNA-seq using next generation high-throughput sequencing technology, exon assembly using bioinformatic tools, and the validation of exon boundaries and expression by quantitative reverse-transcription polymerase chain reaction (qRT-PCR), urea-polyacrylamide gel electrophoresis (UREA-PAGE), and capillary electrophoresis.
RNA-seq is a powerful high throughput sequencing technology involving the generation of a quantitative, genome scale, and single base resolution profile of the transcriptome (Anders et al., 2012; Li et al., 2012; Sanchez-Pla et al., 2012). The details of RNA-seq technology can be found in a recent review (Wang et al., 2009). In general, the mRNA is enriched from a sample of interest followed by the construction of a complementary DNA (cDNA) library using standard reverse-transcription methods. High throughput sequencing is performed using one of several technological platforms, including the illumina genome analyzer, Applied Biosystems (ABI) solid sequencing, and life science’s 454 sequencing. This is an area of rapid improvement where speed, fidelity, and read lengths are increasing dramatically while the overall cost/base is dropping sharply. High throughput sequencing has become a routine method for scientific discovery and advanced clinical diagnosis. Its widespread application has already revolutionized our experimental approaches where visualizing global changes and regulation in gene expression and transcriptome remodeling have become a reality. Indeed, RNA-seq has for the first time made it feasible to catalogue and appreciate the genome wide landscape of the whole transcriptome at single base resolution.
The output of the RNA-seq method is hundreds of millions of RNA sequence reads of about 70 to 100 bases in length. Linking these short reads into a contiguous transcript relies on a sophisticated computational algorithm. In general, the program first needs to map the reads on a particular exon based on matching sequences between the reads and genomic sequences, and then the exon-exon boundary is mapped based on a predicated cDNA database. Finally, all reads associated with a particular gene are combined to generate the total reads for each exon. Therefore, the final profile of each gene contains total reads of each exon at single base resolution. These mapping processes are complicated by a number of issues, including ultra-large datasets and limitations in computational power, repetitive sequences in closely related genes (miss-matching), incomplete genomic databases, sequencing errors, and sensitivity vs. fidelity (Mcintyre et al., 2011; Ozsolak and Milos, 2011). Therefore, to generate a comprehensive expression profile for each exon, it is essential to perform the RNA-seq at sufficient depth. One major advantage of RNA-seq over micro-arrays is the possibility to identify novel, un-annotated exons or transcripts (Daines et al., 2011; Lee et al., 2011; Concha et al., 2012). In a recent study, we have developed two bioinformatic tools, guided transcriptome reconstruction and de novo reconstruction. These tools allow detection of novel exons in known genes and novel transcript clusters (NTCs) (Lee et al., 2011).
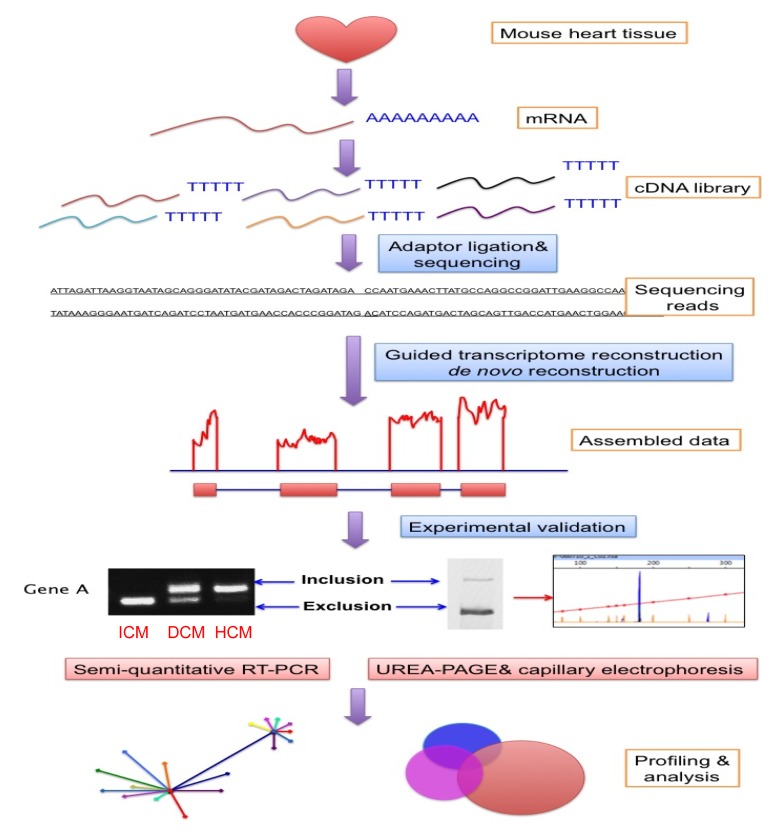
The quantification and specificity of identified known or novel exons should be validated by independent methods, including qRT-PCR. Different transcripts resulting from alternative RNA splicing can be separated by UREA-PAGE and capillary electrophoresis based on size differences. Given the fact that the reads generated from RNA-seq are assembled based on computational analysis, an experimental validation of the findings is always necessary. Indeed, we have identified and confirmed a significant number of differentially expressed exons in normal and diseased hearts by both fluorescent RT-PCR followed by UREA-PAGE and capillary electrophoresis, which show a very high correlation with the bioinformatics prediction (Fig. 2).
Fig. 2.
Workflow of deep RNA-seq on profiling the cardiac transcriptome
ICM: ischemic cardiomyopathy; DCM: dilated cardiomyopathy; HCM: hypertrophic cardiomyopathy
5. Global profiling of alternative RNA splicing in the heart: novel exons
In this study, deep RNA-seq was performed on mRNA samples prepared from adult mouse hearts in basal condition and in failing state as a result of chronic pressure-overload induced by trans-aortic constriction. This is a well-established model system to investigate the pathogenesis of cardiac hypertrophy and heart failure due to mechanical overload, mimicking chronic hypertension in humans.
Among the mRNA transcripts annotated from the RNA-seq data, more than 1 000 novel exons were identified that had not been reported in any published databases. From a selected list of 40 novel exons, 38 (95%) were validated by RT-PCR in mouse heart tissue and all of them were further confirmed by direct DNA sequencing to have the predicted novel exon-exon junctions. The genes containing these novel exons included established regulators in cardiac signaling, mitochondria dynamics, and gene regulation. Many of the novel exons are predicted to have major functional impacts on the parent genes, including mRNA stability, protein truncation, protein activity, and post-translation modification. Using semi-qRT-PCR in human heart failure samples, some of these novel exons showed differential expression patterns in normal or diseased hearts, strongly suggesting that these novel exons may have a functional role in the disease. Therefore, deep RNA-seq revealed a significant number of novel transcripts, which contribute to overall transcriptome complexity in the heart.
6. Differential alternative splicing as a part of global transcriptome remodeling in developing and failing hearts
The onset of heart failure is associated with a significant change in both the quantity and quality of the cardiac transcriptome (Barry et al., 2008; Margulies et al., 2009). Most studies profiling the cardiac transcriptome have employed microarray approaches, which revealed global changes in gene expression in diseased hearts (Asakura and Kitakaze, 2009; Dewey et al., 2011). The features of transcriptome remodeling and the underlying regulatory mechanisms have been the focus of extensive investigation, leading to the identification of a network of responsible transcription factors and co-factors, including myocyte enhancer factor-2 (MEF-2), GATA, and histone deacetylases (HDACs) (Edmondson et al., 1994; Skerjanc et al., 1998; Naya et al., 2002; Backs and Olson, 2006). However, the global transcriptome profile of the alternatively spliced exons in cardiac development and disease remains to be established. RNA-seq analyses of cardiac transcriptome throughout heart development and disease progression hold great promise to address this question. Considering the potential impact of differentially expressed exons on protein function and regulation, alternative RNA splicing may emerge to be an important element in the underlying molecular mechanisms of cardiac lineage commitment, maturation, physiological or pathological responses to stresses. Studies on alternative RNA splicing events, the regulators and the functional consequences at the genome level will open a new frontier for us to explore the fundamental mechanisms of heart disease and potential therapeutic intervention.
Footnotes
Project supported partially by Broad Stem Cell Research Center (BSCRC) Pre-doctoral Fellowship in UCLA, and the National Institutes of Health, USA
References
- 1.Anders S, Reyes A, Huber W. Detecting differential usage of exons from RNA-seq data. Genome Res. 2012 doi: 10.1101/gr.133744.111. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Asakura M, Kitakaze M. Global gene expression profiling in the failing myocardium. Circ J. 2009;73(9):1568–1576. doi: 10.1253/circj.cj-09-0465. [DOI] [PubMed] [Google Scholar]
- 3.Backs J, Olson EN. Control of cardiac growth by histone acetylation/deacetylation. Circ Res. 2006;98(1):15–24. doi: 10.1161/01.RES.0000197782.21444.8f. [DOI] [PubMed] [Google Scholar]
- 4.Barry SP, Davidson SM, Townsend PA. Molecular regulation of cardiac hypertrophy; Int J Biochem Cell Biol; 2008. pp. 2023–2039. [DOI] [PubMed] [Google Scholar]
- 5.Bland CS, Wang ET, Vu A, David MP, Castle JC, Johnson JM, Burge CB, Cooper TA. Global regulation of alternative splicing during myogenic differentiation. Nucleic Acids Res. 2010;38(21):7651–7664. doi: 10.1093/nar/gkq614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buljan M, Chalancon G, Eustermann S, Wagner GP, Fuxreiter M, Bateman A, Babu MM. Tissue-specific splicing of disordered segments that embed binding motifs rewires protein interaction networks; Mol Cell; 2012. pp. 871–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat Rev Mol Cell Biol. 2009;10(11):741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Concha M, Wang X, Cao S, Baddoo M, Fewell C, Lin Z, Hulme W, Hedges D, Mcbride J, Flemington EK. Identification of new viral genes and transcript isoforms during epstein-barr virus reactivation using RNA-seq; J Virol; 2012. pp. 1458–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daines B, Wang H, Wang L, Li Y, Han Y, Emmert D, Gelbart W, Wang X, Li W, Gibbs R, et al. The drosophila melanogaster transcriptome by paired-end RNA sequencing. Genome Res. 2011;21(2):315–324. doi: 10.1101/gr.107854.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dasgupta T, Ladd AN. The importance of celf control: molecular and biological roles of the CUG-BP, Elav-like family of RNA-binding proteins. Wiley Interdiscip Rev RNA. 2012;3(1):104–121. doi: 10.1002/wrna.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.David CJ, Chen M, Assanah M, Canoll P, Manley JL. hnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463(7279):364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de la Grange P, Gratadou L, Delord M, Dutertre M, Auboeuf D. Splicing factor and exon profiling across human tissues; Nucleic Acids Res; 2010. pp. 2825–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dewey FE, Perez MV, Wheeler MT, Watt C, Spin J, Langfelder P, Horvath S, Hannenhalli S, Cappola TP, Ashley EA. Gene coexpression network topology of cardiac development, hypertrophy and failure. Circ Cardiovasc Genet. 2011;4(1):26–35. doi: 10.1161/CIRCGENETICS.110.941757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ding JH, Xu X, Yang D, Chu PH, Dalton ND, Ye Z, Yeakley JM, Cheng H, Xiao RP, Ross J, et al. Dilated cardiomyopathy caused by tissue-specific ablation of SC35 in the heart; EMBO J; 2004. pp. 885–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Edmondson DG, Lyons GE, Martin JF, Olson EN. Mef2 gene expression marks the cardiac and skeletal muscle lineages during mouse embryogenesis. Development. 1994;120(5):1251–1263. doi: 10.1242/dev.120.5.1251. [DOI] [PubMed] [Google Scholar]
- 16.Gang H, Hai Y, Dhingra R, Gordon JW, Yurkova N, Aviv Y, Li H, Aguilar F, Marshall A, Leygue E, et al. A novel hypoxia-inducible spliced variant of mitochondrial death gene Bnip3 promotes survival of ventricular myocytes. Circ Res. 2011;108(9):1084–1092. doi: 10.1161/circresaha.110.238709. [DOI] [PubMed] [Google Scholar]
- 17.Guo W, Schafer S, Greaser ML, Radke MH, Liss M, Govindarajan T, Maatz H, Schulz H, Li S, Parrish AM, et al. RBM20, a gene for hereditary cardiomyopathy, regulates titin splicing. Nat Med. 2012;18(5):766–773. doi: 10.1038/nm.2693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hallegger M, Llorian M, Smith CWJ. Alternative splicing: global insights. FEBS J. 2010;277(4):856–866. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- 19.Honda A, Valogne Y, Bou Nader M, Brechot C, Faivre J. An intron-retaining splice variant of human cyclin A2, expressed in adult differentiated tissues, induces a g1/s cell cycle arrest in vitro. PLoS One. 2012;7(6):e39249. doi: 10.1371/journal.pone.0039249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalsotra A, Wang K, Li PF, Cooper TA. MicroRNAs coordinate an alternative splicing network during mouse postnatal heart development. Genes Dev. 2010;24(7):653–658. doi: 10.1101/gad.1894310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Koshelev M, Sarma S, Price RE, Wehrens XHT, Cooper TA. Heart-specific overexpression of CUGBP1 reproduces functional and molecular abnormalities of myotonic dystrophy type 1. Hum Mol Genet. 2010;19(6):1066–1075. doi: 10.1093/hmg/ddp570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee JH, Gao C, Peng G, Greer C, Ren S, Wang Y, Wang X. Analysis of transcriptome complexity through RNA sequencing in normal and failing murine hearts. Circ Res. 2011;109(12):1332–1341. doi: 10.1161/circresaha.111.249433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li G, Bahn JH, Lee JH, Peng G, Chen Z, Nelson SE, Xiao X. Identification of allele-specific alternative mRNA processing via transcriptome sequencing. Nucleic Acids Res. 2012 doi: 10.1093/nar/gks280. in press [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linke WA, Bucker S. King of hearts: a splicing factor rules cardiac proteins. Nat Med. 2012;18(5):660–661. doi: 10.1038/nm.2762. [DOI] [PubMed] [Google Scholar]
- 25.Margulies KB, Bednarik DP, Dries DL. Genomics, transcriptional profiling, and heart failure. J Am Coll Cardiol. 2009;53(19):1752–1759. doi: 10.1016/j.jacc.2008.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mcintyre L, Lopiano K, Morse A, Amin V, Oberg A, Young L, Nuzhdin S. RNA-seq: technical variability and sampling. BMC Genomics. 2011;12(1):293. doi: 10.1186/1471-2164-12-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naya FJ, Black BL, Wu H, Bassel-Duby R, Richardson JA, Hill JA, Olson EN. Mitochondrial deficiency and cardiac sudden death in mice lacking the MEF2A transcription factor. Nat Med. 2002;8(11):1303–1309. doi: 10.1038/nm789. [DOI] [PubMed] [Google Scholar]
- 28.Ozsolak F, Milos PM. RNA sequencing: advances, challenges and opportunities. Nat Rev Genet. 2011;12(2):87–98. doi: 10.1038/nrg2934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat Genet. 2008;40(12):1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 30.Raghavachari N, Barb J, Yang Y, Liu P, Woodhouse K, Levy D, O′Donnell C, Munson PJ, Kato G. A systematic comparison and evaluation of high density exon arrays and RNA-seq technology used to unravel the peripheral blood transcriptome of sickle cell disease. BMC Med Genomics. 2012;5(1):28. doi: 10.1186/1755-8794-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ramchatesingh J, Zahler A, Neugebauer K, Roth M, Cooper T. A subset of SR proteins activates splicing of the cardiac troponin T alternative exon by direct interactions with an exonic enhancer. Mol Cell Biol. 1995;15(9):4898–4907. doi: 10.1128/mcb.15.9.4898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Refaat MM, Lubitz SA, Makino S, Islam Z, Frangiskakis JM, Mehdi H, Gutmann R, Zhang ML, Bloom HL, Macrae CA, et al. Genetic variation in the alternative splicing regulator RBM20 is associated with dilated cardiomyopathy. Heart Rhythm. 2012;9(3):390–396. doi: 10.1016/j.hrthm.2011.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sanchez-Pla A, Reverter F, Ruiz de Villa MC, Comabella M. Transcriptomics: mRNA and alternative splicing. J Neuroimmunol. 2012;248(1-2):23–31. doi: 10.1016/j.jneuroim.2012.04.008. [DOI] [PubMed] [Google Scholar]
- 34.Skerjanc IS, Petropoulos H, Ridgeway AG, Wilton S. Myocyte enhancer factor 2C and Nkx2-5 up-regulate each other’s expression and initiate cardiomyogenesis in P19 cells. J Biol Chem. 1998;273(52):34904–34910. doi: 10.1074/jbc.273.52.34904. [DOI] [PubMed] [Google Scholar]
- 35.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, et al. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321(5891):956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 36.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang Z, Gerstein M, Snyder M. RNA-seq: a revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10(1):57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warf MB, Berglund JA. MBNL binds similar RNA structures in the cug repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13(12):2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu X, Yang D, Ding JH, Wang W, Chu PH, Dalton ND, Wang HY, Bermingham JR, Jr, Ye Z, Liu F, et al. ASF/SF2-regulated CAMKIIδ alternative splicing temporally reprograms excitation-contraction coupling in cardiac muscle. Cell. 2005;120(1):59–72. doi: 10.1016/j.cell.2004.11.036. [DOI] [PubMed] [Google Scholar]
- 40.Yae T, Tsuchihashi K, Ishimoto T, Motohara T, Yoshikawa M, Yoshida GJ, Wada T, Masuko T, Mogushi K, Tanaka H, et al. Alternative splicing of CD44 mRNA by ESRP1 enhances lung colonization of metastatic cancer cell. Nat Commun. 2012;3:883. doi: 10.1038/ncomms1892. [DOI] [PubMed] [Google Scholar]