Abstract
Initial studies linking Sirtuins to longevity in yeast initiated what is now a rich vein of aging research that is full of promise and fraught with controversy. Missing was a demonstration that enhanced Sirtuin expression extends lifespan in mammals. Now Kanfi et al. provide the evidence but with an interesting plot twist – the lesser known SIRT6 is the longevity factor.
While Sirtuins, a highly conserved class of protein deacetylases, have been linked to longevity in a variety of model organisms and are among the most studied proteins in the context of aging, their role remains controversial. Increased expression of the founding member of the class, yeast SIR2, was reported to extend replicative lifespan over a decade ago and this was followed by reports that overexpression of SIR2 orthologs leads to enhanced longevity in worms and flies 1, 2. More recent reports have called the worm and fly data into question 3, 4 and while increased Sir2 activity may enhance longevity under certain conditions in all three organisms, mechanistic studies have not converged on a specific activity coupled to aging.
Seven mammalian Sir2 homologs (SIRT1-SIRT7) have been identified. A myriad of activities in a range of mammalian tissues have been attributed to SIRT1, a protein most structurally related to yeast Sir2, often linked to protection from the onset of chronic disease states. Yet transgenic mice with increased SIRT1 expression are not long-lived 5, further adding to what is becoming a great science mystery. Until recently, while SIRT1 was being intensely studied, the other Sirtuins were mostly waiting in the wings. In the last few years, however, many of them have been linked to interesting biological functions and, with the recent report by Kanfi et al. 6, SIRT6 assumes a central role in aging.
The primary finding of Kanfi et al. is that transgenic mice overexpressing SIRT6 have enhanced longevity, but only in males 6. While the longevity enhancement is modest (15% increase in median lifespan), the data is rigorous. Similar effects were seen in two different founder transgenic lines, large numbers of mice were included in the study and a mixed genetic background was selected for the study with equal contribution of two long-lived inbred backgrounds. The authors also attempted to address whether the longevity benefit results from retarded aging, as opposed to protection from cancer, a common cause of mortality in mice. While a challenging issue to resolve in a single study, tantalizing evidence is provided that SIRT6 is indeed a modulator of aging. For instance, if longevity is enhanced simply by eliminating the most common type of cancer, lung in these mice, then a post-mortem analysis would yield a skewing of the tumor spectrum. Yet no significant differences were seen in the incidence of different tumors. Moreover, other age-related pathologies were evident in equal proportion. These findings are consistent with a model whereby the aging process is delayed, with animals developing and succumbing to the same diseases later in life (Figure 1). Coupled with earlier findings that SIRT6 overexpression in mice (1) leads to protection from diet-induced obesity and (2) suppresses expression of NF-κB-dependent inflammatory markers, and moreover that SIRT6 expression is induced by caloric restriction in rats, it is fair to say that the deacetylase has certainly entered the realm of top-tier candidate modulatory factors of aging.
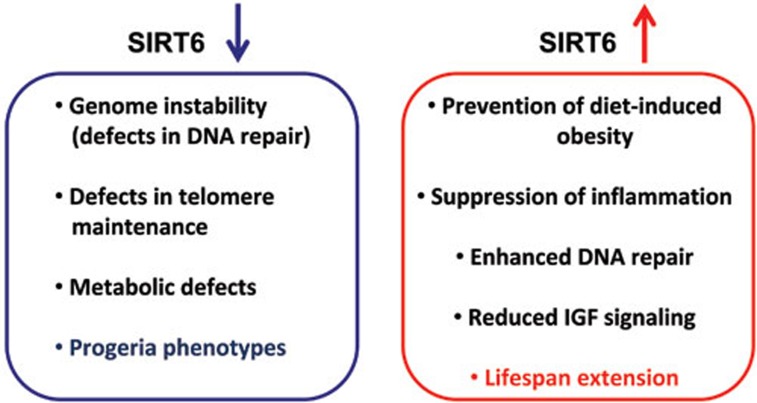
Figure 1.
SIRT6 expression and longevity. Decreased SIRT6 expression leads to genome instability, as well as progeroid phenotypes that resemble accelerated aging. In contrast, increased SIRT6 expression alters cell and physiological phenotypes in a manner that promotes enhanced longevity.
Long debated is whether mouse or human models of progeria, diseases with a rapid onset of a subset of aging pathologies, are truly accelerated aging syndromes 7. One argument against this hypothesis was that the genetic interventions giving rise to progeroid syndromes are non-overlapping with those linked to enhanced longevity. This disconnect is starting to dissolve as genes that go both ways are being identified. p53 is arguably the first example; constitutive p53 activation induces progeria phenotypes 8, whereas enhanced p53 activity under normal regulatory control leads to lifespan extension 9. With the study by Kanfi et al. 6, SIRT6 emerges as a new member of this short list of genes since it was previously shown that SIRT6−/− mice have metabolic defects and rapidly present with abnormalities often associated with aging, such as kyphosis and loss of subcutaneous fat 10. Thus, reduced SIRT6 expression may accelerate aspects of aging whereas enhanced SIRT6 expression may delay them.
A primary question is why increased SIRT6 expression enhances longevity, and several models are possible in addition to regulation of NF-κB. First are the metabolic consequences of enhanced SIRT6 activity, which are further elaborated in Kanfi et al. by examination of metabolic changes during aging of mice on a normal ad lib diet 6. First, Sirt6-transgenic mice are able to maintain healthy glucose metabolism later in life than controls, a feature often associated with longevity. However, both males and females maintain better metabolic function, therefore, this finding, while it may be linked, is separable from the dimorphic effects on lifespan. A further clue comes from the finding that IGF-1 levels are reduced in serum of male but not female overexpressing SIRT6 and that this is coupled to a corresponding increase in IGFBP-1 levels. Interestingly, these changes place the males in line with the females with regard to both factors. The downstream consequences of reduced IGF-1 levels are most apparent in white adipose tissue, which show significant reductions in AKT phosphorylation again only in males. Metabolic changes in fat tissue are increasingly linked to lifespan. These findings raise the intriguing possibility that the enhanced longevity in male Sirt6-transgenic mice occur because they have metabolic profiles more like that of females.
Whole-genome microarray analysis of liver tissue from mice overexpressing SIRT6 raises another intriguing possible mechanism linked to metabolism. First, the biggest gene expression changes were apparent in male transgenic livers and, importantly, many of the observed differences overlap with those of calorie-restricted mice. Since those mice also have reduced serum IGF-1 and increased IGFBP-1 levels 11, gene expression changes could be cause or consequence of changes to the endocrine axis. Moreover, these findings support the hypothesis that SIRT6 overexpression phenocopies aspects of calorie restriction in males. Another possibility that Sirt6-transgenic mice eat less, mimicking calorie restriction in a more direct fashion, can be largely discounted since an early study reported equal food consumption 12.
While SIRT1 is relatively promiscuous in its choice of substrate for deacetylation, finding targets for SIRT6 proved a more challenging endeavor. Recently, however, the identification of histone H3K9 and K56 may explain roles identified for SIRT6 in the DNA damage response and in telomere maintenance 13, 14. Cells from SIRT6−/− mice experience a dramatic elevation in genome instability indicative of defective double strand break (DSB) repair 10 and by contrast, enhanced SIRT6 activity stimulates DSB repair, particularly in response to oxidative stress 15. Deacetylation of H3K56 is linked to enhanced DNA repair, as is mono-ADP ribosylation of PARP, an alternate NAD+-dependent enzymatic activity of SIRT6. SIRT6 also associates with telomere heterochromatin, where it deacetylates both H3K9 and H3K56 13, 16. Reduced SIRT6 expression in primary fibroblasts leads to a range of replication-dependent defects in telomere maintenance, promoting premature senescence. Functions of SIRT6 either at sites of DSBs or at telomeres could underlie both progeroid and/or enhanced longevity phenotypes.
While SIRT1 resembles yeast Sir2 from a structural perspective, it may be SIRT6 that is a closer functional ortholog. In their respective organisms, expression levels of each correlate with longevity. Moreover, both are most closely linked with histone deacetylation and have roles in genome maintenance both at telomeres and in response to DNA damage. Both also may link genome maintenance to nutrient and stress conditions in the cell. A challenge moving ahead will be to link SIRT6 enzymatic functions and cellular properties to the metabolic and longevity phenotypes apparent in the transgenic and knockout mice.
The studies of Kanfi et al. place SIRT6 in a group of genes that when genetically modified can lead to enhanced mammalian longevity. However, they do not rule out roles for other Sirtuins in aging. For instance, while whole body overexpression of SIRT1 appears not to enhance lifespan, given the range of its functions in different tissues, more informative may be the lifespan phenotypes of mice overexpressing SIRT1 in specific tissues. In addition, little is known about other Sirtuins and given interesting metabolic roles especially for those localized to the mitochondria, it would not be surprising if they affect aging as well 2. The role of Sirtuins in aging remains controversial and undoubtedly twists and turns are yet to come, but the report by Kanfi et al. makes the case for Sirtuins and aging far more compelling, at least in the case of SIRT6.
References
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L. Sirtuins, Aging, and Metabolism. Cold Spring Harb Symp Quant Biol. 2011;76:81–90. doi: 10.1101/sqb.2011.76.010629. [DOI] [PubMed] [Google Scholar]
- Viswanathan M, Guarente L. Regulation of Caenorhabditis elegans lifespan by sir-2.1 transgenes. Nature. 2011;477:E1–E2. doi: 10.1038/nature10440. [DOI] [PubMed] [Google Scholar]
- Burnett C, Valentini S, Cabreiro F, et al. Absence of effects of Sir2 overexpression on lifespan in C. elegans and Drosophila. Nature. 2011;477:482–485. doi: 10.1038/nature10296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz D, Munoz-Martin M, Canamero M, et al. Sirt1 improves healthy ageing and protects from metabolic syndrome-associated cancer. Nat Commun. 2010;1:3. doi: 10.1038/ncomms1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Naiman S, Amir G, et al. The sirtuin SIRT6 regulates lifespan in male mice. Nature. 2012;483:218–221. doi: 10.1038/nature10815. [DOI] [PubMed] [Google Scholar]
- Miller RA. 'Accelerated aging': a primrose path to insight. Aging Cell. 2004;3:47–51. doi: 10.1111/j.1474-9728.2004.00081.x. [DOI] [PubMed] [Google Scholar]
- Tyner SD, Venkatachalam S, Choi J, et al. p53 mutant mice that display early ageing-associated phenotypes. Nature. 2002;415:45–53. doi: 10.1038/415045a. [DOI] [PubMed] [Google Scholar]
- Garcia-Cao I, Garcia-Cao M, Martin-Caballero J, et al. "Super p53" mice exhibit enhanced DNA damage response, are tumor resistant and age normally. EMBO J. 2002;21:6225–6235. doi: 10.1093/emboj/cdf595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mostoslavsky R, Chua KF, Lombard DB, et al. Genomic instability and aging-like phenotype in the absence of mammalian SIRT6. Cell. 2006;124:315–329. doi: 10.1016/j.cell.2005.11.044. [DOI] [PubMed] [Google Scholar]
- Swindell WR. Genes and gene expression modules associated with caloric restriction and aging in the laboratory mouse. BMC Genomics. 2009;10:585. doi: 10.1186/1471-2164-10-585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanfi Y, Peshti V, Gil R, et al. SIRT6 protects against pathological damage caused by diet-induced obesity. Aging Cell. 2010;9:162–173. doi: 10.1111/j.1474-9726.2009.00544.x. [DOI] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Berber E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Zwaans BM, Eckersdorff M, Lombard DB. The sirtuin SIRT6 deacetylates H3 K56Ac in vivo to promote genomic stability. Cell Cycle. 2009;8:2662–2663. doi: 10.4161/cc.8.16.9329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Z, Hine C, Tian X, et al. SIRT6 promotes DNA repair under stress by activating PARP1. Science. 2011;332:1443–1446. doi: 10.1126/science.1202723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michishita E, McCord RA, Boxer LD, et al. Cell cycle-dependent deacetylation of telomeric histone H3 lysine K56 by human SIRT6. Cell Cycle. 2009;8:2664–2666. doi: 10.4161/cc.8.16.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]