Abstract
Influenza A virus exerts a large health burden during both yearly epidemics and global pandemics. However, designing effective vaccine and treatment options has proven difficult since the virus evolves rapidly. Therefore, it may be beneficial to identify host proteins associated with viral infection and replication to establish potential new antiviral targets. We have previously measured host protein responses in continuously cultured A549 cells infected with mouse-adapted virus strain A/PR/8/34(H1N1; PR8). We here identify and measure host proteins differentially regulated in more relevant primary human bronchial airway epithelial (HBAE) cells. A total of 3740 cytosolic HBAE proteins were identified by 2D LC–MS/MS, of which 52 were up-regulated ≥2-fold and 41 were down-regulated ≥2-fold after PR8 infection. Up-regulated HBAE proteins clustered primarily into interferon signaling, other host defense processes, and molecular transport, whereas down-regulated proteins were associated with cell death signaling pathways, cell adhesion and motility, and lipid metabolism. Comparison to influenza-infected A549 cells indicated some common influenza-induced host cell alterations, including defense response, molecular transport proteins, and cell adhesion. However, HBAE-specific alterations consisted of interferon and cell death signaling. These data point to important differences between influenza replication in continuous and primary cell lines and/or alveolar and bronchial epithelial cells.
Keywords: Influenza virus, Primary cells, Quantitative proteomics, Host cell response, SILAC
Introduction
Influenza A virus (IAV), a member of the Orthomyxoviridae family, is an enveloped, single-stranded RNA virus with a genome of 8 negative-sense segments, with most encoding a single gene. These eight segments give rise to 10 or 11 distinct proteins depending on the strain (NA, HA, NS1, NS2/NEP, M1, M2, NP, PA, PB1, PB1-F2, and PB2),1 and these have been shown to interact extensively with each other and with host cell proteins throughout the virus lifecycle.2 Some interactions elicit alterations in the host proteome, as exemplified by the virus’s ability to both induce and evade a host immune response,3 to influence autophagy and apoptosis,4−6 and to increase viral protein synthesis while shutting down host protein synthesis.7 Numerous host proteins may also be important for influenza replication, as recently shown in whole genome siRNA screens that identified 100 genes in Drosophila,8 120 genes in U2OS cells,9 as well as 295 genes10 and 287 genes11 in the A549 human carcinoma lung cell line.
Current anti-influenza therapies consist of only two classes of treatments, and these each target a viral protein: zanamivir and oseltamivir are neuraminidase inhibitors, while amantidine and rimantidine target the M2 protein. Recent and currently circulating strains from the 2010–2011 season have remained susceptible to M2 inhibitors; however, resistance to neuraminidase inhibitors is emerging (www.cdc.gov/flu/weekly/). Thus, new targets for which resistance will not quickly be developed are needed, and host cell proteins essential for viral replication represent one option.12
Studying virus–host interactions is increasingly reliant on quantitative proteomic techniques such as 2D-DIGE, isotope-encoded affinity tag (ICAT), and stable isotope labeling of amino acids in cell culture (SILAC). For example, SILAC profiling was used to determine differences in expression between HIV- and mock-infected T-cells13 and the response of several host cell types to hepatitis C infection.14,15 Quantitative proteomics has also been applied to influenza–host interactions, including several studies probing primary macrophages with iTRAQ16 and 2DIGE-MS/MS17 and multiple studies using similar techniques in continuous epithelial cell lines such as AGS,18 MDCK,19−21 A549,22 and Calu-3 cells.23 Several genomic and proteomic studies have also been carried out in influenza-infected macaques.24−26 A novel aspect of the current study is the use of more relevant human primary bronchial epithelial cells, a model that more closely mimics in vivo infection conditions. It has been shown that the response to influenza depends not only on the viral strain27 but also the host species, cell type and location in the lung,28 and the state of cell differentiation.29 While the effect of these different parameters have been characterized in part by measuring secreted cytokines30 and the kinetics of viral replication,27 very little is known about differences in host–virus interactions between different host cell types. Similarly, few studies have compared the effect of IAV infection in continuous and primary cell lines of specific cell types. Therefore, it is important to extend our previous SILAC studies using continuous human lung A549 cells, a type II alveolar epithelial cell, to primary human tracheobronchial airway epithelial (HBAE) cells to compare pathways affected by influenza replication, and identify pathophysiologically relevant responses.
Experimental Procedures
Cells and Viruses
Viruses
Influenza virus strain A/PR/8/34 (H1N1; PR8), an attenuated mouse-adapted strain, was grown in embryonated hens’ eggs from laboratory stocks, after which chorioallantoic fluid was harvested, aliquoted, and titered in MDCK cells by standard procedures.22
Cells and Media
Primary normal human broncho-tracheal epithelial cells were obtained from “healthy donors” by Lonza Inc. and were certified as mycoplasma-, HIV-, HBV-, and HCV-negative. Notably, these cells were capable of >10 cell doublings, which made the six doublings needed for SILAC labeling possible. Cells were cultured in bronchial epithelial growth media (BEGM, Lonza Inc.), which consists of bronchial epithelial basal media (BEBM, Lonza Inc.) supplemented with SingleQuots (Lonza Inc.) containing bovine pituitary extract, hydrocortisone, hEGF, epinephrine, transferrin, insulin, retinoic acid, triiodothyronine, and gentamicin-1000. Cells were maintained as monolayers in 10% CO2 at 37 °C and passaged by trypsinization at 80–90% confluence. For SILAC labeling, cells were grown in SILAC media that consisted of arginine- and lysine-free BEGM supplemented with either C12 (“light”) or C13 (“heavy”) lysine (29 mg/L) (mass difference of 6.0 Da) and C12/N14 (“light”) or C13/N15 (“heavy”) arginine (348 mg/L) (mass difference of 10.0 Da). Light amino acid containing media were prepared from regular stock powder (Sigma, St, Louis MO), and heavy amino acids were obtained from the SILAC Phosphoprotein Identification and Quantification Kit (Invitrogen Canada Inc.; Burlington, Ontario). Separate SILAC labeling and infection experiments were performed four times.
Infection
For SILAC experiments, all cells were grown in a pair of T75 flasks in SILAC media for six cell doublings; after the sixth cell doubling, cells were allowed to reach near confluency. In three replicates C12/N14-light cells were infected with egg-grown PR8 virus that was diluted in gel saline to achieve a multiplicity of infection (MOI) of 7 plaque forming units (PFU) per cell. An equivalent number of C13/N15-heavy cells were mock infected as a control using only gel saline. Previous studies indicate the small amount of egg protein makes no significant measurable contribution to results or conclusions.22 Label swapping was performed for the fourth replicate; the C13/N15 heavy cells were infected, and the C12/N14 light cells were mock infected. For all other infections, cells were grown to near confluency and infected at various MOIs as indicated. In order to synchronize infections, virus- and mock-infected cells were placed at 4 °C for the 1 h virus adsorption, after which inoculum was removed and cells were overlaid with appropriate prewarmed light or heavy SILAC media. Infected and mock-infected cell cultures were then kept at 37 °C until analysis of viral replication and host protein expression. Efficiency of infection was always confirmed by a standard plaque assay as described previously.22
Isolation of Cytosolic Proteins
Twenty-four hours postinfection, cells grown in C12/N14-light or C13/N15-heavy BEGM were collected by brief trypsinization and counted, and then equivalent numbers (roughly 3 × 106 cells) of each group were mixed together. To verify infection status of each culture, aliquots of all separate cultures were saved for virus titration by plaque assay as mentioned above. Mixed cells were washed 3 times in >50 volumes of ice-cold phosphate-buffered saline (PBS) and lysed for cytosolic proteins by adding NP-40 buffer (10 mM TrisHCl pH 7.4, 3 mM CaCl2, 2 mM MgCl2, 0.5% NP-40, 1.1 μM pepstatin A) to the cells and incubating them for 30 min on ice. Nuclei were pelleted at 5000 × g for 10 min and the supernatant was saved as “cytosol”. Both fractions were frozen at −80 °C until further processing.
Protein Digestion
Protein content in the cytosolic fractions were measured using a Protein Assay Kit (Biorad) and bovine serum albumin standards. Three hundred micrograms of each was reduced, to which 6× the volume of 100 mM ammonium bicarbonate was added. Then 100 mM dithiothreitol (DTT) in 100 mM ammonium bicarbonate was added to the peptide mixture, and samples were incubated for 45 min at 60 °C. Iodoacetic acid (500 mM in 100 mM ammonium bicarbonate) was added to each tube for alkylation, and the tubes were incubated for a further 30 min (room temperature, in the dark). Finally, 100 mM DTT was added to quench the excess iodoacetic acid. Samples were digested overnight at 37 °C with 6 μg of sequencing grade trypsin (Promega, Madison, WI) and then stored at −80 °C until further processing.
2D LC–MS/MS and Peptide Identification
Peptide fractionation was carried out using a 2D RP (reversed-phase) high pH–RP low pH peptide system as described previously.31 In short, lyophilized tryptic digests were dissolved in 200 μL of 20 mM ammonium formate pH 10 (buffer A), injected onto a 1 × 100 mm XTerra (Waters, Milford, MA) column and fractionated using a 0.67% acetonitrile per minute linear gradient (Agilent 1100 Series HPLC system, Agilent Technologies, Wilmington, DE) at a 150 μL/min flow rate. Sixty 1-min fractions were collected and concatenated using procedures described elsewhere;31,32 the last 30 fractions were combined with the first 30 fractions in sequential order (i.e., no. 1 with no. 31; no. 2 with no. 32, etc.). Combined fractions were vacuum-dried and redissolved for the second dimension RP separation (0.1% formic acid in water). The second dimension was run on a splitless nanoflow Tempo LC system (Eksigent, Dublin, CA) with 20 μL sample injection via a 300 μm 5 mm PepMap100 precolumn (Dionex, Sunnyvale, CA) and a 100 μm × 200 mm analytical column packed with 5 μm Luna C18(2) (Phenomenex, Torrance, CA). Both eluents A (water) and B (acetonitrile) contained 0.1% formic acid as an ion-pairing modifier. A 0.33% acetonitrile per minute linear gradient (0–30% B) was used for peptide elution, providing a total 2-h run time per fraction in the second dimension.
A QStar Elite mass spectrometer (Applied Biosystems, Foster City, CA) was used in a data-dependent MS/MS acquisition mode. One-second survey MS spectra were collected (m/z 400–1500) followed by MS/MS measurements on the 3 most intense parent ions (80 counts/s threshold, +2 to +4 charge state, m/z 100–1500 mass range for MS/MS), using the manufacturer’s “smart exit” (spectral quality 5) settings. Previously targeted parent ions were excluded from repetitive MS/MS acquisition for 60 s (50 mDa mass tolerance). Spectra were identified using Analyst QS 2.0 (Applied Biosystems) software.
Protein Identification and Quantification
Thirty “.wiff” files from Analyst were submitted simultaneously to Protein Pilot 3.0 (Applied Biosystems) for relative quantification and protein identification using the Paragon algorithm as the search engine. Each MS/MS spectrum was searched against a database of human protein sequences (NCBInr, released March 2008, downloaded from ftp://ftp.ncbi.nih.gov/genomes/H_sapiens/protein/). The search parameters allowed for cysteine modification by iodoacetic acid and biological modifications programmed in the algorithm (i.e., phosphorylations, amidations, semitryptic fragments, etc.). The threshold for detecting proteins (unused protscore (confidence)) in the software was set to 2.0 to achieve 99% confidence, and identified proteins were grouped by the ProGroup algorithm (Applied Biosystems, Foster City, CA) to minimize redundancy. The bias correction option was used to correct for small pipetting errors. The protein sequence coverage was calculated using peptides identified with >95% confidence. A decoy database search strategy (NCBInr Homo sapiens with all protein sequences reversed) was also used to estimate the false discovery rate (FDR), defined as the percentage of reverse proteins identified against the total protein identification. For our data, the estimated FDR was 0.56%, which is low compared to other studies and indicates a very high reliability of the proteins identified.33
For relative quantitation, only peptides unique for a given protein were considered, thus excluding those common to other isoforms or proteins of the same family. Proteins were identified on the basis of having at least one peptide with an ion score above 99% confidence. Among the identified peptides, some of them were excluded from the quantitative analysis for one of the following reasons: (a) The peaks corresponding to the SILAC labels were not detected. (b) The peptides were identified with low identification confidence (<1.0%). (c) Either the same peptide sequence was claimed by more than one protein or more than one peptide was fragmented at the same time because of shared MS/MS spectra. (d) The sum of the signal-to-noise ratio for all of the peak pairs was 6 for the peptide ratios.
Bioinformatics and Statistics
In order to compare multiple biological replicates, protein ratios within each replicate were converted to a z-score that allowed protein ratios to be normalized to the mean and standard deviation of its individual experiment.22 Thus, a protein with a z-score >1.960σ indicates that protein’s differential expression lies outside the 95% confidence level, 2.576σ indicates 99% confidence, and 3.291σ indicates 99.9% confidence; z-scores >1.960 were considered significant. The weighted average z-score was then calculated for each protein found in multiple replicates.
Gi numbers of all significantly regulated proteins were submitted to and analyzed by the DAVID bioinformatic suite at the NIAID, version 6.7,34,35 and gene ontologies were examined with the “FAT” and Panther databases. The gi numbers were also submitted to, and pathways constructed with, Ingenuity Pathway Analysis software (IPA).
Western Blotting
Mock- and influenza-infected HBAE cells were scraped into cold PBS at 24 hpi, pelleted at 5000 × g for 5 min, and lysed with lysis buffer (20 mM Tris pH 7.5, 100 mM NaCl, 0.5% NP-40, 0.5 mM EDTA, 1× antiprotease cocktail (Pierce), 1× phosphatase inhibitor cocktail (Pierce)). Forty micrograms of protein was loaded per lane into SDS-PAGE gels, separated, and transferred to nitrocellulose membranes. Membranes were probed with primary antibodies for viral NP (in-house antibody), viral NS-1 (in-house antibody), SamD9 (Sigma), IFIT1 (Epitomics), E-cadherin (Cell Signaling), GLG1 (Sigma), ISG15 (Rockland), MxB (Santa Cruz), RSAD2 (Abcam), STAT1 (Cell Signaling), β-tubulin (Cell Signaling), PPIA (Epitomics), OASL (Epitomics), β-actin (Sigma), and GAPDH (Santa Cruz) and rabbit HRP-conjugated secondary antibodies. Bands were detected with ECL (Amersham) and the AlphaInnotech FluorChemQ MultiImage III instrument, quantitated using AlphaEase software and virus-to-mock ratios reported without normalization.
Microscopy
HBAE cells were grown to 80% confluence on 25 mm coverslips and infected or mock-infected at MOI = 7. Mock and 0, 12, and 24 h infected cells were fixed in 3% paraformaldehyde for 15 min, permeabilized with 0.3% Triton-X100 in 3% paraformaldehyde for 15 min, and blocked with 1% BSA in PBS. Cells were treated with a primary antibody for NP (made in-house) and a Cy3-conjugated rabbit secondary antibody (Jackson Immuno Research); all antibodies were diluted in 1% BSA and PBS. Coverslips were then mounted onto slides using DAPI-Prolong Gold Antifade, dried, and sealed. Fluorescent images were obtained using a computer-controlled Olympus IX70 microscope (20X objective) equipped with CCD camera and NIS-Element software.
Results
A/PR/8/34 Replicates in Undifferentiated Primary HBAE Cells
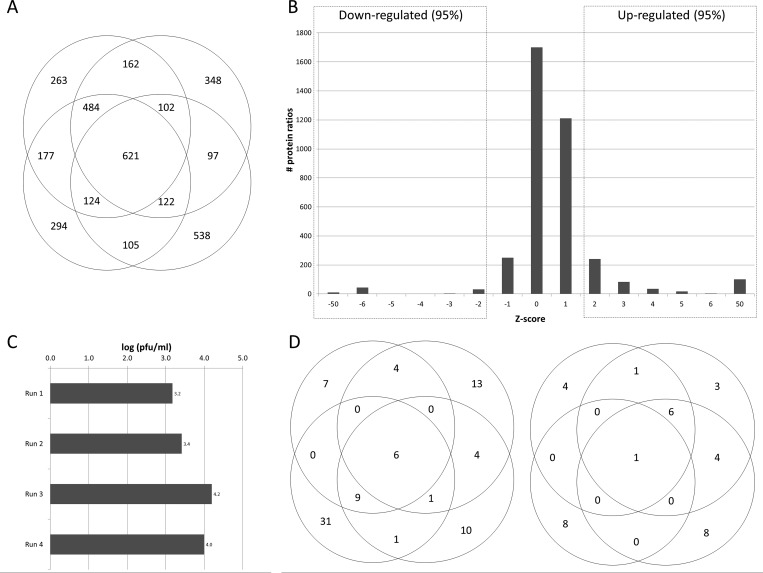
Actively proliferating primary epithelial cell cultures are not commonly used for influenza studies; thus, we initially determined optimal conditions for influenza replication in nondifferentiated HBAE. A low (0.01) and a high (7) MOI were initially chosen to measure the amount of infectious progeny virus released into the supernatant over time. At the low MOI, viral replication was most efficient in MDCK cells, as peak titers reached almost 108 PFU/mL by 48 hpi, whereas maximum titers of 105 PFU/mL were attained by 48 hpi in A549 cells and no detectable virus was recovered from HBAE cells (Figure 1a). At MOI = 7, MDCK and A549 cells reached similar peak titers but at earlier time points, 24 and 31 hpi, respectively; HBAE produced detectable virus by 12 hpi and reached a maximum titer of 105 PFU/mL by 24 hpi (Figure 1b). Influenza replication in HBAE at MOI = 7 was further confirmed by Western blotting for the viral non-structural-1 protein (NS1), which is expressed only during active viral replication. NS1 was clearly expressed in HBAE cells by 6 hpi and increased in abundance at both 12 and 24 hpi (Figure 1b). Additionally, we found that an MOI = 7 was sufficient to demonstrate active and productive infection in approximately 50% of cells by 12 hpi, while >95% of cells demonstrated productive infection by 24 hpi (Figure 1c). For SILAC experiments we therefore chose to study cells 24 h after infection to maximize the amount of time for cellular proteomic changes to occur while retaining active virus replication and minimizing any cytopathic effect (Figure 1d). These data collectively indicate that primary HBAE cells support influenza virus replication. In addition, to verify that HBAE cells could be labeled, we analyzed only heavy-labeled cells using LC–MS/MS and ProteinPilot.36−38 Using the data from ProteinPilot, we determined that 91% of all peptides contained the “C13-label” modification (Supplementary Figure 1). In addition, H:L ratios were reported for each peptide. The C13-containing peptides were highly enriched for high H:L ratios; 66% of these peptides had an H:L ratio >9 (>90% incorporation), and 92% had an H:L ratio >2 (>67% incorporation). In contrast, the peptides that did not contain a C13 label were enriched for peptides with low H:L ratios, e.g., 75% of these peptides had an H:L ratio <1 (<50% incorporation), and virtually all unlabeled peptides belonged to keratins. Therefore, we concluded that the six HBAE cell doublings were sufficient to label the cell proteome.
Figure 1.
Active viral replication in HBAE. (A) HBAE, A549, and MDCK cells were infected at MOI = 0.01 (left) and 7 (right) PFU/cell to monitor and compare efficiency of infectious progeny virus production. Supernatants were collected and titered for progeny virus production by standard plaque assay at indicated time points. (B) HBAE cells were infected with influenza A/PR/8/34 at MOI = 7 PFU/cell, and protein lysates were assayed for accumulation of viral NS-1 protein at 6, 12, and 24 h post infection by Western blotting. (C) HBAE cells were infected for 0, 12, and 24 h at MOI = 7 PFU/cell. After fixation, the percentage of cells infected by virus was shown using immunocytochemistry for the viral nucleoprotein. NP = red; nuclei = blue. (D) Phase-contrast images of HBAE cells at 24 and 48 h post infection and mock infection.
SILAC Identified 3740 Unique Cytosolic HBAE Proteins
We performed four separate replicate experiments using primary human bronchial epithelial cell cultures from the same donor in which the light-labeled culture was infected in three replicates and the heavy-labeled culture was infected for the fourth replicate. This allowed us to demonstrate that the heavy isotopes had no effect on the virus replication. Several proteins were found significantly up-regulated in the light-labeled infections but significantly down-regulated in the heavy-labeled infection. These proteins, which therefore probably represent contaminants, include several keratin species (1, 2, 9), dermcidin, transferrin, thyroid hormone receptor associate protein 3, and nipsnap2 and were excluded from subsequent analysis. Overall, a total of 3740 unique cytoplasmic proteins were detected and quantified; 2282 (61%) of these proteins were detected in 2 or more replicates, 1453 (39%) were found in 3 or more replicates, and 621 (17%) were common to all four replicates (Figure 2a). As these proteins were identified using a human database and rely upon the presence of these proteins in both cultures, no viral proteins were identified during mass spectrometry. To confirm that the samples were infected, supernatants were titered for progeny virus using a standard plaque assay on MDCK cells (Figure 2b).
Figure 2.
Distributions of total, up-regulated, and down-regulated proteins identified. (A) A total of 3740 proteins were identified from four separate biological replicates; 30% of all proteins were found in every run, 53% were found in two or more runs, and 46% were unique to each run. (B) Up- and down-regulated proteins were determined statistically using a frequency distribution curve. In order to compare the four replicate runs, infected:mock ratios for each run were normalized by converting them to z-scores, a measure of deviation from the average. Therefore, positive values represent proteins that are up-regulated in virus-infected cells, and negative values represent down-regulated proteins. To determine which proteins are significantly altered in abundance we used different confidence intervals as indicated. Most proteins were not significantly altered (z-score between −1 and 1); however, a small subset of proteins were strongly up- or down-regulated (z-score >1.96 and <−1.96, respectively; p-value <0.05). Only one of the replicates is shown for clarity. (C) After SILAC-labeled HBAE cells were infected for 24 hpi, the supernatant was collected and titered by plaque assay to demonstrate that the cell monolayer had been infected and had productively produced progeny virus. (D) The number of up- and down-regulated protein pairs identified in each trial at a confidence level of 95% (z-score >1.96). Specific peptides identified, measured, and used to measure up-regulated and down-regulated proteins are listed in Supplementary Table 3.
Comparing the abundance of proteins in infected cultures to uninfected cultures revealed that 97% of all proteins were present in approximately equal amounts (near a 1:1 ratio), thus indicating that IAV infection does not alter the expression level of most proteins in host cells. To determine cutoff values for identifying significantly up- and down-regulated proteins, ratios for infected:uninfected in each trial were first converted to z-scores to normalize each run to its own mean and standard deviation, thus allowing us to compare multiple runs. By converting an average z-score back into an average protein ratio, we determined that a z-score of 1.96 (95% confidence interval) corresponded to a 2-fold change. Similarly, z-scores of 2.58 (99% confidence interval) corresponded to a protein ratio change of 2.5-fold and z-scores of 3.29 (99.9% confidence interval) corresponded to a 3.0-fold change in protein ratio (Figure 2c). Further bioinformatic analyses were primarily performed with the 95% confidence data set but were also compared to more restricted data sets based on 99% and 99.9% confidence intervals (not shown).
Proteins that were identified with only a single peptide were excluded from our data set if they were detected in only a single run; however, any protein identified in multiple trials was included. We also excluded proteins from further analysis if they were not consistently up- or down-regulated between replicate runs. Some proteins identified in the current study were unaltered in at least one replicate and up- or down-regulated in other replicate(s). For these scenarios we grouped ratios into three groups: (a) z-scores −0.09 to 0.9 (“not altered”), (b) ±1.0 to 1.96 (“moderately altered”), or (c) greater than ±1.96 (“highly altered”). We then included only those proteins that had significant values (group c) in at least two runs or one of two runs; additionally, any trial with a nonsignificant value had to have that value still fall within the 80% confidence range (group b) to be considered. Outliers could be found at both the peptide and the protein level such that peptides and proteins that were found only in the virus or mock sample were labeled “9999” or “0000”, respectively. We did not remove any outliers at the peptide level but found that they made up a very small percentage of the peptides in our lists of differentially regulated proteins (the details may be found in Supplementary Table 2) and removing them had minimal impact on the overall protein ratio. For proteins that had a ratio of “9999” and “0” we arbitrarily assigned a z-score of 50 and −50, respectively. These outliers were not included in the statistical analysis but were reincorporated into data sets for further bioinformatic and biological analyses.
In summary, we used stringent parameters to generate a data set of proteins of interest based on 99% confidence in protein identification; protein identification based on 2 or more peptide pairs; 95% confidence interval for altered regulation; and the necessity for proteins to show consistent infected:unifected ratios. In this manner we identified 52 up-regulated proteins and 41 down-regulated proteins from PR8-infected primary HBAE cells (Figure 2c and listed in Tables 1 and 2). The identities, associated z-scores and protein percent coverage of the 52 up-regulated and 41 down-regulated proteins are listed in Supplementary Table 1, and the identities, confidences and other statistical parameters of the specific peptides used for quantitation are listed in Supplementary Table 2. Several of the up-regulated and nonregulated proteins that were identified in the SILAC analysis were confirmed by Western blotting (Figure 3), and most Western blot results confirmed SILAC-determined regulation status. Only one protein, PPIA, that was shown to be up-regulated in SILAC, was not validated using Western blotting. This may be caused by inherent differences in sampling (partially degraded proteins would not be measured by Western blot but their peptides would be detected by MS) or by inherent differences in the different methods' levels of sensitivity.
Table 1. HBAE Proteins Increased >95% Confidencea.
| virus/mock
protein ratio |
no. of peptides used for identification |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| gi accession | symbol | gene name | run 1 | run 2 | run 3 | run 4 | ratio avg | run 1 | run 2 | run 3 | run 4 |
| Cytosolic Proteins Detected More than Once | |||||||||||
| gi|6650772 | PRO1400 | PRO1400 | 100.0 | 100.0 | 100.0 | 18 | 17 | ||||
| gi|33286420 | PKM2 | pyruvate kinase 3 isoform 2 | 100.0 | 100.0 | 100.0 | 81 | 70 | ||||
| gi|5732237 | MLYCD | malonyl coenzyme A decarboxylase | 100.0 | 100.0 | 100.0 | 1 | 1 | ||||
| gi|11321577 | OASL | 2′-5′-oligoadenylate synthetase-like isoform a | 10.0 | 24.5 | 17.2 | 1 | 1 | ||||
| gi|12654159 | IFITM1 | Interferon induced transmembrane protein 1 (9–27) | 20.2 | 100.0 | 20.2 | 3 | 3 | ||||
| gi|19923667 | RSAD2 | Radical S-adenosyl methionine domain containing 2 | 20.9 | 13.9 | 10.0 | 100.0 | 14.9 | 7 | 13 | 8 | 1 |
|
gi|55958476; gi|13436398 |
IFIT3/4 | interferon-induced protein with tetratricopeptide repeats 3/4 | 3.2 | 8.5 | 30.8 | 1.6 | 11.0 | 8 | 8 | 11 | 4 |
| gi|52694752 | GPNMB | glycoprotein (transmembrane) nmb isoform a precursor | 4.1 | 10.7 | 7.4 | 1 | 1 | ||||
|
gi|2996631; gi|7717386 |
MX2 | MX2 | 13.7 | 8.1 | 100.0 | 2.7 | 8.2 | 2 | 3 | 3 | 3 |
|
gi|21751732; gi|7717387 |
MX1 | Myxovirus resistance 1, interferon inducible protein p78 | 7.7 | 3.7 | 10.9 | 4.7 | 6.7 | 3 | 3 | 8 | 4 |
| gi|5834273 | HMGA1 | high mobility group protein-R | 19.7 | 2.1 | 10.9 | 2 | 1 | ||||
| gi|54697154 | MMP1 | matrix metalloproteinase 1 (interstitial collagenase) | 3.5 | 10.1 | 4.5 | 6.0 | 2 | 4 | 1 | ||
|
gi|34192824; gi|55958474 |
IFIT2 | Interferon-induced protein with tetratricopeptide repeats 2 | 7.3 | 5.5 | 8.8 | 2.5 | 6.0 | 7 | 5 | 11 | |
| gi|49574526 | IFIT1 | interferon-induced protein with tetratricopeptide repeats 1 | 6.6 | 3.2 | 4.9 | 4.0 | 4.7 | 6 | 6 | 10 | 3 |
|
gi|14550514; gi|4826774 |
ISG15 | ISG15 ubiquitin-like modifier | 3.6 | 3.4 | 2.7 | 10.8 | 5.1 | 3 | 3 | 3 | 4 |
| gi|34783544 | LGALS7 | LGALS7 protein | 4.8 | 3.6 | 1.6 | 3.3 | 1 | 7 | 1 | ||
|
gi|297097; gi|452647 |
HLA-B/C | HLA-C α chain | 3.8 | 2.4 | 3.1 | 10 | 10 | ||||
| gi|56204179 | GBP1 | Guanylate binding protein 1, interferon-inducible, 67 kDa | 100.0 | 2.9 | 2.9 | ||||||
| gi|39794319 | SPBN | Suprabasin | 2.5 | 3.0 | 2.8 | 1 | 1 | ||||
| gi|14602986 | ACTC1 | Actin, α, cardiac muscle | 1.6 | 4.6 | 100.0 | 3.1 | 44 | 52 | 54 | ||
| gi|54673572 | LYRM7 | Hypothetical protein LOC90624 | 2.9 | 4.1 | 1.4 | 2.8 | 2 | 2 | 3 | ||
| gi|4886493 | SAMHD1 | SAM domain and HD domain 1 | 2.0 | 3.0 | 2.5 | 2 | 1 | ||||
|
gi|13937981; gi|48145531 |
PPIA | Peptidylprolyl isomerase A (cyclophilin A) | 2.3 | 2.1 | 2.2 | 23 | 22 | ||||
| gi|8922707 | TMEM34 | transmembrane protein 34 | 3.0 | 1.8 | 2.4 | 1 | 2 | ||||
| gi|2459624 | TLR2 | Toll-like receptor 2 | 1.6 | 2.5 | 2.6 | 2.2 | 1 | 4 | 1 | ||
| Cytosolic Proteins Detected Only Once | |||||||||||
| gi|56203096 | ACOT7 | brain acyl-CoA hydrolase (BACH) | 100.0 | 100.0 | 8 | ||||||
| gi|15079723 | DBNL | Drebrin-like | 100.0 | 100.0 | 6 | ||||||
| gi|38013966 | TKT | TKT protein | 100.0 | 100.0 | 56 | ||||||
| gi|12053853 | METTL9 | Methyltransferase like 9 | 100.0 | 100.0 | 2 | ||||||
| gi|14549669 | COG3 | vesicle docking protein SEC34 | 100.0 | 100.0 | 2 | ||||||
| gi|485388 | EIF4 | eukaryotic initiation factor 4 | 100.0 | 100.0 | 9 | ||||||
| gi|457262 | YBX1 | Y box binding protein 1 | 100.0 | 100.0 | 4 | ||||||
| gi|55660909 | SEC23B | Sec23 homologue B (S. cerevisiae) | 100.0 | 10.00 | 4 | ||||||
| gi|415849 | ADSS | adenylosuccinate synthetase | 100.0 | 100.0 | 3 | ||||||
| gi|182642 | -- | rapamycin-binding protein | 100.0 | 100.0 | 2 | ||||||
| gi|535179 | BCAM | basal cell adhesion molecule (receptor for laminin) | 100.0 | 100.0 | 2 | ||||||
| gi|5726629 | SNX12 | sorting nexin 12 | 100.0 | 100.0 | 2 | ||||||
| gi|16924221 | ZNF259 | Zinc finger protein 259 | 100.0 | 100.0 | 2 | ||||||
| gi|32479340 | HN1 | hematological and neurological expressed 1 | 100.0 | 100.0 | 2 | ||||||
| gi|55743134 | RPS6KA1 | ribosomal protein S6 kinase, 90 kDa, polypeptide 1 isoform b | 100.0 | 100.0 | 2 | ||||||
| gi|33316810 | OEF1 | OEF1 | 100.0 | 100.0 | 2 | ||||||
| gi|1552516 | -- | trypsinogen C | 65.1 | 65.1 | 7 | ||||||
| gi|11120626 | TYR6 | cationic trypsinogen | 38.9 | 38.9 | 6 | ||||||
| gi|7582292 | eIF4A2 | Eukaryotic translation initation factor 4, A2 | 12.1 | 12.1 | 7 | ||||||
| gi|12001972 | BOLA2 | My016 protein | 14.6 | 14.6 | 3 | ||||||
| gi|48146353 | HSPBP1 | HSPA (heat shock 70 kDa) binding protein, cytoplasmic cochaperone 1 | 68.6 | 68.6 | 2 | ||||||
| gi|52545561 | MTPAP | Mitochondrial poly(A) polymerase | 9.7 | 9.7 | 2 | ||||||
| gi|30511 | HBD | delta globin | 8.4 | 8.4 | 2 | ||||||
| gi|1335273 | NOMO1 | pm5 protein | 7.0 | 7.0 | 25 | ||||||
| gi|18418633 | HBB | mutant β-globin | 5.4 | 5.4 | 2 | ||||||
| gi|999893 | -- | Chain B, Triosephosphate Isomerase (Tim) (E.C.5.3.1.1) Complexed With 2-Phosphoglycolic Acid | 12.9 | 12.9 | 22 | ||||||
| gi|51476968 | STAT1 | Signal transducer and activator of transcription 1 | 3.9 | 3.9 | 7 | ||||||
| gi|9955206 | -- | Chain B, Crystal Structure Of A Rac-Rhogdi Complex | 8.4 | 8.4 | 2 | ||||||
| gi|7671655 | TOMM34 | translocase of outer mitochondrial membrane 34 | 7.8 | 7.8 | 2 | ||||||
| gi|41351087 | CAD | Carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase | 7.5 | 7.5 | 5 | ||||||
| gi|24899174 | SAMD9 | Sterile α motif domain containing 9 | 2.9 | 2.9 | 4 | ||||||
| gi|6573422 | -- | Chain D, Human Glyoxalase I Complexed With S–P- Nitrobenzyloxycarbonylglutathione | 7.1 | 7.1 | 2 | ||||||
| gi|48145879 | CTSH | CTSH | 2.8 | 2.8 | 3 | ||||||
| gi|55960299 | GSN | gelsolin (amyloidosis, Finnish type) | 6.6 | 6.6 | 4 | ||||||
| gi|38174528 | PPP4R1 | Protein phosphatase 4, regulatory subunit 1 | 6.5 | 6.5 | 2 | ||||||
| gi|55663625 | GOT1 | glutamic-oxaloacetic transaminase 1, soluble (aspartate aminotransferase 1) | 6.1 | 6.1 | 5 | ||||||
| gi|182518 | FTL | ferritin light subunit | 5.9 | 5.9 | 2 | ||||||
| gi|1843434 | TXNRD1 | thioredoxin reductase 1 | 5.5 | 5.5 | 4 | ||||||
| gi|15418999 | ANTXR2 | capillary morphogenesis protein-2 | 2.5 | 2.5 | 2 | ||||||
| gi|56204739 | HIST3H2A | histone 3, H2a | 2.4 | 2.4 | 2 | ||||||
| gi|4503445 | TYMP | thymidine phosphorylase (endothelial growth factor) | 4.9 | 4.9 | 3 | ||||||
| gi|40674605 | CAPNS1 | CAPNS1 protein | 2.3 | 2.3 | 7 | ||||||
| gi|30581141 | PSME1 | proteasome activator subunit 1 isoform 2 | 2.3 | 2.3 | 11 | ||||||
| gi|3036787 | HIST1H2BK | histone 1, H2bk | 2.3 | 2.3 | 7 | ||||||
| gi|12803167 | NAP1L1 | Nucleosome assembly protein 1-like 1 | 4.7 | 4.7 | 4 | ||||||
| gi|13623579 | CLPB | caseinolytic peptidase B homologue (E. coli) | 2.3 | 2.3 | 2 | ||||||
| gi|15080241 | PACSIN3 | Protein kinase C and casein kinase substrate in neurons 3 | 2.2 | 2.2 | 7 | ||||||
| gi|4507207 | SRI | sorcin isoform a | 4.5 | 4.5 | 2 | ||||||
| gi|544482 | ALDH1A3 | aldehyde dehydrogenase 6 | 4.3 | 4.3 | 17 | ||||||
| gi|12803089 | NUTF2 | Nuclear transport factor 2 | 4.3 | 4.3 | 2 | ||||||
| gi|55960474 | ANXA7 | annexin A7 | 4.3 | 4.3 | 7 | ||||||
| gi|51895905 | STMN1 | Stathmin1/oncoprotein 18 | 4.2 | 4.2 | 4 | ||||||
| gi|12654831 | PGM3 | Phosphoglucomutase 3 | 4.2 | 4.2 | 2 | ||||||
| gi|13623581 | TXNDC17 | Thioredoxin domain containing 17 | 4.1 | 4.1 | 3 | ||||||
Ranked by average z-score.
Table 2. HBAE Proteins Decreased >95% Confidencea.
| virus/mock
protein ratio |
no. of peptides |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| gi accession | symbol | gene name | run 1 | run 2 | run 3 | run 4 | ratio avg | run 1 | run 2 | run 3 | run 4 |
| Cytosolic Proteins Detected More than Once | |||||||||||
| gi|56205024 | PFDN4 | PFDN4 | 0 | 0 | 0 | 2 | 2 | ||||
| gi|10798851 | FADS2 | fatty acid desaturase 2 | 0.1 | 0.5 | 0.3 | 1 | 2 | ||||
| gi|38511857 | GLG1 | Golgi apparatus protein 1 | 0.3 | 0.2 | 0.3 | 0.4 | 0.3 | 11 | 8 | 13 | 11 |
| gi|1107687 | FAT1 | homologue of Drosophila Fat protein | 0.4 | 0.3 | 0.3 | 6 | 7 | ||||
| gi|14328083 | FDFT1 | Farnesyl-diphosphate farnesyltransferase 1 | 0.3 | 0.3 | 0.4 | 0.3 | 11 | 12 | 13 | ||
| gi|56206185 | SPRR2D | small proline-rich protein 2D | 0.6 | 0.2 | 0.4 | 6 | 2 | ||||
| gi|55662663 | COL12A1 | collagen, type XII, α 1 | 0.3 | 0.4 | 0.4 | 1 | 2 | ||||
| gi|8489095 | TNFRSF10B | Fas-like protein precusor | 0.4 | 0.4 | 0.4 | 4 | 4 | ||||
| gi|40317626 | THBS1 | thrombospondin 1 precursor | 0.4 | 0.3 | 0.5 | 0.4 | 50 | 63 | 55 | ||
| gi|28881898 | F3 | tissue factor | 0.4 | 0.4 | 0.4 | 0.4 | 5 | 5 | 6 | ||
|
gi|34364820; gi|16933542 |
FN1 | Fibronectin 1 | 0.4 | 0.4 | 0.4 | 55 | 75 | ||||
| gi|7243073 | ADAMTS1 | Metalloprotease with thrombospondin type 1 motif, 1 | 0.4 | 0.3 | 0.6 | 0.4 | 2 | 2 | 4 | ||
| gi|9971118 | JAG1 | JAG1 | 0.5 | 0.3 | 0.4 | 1 | 2 | ||||
| gi|1945072 | TNFRSF 10A | cytotoxic ligand TRAIL receptor | 0.5 | 0.4 | 0.4 | 2 | 1 | ||||
| gi|3282161 | TGFBI | Transforming growth factor, β-induced, 68 kDa | 0.5 | 0.4 | 0.4 | 0.4 | 2 | 2 | 2 | ||
| gi|54607033 | INTB4 | integrin β 4 isoform 3 precursor | 0.4 | 0.4 | 0.5 | 0.5 | 49 | 43 | 36 | ||
| gi|12274842 | LAMA5 | Laminin 5 | 0.5 | 0.5 | 0.5 | 7 | 4 | ||||
| gi|55662275 | IGFR2 | Insulin-like growth factor 2 receptor | 0.5 | 0.4 | 0.6 | 0.6 | 0.5 | 16 | 15 | 17 | 14 |
| gi|55960673 | PTPRF | Protein tyrosine phosphatase, receptor type, F | 0.7 | 0.5 | 0.6 | 0.4 | 0.5 | 1 | 3 | 3 | 4 |
| gi|15080220 | C1orf212 | C1orf212 chromosome 1 open reading frame 212 | 0.7 | 0.4 | 0.6 | 0.6 | 2 | 1 | 2 | ||
| gi|46020022 | LAMA3 | laminin α 3b chain | 0.4 | 0.7 | 0.4 | 79 | 58 | ||||
| Cytosolic Proteins Detected Only Once | |||||||||||
| gi|31979223 | RPLP1 | acidic ribosomal phosphoprotein P1 | 0 | 0 | 8 | ||||||
| gi|55957204 | NUP50 | Nucleoporin 50 kDa | 0 | 0 | 2 | ||||||
| gi|13436176 | FADS3 | Fatty acid desaturase 3 | 0 | 0 | 2 | ||||||
| gi|55962101 | EIF4G3 | eukaryotic translation initiation factor 4 gamma, 3 | 0 | 0 | 2 | ||||||
| gi|18568105 | RAB22 | RAB22 | 0 | 0 | 2 | ||||||
| gi|2772927 | TGOLN2 | Trans-Golgi network protein 2 | 0.03 | 0.03 | 2 | ||||||
| gi|48775023 | RELA | RELA protein | 0.3 | 0.3 | 2 | ||||||
| gi|7688699 | RER1 | Retention in endoplasmic reticulum homologue 1 | 0.3 | 0.3 | 3 | ||||||
| gi|52545861 | TRAPPC9 | Trafficking protein particle complex 9 | 0.4 | 0.4 | 3 | ||||||
| gi||4336424 | MCAM | cell surface glycoprotein P1H12 precursor | 0.4 | 0.4 | 3 | ||||||
| gi|6066280 | CROT | peroxisomal carnitine octanoyltransferase | 0.4 | 0.4 | 3 | ||||||
| gi|4505149 | SERPINB7 | serine (or cysteine) proteinase inhibitor, clade B (ovalbumin), member 7 | 0.4 | 0.4 | 2 | ||||||
| gi|7022598 | DARS2 | Aspartyl-tRNA synthetase 2, mitochondrial | 0.4 | 0.4 | 3 | ||||||
| gi|16041779 | SLC39A14 | SLC39A14 protein | 0.4 | 0.4 | 2 | ||||||
| gi|37537242 | RNF149 | Ring finger protein 149 | 0.5 | 0.5 | 2 | ||||||
| gi|34533080 | SH3BP1 | SH3-domain binding protein 1 | 0.5 | 0.5 | 3 | ||||||
| gi|48735299 | SNRPA1 | Small nuclear ribonucleoprotein polypeptide A′ | 0.4 | 0.4 | 3 | ||||||
| gi|13529266 | PSMC6 | Proteasome (prosome, macropain) 26S subunit, ATPase, 6 | 0.4 | 0.4 | 10 | ||||||
| gi|1825562 | PDCD4 | nuclear antigen H731 | 0.5 | 0.5 | 4 | ||||||
Ranked by average z-score.
Figure 3.
Western blotting was performed to confirm SILAC virus:mock ratios. HBAE cells were harvested and lysed with 0.5% NP-40 detergent, nuclei were removed, and cytosolic fractions were dissolved in SDS electrophoresis sample buffer, resolved in 8%, 10%, or 15% mini SDS-PAGE, transferred to PVDF, and probed with various antibodies. Bands were visualized and intensities were measured with an Alpha Innotech FluorChemQ MultiImage III instrument. Molecular weight standards are indicated at left and ratios of each protein (infected divided by mock-infected) are indicated for each protein at right, along with SILAC-measured ratios (far right). Western blot ratios for host proteins that were detected only in infected cells (undetectable in mock) are designated “n.a.”. SILAC ratios are an average of the four replicate runs performed, and Western blotting ratios are from a single experiment in a different donor than was used for SILAC. Bands from Western blotting were quantitated using AlphaEase software and are reported without normalization. *: no viral proteins measured by SILAC because not present in mock-infected samples.
Up-Regulated Proteins Are Associated with Host Cell Defense Responses, Endocytosis, and GTPase Activity
Gene ontology analyses using GO and Panther classification terms were determined for proteins that were up-regulated in response to influenza infection and compared at three different confidence intervals: 95% (z-score >1.96), 99% (z-score >2.58) and 99.9% (z-score >3.29) (Figure 4a). Additionally, canonical pathways and networks that were represented by influenza-altered proteins were constructed with IPA at the 95%, 99%, and 99.9% confidence intervals. Representative pathways and networks are shown only at the 95% interval. Approximately half of the proteins up-regulated in response to influenza infection in HBAE could be attributed to interferon and other known host cell defense responses. These proteins were identified by several gene ontology categories including defense response, immune response, response to virus and defense response to virus (Figure 4a). Collectively, these categories included proteins ISG15, RSAD2, MX1, MX2, IFIT1, IFIT2, IFIT3/4, IFITM1, SAMD9, SAMHD1, OASL, OAS3, TLR2, STAT1, GBP-1, and HLA-B/C. Pathway analyses also identified immune systems and interferon signaling (Figure 5, Supplementary Figure 2a). Proteins belonging to endocytic and intracellular protein trafficking pathways were also indicated as up-regulated (Figure 4a) and included α-actin, MX1, MX2, and PPIA. Related canonical pathways that were identified by IPA include caveolar-mediated endocytosis, viral entry via endocytic pathways, and mechanisms of viral exit from host cells (Supplementary Figure 3b). A third category that was identified among influenza-up-regulated proteins was the presence of GTPase activity, which included proteins MX1, MX2, and GBP1 (Figure 4a).
Figure 4.
Gene ontology (GO) analysis of up- and down-regulated proteins. Lists of (A) up- and (B) down-regulated protein IDs were uploaded into DAVID separately and analyzed for enrichment of categories belonging to biological processes, cellular components, and molecular functions. Additionally, lists of proteins determined at different confidence intervals (95%, 99%, 99.9%) were compared.
Figure 5.
Interactions between up- and down-regulated proteins. Protein IDs and ratios from Tables 1 and 2 (95% confidence interval) were combined and imported into the Ingenuity Pathways Analysis (IPA) tool from which interacting pathways were constructed. Up- and down-regulated proteins are denoted in red and green, respectively; gray proteins indicate that they were detected in our study but not regulated; white proteins interact with many proteins in the network but were not detected in this study. Any known direct connections between these proteins are indicated by solid lines; indirect interactions are not shown here. Networks are titled (A) Infection, Gene Expression, Antimicrobial Processe; (B) Cancer, Dermatological Conditions, Cellular Development; and (C) Connective Tissues, Genetics, CV System Development and Function.
Down-Regulated Proteins Are Associated with Cell Adhesion, Cell Death Signaling, and Lipid Metabolism
Many biological processes were represented among proteins down-regulated at the 95% confidence level. Notably, many of these are not cytosolic proteins but were mainly transmembrane (FADS2, ODZ2, FAT1, F3, GLG1, TNFRSF10A, JAG1, FDFT1, SLC16A2, TNFRSF10B) and extracellular matrix-related proteins (COL12A1, LAMA3, LAMA5, FN1, ADAMTS1, BIGH3) (Figure 4b). Cellular functions that were attributed to these proteins included cell adhesion (COL12A1, FAT1, FN1, GLG1, LAMA3, LAMA5) (Figure 4b) as well as signal transduction (ODZ2, collagen, FAT1, FN, F3, BIGH3, TNFRSF10A, TNFRSF10B, ADAMTS1) (Figure 4a). TRAIL signaling, a process involved in inducing cell death, was also prominently identified (TNFRSF10A, TNFRSF10B) (Figures 4b and 5b, Supplementary Figures 2a and 3d). Third, several enzymes involved in lipid metabolic processes were consistently down-regulated including sterol metabolism (FDFT1) and fatty acid metabolism (FADS2, FADS3) (Figures 4b and 5b, Supplementary Figures 2b and 3d).
Discussion
Our lab has previously used SILAC to study A549 alveolar epithelial cell host pathways that are altered in response to infection by influenza virus A/PR/8/34, a highly attenuated mouse-adapted strain of influenza virus.22 We have now extended this work to include primary human airway epithelial cells, a model that is more closely related to the in vivo situation and that offers opportunity to assess the global relevance of proteomics data obtained using transformed human cell lines. Using a human database, we identified 3740 cytosolic HBAE protein pairs, of which only a small fraction was altered in response to A/PR/8/34 influenza infection. Proteins that were up- and down-regulated were determined statistically using different confidence intervals for which the 95% confidence interval corresponded to a 2-fold or more difference, the 99% limit to at least a 2.5-fold difference, and 99.9% confidence to a 3-fold or greater change. Since many previous studies have used either a 2-fold or 1.5-fold change in protein abundance to create data sets of infection-altered proteins,14,15,22 the cutoff values used here are comparable. As viral proteins are not detected by human database searches, we titered the amount of virus secreted by our SILAC labeled cells at 24 hpi to ensure that viral infection and replication had been successful. It is worth noting that the maximum virus titers produced by HBAE vary from donor to donor. For example, the particular cells used in Figure 1a produced the highest titers we have observed in HBAE, whereas the cells used for the SILAC experiments (Figure 2c) produced considerably lower titers. However, despite these differences, we find robust viral protein expression in these cells from 6 to 72 hpi and can find comparable proteomic expression patterns among HBAE from different donors (data not shown).
The current study consists of the “cytoplasmic” cellular fraction, which includes all NP40 soluble components such as proteins of the cell membrane, cytoplasm, and cytoplasmic organelles, thereby excluding mainly nuclear proteins. We recognize that the nuclear sample is also an important component of the cell to analyze, particularly because influenza virus replicates within the nucleus, and anticipate analyzing these fractions in future studies. However, the primary purpose of the present study was to compare HBAE results to A549 results,22 and the later study had only been performed with the NP-40-soluble fraction.
Up-Regulated Processes
It is well documented that in many cell types interferon expression is a central cellular response to viral infection.39−42 This is thought to be key in developing an antiviral state in both infected and neighboring noninfected cells to limit viral replication.43 Consistent with this paradigm, in our current study up-regulated proteins in HBAE were chiefly enriched for host cell defense responses such as interferon (INF) and the JAK/STAT pathway (Figures 4a and 5a, Supplementary Figures 2a and 3a). Of note, influenza A can also antagonize interferon signaling via the viral NS-1 protein through multiple mechanisms including blocking host translation and sequestering viral RNA in order to prevent its detection by host pathogen recognition processes.43,44 However, the extent of this inhibitory effect is strain-dependent,43,45 and since the NS1 protein produced by the A/PR/8/34 influenza strain is generally thought to be a less potent inhibitor of interferon production than other strains’ NS1 protein,43 this correlates well with our data.
Some of these findings were also unique to HBAE compared to our previous A549 study. Interestingly, neither GO nor IPA identified interferon signaling in PR8-infected A549 cells, whereas it was highly significant in HBAE cells (p-value = 0.001). In addition, while a few interferon-induced proteins (RSAD2, MX1, MX2, and ISG15) were highly up-regulated in both cell types, several (STAT1, RIG-I, SAMD9, SAMHD1, IFIT1, IFIT2, IFIT3) were highly up-regulated in HBAE but unaltered or undetected in A549. Indeed, we were able to confirm with Western blotting that cytosolic STAT1 abundance was increased in HBAE after influenza infection, whereas levels in A549 remained relatively unchanged. SamD9, a molecule downstream of STAT1, was more highly induced in HBAE than A549 after infection, whereas IFIT1 was detected only in HBAE after infection (Figure 6). This suggests that less interferon is produced by A549 than HBAE cells and that this difference is dependent either on the cell type (A549 are alveolar epithelial in origin whereas HBAE are from airways) or properties of the cell line (i.e., primary versus transformed) rather than the virus strain. Notably, a difference in interferon signaling and production could explain the ability of A549, but not HBAE, to support replication of the PR8 virus when infected at a low MOI (0.01 PFU/cell). These observations provide rationale for future studies assessing the precise role(s) of interferon-associated proteins in preventing viral replication in primary epithelia and mechanisms that suppress this effect in transformed or alveolae-derived epithelial cells.
Figure 6.
Comparison of JAK/STAT signaling molecule abundance in A549 and HBAE cells after influenza infection. Cells were harvested and lysed with 0.5% NP-40 detergent, nuclei were removed, and cytosolic fractions were dissolved in SDS electrophoresis sample buffer, resolved in 10% or 12% mini SDS-PAGE, transferred to PVDF, and probed with indicated antibodies. Bands were visualized and intensities were measured with an Alpha Innotech FluorChemQ MultiImage III instrument. Molecular weight standards are indicated at left, and ratios of each protein (infected divided by mock-infected) are indicated for each protein at right. Protein ratios are an average of three experiments each from A549 cells and a single HBAE donor and are reported after normalization to GAPDH.
The interferon-induced proteins commonly up-regulated in both A549 and HBAE (RSAD2, MX1, MX2, and ISG15) were also identified in numerous other influenza studies. For example, myxovirus resistance host proteins Mx1 and Mx2 are up-regulated in a variety of cell models including MDCK20 and A54922,46 cells, as well as in vivo in studies with macaques.26 ISG15 and RSAD2, interferon-induced proteins with anti-influenza activity, were also up-regulated in A549 cells21,22 and primary macrophages.16,47 These proteins have also been identified in proteomics studies of human host cell responses to infection with other viruses such as RSV,48 adenovirus,49 and sindbis virus and HSV.50 Interferon-induced tetratricopeptide repeat proteins (IFIT1–3) have also been found in multiple proteomic studies of influenza involving primary macrophages,16 the polarized Calu-3 airway epithelial cell line23 and animal models.24−26 Interestingly, these three IFIT proteins have been discovered to form an antiviral complex against 5′-triphosphate RNA,51 which is thought to be present during an influenza infection. It is likely that these similar responses to different types of infection reflect the induction of a nonspecific innate immune response against RNA viruses by the host cells.
Down-Regulated Processes
We found 19 proteins that were consistently and strongly down-regulated in primary HBAE cells in response to influenza infection, and these were mainly linked to cell adhesion, death receptor signaling, and lipid metabolism.
Cellular adhesion is necessary for the survival of primary cells, and previous studies have found that a reduction in adhesion molecules precedes and may possibly be instrumental in inducing cell death.52 In epithelial cells adherens junction proteins are essential for cell adhesion and these are largely composed of integrin, cadherin, catenin, and actin complexes.53 Many proteins belonging to these complexes and downstream signaling pathways were detected in our SILAC data among which integrin β 4 was down-regulated 2.4-fold, GLG1 2.5-fold, and THBS1 2.6-fold. Other adhesion-related proteins such as fibronectin, laminin, TGFBI, and TNC were down-regulated >2.0-fold.
However, cell death processes were not identified by any of our analyses, except for the down-regulation of two TRAIL receptors, TNFSFR10A and TNFSFR10B (Figures 4b and 5b). TNFRSF10A, TNFRSF10B and F3 are additionally associated with caspase activity induction, which was similarly indicated as down-regulated (Figure 4b). In contrast, TNFSFR10D was found up-regulated 1.4-fold in PR8-infected A549 cells at the same time point.22 Given that TRAIL signaling is important in clearing influenza-infected epithelial cells, it would be interesting to further investigate the mechanism by which TRAIL-receptors are down-regulated in HBAE.
As cholesterol and lipids are key to maintaining the integrity of the cell membrane and the organization of lipid rafts, it was interesting to find that several proteins related to lipid biosynthesis were down-regulated in response to influenza. For example, a previous study indicated that RSAD2 interferes with cholesterol synthesis and subsequent lipid raft integrity by binding and inhibiting FPPS, an enzyme in the cholesterol biosynthetic pathway.54,55 Our SILAC/HBAE data found RSAD2 highly up-regulated (>3-fold) and FPPS moderately up-regulated (1.5-fold) (Supplementary Figure 1c). Importantly, FPPS is involved not only in the production of cholesterol but also other intermediates, such as isoprenoids FPP and GGPP, which serve as essential activators of GTPase proteins. Our SILAC data identified a number of proteins from this pathway, of which FDFT1 was most significantly altered (2.9-fold down-regulated). Fatty acid synthesis was also affected through a strong downregulation of FADS2 (3.45-fold). A second study has also found that interferon signaling can suppress sterol biosynthesis in macrophages,56 suggesting that this may be a protective response induced by the host cell rather than by the virus. While none of FPPS, FDFT1, nor FADS2 were altered in A549 cells at 24 hpi, IPA indicated other lipid-related pathways as down-regulated in A549 cells including sphingolipid metabolism and VDR/RXR signaling. Collectively, these data suggest that lipid metabolism may be inhibited in epithelial cells during influenza infection.
Conclusion
Overall this proteomics study provides a broad and unbiased profile of cytoplasmic host cell proteins that are up- and down-regulated in response to an H1N1 mouse-adapted influenza virus in primary human airway epithelial cells. While some of our findings correlate with previous proteomic studies involving influenza or other viruses, we have identified many proteins that have not been previously studied with respect to influenza. Thus, our work has expanded the knowledge base for understanding host cell response to influenza infection. Of note, we performed our studies using relevant primary human bronchial epithelial cells, whereas many previous studies have been conducted using transformed cell lines such as alveolar A549 cells or nonlung HEK293 cells. A comparison of functional categories and pathway alterations between our studies with A549 and HBAE identified some common characteristics such as downregulation of cell adhesion and lipid metabolism, as well as upregulation of host defense responses against influenza infection. In contrast, downregulation of TRAIL receptors was specific to HBAE cells. Further validation and investigation into the functions of the candidate proteins we have identified will contribute to understanding novel virus-host interactions and permit integration of this information with previous large-scale analyses of influenza to identify new directions for developing anti-influenza therapies.
Acknowledgments
This work was supported by grant MOP-77759 from the Canadian Institute of Health Research (CIHR) awarded to A.J.H. and CIHR grants PAN-83159, ROP-104906 and MOP-106713 to K.M.C.; by a Small Grant from the Manitoba Institute for Child Health to K.M.C.; by a Manitoba Health Research Council Opportunities Grant awarded to A.J.H. and K.M.C., as well as by CIHR and MHRC graduate studentships awarded to A.L.K. The authors thank Kolawole Opanubi for expert technical assistance, Dr. James House, Director, Animal Sciences for embryonated hens eggs in which some influenza virus stocks were grown, Dr. Ming Yang for anti-NP mAb hybridoma cells, Patty Sauder and Niaz Rahim for anti-influenza virus NS1 monoclonal antibody production, Matthew Stuart-Edwards for developing scripts for bioinformatics analysis. and Dr. Oleg Krokhin for adapting and optimizing the 2D LC–MS/MS platform used for our experiments.
Glossary
Abbreviations
- HBAE
human bronchial airway epithelial cells
- hpi
hours post infection
- IAV
influenza A virus
- INF
interferon
- NS-1
non-structural protein 1
- PR8
influenza strain A/PR/8/34
- SILAC
stable isotope labeling of amino acids in cell culture
Supporting Information Available
This material is available free of charge via the Internet at http://pubs.acs.org.
The authors declare no competing financial interest.
Supplementary Material
References
- Palese P.; Shaw M. L.. Orthomyxoviridae: The Viruses and Their Replication. In Fields Virology, 5th ed.; Lippencott Williams & Wilkins: Philadelphia, 2007; Vol. 2, pp 1647–1689. [Google Scholar]
- Shapira S. D.; Gat-Viks I.; Shum B. O.; Dricot A.; de Grace M. M.; Wu L.; Gupta P. B.; Hao T.; Silver S. J.; Root D. E.; Hill D. E.; Regev A.; Hacohen N. A physical and regulatory map of host-influenza interactions reveals pathways in H1N1 infection. Cell 2009, 139 (7), 1255–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kash J. C.; Goodman A. G.; Korth M. J.; Katze M. G. Hijacking of the host-cell response and translational control during influenza virus infection. Virus Res. 2006, 119 (1), 111–20. [DOI] [PubMed] [Google Scholar]
- Gannage M.; Dormann D.; Albrecht R.; Dengjel J.; Torossi T.; Ramer P. C.; Lee M.; Strowig T.; Arrey F.; Conenello G.; Pypaert M.; Andersen J.; Garcia-Sastre A.; Munz C. Matrix protein 2 of influenza A virus blocks autophagosome fusion with lysosomes. Cell Host Microbe 2009, 6 (4), 367–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig S.; Pleschka S.; Planz O.; Wolff T. Ringing the alarm bells: signalling and apoptosis in influenza virus infected cells. Cell Microbiol. 2006, 8 (3), 375–86. [DOI] [PubMed] [Google Scholar]
- Sumbayev V. V.; Yasinska I. M. Role of MAP kinase-dependent apoptotic pathway in innate immune responses and viral infection. Scand. J. Immunol. 2006, 63 (6), 391–400. [DOI] [PubMed] [Google Scholar]
- Hale B. G.; Randall R. E.; Ortin J.; Jackson D. The multifunctional NS1 protein of influenza A viruses. J. Gen. Virol. 2008, 89 (Pt 10), 2359–76. [DOI] [PubMed] [Google Scholar]
- Hao L.; Sakurai A.; Watanabe T.; Sorensen E.; Nidom C. A.; Newton M. A.; Ahlquist P.; Kawaoka Y. Drosophila RNAi screen identifies host genes important for influenza virus replication. Nature 2008, 454 (7206), 890–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brass A. L.; Huang I. C.; Benita Y.; John S. P.; Krishnan M. N.; Feeley E. M.; Ryan B. J.; Weyer J. L.; van der Weyden L.; Fikrig E.; Adams D. J.; Xavier R. J.; Farzan M.; Elledge S. J. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile virus, and dengue virus. Cell 2009, 139 (7), 1243–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig R.; Stertz S.; Zhou Y.; Inoue A.; Hoffmann H. H.; Bhattacharyya S.; Alamares J. G.; Tscherne D. M.; Ortigoza M. B.; Liang Y.; Gao Q.; Andrews S. E.; Bandyopadhyay S.; De Jesus P.; Tu B. P.; Pache L.; Shih C.; Orth A.; Bonamy G.; Miraglia L.; Ideker T.; Garcia-Sastre A.; Young J. A.; Palese P.; Shaw M. L.; Chanda S. K. Human host factors required for influenza virus replication. Nature 2010, 463 (7282), 813–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlas A.; Machuy N.; Shin Y.; Pleissner K. P.; Artarini A.; Heuer D.; Becker D.; Khalil H.; Ogilvie L. A.; Hess S.; Maurer A. P.; Muller E.; Wolff T.; Rudel T.; Meyer T. F. Genome-wide RNAi screen identifies human host factors crucial for influenza virus replication. Nature 2010, 463 (7282), 818–22. [DOI] [PubMed] [Google Scholar]
- Ludwig S. Targeting cell signalling pathways to fight the flu: towards a paradigm change in anti-influenza therapy. J. Antimicrob. Chemother. 2009, 64 (1), 1–4. [DOI] [PubMed] [Google Scholar]
- Pathak S.; De Souza G. A.; Salte T.; Wiker H. G.; Asjo B. HIV induces both a down-regulation of IRAK-4 that impairs TLR signalling and an up-regulation of the antibiotic peptide dermcidin in monocytic cells. Scand. J. Immunol. 2009, 70 (3), 264–76. [DOI] [PubMed] [Google Scholar]
- Mannova P.; Fang R.; Wang H.; Deng B.; McIntosh M. W.; Hanash S. M.; Beretta L. Modification of host lipid raft proteome upon hepatitis C virus replication. Mol. Cell. Proteomics 2006, 5 (12), 2319–25. [DOI] [PubMed] [Google Scholar]
- Kang S. M.; Shin M. J.; Kim J. H.; Oh J. W. Proteomic profiling of cellular proteins interacting with the hepatitis C virus core protein. Proteomics 2005, 5 (8), 2227–37. [DOI] [PubMed] [Google Scholar]
- Lietzen N.; Ohman T.; Rintahaka J.; Julkunen I.; Aittokallio T.; Matikainen S.; Nyman T. A. Quantitative subcellular proteome and secretome profiling of influenza A virus-infected human primary macrophages. PLoS Pathog. 2011, 7 (5), e1001340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohman T.; Rintahaka J.; Kalkkinen N.; Matikainen S.; Nyman T. A. Actin and RIG-I/MAVS signaling components translocate to mitochondria upon influenza A virus infection of human primary macrophages. J. Immunol. 2009, 182 (9), 5682–92. [DOI] [PubMed] [Google Scholar]
- Liu N.; Song W.; Wang P.; Lee K.; Chan W.; Chen H.; Cai Z. Proteomics analysis of differential expression of cellular proteins in response to avian H9N2 virus infection in human cells. Proteomics 2008, 8 (9), 1851–8. [DOI] [PubMed] [Google Scholar]
- Zhang L.; Zhang X.; Ma Q.; Ma F.; Zhou H. Transcriptomics and proteomics in the study of H1N1 2009. Genomics, Proteomics Bioinf. 2010, 8 (3), 139–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vester D.; Rapp E.; Gade D.; Genzel Y.; Reichl U. Quantitative analysis of cellular proteome alterations in human influenza A virus-infected mammalian cell lines. Proteomics 2009, 9 (12), 3316–27. [DOI] [PubMed] [Google Scholar]
- Liu H. C.; Hicks J.; Yoo D. Proteomic dissection of viral pathogenesis. Dev. Biol. (Basel) 2008, 132, 43–53. [DOI] [PubMed] [Google Scholar]
- Coombs K. M.; Berard A.; Xu W.; Krokhin O.; Meng X.; Cortens J. P.; Kobasa D.; Wilkins J.; Brown E. G. Quantitative proteomic analyses of influenza virus-infected cultured human lung cells. J. Virol. 2010, 84 (20), 10888–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C.; Bankhead A.; Eisfeld A. J.; Hatta Y.; Jeng S.; Chang J. H.; Aicher L. D.; Proll S.; Ellis A. L.; Law G. L.; Waters K.; Neumann G.; Katze M. G.; McWeeney S.; Kawaoka Y. Host regulatory network response to infection with highly pathogenic H5N1 avian influenza virus. J. Virol. 2011, 85, 10955–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zinman G.; Brower-Sinning R.; Emeche C. H.; Ernst J.; Huang G. T.; Mahony S.; Myers A. J.; O’Dee D. M.; Flynn J. L.; Nau G. J.; Ross T. M.; Salter R. D.; Benos P. V.; Bar Joseph Z.; Morel P. A. Large scale comparison of innate responses to viral and bacterial pathogens in mouse and macaque. PLoS One 2011, 6 (7), e22401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J. N.; Palermo R. E.; Baskin C. R.; Gritsenko M.; Sabourin P. J.; Long J. P.; Sabourin C. L.; Bielefeldt-Ohmann H.; Garcia-Sastre A.; Albrecht R.; Tumpey T. M.; Jacobs J. M.; Smith R. D.; Katze M. G. Macaque proteome response to highly pathogenic avian influenza and 1918 reassortant influenza virus infections. J. Virol. 2010, 84 (22), 12058–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baas T.; Baskin C. R.; Diamond D. L.; Garcia-Sastre A.; Bielefeldt-Ohmann H.; Tumpey T. M.; Thomas M. J.; Carter V. S.; Teal T. H.; Van Hoeven N.; Proll S.; Jacobs J. M.; Caldwell Z. R.; Gritsenko M. A.; Hukkanen R. R.; Camp D. G. 2nd; Smith R. D.; Katze M. G. Integrated molecular signature of disease: analysis of influenza virus-infected macaques through functional genomics and proteomics. J. Virol. 2006, 80 (21), 10813–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scull M. A.; Gillim-Ross L.; Santos C.; Roberts K. L.; Bordonali E.; Subbarao K.; Barclay W. S.; Pickles R. J. Avian Influenza virus glycoproteins restrict virus replication and spread through human airway epithelium at temperatures of the proximal airways. PLoS Pathog. 2009, 5 (5), e1000424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibricevic A.; Pekosz A.; Walter M. J.; Newby C.; Battaile J. T.; Brown E. G.; Holtzman M. J.; Brody S. L. Influenza virus receptor specificity and cell tropism in mouse and human airway epithelial cells. J. Virol. 2006, 80 (15), 7469–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. W.; Yuen K. M.; Yu W. C.; Ho C. C.; Nicholls J. M.; Peiris J. S.; Chan M. C. Influenza H5N1 and H1N1 virus replication and innate immune responses in bronchial epithelial cells are influenced by the state of differentiation. PLoS One 2010, 5 (1), e8713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson C. I.; Barclay W. S.; Zambon M. C.; Pickles R. J. Infection of human airway epithelium by human and avian strains of influenza a virus. J. Virol. 2006, 80 (16), 8060–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer V.; Yamchuk A.; Cortens J.; Sousa S.; Ens W.; Standing K. G.; Wilkins J. A.; Krokhin O. V. Sequence-specific retention calculator. A family of peptide retention time prediction algorithms in reversed-phase HPLC: applicability to various chromatographic conditions and columns. Anal. Chem. 2007, 79 (22), 8762–8. [DOI] [PubMed] [Google Scholar]
- Dwivedi R. C.; Spicer V.; Harder M.; Antonovici M.; Ens W.; Standing K. G.; Wilkins J. A.; Krokhin O. V. Practical implementation of 2D HPLC scheme with accurate peptide retention prediction in both dimensions for high-throughput bottom-up proteomics. Anal. Chem. 2008, 80 (18), 7036–42. [DOI] [PubMed] [Google Scholar]
- Ho Y. S.; Wang Y. J.; Lin J. K. Induction of p53 and p21/WAF1/CIP1 expression by nitric oxide and their association with apoptosis in human cancer cells. Mol. Carcinog. 1996, 16 (1), 20–31. [DOI] [PubMed] [Google Scholar]
- Huang da W.; Sherman B. T.; Lempicki R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4 (1), 44–57. [DOI] [PubMed] [Google Scholar]
- Dennis G. Jr.; Sherman B. T.; Hosack D. A.; Yang J.; Gao W.; Lane H. C.; Lempicki R. A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4 (5), P3. [PubMed] [Google Scholar]
- Abedin M. J.; Wang D.; McDonnell M. A.; Lehmann U.; Kelekar A. Autophagy delays apoptotic death in breast cancer cells following DNA damage. Cell Death Differ. 2007, 14 (3), 500–10. [DOI] [PubMed] [Google Scholar]
- Arcangeletti M. C.; De Conto F.; Ferraglia F.; Pinardi F.; Gatti R.; Orlandini G.; Covan S.; Motta F.; Rodighiero I.; Dettori G.; Chezzi C. Host-cell-dependent role of actin cytoskeleton during the replication of a human strain of influenza A virus. Arch. Virol. 2008, 153 (7), 1209–21. [DOI] [PubMed] [Google Scholar]
- Arumugam E.; Pereira J.; Gilbert P. M. Reconstruction after skin sparing mastectomy. Ann. R. Coll. Surg. Engl. 2005, 87 (4), 314.; author reply 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu A. C.; Barr I.; Hansbro P. M.; Wark P. A. Human Influenza is More Effective than Avian Influenza at Antiviral Suppression in Airway Cells. Am. J. Respir. Cell Mol. Biol. 2010, 44, 906–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapp S.; Derby N. R.; Singer R.; Shaw A.; Williams V. G.; Turville S. G.; Bess J. W. Jr.; Lifson J. D.; Robbiani M. Double-stranded RNA analog poly(I:C) inhibits human immunodeficiency virus amplification in dendritic cells via type I interferon-mediated activation of APOBEC3G. J. Virol. 2009, 83 (2), 884–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy J. A.; Duerst R. J.; Smith T. J.; Morrison L. A. Herpes simplex virus type 2 virion host shutoff protein regulates alpha/beta interferon but not adaptive immune responses during primary infection in vivo. J. Virol. 2003, 77 (17), 9337–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F. F.; Zhang Y. B.; Liu T. K.; Liu Y.; Sun F.; Jiang J.; Gui J. F. Fish virus-induced interferon exerts antiviral function through Stat1 pathway. Mol. Immunol. 2010, 47 (14), 2330–41. [DOI] [PubMed] [Google Scholar]
- Haye K.; Burmakina S.; Moran T.; Garcia-Sastre A.; Fernandez-Sesma A. The NS1 protein of a human influenza virus inhibits type I interferon production and the induction of antiviral responses in primary human dendritic and respiratory epithelial cells. J. Virol. 2009, 83 (13), 6849–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mibayashi M.; Martinez-Sobrido L.; Loo Y. M.; Cardenas W. B.; Gale M. Jr.; Garcia-Sastre A. Inhibition of retinoic acid-inducible gene I-mediated induction of beta interferon by the NS1 protein of influenza A virus. J. Virol. 2007, 81 (2), 514–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale B. G.; Barclay W. S.; Randall R. E.; Russell R. J. Structure of an avian influenza A virus NS1 protein effector domain. Virology 2008, 378 (1), 1–5. [DOI] [PubMed] [Google Scholar]
- Geiss G. K.; Salvatore M.; Tumpey T. M.; Carter V. S.; Wang X.; Basler C. F.; Taubenberger J. K.; Bumgarner R. E.; Palese P.; Katze M. G.; Garcia-Sastre A. Cellular transcriptional profiling in influenza A virus-infected lung epithelial cells: the role of the nonstructural NS1 protein in the evasion of the host innate defense and its potential contribution to pandemic influenza. Proc. Natl. Acad. Sci. U.S.A. 2002, 99 (16), 10736–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S. M.; Gardy J. L.; Cheung C. Y.; Cheung T. K.; Hui K. P.; Ip N. Y.; Guan Y.; Hancock R. E.; Peiris J. S. Systems-level comparison of host-responses elicited by avian H5N1 and seasonal H1N1 influenza viruses in primary human macrophages. PLoS One 2009, 4 (12), e8072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munday D.; Emmott E.; Surtees R.; Lardeau C. H.; Wu W.; Duprex W. P.; Dove B. K.; Barr J. N.; Hiscox J. A. Quantitative proteomic analysis of A549 cells infected with human respiratory syncytial virus. Mol. Cell. Proteomics 2010, 9, 2438–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H.; Boije H.; Granberg F.; Pettersson U.; Svensson C. Activation of the interferon-induced STAT pathway during an adenovirus type 12 infection. Virology 2009, 392 (2), 186–95. [DOI] [PubMed] [Google Scholar]
- Lenschow D. J.; Lai C.; Frias-Staheli N.; Giannakopoulos N. V.; Lutz A.; Wolff T.; Osiak A.; Levine B.; Schmidt R. E.; Garcia-Sastre A.; Leib D. A.; Pekosz A.; Knobeloch K. P.; Horak I.; Virgin H. W. t. IFN-stimulated gene 15 functions as a critical antiviral molecule against influenza, herpes, and Sindbis viruses. Proc. Natl. Acad. Sci. U.S.A. 2007, 104 (4), 1371–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichlmair A.; Lassnig C.; Eberle C. A.; Gorna M. W.; Baumann C. L.; Burkard T. R.; Burckstummer T.; Stefanovic A.; Krieger S.; Bennett K. L.; Rulicke T.; Weber F.; Colinge J.; Muller M.; Superti-Furga G. IFIT1 is an antiviral protein that recognizes 5′-triphosphate RNA. Nat. Immunol. 2011, 12 (7), 624–30. [DOI] [PubMed] [Google Scholar]
- Ling Y.; Zhong Y.; Perez-Soler R. Disruption of cell adhesion and caspase-mediated proteolysis of beta- and gamma-catenins and APC protein in paclitaxel-induced apoptosis. Mol. Pharmacol. 2001, 59 (3), 593–603. [DOI] [PubMed] [Google Scholar]
- Suzanne M.; Steller H. Letting go: modification of cell adhesion during apoptosis. J. Biol. 2009, 8 (5), 49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X.; Hinson E. R.; Cresswell P. The interferon-inducible protein viperin inhibits influenza virus release by perturbing lipid rafts. Cell Host Microbe 2007, 2 (2), 96–105. [DOI] [PubMed] [Google Scholar]
- Waheed A. A.; Freed E. O. Influenza virus not cRAFTy enough to dodge viperin. Cell Host Microbe 2007, 2 (2), 71–2. [DOI] [PubMed] [Google Scholar]
- Blanc M.; Hsieh W. Y.; Robertson K. A.; Watterson S.; Shui G.; Lacaze P.; Khondoker M.; Dickinson P.; Sing G.; Rodriguez-Martin S.; Phelan P.; Forster T.; Strobl B.; Muller M.; Riemersma R.; Osborne T.; Wenk M. R.; Angulo A.; Ghazal P. Host defense against viral infection involves interferon mediated down-regulation of sterol biosynthesis. PLoS Biol. 2011, 9 (3), e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.