Abstract
Mitochondria are able to modulate cell state and fate during normal and pathophysiologic conditions through a nuclear-mediated mechanism collectively termed as a retrograde response. Our previous studies in Drosophila melanogaster have clearly established that progress through the cell cycle is precisely regulated by the intrinsic activity of the mitochondrion by specific signaling cascades mounted by the cell. As a means to further our understanding of how mitochondrial energy status affects nuclear control of basic cell decisions, we have employed Affymetrix microarray-based transcriptional profiling of Drosophila S2 cells knocked down for the gene encoding subunit Va of the complex IV of the mitochondrial electron transport chain. The profiling data identify transcriptional upregulation of glycolytic genes, and metabolic studies confirm this increase in glycolysis. The data provide a model of the shift of metabolism from a predominately oxidative state toward a predominately aerobic glycolytic state mediated through transcriptional control. The transcriptional changes alter many signaling systems, including p53, insulin, hypoxia-induced factor α, and conserved mitochondrial retrograde responses. This rich dataset provides many novel targets for further understanding the mechanism whereby the mitochondrion manages energy substrate disposition and directs cellular fate decisions.
Keywords: electron transport, glycolysis, cytochrome oxidase Va, lactate, microarray
Mitochondria are dynamic cellular organelles that act as metabolic hubs to integrate diverse cell extrinsic and intrinsic signals that modulate cell proliferation, differentiation, and death (DiMauro and Schon 2003; Ott et al. 2009; Mandal et al. 2011;). The Drosophila mitochondrial genome encodes only 13 proteins (Garesse and Kaguni 2005), with the vast majority of proteins involved in mitochondrial structure and function encoded by the nuclear genome. Therefore, proper communication between mitochondria and the nucleus is essential for maintaining cellular homeostasis. As mitochondrial biogenesis is completely dependent on the nuclear genome, much attention has been paid to understanding anterograde regulation, the mechanism by which information and materials are transferred from the nucleus and cytoplasm to the mitochondria. However, recent studies in diverse organisms have uncovered a unique process of retrograde regulation by which mitochondria exert specific effects on nuclear function and thereby modulate cellular function under normal and pathophysiological conditions. Although the phenomenon of mitochondrial retrograde regulation is conserved from yeast to humans, the molecular mechanisms underlying the process vary across phyla (Butow and Avadhani 2004).
In budding yeast, the organism most investigated for mitochondrial retrograde signaling, a group of transcription factors known as retrograde (RTG) proteins are involved in transducing a mitochondrial dysfunction signal to the nucleus (Liu and Butow 2006). Through intranuclear translocation, the RTG proteins induce the transcription of specific target genes, which in turn modulate mitochondrial function. A primary function of the RTG target genes is to maintain glutamate supplies to meet biosynthetic needs, as glutamate through the amine derivative glutamine provides all the nitrogen used in biosynthetic reactions. Retrograde regulation in higher plants, as observed in Brassica juncea, involves the MAPK signaling pathway in modulating the expression of nuclear genes associated with cytoplasmic male sterility (Yang et al. 2008). Signaling from mitochondria to nucleus has also been evidenced in mammalian cells. Using C2C12 skeletal myoblasts, mitochondrial stress was found to increase intracytoplasmic calcium ion levels and subsequently activate calcineurin (Biswas et al. 1999). In a model of cancer, osteosarcoma cells depleted of mitochondria were observed to have increased inosine 5′-monophosphate dehydrogenase type 2 and ubiquinol cytochrome-c reductase core protein I proteins as a response to mitochondrial depletion (Kulawiec et al. 2006). This increase in protein production was returned to baseline wild-type levels with repletion of mitochondria through cybrid formation, indicating continuous monitoring and a reversible feedback control.
In recent years, our studies with the genetically tractable organism Drosophila melanogaster led to the identification of two independent retrograde signaling pathways that are activated upon mitochondrial dysfunction and impose a block in G1–S progression during the cell cycle (Mandal et al. 2005; Liao et al. 2006). Molecular genetics analyses revealed that cells mutant for the gene encoding Cytochrome c oxidase subunit Va (CoVa ) of complex IV of the electron transport chain specifically activate a retrograde signaling pathway that involves both AMP-activated protein kinase and p53. The activated p53 leads to transcriptional activation of archipelago, the F-box protein responsible for specific ubiquitinylation of CyclinE (Mandal et al. 2010). The targeting of CyclinE results in proteasomal degradation and thereby imposes a block in G1–S progression. Interestingly, despite a significant drop in cellular ATP level, the CoVa mutant cells do not apoptose, undergo normal differentiation, and can even send axonal projections to the brain. This suggests that apart from activating a G1 cell-cycle checkpoint, retrograde signaling in CoVa mutant cells also modulates nuclear gene expression to support cell survival and activity in an altered metabolic condition. To better understand the genome-wide response to mitochondrial dysfunction, we have employed Affymetrix 3′ gene expression microarrays to define the transcriptional changes in Drosophila S2 cells knocked down for CoVa as a follow-up of our initial mechanistic studies.
The transcriptional profiling experiments described herein reveal that with loss of CoVa by RNA interference (RNAi) there is upregulation of glycolytic genes, thereby demonstrating a shift from oxidative phosphorylation to aerobic glycolysis. A systems biologic interpretation of the most highly differentially expressed genes reveals a portrait of the cellular response to abrogation of electron transport function and identifies the specific genes the cell uses to acquire glucose, control the metabolism through the glycolytic pathway, and generate and dispose of metabolites. Conserved signaling pathways are also found within this data. These responses include the action of p53, insulin, hypoxia-induced factor α (Hifα), stress oxidant responses, and other conserved mitochondrial retrograde signals. This transcriptional data therefore supplies important models of cell-cycle control, energy management, and conserved mitochondrial retrograde responses.
Materials and Methods
CoVa RNA interference in S2 cells and microarray expression profiling
RNAi using a sequence specific to CoVa was performed in Drosophila S2 cells as previously described (Mandal et al. 2005). A GFP sequence not found in the Drosophila genome was used as an experimental control. A DNA template for in vitro transcription was amplified using CoVa primer sequences TAATACGACTCACTATAGGCTGCTACTCGTAA (forward) and TAATACGACTCACTATAGGGTACTTCGTA (reverse); GFP primer sequences TAATACGACTCACTATAGGGAGTGAA (forward) and TAATACGACTCACTATAGGGAGCTTC (reverse). A plasmid containing the GFP coding sequence was kindly provided by Dr. Arnold Berk of the Molecular Biology Institute at UCLA. Using the DNA template amplified from either CoVa or GFP, interfering RNA specific for CoVa and GFP were in vitro transcribed using Megascript T7 RNA polymerase (Ambion, Austin, TX). Cultured Drosophila S2 cells were transfected with 20 μg of RNA specific for either GFP or CoVa using calcium phosphate. In the pilot experiment, cells were harvested at 48 and 168 hr after transfection. In the second and third replicates, cells were harvested at 72, 96, 120, and 168 hr after transfection for a total of three independent time course experiments. At the time of cell collection, low-speed centrifugation and washing in PBS was performed. Total RNA was extracted using trizol (Invitrogen; Carlsbad, CA) as per the manufacturer's protocol. RNA cleanup using the mini RNEASY column was performed as per the manufacturer's protocol (Qiagen; Valencia, CA). RNA quality was ensured by spectrophotometric absorption at 260nm/280nm, as well as by the Agilent Bioanalyzer, which ensured integrity of the small and large ribosomal subunits and lack of degradation (Agilent; Santa Clara, CA).
Total RNA (1 μg) was used to generate microarray probes by standard Affymetrix protocol (Enzo Diagnostics; Farmingdale, NY), which were hybridized to the Affymetrix Drosophila genome 2 arrays (Affymetrix; Santa Clara, CA). The Gene Chip Operating System was used to define absent/present calls and generate cel files using the default settings. Data files (cel) and corresponding text files were uploaded into the dCHIP program and normalized to the median intensity array. Pairwise comparisons were made between the GFP controls and the CoVa RNA–interfered S2 cells.
Quantitative reverse-transcription polymerase chain reaction
Phosphofructokinase (Pfk), phosphoglycerate kinase (Pgk), and the Drosophila homolog of lactate dehydrogenase, Ecdysone-inducible gene L3 (Impl3), were evaluated by quantitative reverse-transcription polymerase chain reaction (qRT-PCR). RNA (1 μg) from the 72 hr time point was evaluated by Super Script Platinum SYBR green One Step q-PCR according to manufacturer's protocol (Invitrogen) using an ABI 7500 thermocycler with the following parameters: 50 C for 10 min, 95 C for 5 min, then 40 cycles of 95 C for 15 sec and 60 C for 60 sec. Data were normalized to Rpl10, and relative quantification to the GFP sample was made using the ΔΔCt method (Livak and Schmittgen 2001). Rpl10 was used as an amplification control and was selected from a survey of the microarray data for genes with the lowest coefficient of variation. The sequences of the primers are as follows: Impl3 forward, ATGGCATTGACAAGGATGTG; Impl3 reverse, GACATGATGTTGGCGGACTT; Pfk forward, AGACGATGGGTGGCTACTGT; Pfk reverse, GGCCATGTGGTAGACATCCT; Pgk forward, AATTGTCGCTGCCTTGGATA; Pgk reverse, GGTGCCAGGGTGTACTTGAT; Rpl10 forward, AAGAAGGTGCTCTGCCTGTC; Rpl10 reverse, CGCACATTCTGCCAGTTCT. Statistical significance was evaluated by Fisher's protected least significant difference using StatView software version 5 (SAS, Cary, NC).
Lactate measurement
The conditioned media from the S2 cells was collected at 168 hr after either GFP (control) or CoVa RNAi. Lactate measurements were made in triplicate using an enzyme-linked ultraviolet absorption method (Raisio Diagnostics, Rome, Italy) calibrated to known standards. Statistical significance was evaluated by Fisher's protected least significant difference.
Quantification of glycolysis through the extracellular acidification rate of S2 cell cultures
S2 cells were cultured in Schneider's media, and CoVa and GFP knockdown was performed using small interfering RNA as previously described (Mandal et al. 2005). Cells were subcultured every 3 days, and daily cell counts were assayed. One hundred thirty-two hours after RNAi treatment, coincident with the growth plateau seen in CoVa knockdown cells and with the observed shift to glycolytic gene expression found in the microarray analysis, the cells were counted and seeded into the proprietary Seahorse culture plate for a projected density of 50,000 cells per milliliter after an overnight rest. The following morning (144 hr), the cell media was decanted and replaced with DMEM low-glucose media (glucose 2.5 mM) for 5 hr. Real-time measurements of the media pH was made as an index of the cellular response to a glucose load (25 mM), followed by a bolus of 2-deoxyglucose (2dG, 225 mM) as a specific inhibitor of glycolysis in the extracellular flux (XF) instrument (Seahorse Bioscience; Chicopee, MA). The resulting graph of extracellular pH over time indicates the glycolytic capacity of the cells, and using the area under this curve (AUC), direct comparison of the GFP controls with the CoVa knockdown cells was made after correcting for cell count, which was performed immediately after the extracellular flux measurements were recorded. The experiment was performed independently three times, and statistical significance for each experiment was evaluated by t-test.
Results
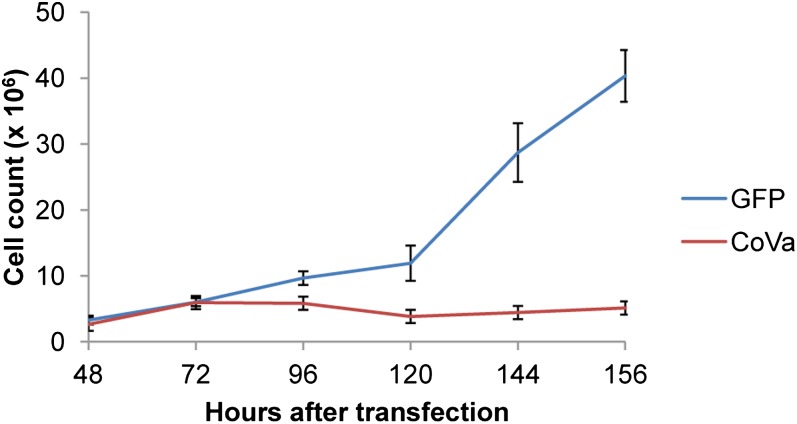
Growth of Drosophila S2 cells is slowed when RNAi is used to abolish CoVa transcription (Figure 1), consistent with both in vivo and in vitro prior studies from our laboratory (Mandal et al. 2005). S2 cells transfected with a GFP control retain proliferative capacity after transfection, whereas CoVa knockdown cells display slowed growth by 96 hr after transfection. Using these growth kinetics, we selected 72, 96, 120, and 168 hr as time points to perform microarray expression profiling. Microarray signal intensity of CoVa transcripts revealed a knockdown that paralleled the cell growth kinetics, with a mean knockdown of 70% at the termination of the experiment (supporting information, Figure S1). The expression profile time series was performed in triplicate and is available at GEO accession number GSE32912.
Figure 1.
Proliferation profile of Drosophila S2 cells treated with either GFP or CoVa RNAi. S2 cells were treated with either GFP (control, blue) or CoVa interfering RNA (red). After initial rounds of mitosis, CoVa RNAi–treated cells slow down and stop dividing. Error bars indicate standard deviation.
Transcriptional profiling identifies groups of genes that are coordinately and consistently altered by loss of electron transport function through CoVa RNAi
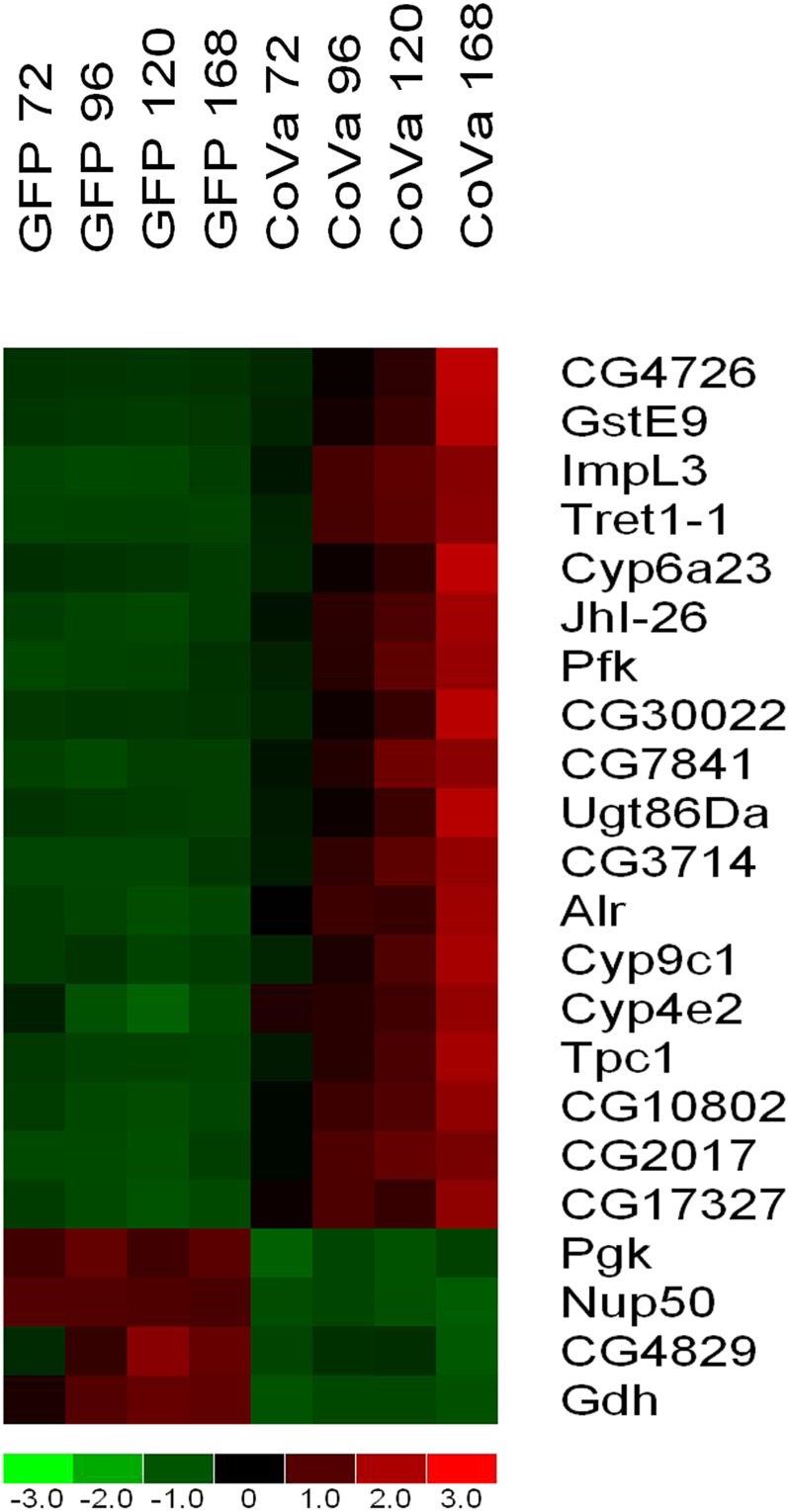
To identify the genes most reliably changed by loss of CoVa, a stringent pairwise comparison was made between all control time points and all CoVa time points using all three independent time course experiments and the following comparison criteria: a minimum of 2-fold or greater difference below a 90% confidence bound; absolute difference greater than 500 (which is greater than 5-fold higher than the noise floor); and a P value less than 0.05 using a Welch-modified two-sample t-test. This identified 25 probesets consisting of 22 genes consistently and robustly altered by loss of CoVa expression (Figure 2). Of these 22 significantly and differentially expressed genes, 18 are upregulated and 4 are downregulated. To identify the effect of time in culture, a repeated measures analysis of variance was performed. This analysis identified that 19 of these 22 genes are statistically significantly different, indicating that time in culture after initial knockdown was related to the degree of expression changes (P range from 1 × 10−5 to 0.03). Three genes, Tret1-1, Nup50, and CG4829, were not found to be significantly different using a repeated measures analysis of variance, indicating that the abrupt, profound, and unwavering changes in their gene expression are not correlated to time in culture after initial knockdown. Despite the lack of significance by the repeated measures test, the expressions of these three genes are exquisitely correlated to CoVa knockdown and serve as time-independent markers of electron transport function. A less stringent comparison was made using the following criteria: a minimum of 1.5 times or greater difference below a 90% confidence bound; absolute difference greater than 200; and a P value less than 0.05 using a Welch-modified two-sample t-test (see Table S1). This less stringent comparison identified 142 probesets to be differentially expressed, with 111 probesets upregulated and 31 probesets downregulated.
Figure 2.
Microarray expression profiling identifies genes differentially expressed by CoVa knockdown in Drosophila S2 cells. Using microarray-based transcriptional profiling, 22 genes are identified to be differentially expressed as a result of CoVa knockdown in Drosophila S2 cells (see text for comparison criteria). Listed from left to right are the time-course samples of S2 cells with GFP indicating control and CoVa indicating CoVa knockdown. The number following the identifier is the number of hours after RNA interference was initiated. Listed from top to bottom are genes most highly upregulated to most highly downregulated. The color map bar on the bottom of the figure displays fold change of the gene expression, with red indicating a fold change of 1 or more, and green indicating a fold change of −1 or less. For a description of the specific genes, see the Results and Discussion sections.
Glycolysis gene expression is strongly upregulated in response to CoVa RNAi
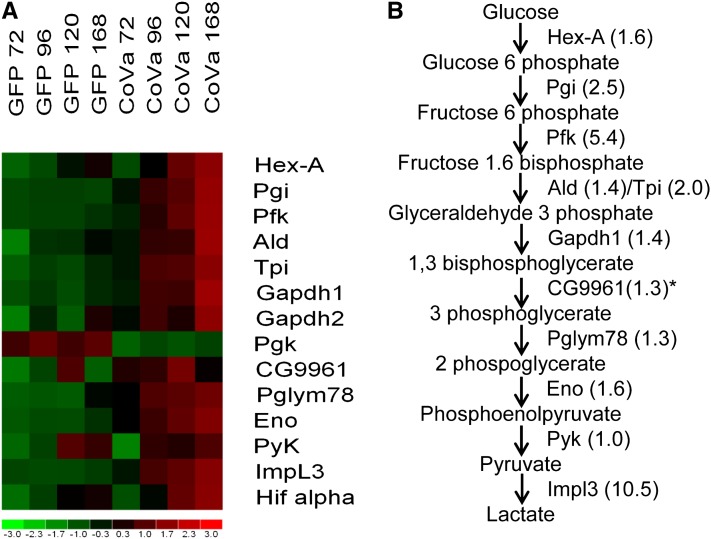
The majority of genes identified in the stringent comparison are novel targets, as most are uncharacterized or possess a CG identifier. The 22 stringently identified genes were evaluated with the online software Database for Annotation, Visualization and Integrated Discovery of the National Institutes of Allergy and Infectious Diseases; this software identifies groups of genes that are overrepresented using an unbiased algorithm based upon the Gene Ontology classification (Huang da et al. 2009). This analysis identified glycolysis genes to be significantly overrepresented with loss of CoVa (software settings: functional annotation, gene list, AFFYMETRIX_3PRIME_IVT_ID, genelist). Overrepresentation of glycolysis genes was found using either the complete Drosophila genome as a background list (all 18,769 probesets found on the Affymetrix Drosophila Genome 2 microarray; P < 0.0007) or using an abbreviated background list containing probesets found to have a Present call in 100% of the samples (6670 probe sets; P < 0.03). We therefore examined the expression profile of all enzymes of the glycolytic pathway from hexokinase to lactate dehydrogenase (Figure 3). Although lactate dehydrogenase (Impl3) is not classically considered a member of the glycolysis pathway, it is required to dispose of pyruvate and allow continued metabolic substrate flux through glycolysis, particularly when disposal of pyruvate through the citric acid cycle and oxidative phosphorylation is impaired. Glycolytic genes are upregulated in response to CoVa knockdown. Interestingly, the described Drosophila Pgk is highly downregulated. Using an in silico approach, the Drosophila genome was surveyed for sequences similar to Pgk, and CG9961 was identified to have a high sequence similarity to Pgk. Pgk and CG9961 are located in tandem on Drosophila chromosome 2L. The transcripts of Pgk and CG9961 share 67% of their nucleotide sequence and 63% of their amino acid sequence. When similar amino acid residues are accounted for, the two proteins share 76% similarity using the basic local alignment search tool (BLAST) for protein pairwise comparison (Altschul et al. 1990). The microarray data reveals that when CoVa expression is downregulated, the expression of CG9961 is upregulated. Henceforth, we refer to CG9961 as Drosophila phosphoglycerate kinase 2 (dPGK2).
Figure 3.
Glycolytic gene expression is increased as a response to CoVa knockdown in Drosophila S2 cells. The microarray data were collated for glycolytic gene expression. (A) The heat map of glycolytic genes. (B) The glycolysis pathway, enzymes, and fold changes at 168 hr after CoVa knockdown. The heat map displays from top to bottom the glycolytic genes arranged by their position in the glycolytic pathway, with the gene at the bottom of the figure Hifα, a transcription factor known to transcriptionally control glycolytic gene expression. Listed from left to right are the time course samples of S2 cells, with GFP indicating control and CoVa indicating CoVa knockdown. The number following the identifier is the number of hours after RNA interference was initiated. The color map bar on the bottom of the figure displays fold change of the gene expression, with red indicating a fold change of 1 or more, and green indicating a fold change of −1 or less. The specific genes from top to bottom are Hexokinase-A (Hex-A), Phosphoglucose isomerase (Pgi), Pfk, Aldolase (Ald), Triose phosphate isomerase (Tpi), Glyceraldehyde 3 phosphate dehydrogenase 1 (Gapdh1), Glyceraldehyde 3 phosphate dehydrogenase 2 (Gapdh2), Pgk, CG9961, Phosphoglyceromutase (Pglym78), Enolase (Eno), Pyruvate kinase (PyK), Impl3, and Hifα. *The fold change of CG9961, the alternate Pgk equivalent that is increased by CoVa knockdown, is displayed.
Upregulation of glycolytic genes prompted us to examine the expression of Hifα, the transcription factor known to be responsible for controlling glycolytic gene transcription (Figure 3)(Semenza et al. 1994). The microarray signal intensity of Hifα is increased by a mean of 50% as a response to CoVa knockdown, with a graded increase proportional to the time in culture after initial knockdown. When the microarray signal intensities of all of the GFP controls are compared with all CoVa samples there is a nonsignificant statistical trend toward increasing Hifα expression in the CoVa knockdown cells (P < 0.06). This difference peaks at the 168 hr time point, with a 70–90% increase in Hifα expression (P < 0.02).
Glycolytic capacity of S2 cells is greatly increased by CoVa RNAi
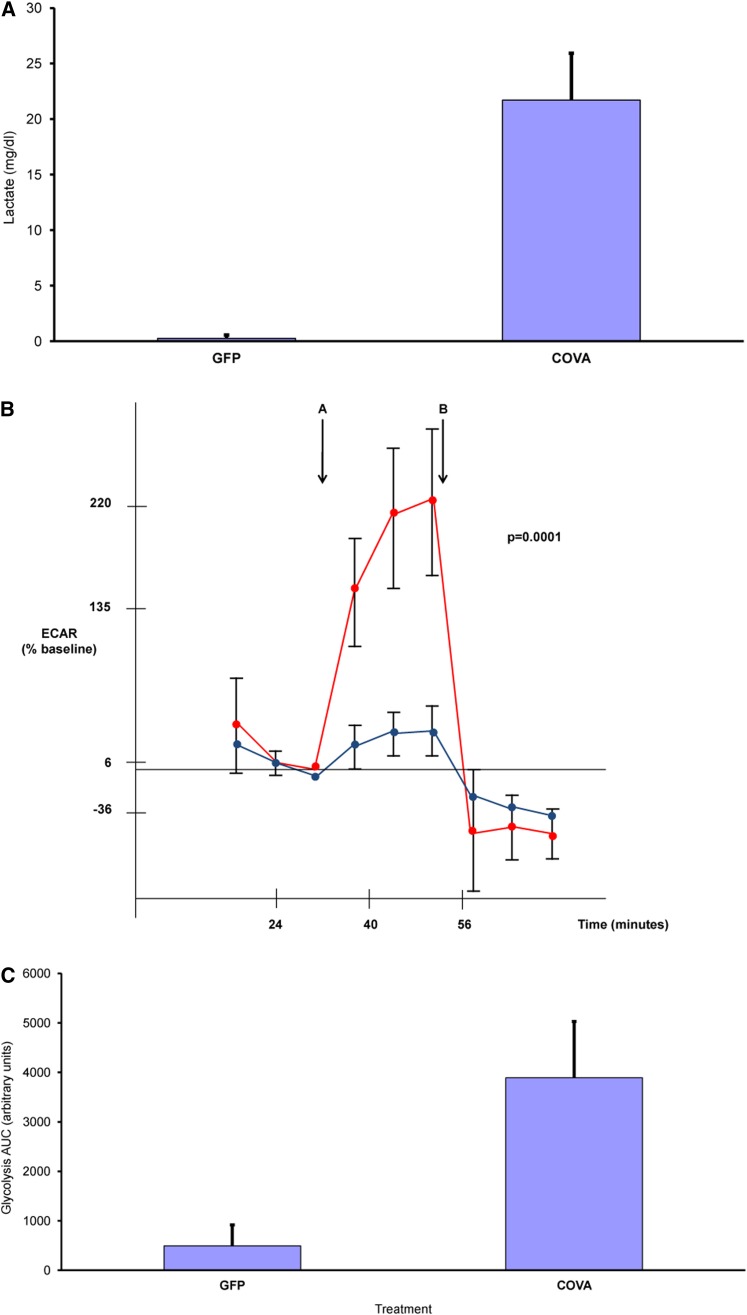
To confirm the microarray results and evaluate the glycolytic capacity of CoVa knockdown cells, three additional confirmatory experiments were performed. First, qRT-PCR for selected glycolysis genes confirmed the microarray results (Table S2). qRT-PCR for Pfk, Pgk, and Impl3 revealed significant differences between GFP controls and CoVa knockdown, and it paralleled the microarray results. The qRT-PCR results correlate to the microarray findings with a correlation coefficient of 0.9. Second, as aerobic glycolysis should result in increased lactate production, the lactate content of the conditioned S2 cell media would be an indirect assay for the glycolytic capacity of the cells. A lactate-specific spectrophotometric assay showed that the lactate concentration increased upon CoVa knockdown (Figure 4A). The conditioned S2 media concentration of lactate was 0.3 mg/dl in the GFP controls compared with 21.7 mg/dl in the CoVa knockdown (P < 0.009). Third, to conclusively assay the glycolytic capacity of the S2 cell cultures, metabolic flux studies using the Seahorse XF24 multiparameter analyzer was performed using glucose and 2dG as a specific inhibitor of glycolysis with real-time measurement of the pH of the extracellular media.
Figure 4.
Abrogation of mitochondrial electron transport function through CoVa knockdown results in increased glycolysis and lactate production in Drosophila S2 cells. (A) Media concentrations of lactate are increased in response to CoVa knockdown in Drosophila S2 cells (P < 0.009). (B) Glycolysis rates are increased in Drosophila S2 cells when CoVa expression is knocked down. Media pH measurements were recorded in response to a bolus of glucose (point A) through administration of 2dG, a specific inhibitor of glycolysis (point B). The AUC is proportional to the cumulative glycolysis occurring in the cells. GFP controls are in blue and CoVa downregulated cells are in red. The extracellular acidification profile shown is from one of three independently performed experiments. (C) CoVa downregulated cells metabolize glucose through glycolysis at a rate 7.9 times the GFP controls (P < 0.0001).
Cell culture of S2 cells was performed with CoVa knockdown as per the same protocol as the microarray experiments. At 144 hr after transfection, the metabolic flux studies were performed after placement of the cells in a 5 hr treatment of low glucose-containing medium (Figure 4B). When the glucose-limited cells were exposed to a bolus of glucose (arrow A), there was increased extracellular acidification found in the CoVa knockdown cells compared with the GFP controls. As the CoVa knockdown cells acidified the extracellular media to a greater extent, this suggests increased lactate production through increased glycolysis, with the end product of lactate being transferred to the extracellular space in CoVa-deficient cells. To specifically abrogate glycolysis, 2dG was then applied to the cells (arrow B). 2dG is an analog of glucose that is unable to undergo glycolysis. 2dG stopped the extracellular acidification of both the GFP controls and CoVa, but to a much larger degree in the CoVa knockdown cells. The AUC of the extracellular acidification rate from the time point of glucose injection through the 2dG treatment is proportional to the level of glycolysis occurring in the cells. CoVa-deficient cells had a 7.9-fold increase in the glycolysis AUC found compared with the CoVa knockdown cells (P < 0.0001)(Figure 4C). We conclude from these experiments that glycolysis is increased in response to abrogation of complex IV function through CoVa RNAi.
Discussion
We have developed a rich dataset describing the transcriptional changes that are associated with RNAi-induced gene knockdown of CoVa in Drosophila S2 cells using microarray-based transcriptional profiling. The selected time points were designed to capture the cell-cycle arrest induced by loss of activity of the fourth complex of the electron transport chain to illuminate the mechanisms operative behind retrograde mitochondrial signaling and cell growth slowing (Mandal et al. 2005). The most striking aspect of these data is the transcriptional upregulation of glycolytic genes, which results in increased glycolytic capacity as confirmed in metabolic studies (Figures 3 and 4, respectively). Further scrutiny of the differentially expressed gene list clearly demonstrates the mechanism by which the cell directs energy disposition and controls cellular function.
A systems biologic analysis outlines the specific genes responsible for energy substrate management in time of stress
A systems biologic analysis of the 21 highest differentially expressed genes describes a dominant metabolic pathway defined by the acquisition of glucose from both extracellular and intracellular stores, utilization of the obtained glucose through the glycolytic pathway, and disposal of reducing equivalents to lactate production (Figure 5). This analysis contains 21 of the 22 genes identified and has excluded CG7841 as it is completely uncharacterized.
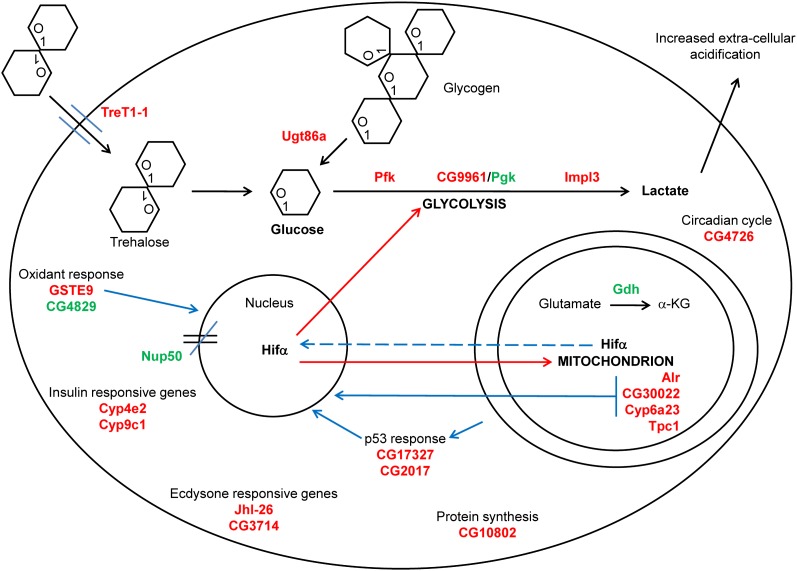
Figure 5.
The 21 genes most differentially expressed in response to CoVa knockdown in Drosophila S2 cells identifies metabolic pathways and mitochondrial retrograde signals. After knockdown of CoVa, energy substrate disposition is dominated by the acquisition of glucose from both extracellular sources, such as from trehalose, and from intracellular sources, such as glycogen. The glucose is then metabolized through glycolysis and results in increased lactate production and the direction of reducing equivalents away from the mitochondrion by downregulation of glutamate dehydrogenase. The cellular response to electron transport deficiency also leads to the induction of specific mitochondrial retrograde signals. These signals include experimentally validated targets, including the mitochondrial proteins Alr and CG30022, as well as evidence of a p53-response, also experimentally validated, through the upregulation of the genes CG17327 and CG2017. Hifα provides a biologically relevant mitochondrial retrograde signal, as it is known to continuously sense ambient oxygen tension, the primary electron acceptor of the oxidative function of the mitochondrion. In addition, it is known to control glycolytic and cytochrome gene expression. Other pathways identified in the transcriptional response to CoVa knockdown include the ecdysone response, circadian cycling, new protein synthesis, and the closing of nuclear pores by affecting the importin-alpha and importin-beta nuclear pore complex. For a discussion of the specific genes, see the Results and Discussion sections. Genes in red indicate an upregulated gene, and genes in green indicate a downregulated gene. Solid black lines indicate a metabolic pathway, solid blue lines indicate an experimentally validated mitochondrial retrograde signal, dashed blue lines indicate a suspected mitochondrial retrograde signal, and red lines indicate genes whose expression is known to be controlled by Hifα (Semenza et al. 1994).
Trehalose, a disaccharide of glucose molecules linked at the first carbon position, is the primary circulating carbon source in insects. Trehalose transporter 1-1 (Tret1-1) is highly upregulated in response to CoVa knockdown, thereby providing a mechanism by which trehalose may be drawn into the cell. In addition, Ugt86a, a UDP-glycosyl transferase with possible glycogen phosphorylase activity, may be able to mobilize glucose from intracellular glycogen stores, further providing glucose for glycolysis. The key glycolytic enzymes found to be upregulated by CoVa knockdown include Pfk and the Drosophila lactate dehydrogenase Impl3. The expression profiling also identifies a novel transcript, dPGK2, which encodes an alternative stress-induced enzyme containing homology to Pgk and serves the activity of Pgk during abrogation of electron transport function. Early attempts at characterizing the Drosophila Pgk identified at least three different electrophoretic variants (Chew and Cooper 1973), and subsequent studies using RNase protection assay and primer extension found multiple transcript clusters (Roselli-Rehfuss et al. 1992). Multiple isoforms of Pgk are found in Drosophila, indicating a regulatory mechanism similar to mammals. Future directed studies will clarify the role of dPgk2 in times of mitochondrial stress.
Disposition of metabolic substrate to the glycolytic pathway is coordinately linked to substrate disposition away from the mitochondrion by modulating the production of the key anaplerotic intermediate α-ketoglutarate. There is likely decreased α-ketoglutarate production in the mitochondrion as the expression of glutamate dehydrogenase (Gdh), a primary mechanism to produce α-ketoglutarate in the mitochondrion, is rapidly and profoundly decreased. We therefore conclude that the electron transport deficiency caused by CoVa RNAi leads to a transcriptional response characterized by the preferential utilization of glucose and the shunting of metabolic precursors away from the oxidative function of the mitochondria mediated through the decreased expression of Gdh.
Evidence of conserved mitochondrial retrograde signals are identified within the systems biologic analysis
Our previous genetic interaction studies had implicated p53 as being operative in the growth arrest associated with loss of CoVa (Mandal et al. 2005). CG17327 and CG2017 are two genes responsive to p53 (Akdemir et al. 2007) and are increased in response to CoVa RNAi (Figure 5). CG17327 is an aminoacyl- tRNA hydrolase, and CG2017 is a GTP binding protein acting as a possible protein synthesis factor. Interestingly, both of these genes were also identified in a screen for Drosophila oxidant stress employing hyperoxia (Gruenewald et al. 2009).
The gene Augmenter of liver regeneration (Alr), which contains both sulfhydryl oxidase and cytochrome c reductase activities (Thirunavukkarasu et al. 2008), is upregulated by CoVa RNAi. Alr maintains mitochondria in a rudimentary ultra-structure network that is associated with low oxidative capacity and is thought to be one mechanism that maintains pluripotency in a murine hematopoietic stem cell model (Todd et al. 2010; Wilkerson and Sankar 2011). In a separate model of mitochondrial dysfunction employing the Drosophila mutant total knock out, CG30022, a mitochondrial hydrolase, and Cyp6a23 were identified to be upregulated (Fernandez-Ayala et al. 2010). Both CG30022 and Cyp6a23 are found to be upregulated in response to CoVa RNAi and therefore are part of a conserved response to mitochondrial dysfunction (Figure 5).
Glutathione S-transferases (GST) carry out a wide range of cellular functions, including the removal of reactive oxygen species, regeneration of S-thiolated proteins and antioxidants (Dixon et al. 2011), and catalysis of the conjugation of reduced glutathione to both endogenous compounds as well as exogenous xenobiotics (Sheehan et al. 2001). GstE9 is a cytosolic protein of the GST epsilon class previously described to be found in response to an oxidant stress (Li et al. 2008), as well as part of a larger transcriptional network associated with oxidative phosphorylation genes (Pile et al. 2003).
As Impl3 is one of the consistently highest upregulated genes by CoVa RNAi, examination of the known literature on lactate dehydrogenase regulation, coupled with an analysis of the lactate dehydrogenase 5′ regulatory region, identifies Hifα to be an important arbiter of Impl3 expression (Bruick and Mcknight 2001). A known function of Hifα is to sense the ambient levels of oxygen, the final electron acceptor of the electron transport chain in the mitochondria. In addition, it is known that Hifα controls the expression of both glycolytic and cytochrome oxidase genes (Semenza et al. 1994; Fukuda et al. 2007). Hifα therefore provides an attractive candidate for mitochondrial retrograde signaling, as it senses the primary substrate reduced by the mitochondria, controls glycolytic substrate flux via the transcriptional control of glycolysis, and controls the expression of the cytochrome oxidase genes. A plausible model would therefore incorporate Hifα as a central player in mitochondrial retrograde signaling, most especially in times of mitochondrial dysfunction (Figure 5). It has long been known that inhibition of complex IV through pharmacologic means results in upregulation of glycolytic genes (Simon 1953). Examination of our profiling data through coregulated gene network analysis reveals that most of the genes responding to CoVa RNAi are correlated to Impl3, which suggests Hifα is a central gene directing the transcriptional response. When the stringent gene list describing the response to CoVa knockdown (Figure 2) was examined for the Hifα binding sites within the 500 base pairs surrounding the transcriptional start site of the gene, 19 of the 22 genes possess at least one high similarity Hifα binding site (Table S3). Further, the microarray data reveal transcriptional upregulation of Hifα (Figure 3). This is interesting in that most literature regarding Hifα has focused on the post transcriptional stabilization of Hifα, usually through the action of hydroxy-prolyl hydroxylase (Bruick and Mcknight 2001). Adding more complexity to the regulation of Hif1α is the recent report that Hif1α may be stabilized by alternative methods such as neddylation (Ryu et al. 2011), a mechanism of stabilization that requires the presence of reactive oxygen species. Traditionally the stabilization of Hif1a has been thought to require reactive oxygen species (Chandel et al. 2000); however, Hif1α may be stabilized in a reactive oxygen species–independent fashion (Chua et al. 2010). As our prior observation has revealed low reactive oxygen species generation with CoVa RNAi in vivo, we concur that stabilization of Hif1α may occur in the absence of reactive oxygen species. Undoubtedly, the regulation of Hif1α action is achieved through multiple mechanisms and will not be the same under all biologic conditions. Regardless of the specific mechanism by which Hifα mRNA is increased in response to CoVa knockdown, as glycolytic genes are found to be upregulated and Hifα binding sites are identified in the majority of the top differentially regulated genes, we conclude that Hifα is likely to play a key role in mitochondrial retrograde signaling in times of mitochondrial dysfunction.
We conclude that the most likely mitochondrial retrograde signals identified in our expression profiling experiments are p53, alr, CG30022, Cyp6a23, GstE9, and Hifα (Figure 5). These identified genes may be the mitochondrial retrograde signal itself or an output from a retrograde signal system. As these genes have been identified in other biologic systems, their importance in mitochondrial regulation of the nucleus must be emphasized. Further directed studies will clarify the roles that these genes play in modulating nuclear function.
Cytochrome p450 proteins, developmental cues, nuclear pores, and redox generators are identifiable within the systems biologic analysis
Cytochrome p450 (CYP450) proteins comprise a large group of heme-containing proteins that predominantly perform oxidation reactions. Targets of CYP450 proteins include both endogenous molecules, such as steroid hormones and lipids, as well as exogenous compounds, such as xenobiotics. A transcriptional response that directs detoxifying xenobiotics may be a conserved reaction to counter the threat posed by xenobiotics, compounds that often have the electron transport chain as their target. The expressions of Cyp4e2 and Cyp9c1 are increased in response to CoVa RNAi, and interestingly, both of these proteins have been identified in a screen for class O Forkhead box protein function (Junger et al. 2003) and are thought to be increased in response to decreased insulin signaling (Figure 5).
Ecdysone is a steroidal prohormone that is involved in larval maturation and directs stage-dependent gene expression. Jhl-26 (Dubrovsky et al. 2000; Palli 2009) and CG3714 (Beckstead et al. 2005) are known to be responsive to ecdysone and are both increased in response to CoVa knockdown. These two genes may therefore serve as potentially important links between energy status and developmental stage (Figure 5).
The nuclear pore is a large specialized multiprotein channel that connects the cytoplasm to the nucleus and is composed of nucleoporins. Nucleoporin 50 kD (Nup50) acts as a cofactor for the importin-alpha and importin-beta heterodimer, which allows for transportation of nuclear-targeted proteins through the nuclear pore complex (Lindsay et al. 2002). An example of a signaling molecule dependent upon nuclear pores and the importin-alpha and importin-beta system is the mothers against decapentaplegic class of protein of the transforming growth factor beta signaling cascade. Therefore, signaling molecules may be excluded from the nucleus as a direct result of cellular energy stores (Figure 5).
The last two genes identified in the stringent analysis include thiamine pyrophosphate carrier protein 1 (Tpc1), a mitochondrial thiamine transporter (Iacopetta et al. 2010), and CG4829, a protein-glutamine-gamma glutamyl transferase, genes that are upregulated and downregulated, respectively, in response to CoVa RNAi (Figure 5). Gamma glutamyl transferases catalyze the transfer of glutamyl groups from reduced glutathione to water, proteins, and amino acids. In the process of this catalysis, hydrogen peroxide may be released, and thus, gamma glutamyl transferases have been shown to be a significant source of cellular oxidants (Maellaro et al. 2000).
Summary
To conclude, transcriptional profiling of CoVa RNAi reveals the plasticity of the Drosophila cell to respond to experimental manipulation of the electron transport chain, and it provides molecular mechanisms for understanding how energy substrate management is controlled in times of stress. Through these studies, we witness the response of the transcriptional profile to experimentally induced electron transport deficiency and are able to discern retrograde mitochondrial signals and outputs. Understanding the mechanism through which the mitochondrion orchestrates a nuclear response and is able to manage the disposition of energy precursors may help to develop novel therapeutics for cancer, as well as to better understand the metabolic program of rapidly dividing cells that preferentially utilize glucose as their fuel source.
Supplementary Material
Acknowledgments
W.A. Freije graciously acknowledges the salary support received from the faculty plan of the Department of Obstetrics and Gynecology, as well as the support of Professor Gautam Chaudhuri, Chair of the Department of Obstetrics and Gynecology. We are grateful for the 5′ analysis made by Kelvin Zhang, the editing critique by Kevin Jones, the repeated measures analysis of variance performed by the Statistical Computing Group of the UCLA Academic Technology Services, and discussions with Preeta Guptan. This study was supported by a grant from the National Institutes of Health (R01-EY-08152 to U. Banerjee).
Footnotes
Communicating editor: J. A. Birchler
Literature Cited
- Akdemir F., Christich A., Sogame N., Chapo J., Abrams J. M., 2007. p53 directs focused genomic responses in Drosophila. Oncogene 26: 5184–5193 [DOI] [PubMed] [Google Scholar]
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., 1990. Basic local alignment search tool. J. Mol. Biol. 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Beckstead R. B., Lam G., Thummel C. S., 2005. The genomic response to 20-hydroxyecdysone at the onset of Drosophila metamorphosis. Genome Biol. 6: R99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas G., Adebanjo O. A., Freedman B. D., Anandatheerthavarada H. K., Vijayasarathy C., et al. , 1999. Retrograde Ca2+ signaling in C2C12 skeletal myocytes in response to mitochondrial genetic and metabolic stress: a novel mode of inter-organelle crosstalk. EMBO J. 18: 522–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick R. K., McKnight S. L., 2001. A conserved family of Prolyl-4-Hydroxylases that modify HIF. Science 294: 1337–1340 [DOI] [PubMed] [Google Scholar]
- Butow R. A., Avadhani N. G., 2004. Mitochondrial signaling: the retrograde response. Mol. Cell 14: 1–15 [DOI] [PubMed] [Google Scholar]
- Chandel N. S., McClintock D. S., Feliciano C. E., Wood T. M., Melendez J. A., et al. , 2000. Reactive oxygen species generated at mitochondrial complex III stabilize hypoxia-inducible factor-1alpha during hypoxia: a mechanism of O2 sensing. J. Biol. Chem. 275: 25130–25138 [DOI] [PubMed] [Google Scholar]
- Chew G. K., Cooper D. W., 1973. Phosphoglycerate kinas polymorphism in Drosophila. Biochem. Genet. 8: 267–270 [DOI] [PubMed] [Google Scholar]
- Chua Y. L., Dufour E., Dassa E. P., Rustin P., Jacobs H. T., et al. , 2010. Stabilization of hypoxia-inducible factor-1alpha protein in hypoxia occurs independently of mitochondrial reactive oxygen species production. J. Biol. Chem. 285: 31277–31284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMauro S., Schon E. A., 2003. Mitochondrial respiratory-chain diseases. N. Engl. J. Med. 348: 2656–2668 [DOI] [PubMed] [Google Scholar]
- Dixon D. P., Steel P. G., Edwards R., 2011. Roles for glutathione transferases in antioxidant recycling. Plant Signal. Behav. 6: 1223–1227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubrovsky E. B., Dubrovskaya V. A., Bilderback A. L., Berger E. M., 2000. The isolation of two juvenile hormone-inducible genes in Drosophila melanogaster. Dev. Biol. 224: 486–495 [DOI] [PubMed] [Google Scholar]
- Fernandez-Ayala D. J., Chen S., Kemppainen E., O'Dell K. M., Jacobs H. T., 2010. Gene expression in a Drosophila model of mitochondrial disease. PLoS ONE 5: e8549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda R., Zhang H., Kim J. W., Shimoda L., Dang C. V., et al. , 2007. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 129: 111–122 [DOI] [PubMed] [Google Scholar]
- Garesse R., Kaguni L. S., 2005. A Drosophila model of mitochondrial DNA replication: proteins, genes and regulation. IUBMB Life 57: 555–561 [DOI] [PubMed] [Google Scholar]
- Gruenewald C., Botella J. A., Bayersdorfer F., Navarro J. A., Schneuwly S., 2009. Hyperoxia-induced neurodegeneration as a tool to identify neuroprotective genes in Drosophila melanogaster. Free Radic. Biol. Med. 46: 1668–1676 [DOI] [PubMed] [Google Scholar]
- Huang da W., Sherman B. T., Lempicki R. A., 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Iacopetta D., Carrisi C., De Filippis G., Calcagnile V. M., Cappello A. R., et al. , 2010. The biochemical properties of the mitochondrial thiamine pyrophosphate carrier from Drosophila melanogaster. FEBS J. 277: 1172–1181 [DOI] [PubMed] [Google Scholar]
- Junger M. A., Rintelen F., Stocker H., Wasserman J. D., Vegh M., et al. , 2003. The Drosophila Forkhead transcription factor FOXO mediates the reduction in cell number associated with reduced insulin signaling. J. Biol. 2: 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulawiec M., Arnouk H., Desouki M. M., Kazim L., Still I., et al. , 2006. Proteomic analysis of mitochondria-to-nucleus retrograde response in human cancer. Cancer Biol. Ther. 5: 967–975 [DOI] [PubMed] [Google Scholar]
- Li H. M., Buczkowski G., Mittapalli O., Xie J., Wu J., et al. , 2008. Transcriptomic profiles of Drosophila melanogaster third instar larval midgut and responses to oxidative stress. Insect Mol. Biol. 17: 325–339 [DOI] [PubMed] [Google Scholar]
- Liao T. S., Call G. B., Guptan P., Cespedes A., Marshall J., et al. , 2006. An efficient genetic screen in Drosophila to identify nuclear-encoded genes with mitochondrial function. Genetics 174: 525–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay M. E., Plafker K., Smith A. E., Clurman B. E., Macara I. G., 2002. Npap60/Nup50 is a tri-stable switch that stimulates importin-alpha:beta-mediated nuclear protein import. Cell 110: 349–360 [DOI] [PubMed] [Google Scholar]
- Liu Z., Butow R. A., 2006. Mitochondrial retrograde signaling. Annu. Rev. Genet. 40: 159–185 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)). Method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Maellaro E., Dominici S., Del Bello B., Valentini M. A., Pieri L., et al. , 2000. Membrane gamma-glutamyl transpeptidase activity of melanoma cells: effects on cellular H(2)O(2) production, cell surface protein thiol oxidation and NF-kappa B activation status. J. Cell Sci. 113: 2671–2678 [DOI] [PubMed] [Google Scholar]
- Mandal S., Guptan P., Owusu-Ansah E., Banerjee U., 2005. Mitochondrial regulation of cell cycle progression during development as revealed by the tenured mutation in Drosophila. Dev. Cell 9: 843–854 [DOI] [PubMed] [Google Scholar]
- Mandal S., Freije W. A., Guptan P., Banerjee U., 2010. Metabolic control of G1-S transition: cyclin E degradation by p53-induced activation of the ubiquitin-proteasome system. J. Cell Biol. 188: 473–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal S., Lindgren A. G., Srivastava A. S., Clark A. T., Banerjee U., 2011. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells 29: 486–495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ott M., Norberg E., Zhivotovsky B., Orrenius S., 2009. Mitochondrial targeting of tBid/Bax: a role for the TOM complex. [quest] Cell Death Differ. 16: 1075–1082 [DOI] [PubMed] [Google Scholar]
- Pile L. A., Spellman P. T., Katzenberger R. J., Wassarman D. A., 2003. The SIN3 deacetylase complex represses genes encoding mitochondrial proteins: implications for the regulation of energy metabolism. J. Biol. Chem. 278: 37840–37848 [DOI] [PubMed] [Google Scholar]
- Palli S. R., 2009. Recent advances in the mode of action of juvenile hormones and their analogs, pp. 111–129 in Biorational Control of Arthropod Pests, edited by I. Ishaaya and A. R. Horowitz. Springer, Dordrecht, The Netherlands
- Roselli-Rehfuss L., Ye F., Lissemore J. L., Sullivan D. T., 1992. Structure and expression of the phosphoglycerate kinase (Pgk) gene of Drosophila melanogaster. Mol. Gen. Genet. 235: 213–220 [DOI] [PubMed] [Google Scholar]
- Ryu J. H., Li S. H., Park H. S., Park J. W., Lee B., et al. , 2011. Hypoxia-inducible factor alpha subunit stabilization by NEDD8 conjugation is reactive oxygen species-dependent. J. Biol. Chem. 286: 6963–6970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semenza G. L., Roth P. H., Fang H. M., Wang G. L., 1994. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J. Biol. Chem. 269: 23757–23763 [PubMed] [Google Scholar]
- Sheehan D., Meade G., Foley V. M., Dowd C. A., 2001. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 360: 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon E. W., 1953. Dinitrocresol, cyanide, and the pasteur effect in yeast. J. Exp. Bot. 4: 393–402 [Google Scholar]
- Thirunavukkarasu C., Wang L. F., Harvey S. A., Watkins S. C., Chaillet J. R., et al. , 2008. Augmenter of liver regeneration: an important intracellular survival factor for hepatocytes. J. Hepatol. 48: 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd L. R., Damin M. N., Gomathinayagam R., Horn S. R., Means A. R., et al. , 2010. Growth factor erv1-like modulates Drp1 to preserve mitochondrial dynamics and function in mouse embryonic stem cells. Mol. Biol. Cell 21: 1225–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkerson D. C., Sankar U., 2011. Mitochondria: a sulfhydryl oxidase and fission GTPase connect mitochondrial dynamics with pluripotency in embryonic stem cells. Int. J. Biochem. Cell Biol. 43: 1252–1256 [DOI] [PubMed] [Google Scholar]
- Yang J., Zhang M., Yu J., 2008. Mitochondrial retrograde regulation tuning fork in nuclear genes expressions of higher plants. J. Genet. Genomics 35: 65–71 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.