Dear Editor,
Receptor-like kinases (RLKs) constitute the major family of cell surface-associated receptors in plants and play essential roles in perceiving extracellular signals1. Over two hundred members of the largest subfamily of RLKs that contain leucine-rich repeat extracellular domains (LRR-RLK) are found in Arabidopsis, among which BRI1-associated kinase 1 (BAK1) is one of the best studied. BAK1 was initially identified based on its association with the LRR-RLK BRI1, which perceives brassinosteroid, an important hormone that regulates a wide range of developmental and physiological processes in plants2. BAK1 also serves as a co-receptor for several other LRR-RLKs that perceive pathogen-associated molecular patterns (PAMPs), including flagellin-sensing 2 (FLS2) and elongation factor EF-Tu receptor3,4, and is therefore required for the innate immunity of plants. The reciprocal phosphorylation on the cytoplasmic domains of BAK1 and the ligand-binding RLKs in the complex is a prerequisite for the full activation of the receptor kinase5. The crystal structure of BAK1 complexed with AvrPtoB, which is an effector secreted by Pseudomonas syringae pv. Tomato to suppress PAMP-triggered immunity, recently revealed the mechanism by which BAK1 activity is inhibited6. However, the mechanism for BAK1 activation, particularly the impact of phosphorylation of key residues on BAK1 activation, remains unclear.
In the present study, we report the crystal structure of the fully phosphorylated BAK1 cytoplasmic domain (BAK1-CD) (residues 259-583) complexed with an ATP analog (Figure 1, Supplementary information, Figures S1 and S2). Combined with quantitative mass spectrometry (MS) analysis of the impact of specific phosphorylated residues on BAK1 activation and further phosphorylation of FLS2, our structure indicates the essential roles of the phosphorylation in BAK1 activation; in particular, the phosphorylations of T450 and T455 have stronger functional impact than other phosphorylated residues by stabilizing key motifs of the BAK1 kinase.
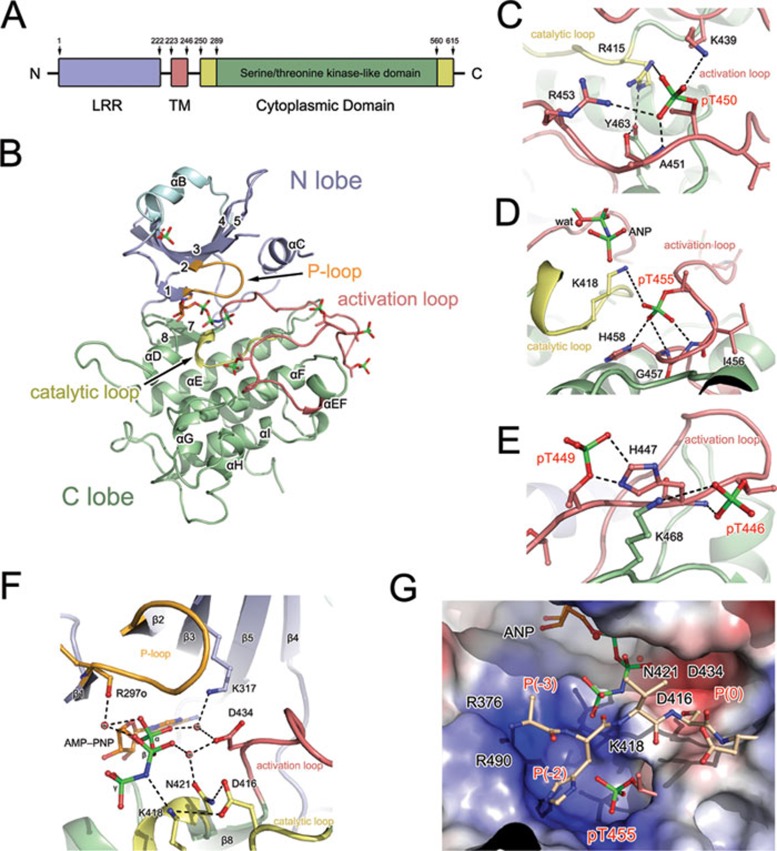
Figure 1.
Structure of the cytoplasmic domain of BRI1-associated kinase 1 of Arabidopsis thaliana. (A) Schematic diagram of the molecular structure of BAK1. Leucine-rich repeat (LRR), transmembrane (TM), and kinase domains (KD) are colored blue, red, and green, respectively. The accessory portion of BAK1-CD is colored yellow. The residue numbers are indicated. (B) Overall structure of BAK1-CD in complex with AMP-PNP. The polypeptide of BAK1-CD is shown as a cartoon representation, and the bound AMP-PNP molecule is presented as a colored stick. The N-terminal extension, N-lobe, activation loop, catalytic loop, and C-lobe are colored cyan, light blue, red, yellow, and pale green, respectively. The phosphorylated residues are displayed as colored sticks. (C-E) The phosphorylated residues contribute to the stabilization of the activation loop conformation. The interaction networks formed by pT450 (C), pT455 (D), and pT446 and pT449 (E) are highlighted. The key motifs of BAK1-CD are shown using the same color scheme as that in panel B. The red sphere represents the solvent molecule (also marked by “wat”). The dashed lines denote the hydrogen bonding interactions. (F) Catalytic machinery involved in ATP hydrolysis. Structure of the BAK1 kinase catalytic machinery. The red spheres labeled with “w” represent the water molecules involved in ATP binding. The dashed lines denote the hydrogen bonding interactions. (G) Substrate-binding pocket of BAK1-CD. The substrate-binding site of BAK1-CD is shown as a potential surface representation. A substrate peptide modeled according to the transphosphorylation site S1014 of FLS2 is shown in the binding pocket. The key residues, which are potentially crucial for substrate binding, are shown. The P(0), P(-2), and P(-3) sites of the substrate are labeled.
The core BAK1 kinase domain has the canonical bilobal structural architecture of a protein kinase family with N- and C-lobes. The extra N-terminal portion (G272-N291) forms a long extension, followed by a short helix (αB), which caps the top of the twisted β-sheets in the N-lobe. The C-lobe is predominantly α-helical, containing eight α-helices and two β-strands. The activation loop (D434-E462), which is restricted by the highly conserved DGF and APE motifs, attaches to the N-lobe and presents a fully extended architecture like that in other known active kinase structures7,8. A well-ordered adenylyl imidodiphosphate (AMP-PNP) molecule binds with the canonical ATP-binding pocket. No cation is present in the structure, even though sufficient amounts of magnesium have been supplied in the purification and crystallization buffers.
A key feature of the kinase activation loop is the localization of phosphorylated residues that mediate the kinase functions. The activation loop of BAK1 contains four phosphorylation sites, namely, T446, T449, T450, and T455, that are essential for its biological functions5. These sites were evidently shown by the electron density and the specific localization was verified via MS analysis, in particular, the phosphorylation of T455 was first identified using crystallographic data (Supplementary information, Table S1 and Figure S3). All phosphorylated residues form strong hydrogen bonds or salt bridges with residues in the activation loop, the catalytic loop, or the C-lobe through their phosphate moiety, indicating their important role in determining the correct conformation of BAK1-CD. Phosphorylated T450 (pT450) is oriented toward the kinase core and is trapped in a positively charged pocket formed by R415-K439-R453 (Figure 1C). The interaction between the phosphate moiety of pT450 and R415, which is located at the catalytic loop and precedes the catalytic base D416, appears to play a key role in stabilizing the catalytic loop. R415 also forms a hydrogen bond with Y463, which is clamped by R415 and A451. This observation provides a structural explanation for the complete abolishment of BAK1 autophosphorylation by the Y463F mutation in vitro9. pT450 also interacts with K439 and R453, which are located on the two sides of the activation loop, and presumably facilitate the correct conformation of the activation loop. All these interactions with pT450 contribute to the correct orientation of and electrostatic environment for the catalytic base, as well as the proper active orientation of the αC helix. Phosphorylated T455 (pT455) is recognized as one of the most important phosphorylated sites for BAK1 functions5. In the BAK1-CD structure, pT455 forms hydrogen bonds with the nitrogen atom of G457 and the imidazole group of H458 through its phosphate group, thereby enhancing the stability of the activation loop (Figure 1D). Moreover, the hydrogen-bonding interaction between pT455 and K418 indicates that pT455 is also involved in bridging the catalytic and activation loops to promote kinase activity. The phosphate moieties of pT446 and pT449 fold outward from the kinase core and interact with the backbone amino group and imidazole moiety of H447 to stabilize the conformation of the activation loop (Figure 1E). These structural observations suggest a critical role of the phosphorylated residues in BAK1 activity by stabilizing the conformation of its key motifs, given that the correct conformation of the activation and catalytic loops is essential for the kinase activity10.
A well-positioned ATP is one of the crucial features of a kinase catalytic machinery. In the BAK1-CD-AMP-PNP binary complex structure, L295, Y363-M366, and L423 form a deep cavity to clamp the adenine group of AMP-PNP (Supplementary information, Figure S4). The functional importance of the ATP-binding motif is demonstrated by the complete loss of autophosphorylation activity upon mutation of the Y363 residue (Supplementary information, Figure S5). A glycine-rich nucleotide-binding loop (R297-K302), called the P-loop, contributes to the stabilization of the bound AMP-PNP by interacting with the phosphate groups of AMP-PNP. One solvent-mediated hydrogen bond is observed between the backbone carbonyl atom of R297 and the β-phosphate (Figure 1F). A similar observation on the lack of interaction between the bound ATP or AMP-PNP molecule and the P-loop was reported in the crystal structure of insulin receptor tyrosine kinase (IRK)10 (Supplementary information, Figure S6D). By contrast, the P-loops of the cAMP-dependent protein kinase (cAPK) (Supplementary information, Figure S6B) and the γ-subunit of phosphorylase kinase (PHK) (Supplementary information, Figure S6C) significantly contribute to the stabilization of the β- and γ-phosphates of ATP11. This phenomenon indicates the variations in the stabilization of ATP binding by the P-loop of different kinase family members. In addition to the interactions with the P-loop, two linkages involving the α-phosphate are formed with D434 of the activation loop and K317.
The catalytic base D416, together with K418 and D434, is found above the γ-phosphate group of AMP-PNP at the top of a pyramid-shaped interaction network. Notably, no cations are bound to the catalytic base D416 at the active center even though a sufficient amount of magnesium has been supplied (Supplementary information, Figure S7). Instead, two solvent molecules replace the metal ions and mediate the interactions between AMP-PNP and the catalytic residues. In the structures of cAPK, PHK, and IRK, the bound metal ions form direct bonds with the phosphate groups of ATP and the catalytic residues. These observations suggest that the mechanisms for the catalytic activity of BAK1 be less similar with other kinases.
The conservation of the sequence and structural features between BAK1-CD and IRK12 and PHK13 indicates similar characteristics of the substrate-binding sites. In our study, FLS2 was phosphorylated by BAK1-CD in vitro (Supplementary information, Table S2A). Hence, the sequence around one transphosphorylation residue, S1014 of FLS2 (1011AHVSD1015), was used to build a BAK1-CD complex model. S1014 of FLS2 is located at the P(0) position, and the P(-3) to P(-1) sites fit the substrate pocket shape well in terms of complementarity and interactions. pT455 is positioned at the P(-2) site of the BAK1-CD substrate-binding site, and the phosphate of pT455 is likely to form hydrophilic connections with the substrate peptide (Figure 1G). Although this threonine residue is highly conserved in serine/threonine kinases, the direct structural evidence for the phosphorylation of this residue is only observed in BAK1-CD. Previous results have revealed that the substitution of K317 in the catalytic core and the mutation of T455 can significantly impair or abolish the transphosphorylation activity of BAK1. In the BAK1-CD-AMP-PNP structure, K317 interacts with the α-phosphate of AMP-PNP through solvent-mediated hydrogen bonding; however, this interaction occurs far from the substrate-binding groove, which is consistent with the previously reported result that mutations at K317 did not affect the binding affinity of BAK1 to BRI12.
We further examined the site-specific impact of the phosphorylated residues on BAK1 activation using quantitative MS-based assays with high mass accuracy measurements (< 10 ppm) (Supplementary information, Table S3 and Figure S8) to quantitatively evaluate their contributions and determine if phosphorylation occurs in an interdependent manner. Eleven residues in the BAK1-CD catalytic or activation loop were point mutated, including all four threonine residues in the activation loop that undergo autophosphorylation and transphosphorylation (Supplementary information, Table S4). Given their central roles in enzymatic catalysis, substitutions of D416 and N421 with alanine prevented phosphorylation on all six phosphorylated sites in BAK1-CD. The T450A and R415A mutants also lost nearly all phosphorylation activity at different sites, as can be inferred from the critical roles of pT450 and R415 in bridging the catalytic and activation loops (Figure 1C). The substitution of another structurally critical residue, T455, with alanine led to an almost complete disruption of the phosphorylation activity. The slightly retained phosphorylation levels on S290 and T312 in the T455A mutant suggest the less important functions of these two residues compared with those of the other four in the activation loop. The T446A mutant showed a more profound effect in reducing the overall autophosphorylation levels than T449A, even though both of the phosphorylated residues interact with H447 and presumably stabilize the conformation of the activation loop. This difference in effect may have stemmed from the additional role of pT446 in the interaction with K468. The H458A mutant eliminated the phosphorylation activity across all six S/T sites. By contrast, the H447A mutant in the activation loop retained a significant portion of phosphorylation on most of the sites examined. The mutants on the residues adjacent to the activation loop (K439A and R453A) retained most of the phosphorylation activity on certain sites, indicating their subtler role in regulating the phosphorylation activity than key residues in the activation loop.
FLS2 forms a complex with BAK1 in vivo to trigger the innate immunity of plants when flagellin is detected. We chose this protein to examine further the roles of BAK1 phosphorylation in mediating the activation of an immunity-related LRR-RLK. FLS2-CD showed distinct phosphorylation signals on several sites of the juxtamembrane domain and CD regions upon treatment with BAK1-CD (Supplementary information, Table S2A and Figure S9). This is the first report of in vitro transphosphorylation of FLS2 by BAK1. Moreover, the substitution of D416 in BAK1 resulted in the complete loss of phosphorylation activity toward FLS2, which is consistent with the central role of D416 in kinase reactions (Supplementary information, Table S2B). Mutations of T450 and T455 to alanine eliminated the FLS2 phosphorylation signals, thereby emphasizing the essential roles of these two residues in regulating the transphosphorylation activity of BAK1. By contrast, the T449A and T446A mutants retained most of their transphosphorylation activity toward FLS2; this result is consistent with that of their autophosphorylation activity, which was also partially retained. The transphosphorylation activity of the T449A mutant was significantly higher than that of T446A, which is consistent with the difference in their autophosphorylation activities. The different activities of the two mutants support our structure-derived hypothesis that pT446 plays an additional role in the overall folding of BAK1-CD.
In summary, our study has provided a structural basis for understanding the essential roles of BAK1 phosphorylation in its kinase functions. The crystal structure indicates that certain phosphorylated residues (T450 and T455) have stronger functional effects than other phosphorylated residues by interacting with both the catalytic and activation loops to achieve a conformational stability. The distinct impact of specific phosphorylated residues on BAK1 activation and further phosphorylation of FLS2 was determined via quantitative MS analysis. The phosphorylation of BAK1 in planta is likely initiated by the interaction with its partners, such as BRI1 or FLS2. Our study has provided clear structural evidence to support the previous speculation5 that the initial phosphorylation of the BAK1 activation loop locks BAK1 kinase in the active conformation. BAK1 in turn phosphorylates and fully activates the ligand-binding RLK, which phosphorylates downstream substrates for signal transduction and the development of the innate immune responses of plants14,15.
Acknowledgments
We would like to thank the staff of the Beijing Synchrotron Radiation Facility and Shanghai Synchrotron Radiation Facility for their generous help on diffraction data collection, and Dr Tae-Wuk Kim for providing Supplementary information, Figure S5. This work was supported by the National Natural Science Foundation of China (31000332, 31100208, and 31170782) and partly by a grant from the Herman Frasch Foundation (HF07 639 to Z-Y W).
Footnotes
(Supplementary information is linked to the online version of the paper on the Cell Research website.)
Supplementary Material
Structural basis for the impact of phosphorylation on the activation of plant receptor-like kinase BAK1
References
- Tang W, Deng Z, Wang ZY. Proteomics shed light on the brassinosteroid signaling mechanisms. Curr Opin Plant Biol. 2011;13:27–33. doi: 10.1016/j.pbi.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Wen J, Lease KA, Doke JT, Tax FE, Walker JC. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110:213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- Chinchilla D, Zipfel C, Robatzek S, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448:497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- Zipfel C, Kunze G, Chinchilla D, et al. Perception of the bacterial PAMP EF-Tu by the receptor EFR restricts Agrobacterium-mediated transformation. Cell. 2006;125:749–760. doi: 10.1016/j.cell.2006.03.037. [DOI] [PubMed] [Google Scholar]
- Wang X, Kota U, He K, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15:220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- Cheng W, Munkvold KR, Gao H, et al. Structural analysis of Pseudomonas syringae AvrPtoB bound to host BAK1 reveals two similar kinase-interacting domains in a type III effector. Cell Host Microbe. 2011;10:616–626. doi: 10.1016/j.chom.2011.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR, Wei L, Ellis L, Hendrickson WA. Crystal structure of the tyrosine kinase domain of the human insulin receptor. Nature. 1994;372:746–754. doi: 10.1038/372746a0. [DOI] [PubMed] [Google Scholar]
- Mol CD, Lim KB, Sridhar V, et al. Structure of a c-kit product complex reveals the basis for kinase transactivation. J Biol Chem. 2003;278:31461–31464. doi: 10.1074/jbc.C300186200. [DOI] [PubMed] [Google Scholar]
- Oh MH, Wang X, Wu X, Zhao Y, Clouse SD, Huber SC. Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc Natl Acad Sci USA. 2010;107:17827–17832. doi: 10.1073/pnas.0915064107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Hubbard SR. Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat Rev Mol Cell Biol. 2004;5:464–471. doi: 10.1038/nrm1399. [DOI] [PubMed] [Google Scholar]
- Bossemeyer D, Engh RA, Kinzel V, Ponstingl H, Huber R. Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24) EMBO J. 1993;12:849–859. doi: 10.1002/j.1460-2075.1993.tb05725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard SR. Crystal structure of the activated insulin receptor tyrosine kinase in complex with peptide substrate and ATP analog. EMBO J. 1997;16:5572–5581. doi: 10.1093/emboj/16.18.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abuhammad AM, Lowe ED, Fullam E, Noble M, Garman EF, Sim E. Probing the architecture of the Mycobacterium marinum arylamine N-acetyltransferase active site. Protein Cell. 2010;1:384–392. doi: 10.1007/s13238-010-0037-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang W, Kim TW, Oses-Prieto JA, et al. BSKs mediate signal transduction from the receptor kinase BRI1 in Arabidopsis. Science. 2008;321:557–560. doi: 10.1126/science.1156973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TW, Wang ZY. Brassinosteroid signal transduction from receptor kinases to transcription factors. Annu Rev Plant Biol. 2010;61:681–704. doi: 10.1146/annurev.arplant.043008.092057. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Structural basis for the impact of phosphorylation on the activation of plant receptor-like kinase BAK1