Abstract
During development, angiogenesis occurs as a controlled series of events leading to neovascularization that supports changing tissue requirements. Several pro- and antiangiogenic factors orchestrate a complex, dynamic process to allow initial sprouting and invasion, subsequent pruning and remodeling, and finally maturation and survival of blood vessels. In the last decade, a new class of small RNA molecules termed micro-RNAs (miRs) have emerged as key regulators of several cellular processes including angiogenesis. Micro-RNAs such as miR-132, miR-126, miR-296, miR-145, and miR-92a have been shown to play pro- and antiangiogenic roles in the vasculature of both endothelial cells and perivascular cells. However, in pathological situations such as cancer or inflammation, the same angiogenic signaling pathways and miRs are dysregulated and exploited, typically resulting in poorly organized vessels with leaky and tortuous properties. This article is a brief overview of specific miRs that have been reported to play a role in the vasculature. The authors explore emerging principles that suggest miRs insulate cellular processes from external perturbations and provide robustness to biological systems in the context of angiogenesis.
Keywords: miR-132, angiogenic switch, endothelial quiescence
Introduction
An adult human has about 1 to 10 × 1012 endothelial cells lining the blood vessels with only about 1 in 10,000 undergoing cell cycle at any given time.1,2 This fraction can dramatically increase during specific physiological requirements such as wound healing and tissue repair or pathological conditions such as tumorigenesis. The quiescent endothelium resumes an active proliferation program that results in growth of new blood vessels governed by a complex milieu of growth factors and signaling networks to generate new blood vessels in a process broadly termed angiogenesis. Some of these proliferation programs and signaling networks are fundamental processes that shape the development of vasculature in the embryo and are reactivated and rewired during pathological neovascularization.3 Although pro- and antiangiogenic receptors and signaling networks have been well characterized, the molecular mechanisms that enable the endothelial cells to reverse their quiescence and reenter the cell cycle rapidly remain poorly understood. In the past decade or so, it has become increasingly clear that small noncoding RNAs, particularly micro-RNAs (miRs), in the genome rapidly respond to stimuli and facilitate physiological processes including angiogenesis. Emerging evidence points to miRs as “agents of robustness” that help organisms and cells maintain their phenotypes by buffering against variations both internal and external. This review will focus on a few examples from endothelial biology in which the role of miRs seems to provide a critical threshold to facilitate or inhibit angiogenic responses.
A Brief Primer on miRs
miRs are small 18- to 25-nucleotide sequences most often found in the intronic or intergenic regions. They are transcribed by RNA polymerase II, processed into pri-miRs by an RNAse Type III enzyme Drosha, and exported out of the nucleus by exportin-5 and then cleaved by another RNAse Dicer.4 In a process that is not entirely clear, one of the strands of the miR duplex selectively assembles an miR-induced silencing complex (miRISC) involving members of the Argonaute protein family (AGO). The miR strand then binds to target mRNA’s 3′ untranslated regions (UTRs), often forming contiguous base-pairing in the “seed sequence” between nucleotides 2 and 10 of the miR and partial base- pairing with other nucleotides of the miR. This triggers the recruitment of a host of other proteins that can modify RNA molecules, and the target mRNA is deadenylated, decapped, and degraded by exonucleases. In general, it is thought that miR-mediated repression of translation contributes to a decrease in protein output that often correlates with the decrease in mRNA levels of the targets.5
Global Loss of miRs Affects Development and Function of the Vasculature
The role of miRs in cardiovascular development was immediately apparent with the generation of Dicer hypomorphic mice. These mice had severe vascular deformation in the embryo and yolk sac and died at E12.5-E14.5.6 The knockdown of both maternal and zygotic Dicer in zebrafish also resulted in pericardial edema and lack of proper circulation.7 Suarez et al 8 reported that the knockdown of Dicer in human endothelial cells affected several critical target genes important for endothelial function, suggesting that miR dysregulation impairs endothelial biology. Subsequently, selective ablation of Dicer in mouse endothelium also led to significant defects in postnatal angiogenesis in response to a variety of stimuli such as growth factors, ischemia, and wound healing.9 Recently, the same group has shown that deletion of Dicer in smooth muscle cells results in late embryonic lethality associated with extensive internal hemorrhage.10 Inducible, smooth muscle cell specific deletion of Dicer in mice postnatally led to a significant loss of contractile function, vascular contractile function, smooth muscle cell (SMC) differentiation, and vascular remodeling.11 Taken together, the profound phenotypes seen upon loss of Dicer in the vascular compartment suggest that miRs regulate fundamental processes that shape both structure and function of the vasculature.
Specific miRs Implicated in Angiogenesis
Several excellent reviews have comprehensively addressed the different miRs that play a role in angiogenesis and cardiovascular development.12,13 Therefore, we will provide a few illustrative examples of specific miRs and present a broad evolutionary perspective on the emergence of miRs as an adaptation to insulate cardiovascular systems from external stress.
miR-1/miR-133 in Cardiovascular Development
miR-1 and miR-133 were among the first miRs to be characterized as regulators of muscle proliferation and differentiation both in cardiac and skeletal muscles.14 Zhao et al 15 showed that overexpression of miR-1 in mice results in developmental lethality with cardiovascular failure due to the dysregulation of the transcription factor Hand2. The same group followed up with loss-of-function studies in miR-1-2 knockout mice, showing that miR-1 was critical for cardiogenesis, cardiac conduction, and cell cycle.16 Similarly, miR-133, which is co-transcribed with miR-1, has been shown to be responsible for regulating cardiomyocyte hypertrophy through regulation of target genes such as RhoA and Cdc42.17 Interestingly, both these miRs have been shown to regulate muscle gene expression in zebrafish models,18,19 with miR-133 particularly involved as a negative regulator of appendage regeneration.
miR-143/miR-145 in Vascular Smooth Muscle Biology
Several groups have shown miR-143 and miR-145 to play fundamental roles in vascular smooth muscle cell function.20-23 These miRs are co-transcribed in response to serum response factor and target a network of transcription factors including Kruppel-like factor (KLF) genes, Elk1, and cytoskeletal proteins to regulate the proliferation, differentiation, contraction, and migration of smooth muscle cells.24 Although mice without either miR-145 or both miR-143 and miR-145 show normal development, they have significantly less neointima formation after vascular injury. Also, these mice have a decrease in blood pressure and enhanced development of atherosclerotic lesions. Taken together, these observations point to the role of miR-143/145 in smooth muscle cell responses to insults and injuries in the vasculature.
miR-126 as a Regulator of VEGF Signaling in the Endothelium
Targeted deletion of miR-126 causes a loss of vascular integrity in mice and zebrafish during development and leads to defective angiogenesis.25 Mice that survive the loss of miR-126 during development show defective neovascularization following myocardial infarction. It has been shown that miR-126 inhibits sprout-related protein SPRED1, a negative inhibitor of VEGF signaling.26 Recent work has shown that blood flow–induced upregulation of miR-126 by a mechanosensitive transcription factor Klf2 in endothelial cells activated VEGF signaling pathways and led to sprouting and remodeling of the aortic arch in developing zebrafish.27 These studies highlight a unique role for an miR in translating physical or mechanical stress into a biological response.
miR-132 as a Genomic “First Responder” to Activation
miR-132 is encoded on human chromosome 17 and is transcribed by the transcription factor cAMP-response element binding protein (CREB) in multiple cell types.28 We recently characterized miR-132 as a key regulator of pathological neovascularization that affects endothelial activation (Fig. 1) by downregulation of p120RasGAP.29 Angiogenic growth factors such as VEGF, bFGF, and conditioned media from a variety of tumors lead to phosphorylation of CREB and rapid transcription of miR-132 that peaks about 3 to 6 hours after activation. Using gain and loss-of-function studies, we showed that miR-132 affected endothelial proliferation, tube formation in vitro, and both developmental and pathological angiogenesis in vivo. We were able to demonstrate that there is a reciprocal relati onship between miR-132 and p120RasGAP during vascular hyperproliferation, including hemangiomas and tumor angiogenesis. Lagos et al 30 reported that miR-132 is upregulated with similar kinetics and plays a key role in lymphatic endothelial cells during viral infections. Our observations highlight that miR-132 not only is among the early response genes in endothelial activation but also is a critical regulator of the downstream events that control endothelial proliferation, tube formation, and angiogenesis in vivo.
Figure 1.
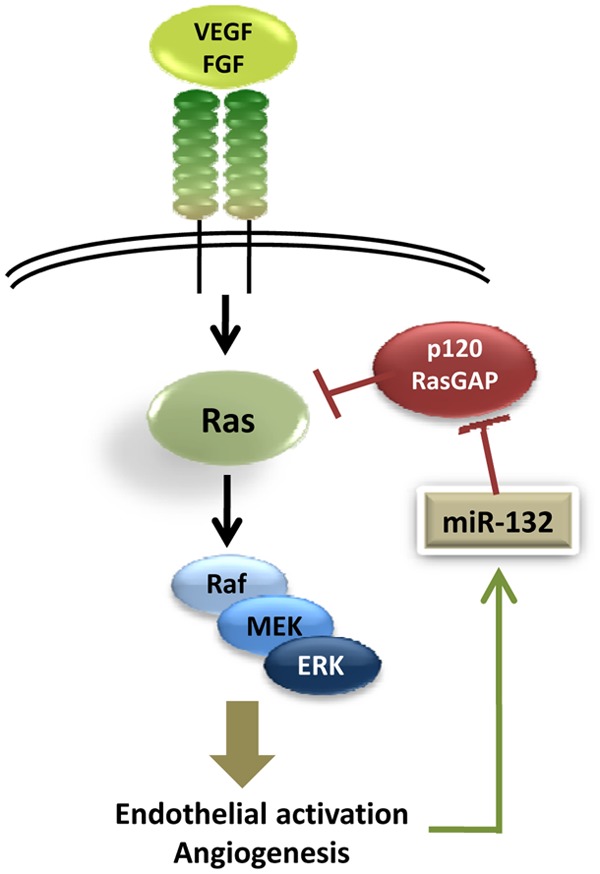
Micro-RNA (miR)-132/p120RasGAP as an angiogenic switch. miR-132 is upreg- ulated by growth factor signaling in endothelial cells and downregulates p120RasGAP levels. This in turn stabilizes active Ras and amplifies endothelial activation leading to angiogenesis. VEGF = vascular endothelial growth factor; FGF = fibroblast growth factor; MEK = MAP kinase or ERK kinase; ERK = extracellular signal-regulated kinase.
miRs That Affect Angiogenic Signaling Cascades Indirectly
miR-296 has been shown to be induced in brain endothelial cells during glioma progression.31 miR-296 directly decreased the levels of hepatocyte-growth factor regulated tyrosine kinase substrate (HGS) and indirectly affected the levels of VEGFR2 and PDGFRβ by altering their trafficking to the cell surface. Hypoxia has been shown to trigger miR-424 expression, which leads to degradation of an ubiquitin ligase scaffold protein cullin-2. Degradation of cullin-2 prevents HIF-1α downregulation and facilitates a cascade of proangiogenic signaling.32
miR-17~92 Cluster as Intrinsic Antiangiogenic miRs in the Endothelium
miR-17~92 is a well-characterized group of miRs that are transcribed as a single polycistronic RNA. Several studies have reported that this cluster of miRs functions as an oncogene and drives key physiological responses during development and disease.33 Bonauer et al 34 first reported that miR-17~92 cluster is highly expressed in human endothelial cells and plays a critical role as a negative regulator of angiogenesis, partly through targeting the integrin subunit α5. Those investigators also demonstrated that the loss of miR-92a improved functional recovery after myocardial infarction and limb ischemia by enhancing blood vessel growth. miR-17 has been proposed as another key antiangiogenic miR in endothelial cells putatively through its targeting of Janus kinase1 (JAK1).35
This representative list of miRs that regulate different aspects of endothelial/smooth muscle growth and differentiation highlights the biological complexity and perhaps redundancy in the myriad network of factors that shape the structure and function of blood vessels. Although the presence of pro- and antiangiogenic factors has been appreciated for decades, it is becoming clear that miRs provide a unique layer of checks and balances because of their ability to alter biological responses both on a broader and subtler scale.
miRs as Canalization Factors during Development
Decades ago, C.H. Waddington36 introduced the term canalization of development to describe genetic robustness/buffering, where the phenotype of an organism is protected from external perturbations. Over these decades, we have acquired a much better understanding of what specific mechanisms confer genetic robustness during development and disease.37,38 Also, recent work has considerably expanded the repertoire of perturbations that cells and organisms experience. For instance, the process of angiogenesis is susceptible to various external stimuli such as oxygen concentrations, growth factor gradients, and the presence of inhibitors of angiogenesis. There are also internal fluctuations in cells that contribute to the variance, such as transcriptional activity, gene dosage, and leaky transcription. An endothelial cell needs to assess these diverse inputs and respond appropriately by staying quiescent or set off a cascade of programs triggering proliferation, migration, and tube formation to form a new blood vessel. Multiple intrinsic mechanisms facilitate this decision making in the endothelial cells. First, there are feedback and feedforward loops (e.g., Notch pathways) that set a threshold for endothelial activation. Second, multiple signaling networks and integrin pathways (e.g., crosstalk between VEGFR and integrinβ3) regulate angiogenesis, providing a degree of redundancy that ensures responses occur only in the right biological contexts. Third, genetic and posttranslational mechanisms provide an additional layer of immunity, preventing small changes at the genetic level influencing the fate of the quiescent endothelium (e.g., methylation of promoters of endogenous inhibitors of angiogenesis). In addition to these mechanisms, recent evidence indicates that miRs provide robustness and buffering in several processes including angiogenesis.39 It can be argued that miRs can either decrease the residual activation to strengthen endothelial quiescence in a coherent feedforward loop or ensure the appropriate activation by restricting the response to a certain range of growth factor/nutrient/oxygen concentration in an incoherent feedforward loop. These highly coordinated networks are often disrupted or diverted to promote hyperproliferative responses during pathological angiogenesis.
Some of the early evidence for the role of miRs in canalization of development came from studies in Drosophila. miR-1, described earlier as a critical regulator of cardiovascular development, is expendable for normal muscle differentiation in Drosophila but is critical during the stressful “rapid growth” phase.40 The loss of miR-1 appears to destabilize the differentiation program, making it vulnerable to external perturbations. Similarly, miR-7 functions in a complex Notch-EGFR feedback and feedforward loop to insulate Drosophila photoreceptor determination from temperature fluctuations.41 More recently, Staton et al 42 demonstrated that migration of germ cells in zebrafish was buffered from fluctuations of SDF1 chemokine levels by miR-430. Our observations with miR-132 regulation of p120RasGAP suggest the possibility that miR-132 might act as a buffer by setting the threshold for growth factor mediated activation of endothelial cells.
miRs as Modulators of Morphological Complexity during Evolution
The circulatory system is one of the first organs to develop in an embryo so that organogenesis can be sustained with the delivery of oxygen and nutrients. During early development, blood vessels arise de novo from endothelial precursor cells (angioblasts) that form primitive capillary networks in a process broadly termed vasculogenesis.43 These early capillaries can then sprout and branch into a capillary network in a process often referred to as angiogenesis.44 The need to deliver nutrients and oxygen in complex multicellular organisms that had increasingly dense tissue architecture drove the evolution of vasculature. Interestingly, some features of vertebrate angiogenesis such as tube formation and sprouting are also seen in insect trachea.45 In fact, VEGF orthologs have been identified in Drosophila, where they play a critical role in blood cell migration.46 The vertebrate circulatory system gained a degree of complexity by the development of a specialized tubular system consisting of endothelial cells, adding a smooth muscle component to the vascular wall and innervation of the muscle tissue to place the vascular tone under the control of the sympathetic nervous system. Because the acquisition of such complexity might coincide with the evolution of canalization factors, it has been postulated that miRs are active modulators of morphological complexity. Indeed, miR genes are continuously being added to metazoan genomes, whereas there appears to be no discernible loss of miR genes.47 Some of the miRs that we described earlier have also been discovered in ancient species. For instance, miR-132 has been discovered in sea lampreys and sharks.48 miR-1/miR-133 cluster has been identified as a co-transcribed miR in deuterostomes.49 Christodoulou et al 50 recently explored this in more detail and reported that miR evolution and establishment of tissue identities were closely interlinked in bilaterian evolution. Therefore, it is plausible that miRs such as miR-132 were selected during the course of evolution as modulators of morphogenesis. Not surprisingly, mammalian vascular and nervous systems, given their common morphological traits and signaling modules,51 also share common miR regulators including miR-132.28,52
Pathological Angiogenesis Recapitulates Developmental Angiogenesis
Pathological neovascularization shares fundamental mechanisms with developmental angiogenesis at multiple levels, including receptor signaling cascades, migration, invasion, proliferation, and tube formation.53 A developing tumor begins to secrete angiogenic factors partly in response to hypoxia. This leads to the activation of quiescent endothelial cells that proliferate, migrate, and establish a robust capillary network. However, in contrast to normal vasculature, pathological neovascularization in tumors results in leaky immature vessels with poor pericyte coverage. This phenomenon of a small dormant tumor acquiring a vascular network has been historically referred to as the angiogenic switch.54 The molecular mechanisms that drive tumor angiogenesis now include a host of miRs that act on different cell types and pathways but in the end facilitate the establishment of a robust vascular network.55 Tumors or inflammatory stimuli such as cytokines significantly influence angiogenesis by transcribing proangiogenic miRs and inhibiting antiangiogenic miRs in the endothelium, a process that overcomes the canalization of angiogenesis. Therefore, it is likely that restoration of antiangiogenic miRs or blockade of proangiogenic miRs might potentially function in a synergistic manner with other therapies by resetting the activation threshold of the endothelium.
Conclusions
Since the discovery of miRs, we have accumulated a large body of knowledge about their transcription, processing, and expression in tissues and organisms during development and disease. Target prediction programs and high-throughput genomics/proteomics methods to identify miR targets are being refined. Ongoing efforts are focused on understanding the functions of different miRs in a context-dependent manner. Although miRs represent a unique, potentially exciting therapeutic agent for targeting angiogenesis, uncovering the biological role of miRs holds the clues to understanding the equilibrium between quiescence and activation of the endothelium. Such understanding will ultimately benefit and inform the development of miR therapeutic agents for treatment of pathological neovascularization across several human diseases, including cancer.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: Work in the Cheresh laboratory is funded by NCI and NHLBI. S.A. is supported by a postdoctoral fellowship award (09POST2040038) by American Heart Association.
References
- 1. Engerman RL, Pfaffenbach D, Davis MD. Cell turnover of capillaries. Lab Invest. 1967; 17(6):738-43 [PubMed] [Google Scholar]
- 2. Hobson B, Denekamp J. Endothelial proliferation in tumours and normal tissues: continuous labelling studies. Br J Cancer. 1984;49(4):405-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473(7347):298-307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116(2): 281-97 [DOI] [PubMed] [Google Scholar]
- 5. Huntzinger E, Izaurralde E. Gene silencing by microRNAs: contributions of translational repression and mRNA decay. Nat Rev Genet. 2011;12(2):99-110 [DOI] [PubMed] [Google Scholar]
- 6. Yang WJ, Yang DD, Na S, Sandusky GE, Zhang Q, Zhao G. Dicer is required for embryonic angiogenesis during mouse development. J Biol Chem. 2005;280(10):9330-5 [DOI] [PubMed] [Google Scholar]
- 7. Giraldez AJ, Cinalli RM, Glasner ME, et al. MicroRNAs regulate brain morphogenesis in zebrafish. Science. 2005;308(5723):833-8 [DOI] [PubMed] [Google Scholar]
- 8. Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100(8):1164-73 [DOI] [PubMed] [Google Scholar]
- 9. Suarez Y, Fernandez-Hernando C, Yu J, et al. Dicer-dependent endothelial microRNAs are necessary for postnatal angiogenesis. Proc Natl Acad Sci U S A. 2008;105(37):14082-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Albinsson S, Suarez Y, Skoura A, Offermanns S, Miano JM, Sessa WC. MicroRNAs are necessary for vascular smooth muscle growth, differentiation, and function. Arterioscler Thromb Vasc Biol. 2010;30(6):1118-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Albinsson S, Skoura A, Yu J, et al. Smooth muscle miRNAs are critical for post-natal regulation of blood pressure and vascular function. PLoS ONE. 2011;6(4):e18869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang S, Olson EN. Angiomirs—key regulators of angiogenesis. Curr Opin Genet Dev. 2009;19(3):205-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bonauer A, Boon RA, Dimmeler S. Vascular microRNAs. Curr Drug Targets. 2010;11(8):943-9 [DOI] [PubMed] [Google Scholar]
- 14. Chen JF, Mandel EM, Thomson JM, et al. The role of microRNAs-1 and microRNAs-133 in skeletal muscle proliferation and differentiation. Nat Genet. 2006;38(2):228-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zhao Y, Samal E, Srivastava D. Serum response factor regulates a muscle-specific microRNAs that targets hand2 during cardiogenesis. Nature. 2005;436(7048):214-20 [DOI] [PubMed] [Google Scholar]
- 16. Zhao Y, Ransom JF, Li A, et al. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking miRNAs-1-2. Cell. 2007;129(2):303-17 [DOI] [PubMed] [Google Scholar]
- 17. Care A, Catalucci D, Felicetti F, et al. MicroRNAs-133 controls cardiac hypertrophy. Nat Med. 2007;13(5):613-8 [DOI] [PubMed] [Google Scholar]
- 18. Mishima Y, Abreu-Goodger C, Staton AA, et al. Zebrafish Mir-1 and Mir-133 shape muscle gene expression and regulate sarcomeric actin organization. Genes Dev. 2009;23(5):619-32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yin VP, Thomson JM, Thummel R, Hyde DR, Hammond SM, Poss KD. Fgf-dependent depletion of microRNAs-133 promotes appendage regeneration in zebrafish. Genes Dev. 2008;22(6):728-33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boettger T, Beetz N, Kostin S, et al. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119(9):2634-47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cordes KR, Sheehy NT, White MP, et al. Mir-145 and Mir-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Elia L, Quintavalle M, Zhang J, et al. The knockout of Mir-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: correlates with human disease. Cell Death Differ. 2009;16(12):1590-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Xin M, Small EM, Sutherland LB, et al. MicroRNAs Mir-143 and Mir-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23(18):2166-78 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rangrez AY, Massy ZA, Metzinger-Le Meuth V, Metzinger L. Mir-143 and Mir-145: molecular keys to switch the phenotype of vascular smooth muscle cells. Circ Cardiovasc Genet. 2011;4(2):197-205 [DOI] [PubMed] [Google Scholar]
- 25. Fish JE, Santoro MM, Morton SU, et al. Mir-126 regulates angiogenic signaling and vascular integrity. Dev Cell. 2008;15(2):272-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wang S, Aurora AB, Johnson BA, et al. The endothelial-specific microRNAs Mir-126 governs vascular integrity and angiogenesis. Dev Cell. 2008;15(2):261-71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nicoli S, Standley C, Walker P, Hurlstone A, Fogarty KE, Lawson ND. MicroRNAs-mediated integration of haemodynamics and vegf signalling during angiogenesis. Nature. 2010;464(7292):1196-200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vo N, Klein ME, Varlamova O, et al. A camp-response element binding protein-induced microRNAs regulates neuronal morphogenesis. Proc Natl Acad Sci U S A. 2005;102(45):16426-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Anand S, Majeti BK, Acevedo LM, et al. MicroRNAs-132-mediated loss of p120rasgap activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16(8):909-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lagos D, Pollara G, Henderson S, et al. Mir-132 regulates antiviral innate immunity through suppression of the p300 transcriptional co-activator. Nat Cell Biol. 2010;12(5):513-9 [DOI] [PubMed] [Google Scholar]
- 31. Wurdinger T, Tannous BA, Saydam O, et al. Mir-296 regulates growth factor receptor overexpression in angiogenic endothelial cells. Cancer Cell. 2008;14(5):382-93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ghosh G, Subramanian IV, Adhikari N, et al. Hypoxia-induced microRNAs-424 expression in human endothelial cells regulates Hif-alpha isoforms and promotes angiogenesis. J Clin Invest. 120(11):4141-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mendell JT. Myriad roles for the Mir-17-92 cluster in development and disease. Cell. 2008;133(2):217-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bonauer A, Carmona G, Iwasaki M, et al. MicroRNAs-92a controls angiogenesis and functional recovery of ischemic tissues in mice. Science. 2009;324(5935):1710-3 [DOI] [PubMed] [Google Scholar]
- 35. Doebele C, Bonauer A, Fischer A, et al. Members of the microRNAs-17-92 cluster exhibit a cell-intrinsic antiangiogenic function in endothelial cells. Blood. 2010;115(23):4944-50 [DOI] [PubMed] [Google Scholar]
- 36. Waddington CH. Canalization of development and genetic assimilation of acquired characters. Nature. 1959;183(4676):1654-5 [DOI] [PubMed] [Google Scholar]
- 37. Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467(7312):167-73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kitano H. Biological robustness. Nat Rev Genet. 2004;5(11):826-37 [DOI] [PubMed] [Google Scholar]
- 39. Hornstein E, Shomron N. Canalization of development by microRNAs. Nat Genet. 2006;38 Suppl:S20-4 [DOI] [PubMed] [Google Scholar]
- 40. Sokol NS, Ambros V. Mesodermally expressed drosophila microRNAs-1 is regulated by twist and is required in muscles during larval growth. Genes Dev. 2005;19(19):2343-54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Li X, Cassidy JJ, Reinke CA, Fischboeck S, Carthew RW. A microRNAs imparts robustness against environmental fluctuation during development. Cell. 2009;137(2):273-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Staton AA, Knaut H, Giraldez AJ. MiRNAs regulation of Sdf1 chemokine signaling provides genetic robustness to germ cell migration. Nat Genet. 2011;43(3):204-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Risau W, Flamme I. Vasculogenesis. Annu Rev Cell Dev Biol. 1995;11:73-91 [DOI] [PubMed] [Google Scholar]
- 44. Carmeliet P. Mechanisms of angiogenesis and arteriogenesis. Nat Med. 2000;6(4):389-95 [DOI] [PubMed] [Google Scholar]
- 45. Lubarsky B, Krasnow MA. Tube morphogenesis: making and shaping biological tubes. Cell. 2003;112(1):19-28 [DOI] [PubMed] [Google Scholar]
- 46. Cho NK, Keyes L, Johnson E, et al. Developmental control of blood cell migration by the Drosophila VEGF pathway. Cell. 2002;108(6):865-76 [DOI] [PubMed] [Google Scholar]
- 47. Wheeler BM, Heimberg AM, Moy VN, et al. The deep evolution of metazoan microRNAs. Evol Dev. 2009;11(1):50-68 [DOI] [PubMed] [Google Scholar]
- 48. Heimberg AM, Sempere LF, Moy VN, Donoghue PC, Peterson KJ. MicroRNAs and the advent of vertebrate morphological complexity. Proc Natl Acad Sci U S A. 2008;105(8):2946-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Campo-Paysaa F, Semon M, Cameron RA, Peterson KJ, Schubert M. MicroRNAs complements in deuterostomes: origin and evolution of microRNAs. Evol Dev. 2011;13(1):15-27 [DOI] [PubMed] [Google Scholar]
- 50. Christodoulou F, Raible F, Tomer R, et al. Ancient animal microRNAs and the evolution of tissue identity. Nature. 2010;463(7284):1084-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Tam SJ, Watts RJ. Connecting vascular and nervous system development: angiogenesis and the blood-brain barrier. Annu Rev Neurosci. 2010;33:379-408 [DOI] [PubMed] [Google Scholar]
- 52. Magill ST, Cambronne XA, Luikart BW, et al. MicroRNAs-132 regulates dendritic growth and arborization of newborn neurons in the adult hippocampus. Proc Natl Acad Sci U S A. 2010;107(47):20382-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Plate KH, Breier G, Risau W. Molecular mechanisms of developmental and tumor angiogenesis. Brain Pathol. 1994;4(3):207-18 [DOI] [PubMed] [Google Scholar]
- 54. Bergers G, Benjamin LE. Tumorigenesis and the angiogenic switch. Nat Rev Cancer. 2003; 3(6):401-10 [DOI] [PubMed] [Google Scholar]
- 55. Fish JE, Srivastava D. MicroRNAs: opening a new vein in angiogenesis research. Sci Signal. 2009;2(52):pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]


