Abstract
Cytoprotective functions of a 20S proteasome activator were investigated. Saccharomyces cerevisiae Blm10 and human 20S proteasome activator 200 (PA200) are homologs. Comparative genome-wide analyses of untreated diploid cells lacking Blm10 and growing at steady state at defined growth rates revealed downregulation of numerous genes required for accurate chromosome structure, assembly and repair, and upregulation of a specific subset of genes encoding protein-folding chaperones. Blm10 loss or truncation of the Ubp3/Blm3 deubiquitinating enzyme caused massive chromosomal damage and cell death in homozygous diploids after phleomycin treatments, indicating that Blm10 and Ubp3/Blm3 function to stabilize the genome and protect against cell death. Diploids lacking Blm10 also were sensitized to doxorubicin, hydroxyurea, 5-fluorouracil, rapamycin, hydrogen peroxide, methyl methanesulfonate, and calcofluor. Fluorescently tagged Blm10 localized in nuclei, with enhanced fluorescence after DNA replication. After DNA damage that caused a classic G2/M arrest, fluorescence remained diffuse, with evidence of nuclear fragmentation in some cells. Protective functions of Blm10 did not require the carboxyl-terminal region that makes close contact with 20S proteasomes, indicating that protection does not require this contact or the truncated Blm10 can interact with the proteasome apart from this region. Without its carboxyl-terminus, Blm10(−339aa) localized to nuclei in untreated, nonproliferating (G0) cells, but not during G1 S, G2, and M. The results indicate Blm10 functions in protective mechanisms that include the machinery that assures proper assembly of chromosomes. These essential guardian functions have implications for ubiquitin-independent targeting in anticancer therapy. Targeting Blm10/PA200 together with one or more of the upregulated chaperones or a conventional treatment could be efficacious.
Keywords: 20S proteasome activator, BLM10/PA200, UBP3/BLM3, DNA damage, molecular chaperones
Aggregated, unfolded, misfolded, and nonfunctional proteins accumulate in many human diseases, such as cancers (Coughlan and Brodsky 2003; Scott and Frydman 2003; Huo 2010; Nagaraj et al. 2010). Proteasomes degrade such proteins, along with those otherwise damaged or altered or no longer needed (Dobson 2003; Goldberg 2003; Demartino and Gillette 2007; Rosenzweig and Glickman 2008). These multicatalytic proteinase complexes conduct the preponderance of intracellular protein degradation and dispense with DNA-damaging agents or other toxic compounds in cells. Selective inhibition of proteasomes in cancer cells is an anticancer treatment strategy whose efficacy lies in blocking metabolic functions, inducing apoptosis, and sensitizing malignant cells and tumors to chemotherapeutic agents and radiation (Voorhees and Orlowski 2006; Orlowski and Kuhn 2008; Yang et al. 2009).
Proteolytic activities of proteasomes occur inside 20S multisubunit core particles, of which homologous human proteasome activator 200 (PA200) and yeast Blm10 are activators and regulatory proteins (Schmidt et al. 2005b; Főrster and Hill 2006; Finley 2009; Stadtmueller and Hill 2011; Savulescu and Glickman 2011; Lopez et al. 2011; Dange et al. 2011). We found PA200 widely distributed in adult human tissues but not fetal tissues (Febres et al. 2001), and others found it widely distributed in mouse tissues (Ustrell et al. 2002) and required for normal spermatogenesis (Khor et al. 2006). The yeast and human proteins share 17% sequence identity (Ustrell et al. 2002; Ortega et al. 2005; Iwanczyk et al. 2006). It was actually the divergent sequences that led to the prediction that PA200 and Blm10 may perform different in vivo roles (Főrster and Hill 2006).
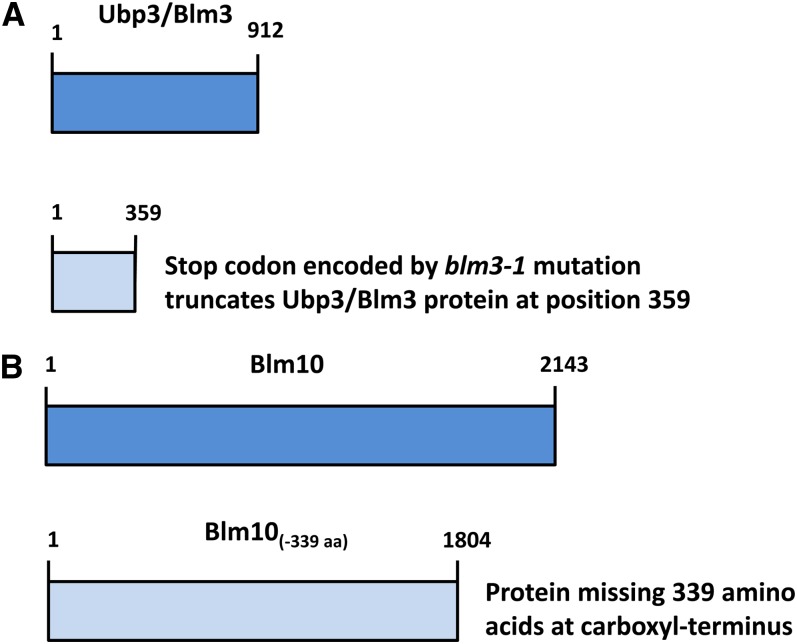
Blm10 was first discovered as a multicopy suppressor (Febres et al. 2001; Doherty et al. 2004) of the hypersusceptibilities to killing by anticancer bleomycins and structurally related phleomycins that are conferred by the blm3-1 mutation (Moore 1991). This nonsense mutation in the UBP3/BLM3 (human Ubp10/Usp10) ubiquitin-specific protease gene truncates upstream of the ubiquitin hydrolase domain (McCullock et al. 2006) (Figure 1). In addition to bleomycin and phleomycin (Moore 1991; Febres et al. 2001; McCullock et al. 2006), the blm3-1 mutation confers hypersusceptibilities to lethal effects of gamma irradiation and hydrogen peroxide (Moore 1991) and canavanine, hydroxyurea, and growth at 37° (McCullock et al. 2006). It was proposed that Ubp3 promotes protein stability by deubiquitinating misfolded proteins, permitting their refolding and function (Brew and Huffaker 2002). Genetic interaction data suggest a role for Ubp3 in transcriptional elongation (McCullock et al. 2006). It was suggested that Ubp3 physically interacts with the 26S proteasome and the Rad4 protein to facilitate degradation of Rad4 and suppression of DNA nucleotide excision repair (Mao and Smerdon 2010).
Figure 1 .
Truncations of the Blm10 and Ubp3/Blm3 proteins as described in the text. Dark blue indicates full-length proteins; light blue: truncated proteins.
As energy-independent 20S proteasome activators, Blm10 and PA200 do not require ATPases and ubiquitinated substrates for activation (Ustrell et al. 2002; Schmidt et al. 2005a). Structural and biochemical properties of Blm10/PA200 were recently reviewed (Stadtmueller and Hill 2011; Savulescu and Glickman 2011; Lopez et al. 2011; Dange et al. 2011). Electron microscopy (Schmidt et al. 2005a; Iwanczyk et al. 2006) and crystal structure (Sadre-Bazzaz et al. 2010) show Blm10 docks onto the axial end of the core particle cylinder, allowing it to regulate the state of the core particle channel. Active gate opening by Blm10 engages its carboxyl-terminus with the core particle (Dange et al. 2011). In proteasome assembly and maturation, Blm10 associates with nascent and de novo synthesized 20S core particles (Fehlker et al. 2003); caps the core particle in its association with stable, mature complexes (Schmidt et al. 2005a); and binds to preactivated core particles (Lehmann et al. 2008). The protein is detected in association with mature proteasomes (Schmidt et al. 2005a; Iwanczyk et al. 2006), and half (Li et al. 2007; Marques et al. 2007) and full (Fehlker et al. 2003; Li et al. 2007; Marques et al. 2007) precursor complexes. Although initial computer modeling of the predicted Blm10 amino acid sequence led to its classification as a potential membrane transport protein containing seven to 10 transmembrane domains (Febres et al. 2001), these are now known to be HEAT domains (Kajava et al. 2004). HEAT repeat proteins have a characterized solenoid structure that facilitates Blm10 binding to the core particle surface, wrapping around the core particle, and looping into the catalytic chamber to interact with core components. PA200 attaches to the α-ring surface in a defined conformation, coming into contact with all subunits except α7 (Glickman and Raveh 2005; Ortega et al. 2005).
Although strains with the BLM10 gene deleted are hypersusceptible to the lethal effects of bleomycin and phleomycin (Febres et al. 2001; Doherty et al. 2004; Schmidt et al. 2005a), no evidence exists that Blm10 or PA200 performs a direct role in DNA repair. PA200 was previously reported to be involved in DNA repair based on the change of finely punctated patterns of PA200 in HeLa nuclei to foci after gamma irradiation but not after hydrogen peroxide or ultraviolet light treatments (Ustrell et al. 2002). It is now known that this form of cellular PA200 is found associated with proteasomes and that PA200 in association with proteasomes, rather than independently, accumulates on chromatin after ionizing irradiation (Blickwedehl et al. 2007). Consistent with these findings, it is known that proteasomes in yeast associate with sites of DNA double-strand breaks (Krogan et al. 2004).
The purpose of the current studies was to investigate some of the properties conferred by the BLM10 gene. Because of the valuable biology that can be obtained from the comprehensive and simultaneous analyses of thousands of genes, we used the technologies of systems biology to measure and compare global gene expression in cells with and without Blm10. To obtain this systems-level understanding, we sought to identify differentially coexpressed genes and discover some of the interactive networks and pathways affected by the loss of the activator irrespective of whether the regulated genes may be directly or indirectly due to the absence of Blm10. We grew strains under steady-state growth conditions in chemostats to avoid confounding changes in gene expression caused by changes in growth rates between normal and mutant strains (Brauer et al. 2008). We followed up the comparative genomic analyses with comparative studies of global chromosomal integrity. We investigated a potential role for Blm10 or Ubp3/Blm3 in maintaining chromosomal integrity after DNA damage and a potential role of Blm10 in protecting against agents with different mechanisms of action. In protein localization experiments, we constructed a YFP-Blm10 fusion protein and used it to track Blm10 localization during the cell cycle and before and after DNA damage. Functions of the Blm10 carboxyl-terminus in protection and proper localization were also examined. Blm10 activation or regulation of the 20S proteasome is shown to be an important step in properly assembling chromosomes. The findings provide important new insights into the molecular mechanisms of protection and genomic stability conferred by Blm10 and suggest Blm10/PA200 inhibition could hold promise as a novel approach to cancer treatment.
Materials and Methods
Strains, plasmids, and primers
Yeast strains, plasmids, and primers used in these studies are listed, respectively, in Table 1, Table 2, and Table 3. Transformations and plasmid isolations were adapted from published methods (Rose and Broach 1991; Febres et al. 2001; Gietz and Woods 2002). For chemostat experiments, pulsed-field gel electrophoretic analyses, survival, and susceptibility experiments, we used diploid strains to avoid suppressors that could mask or modify mutant phenotypes.
Table 1. Yeast strains.
| Strain | Genotype | Source |
|---|---|---|
| BESY54 | derived from M1452-98B. ade2-40 ilv1-92 trp1-1 ura3-1 YFP-BLM10 SPC42-CFP | Eric Muller |
| BMA-8A | MATa ura3-52 trp1Δ63 leu2Δ his3Δ200 gal2 | Agnes Baudin |
| CM1452-98B | MATα ura3-52 ade2-40 leu2-3 ilv1-92 | This laboratory |
| CM1469-5A | MATα ilv1-92 his3-11 or his3-15 leu2-3, 112 trp1-1 ura3-1 | This laboratory |
| CM1469-5C | MATa ade2-40 or ade2-1 ura3-1 ilvl-92 trp5-12 ura3-1 blm3-1 | This laboratory |
| CM1522-9B | MATa ura3-52 trp1Δ63 leu2Δ his3Δ200 blm10Δ::HIS3 | This laboratory |
| CM-1526 | MATa/MATα ade2-40/ade2-40 trp1-1/trp1-1 HIS3/his3-11 LEU2/leu2-3 ura3-1/ura3-1 ILV1/ilv1-92 blm3-1/blm3-1 | This laboratory |
| CM-1527 | MATa/MATα LEU2/leu2-3 ILV1/ilv1-92 HIS3/his3-11 ade2-40/ade2-40 TRP1/trp1-1 | This laboratory |
| CM-1528 | MATa/MATα LEU2/leu2-3 ade2-40/ade2-40 trp1-1/trp1-1 URA3/ura3-1 ILV1/ilv1-92 BLM3/blm3-1 | This laboratory |
| CM-1529 | MATa/MATα LEU2/leu2-3 URA3/ura3-1 ILV1/ilv1-92 ade2-40/ade2-40 trp1-1/trp1-1 | This laboratory |
| CM1530-1A | MATa ura3-52 trp1Δ63 leu2Δ his3Δ200 gal2 blm10Δ::HIS3 | This laboratory |
| CM-1531 | MATa/MATα trp1/trp1 leu2/leu2 ura3/ura3 blm10Δ::HIS3/ blm10Δ::HIS3 | This laboratory |
| CM1531-1B | MATα ura3-52 ade2-40 leu2-3 ilv1-92 BLM10::YFP | This laboratory |
| DBY9500 (CEN.PK) | MATa/MATα MAL2-8C/ MAL2-8C | Maitreya Dunham |
| EJ758 | MATa his3-D200 leu2-3,112 ura3-52 pep4::URA3 | Eric Phizicky, Elizabeth Jones |
Table 2. Plasmids.
| Plasmid Name | Experiment | Parental Vector | Escherichia coli Selection | Yeast Selection | Tag | Insert |
|---|---|---|---|---|---|---|
| pRS303 | Deletion | pBluescript | Amp | HIS3 | None | HIS3 |
| pDH22 | Localization | Amp | Kanr | None | Kan and YFP | |
| pSH47 | Localization | Amp | URA3 | None | Cre recombinase | |
| pUB23 | β-galactosidase | URA3 | ||||
| pYEX 4T-1B | GST | pYEULC | Amp | URA3 | GST | BLM10 |
GST, Glutathione-S-transferase.
Table 3. Oligonucleotide primers.
| Primer Name | Primer Use | Length (Bases) | Sequence 5′-3′ |
|---|---|---|---|
| 1 | Sequencing | 20 | ATATGCCGCAGACGGAAGAC |
| 2 | Sequencing | 20 | ATATAAGACTGAAAGTCATG |
| 3 | Sequencing | 21 | GCCTATCGTTACATCCGTTGT |
| 4 | Sequencing | 21 | AGTAATTCGGTTTATTGTGAT |
| 5 | Sequencing | 19 | CAAAGAACAAATCAAAAGA |
| 6 | Sequencing | 20 | TCAGTGGCACGTACCTTCTA |
| 7 | Sequencing | 21 | CTTCATTGACGTTGATTTCCT |
| 8 | Sequencing | 22 | CAAAAAGAAAAAGCGTGAGTAC |
| 9 | Sequencing | 22 | AAAGCTCAATTTACGTGAGAAT |
| 10 | Sequencing | 22 | GTTGGTATTTGATCACCCATAC |
| 11 | Sequencing | 21 | GTTCGTGCGGCATCCATTTTG |
| PA-05 | Sequencing/deletion verification/YFP verification | 30 | GCGCGGTACCATTACGCAGAATAATCTATG |
| YFP-up | YFP cassette | 94 | TTCAATTGGGATAAGGTCTTGTTAGTAATGGGAAT |
| GGGTGATTTGATATCATCGTCATTGTTAGCGGTCAT | |||
| TTTGTACAATTCATCCATACCATG | |||
| YFP-down | YFP cassette | 82 | TTGCATACATAAACTTTATCATTGTTCGTTAGCTAG |
| CTTTGCACATTAATTTTTCGATTTGTTACCGCCACGG | |||
| CCGCCAGGG | |||
| Deletion primer 1 | Replacement cassette | 72 | ATGATCTCAAACTGCTTCTTAATATAGGCATCCAC |
| CTTTTCTGGGACGCTTTTTACTCTTGGCCTCCTCTAG | |||
| Deletion primer 2 | Replacement cassette | 68 | CAAATCTACATGTATATACAGATCTATACAGCAA |
| TTATAGGATATCTTTCGTTCAGAATGACACG | |||
| HIS3 R | Verify deletion | 21 | CAGACAATCAACGTGGAGGGT |
| NF | Sequencing | 22 | ATTCCCATTACTAACAAGACCT |
| NR | Sequencing | 22 | ATCGCAATATAAAGATTAACTA |
| L1F | Sequencing | 22 | AATCTTATATTGCGATCAGCTC |
| L1R | Sequencing | 24 | GATATGATAAGATAGGGCACAAC |
| L2F | Sequencing | 24 | GGGATTTTTACTGATGATCAAATG |
| L2R | Sequencing | 25 | GATATGATAATGATAGGGCACAAC |
| L3F | Sequencing | 24 | TGTTTAACTTCTTTTTGTCACGAA |
| L3R | Sequencing | 25 | GATAGGAATGAAAGCGGCTATAGA |
| L4F | Sequencing | 24 | AACCTCATCAACGGTATTGTATCT |
| L4R | Sequencing | 24 | TATTTCGGTTGTACATAGAGTTGC |
| L5F | Sequencing | 25 | ACTCTATGTACAACCGAAATAACTG |
| L5R | Sequencing | 21 | AAATATCAATCTGCCGATGTC |
| L6F | Sequencing | 25 | AGTGTATGTGTCATTTCCGATCAAG |
| L6R | Sequencing | 23 | CATATTCAGTTCGCAGAAACCAG |
| CF | Sequencing | 24 | TCATCTGGTTTCTGCGAACTGAAT |
| CR | Sequencing | 25 | GTTAGCGACAGCTGGCGAACCTGA |
Deletion of BLM10
Using polymerase chain reaction (PCR) deletion and replacement methods (Baudin et al. 1993), chromosomal BLM10 was deleted in haploid strains by PCR amplification of the HIS3 replacement cassette in plasmid pRS303 (kindly provided by Dr. Susan Henry), transformation of the PCR product into strain BMA-8A (kindly donated by Dr. Agnes Baudin), selection of HIS3+ recombinants, and verification by PCR of the correct gene deletion using oligonucleotides complementary to BLM10 flanking sequences (Table 3). Segregants from heterozygous diploids sporulated in 0.25% yeast extract, 0.1% glucose, 0.98% potassium acetate, and 40 mg/mL uracil, tryptophan, and leucine were intercrossed.
Chemostats
Detailed procedures for chemostat setup, temperature and pH probes, glucose-limited medium, aeration, daily monitoring, data acquisition, harvests, sample processing, and RNA preparations were followed as described at http://dunham.gs.washington.edu/protocols.shtml. Chemostats with individually calibrated pH and temperature probes were established in 500-mL fermenter vessels (Sixfors; Infors AG, Bottmingen, Switzerland) containing 300-mL cultures. They were stirred at 400 rpm with 5 L per minute of humidified and filtered air. Equivalent population growth (dilution) rates and doubling times were confirmed by measuring effluent volumes over time, microscopic examination, and cell counts.
Microarrays
RNA was isolated, labeled, and hybridized according to the instructions supplied by Agilent. Reference RNA was isolated from diploid strain DBY9500 (CEN.PK; Mal2-8C/Mal2-8C), labeled, and used in all hybridization samples. Cells had been grown to steady state in a chemostat and kindly provided by Dr. Maitreya Dunham.
Analyses of gene expression data
Mean expression values were calculated among replicated experiments and microarrays. Data were analyzed using the Agilent Scanner and Feature Extraction software, Princeton University MicroArray database (Gollub et al. 2003), Gene Ontology Local Exploration Map (GOLEM) software (Sealfon et al. 2006), Saccharomyces Genome Database (SGD 2010), and bioPIXIE (biological Process Inference from eXperimental Interaction Evidence) software (Myers et al. 2005). Results are presented for dilution rate = 0.06 hr−1 (doubling time = ∼12 hr) and were comparable for dilution rate = 0.12 hr−1 (doubling time = ∼6 hr).
β-galactosidase assays
Strains CM1469-5A (BLM10, UBP3/BLM3), CM1522-9B (blm10Δ, UBP3/BLM3), and CM1469-5C (BLM10, blm3-1) are auxotrophic for uracil. They were grown in synthetic selective media lacking uracil to select the URA3 gene on plasmid pUB23 (kindly provided by Dr. Alfred Goldberg). After induction of the fusion protein by galactose, β-galactosidase was assayed as published (Rose and Botstein 1983).
Pulsed-field gel electrophoreses and survival
Pulsed-field gel electrophoresis and cell survival were measured in parallel in each experiment as previously described (Moore et al. 2000). Phleomycin was supplied and prepared as previously described (Moore 1982, 1988). Concentrations were determined using the Beer-Lambert equation (OD245 /1.6 × 10−2). Liquid-holding (LH) recovery was measured under nondividing, non-replicating conditions maintaining full viability (Moore et al. 2000).
Susceptibility tests
Fresh cells were harvested from overnight cultures grown in standard, nonsynthetic complete medium (YPAD; Moore 1982) with aeration at 30° to 5 × 108 cells/mL, washed twice with deionized water to remove growth medium, and resuspended at 5 × 107 cells/mL in deionized water. Five microliters of several dilutions were pipetted on YPAD plates, prepared the preceding day, with varying dilutions of 1% methyl methanesulfonate (Sigma-Aldrich), hydrogen peroxide (3% stabilized, pharmaceutical grade), 0.02 M 5-fluorouracil (3 mg/mL; City Chemical), 5 M hydroxyurea (0.38 g/mL; Sigma-Aldrich), 0.001 M doxorubicin chlorhydrate (adriamycin, Tecoland; 0.58 mg/mL DMSO [Fisher Scientific]), rapamycin/sirolimus (Tecoland; 0.09 mg/mL), or calcofluor white (American Cyanamid). Plates were incubated at 30° for up to 4 days.
Yellow fluorescent protein (YFP) fusion and fluorescence microscopy
YFP was fused to the N-terminus of Blm10 as previously described (Prein et al. 2000). The fusion complements a Blm10 deletion mutant as judged by its total relief of hypersensitivity to phleomycin. CM1452-98B (YFP-Blm10) and BESY54 (YFP-Blm10, Spc42-CFP) were grown on nonsynthetic complete medium and imaged using a DeltaVision microscopy system from Applied Precision (Issaquah, WA). The system incorporates an Olympus IL-70 microscope, a u-plan-apo 100× oil objective (1.35NA), a CoolSnap HQ digital camera from Roper Scientific (Tucson, AZ), and optical filter sets from Omega Optical (Brattleboro, VT). Live cells were imaged on thin pads of medium containing 1% agarose (Muller et al. 2005). Images were analyzed and quantitated using the program Fluorcal, an integrated set of Matlab scripts designed for the automated selection and analysis of regions of interest within images obtained by fluorescence microscopy (Shimogawa et al. 2010). Phleomycin D1 was purchased from Invitrogen (Life Technologies, Carlsbad, CA) in the formulation of Zeocin.
Glutathione-S-transferase (GST) fusion, immunofluorescence, and quantitative expression of Blm10(-339aa)
Truncated Blm10-GST under control of the CUP1 promoter was purified from a pool of slightly modified plasmid pYEX 4T-1 (kindly provided by Dr. Eric Phizicky; Martzen et al. 1999), confirmed by DNA sequencing, and transformed into blm10Δ cells. To localize Blm10-GST, cells were grown to early exponential phase (106−107 cells/mL) in liquid YPAD containing copper (50 μM) to induce expression of fusion protein. Cells were fixed by adding 3.7% formaldehyde directly to the medium, incubated for at least 1 hr, converted to spheroplasts in 1 mL of 50 μL/mL Zymolyase 100T in 0.1 M potassium phosphate (pH 7.5) with 2 μL/mL 2-mercaptoethanol, gently pelleted, and treated with Alexa Fluor 488 anti-GST conjugate antibody (Molecular Probes) or 4′,6-diamidino-2-phenylindole (DAPI, 1 mg/mL in water). Cells were observed using a Zeiss fluorescent microscope at 100× with FITC and DAPI filters.
To quantitate growth and survival, copper and phleomycin were added to fresh cells grown overnight in YPAD, washed, and resuspended in YPAD at 5 × 106 cells/mL. Cells were counted and plated on YPAD at each time point to determine viability.
Results
Genomic instability signature of downregulated genes
To determine how cells alter their gene expression in the absence of the Blm10 protein, we studied microarray-based global gene expression for the entire genome (∼6000 genes) in normal (BLM10/BLM10) and mutant (blm10Δ/ blm10Δ) diploids in the context-specific environments of chemostats. Diploids of both genotypes were grown in continuous steady-state at the same controlled growth rates (Brauer et al. 2005; Kubitschek 1970). In these steady-state growth conditions, the growth rate is determined by the medium flow rate and therefore identical between normal and mutant cultures. This was an important part of the experimental design because we found blm10Δ/blm10Δ cells grow slowly outside of chemostats in some media in comparison with BLM10Δ/BLM10Δ cells of the same genetic background.
To our surprise, many genes specifically encoding proteins required for proper chromosome organization, assembly, function, repair, and progression through the cell cycle were downregulated to different extents in blm10Δ/blm10Δ cells. Some of these are shown in Table 4, along with their human homologs. Among these were genes encoding 18 DNA packaging, nucleosome organization, and chromatin assembly proteins; nine DNA damage and checkpoint proteins; seven DNA repair proteins; eight transcription factors; and 34 proteins assuring correct numbers of chromosomes segregate during cell division. Twenty-five additional genes involved in nucleosome modeling and DNA packaging were downregulated more than 25% (supporting information, Table S1). In addition, the recombinational repair gene, RAD51, was downregulated 62%, and all six remaining YRF1 helicase genes were downregulated 76–91% (Table S1).
Table 4. Some of the significantly downregulated genes critical for chromosomal integrity.
| Genes | |
|---|---|
| Chromosome organization and function | |
| DNA packaging, nucleosome organization, chromatin assembly or dissassembly, nucleosome and chromatin remodeling, chromatin organization and modification | ACS2 2.8, HST3 2.3, HHF1 2.2, HTZ1 2.2, HHO1 2.1, NHP6A 2.1, HTB1 2.0, HTB2 2.0 |
| DNA repair: double-strand break repair, mismatch repair, postreplication repair | RAD59 2.4, MCK1 2.3, NHP6A 2.1, HTB1 2.0, PSY3 1.8, MEC3 1.6, DUN1 1.5 |
| Mitotic DNA recombination, telomere arrangement and maintenance | RAD59 2.4, HHO1 2.1, YRF1-3 2.0, YRF1-6 2.0, PSY3 1.8, MEC3 1.6 |
| Regulation and progression of cell cycle, cyclin-dependent protein kinase activity | CLB2 3.3, PCL9 3.2, SIC1 2.6, CLB1 2.0, MEC3 1.6 |
| G1/S transition of mitotic cell cycle | PCL9 3.2, SIC1 2.6 |
| Mitotic spindle assembly and chromosome segregation | CIN8 2.4 |
| Spindle pole body separation, microtubule cytoskeleton organization, G2/M transition | CLB2 3.3, CLB1 2.0, HSL1 1.9 |
| Chromatin silencing and negative regulation of gene expression, epigenetic | HTZ1 2.2, MEC3 1.6 |
| Meiotic DNA replication, meiotic DNA recombination | RIM4 2.2, MUM2 2.1 |
| Checkpoints | |
| Genome integrity, DNA damage sensor | MEC1 1.6, MEC3 1.6, SGS1 1.6, DUN1 1.5 |
| DNA replication, gap repair of damaged DNA | TOF1 1.5 |
| Septin | HSL1 1.9, GIN4 1.7 |
| Spindle checkpoint activation, protects sister chromatid cohesion in mitosis | PHB2 1.6 |
| Meiotic recombination | FPR3 1.7 |
| Cytokinesis | |
| Cytokinesis, cell division | DSE2 5.4, SUN4 5.3, DSE1 4.6, CTS1 4.1, SCW11 3.7, CHS2 3.5, BUD9 3.2, EGT2 2.8, HOF1 2.5, MYO1 2.3, RAX1 2.3, IQG1 2.2, HSL1 1.9 |
| Transcription factors | |
| G1 cell-cycle progression, cyclin-dependant kinase target | SFG1 3.6 |
| Activates expression of early G1-specific genes | ACE2 2.6 |
| Activates transcription of genes expressed at M/G1 and G1, activates cyclin Pcl9 | SWI5 2.5 |
| Response to DNA damage stimulus, expression highest in G1 | TOS4 1.8 |
| Response to DNA damage stimulus, progression from G1 to S and G2 to M | CKA2 1.6 |
| Involved in directing transcription of genes by RNA polymerases, I, II, and III | SPT15 1.5 |
| RNA polymerase II initiation and elongation | TFG2 1.5 |
| Calcineurin B; calcineurin regulates stress-response transcription factor Crz1; human protein participates in apoptosis and other signaling pathways | CNB1 1.5 |
Genes are listed in each subgroup or group in order of their fold decreased expression. Essential genes are underlined. Yeast genes in bold have the following human homologs: ACS (ACS2), ATR (MEC1), BLM and WRN (SGS1), BRSK2 (HSL1), CCNB1 (CLB1), CCNB2 (CLB2), CHEK2 (DUN1), CSNK2A1 (CKA2), FPR3 (FPR3), GSK3 family (MCK1), H1F0 (HHO1), H2AFV, (HTZ1), HIST1H2BH (HTB1), HIST1H2BO (HTB2), HIST1H4N (HHF1), HMGB1/HMG1 (NHP6A), Hus1 (MEC3), IQGAP1 (IQG1), Kip1 (SIC1), and MYH11 (MYO1), PHB2 (PHB2), PPP3R2 (CNB1), RAP30 (TFG2), TBP (SPT15).
While striking, the downregulation of all of these genes may not necessarily decrease protein expression in all instances after posttranscriptional and posttranslational modification, or actually be a direct rather than indirect consequence in all cases by the absence of Blm10. Moreover, the actual levels of downregulation may not be strictly quantitative. However, what is most important and remarkable is the unique and composite signature defined by the simultaneous downregulation of the genes. As a whole, the results suggest a vulnerability of cells without Blm10.
Chaperone-mediated, protein-folding signature of upregulated genes
We reasoned that mutant cells may compensate by upregulating particular pathways or genes. To evaluate this possibility, we first submitted all genes and open reading frames (ORFs) upregulated greater than fourfold in blm10Δ/blm10Δ cells (gray nodes in Figure 2) to bioPIXIE (Myers et al. 2005) to query whether any of the genes colocalized near each other in a functional network, and if so, what other genes colocalized with them.
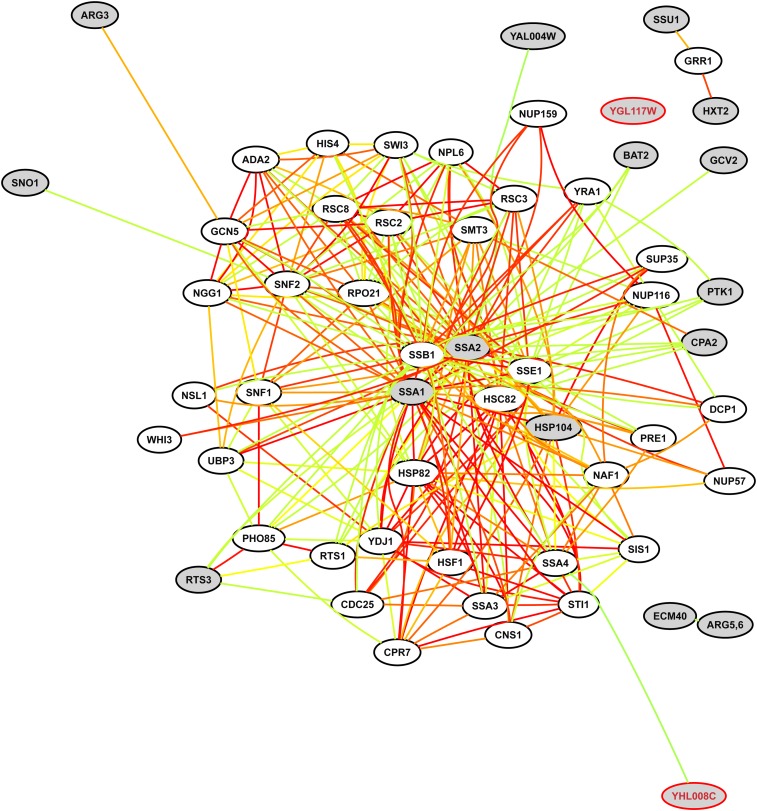
Figure 2 .
Pathway analysis of coordinated upregulation without Blm10. Gray nodes are genes and ORFs upregulated fourfold to eightfold in blm10Δ/blm10Δ mutant cells relative to BLM10/BLM10 cells. YAL004W is a dubious ORF, and YGL117W and YHL008C are uncharacterized ORFs (SGD Project 2010). Confidence-weighted pairwise linkages between genes are color-coded (Myers et al. 2005): red (highest confidence), orange, yellow, green (lowest confidence).
The most relevant part of the functional network surrounding the upregulated genes was extracted by bioPIXIE and is shown in Figure 2. Most central and geometrically close in the integrated functional network lie SSA1- and SSA2-encoded chaperone proteins, homologs of yeast and human members of the heat shock protein 70 multigene family, together with HSP104 of the Hsp100 family. Ssa1 and Ssa2 are novel G2/M checkpoint proteins that copurify with the DNA damage-dependent checkpoint protein, Rad9, and function after ultraviolet light irradiation in phosphorylating Rad9 and another checkpoint protein, Rad53 (Gilbert et al. 2003). It is proposed that the chaperone activities of Ssa1 and Ssa2 remodel the large Rad9 complex to a smaller Rad53 activating complex after genomic insult (Van Den Bosch and Lowndes 2004).
By far, the highest enrichment bioPIXIE measured in the whole network (P value 4 × 10−9) comprised more than 19% of all genes (Table 5). This specific subset encodes protein-folding chaperones, nine of which were upregulated twofold to sevenfold in mutant cells (Table 6). Those that physically interact with Blm10 (SGD Project 2010) are indicated in Table 6. Eight additional protein-folding genes were enriched (GOLEM, P value 5 × 10−10) among 144 genes upregulated twofold or more, and are listed in Table 7 with other protein-folding genes regulated ≥1.5 fold. As a group, the encoded molecular chaperones in Figure 2 and Tables 6 and 7 assist to properly fold and assemble nascent polypeptide chains or refold previously denatured or aggregated proteins. Equally important, they interact with partially folded or unfolded protein subunits to stabilize, translocate, or degrade them.
Table 5. Gene ontology terms enriched in the gene network shown in Figure 2.
| Go term | Cluster Frequency | Genome Frequency | P Value | Genes |
|---|---|---|---|---|
| Protein folding | 11/57 19.3% | 70 / 6471 1.1% | 4.10−9 | SSA4, STI1, SSA2, SSE1, SIS1, HSC82, HSP82, SSA3, SSA1, HSP104, CNS1 |
| Arginine biosynthesis | 4/57 7.0% | 10 / 6471 0.2% | 3.42−4 | ARG3, CPA2, ARG5, 6, ECM40 |
| Cellular physiological process | 55/57 96.5% | 4689/6471 72.5% | 7.83−4 | SMT3, SSA4, SSB1, HIS4, GCV2, NUP57, NSL1, WHI3, ARG3, STI1, HSF1, CPA2, RTS3, RSC8, DCP1, SSA2, SNO1, ADA2, YDJ1, RTS1, RSC3, SSE1, NUP159, SUP35, YRA1, SWI3, NPL6, NUP116, SNF1, SIS1, GRR1, PTK1, CDC25, PRE1, HSC82, SSU1, PHO85, UBP3, NGG1, HSP82, BAT2, NAF1, RPO21, ARG5, 6, SSA3, SSA1, ECM40, SNF2, HXT2, HSP104, YHL008C, GCN5, TRP5, CNS1, RSC2 |
| Cellular process | 55/57 96.5% | 4728/6471 73.1% | 1.19−3 | SMT3, SSA4, SSB1, HIS4, GCV2, NUP57, NSL1, WHI3, ARG3, STI1, HSF1, CPA2, RTS3, RSC8, DCP1, SSA2, SNO1, ADA2, YDJ1, RTS1, RSC3, SSE1, NUP159, SUP35, YRA1, SWI3, NPL6, NUP116, SNF1, SIS1, GRR1, PTK1, CDC25, PRE1, HSC82, SSU1, PHO85, UBP3, NGG1, HSP82, BAT2, NAF1, RPO21, ARG5, 6, SSA3, SSA1, ECM40, SNF2, HXT2, HSP104, YHL008C, GCN5, TRP5, CNS1, RSC2 |
Table 6. Regulation of protein-folding molecular chaperones encoded by genes in Figure 2.
| Gene | Fold Change | Encoded Chaperone Activity | Human Homolog or Domain |
|---|---|---|---|
| SSA2 | ↑7.1 | Hsp70 family member, member of Rad9 DNA-checkpoint complex | Hsp70 |
| SSA1a | ↑4.2 | Hsp70 family, chaperone complex with ADJ1, protein refolding, member of Rad9 DNA-checkpoint complex | Hsp70 |
| HSP104a | ↑4.0 | Hsp100 family, acts in conjunction with Ssa1 and Ydj1 (Hsp40), protein refolding | |
| SSA3 | ↑3.8 | Hsp70 family member | HSPA1AB |
| HSP82 | ↑3.3 | Hsp90 isoform, associates with Cpr6, Sti1, Cns1, Hch1, Aha1, Sse1, nascent chain folding, protein refolding, proteasome assembly | HSP90AB1 |
| SIS1 | ↑2.7 | HSP40 (DNAJ) co-chaperone, interacts with Ssa1 | DNAJB1 [HSP40] |
| SSE1 | ↑2.6 | Hsp70 family member, component of Hsp90 chaperone complex, protein refolding | HSPA4 |
| HSC82a | ↑2.3 | Hsp90 isoform, associates with Sti1, Cns1, Cpr6, Hch1, Aha1, Sse1, nascent chain folding, protein refolding, proteasome assembly | HS90AB1 |
| STI1 | ↑1.9 | Hsp90 co-chaperone, interacts with Ssa and Hsp70 chaperones | STIP1 |
| SSA4 | ↑1.2 | Hsp70 family member | HSPA8 |
| CNS1b | ↓1.7 | Hsp90 co-chaperone, binds Hsp82 and Ssa1 | TTC4 |
Chaperone physically interacts with Blm10.
Essential gene.
Table 7. Additional protein-folding genes regulated ≥1.5-fold.
| Gene | Fold Change | Encoded Chaperone Activity | Human Homolog or Domain |
|---|---|---|---|
| HSP78 | ↑3.7 | Hsp100 family, mitochondrial homolog of Hsp104, protein refolding | Clp/Hsp100 |
| HSP26 | ↑2.9 | Small molecular chaperone | |
| AHA1 | ↑2.6 | Co-chaperone, binds Hsp82, activates Hsp90, similar to Hch1 | AHSA1 |
| HCH1 | ↑2.6 | Co-chaperone, binds and activates Hsp90 | |
| APJ1 | ↑2.4 | HSP40 (DNAJ) family, regulates Hsp70 activity, genetically interacts with Ydj1 | Contains a DNAJ domain |
| HSP42 | ↑2.3 | Small molecular chaperone | |
| CPR6 | ↑2.3 | Binds Hsp82, protein refolding | PPID |
| CIN1 | ↑2.1 | Tubulin-folding factor | TBCD |
| HSP31 | ↑1.9 | DJ-1/Pfpl family, amino acid substitution in DJ-1 associated with early-onset Parkinson's | DJ-1 |
| YDJ1 | ↑1.8 | Hsp40 (DNAJ), Ssa1 co-chaperone, regulates Hsp70 and Hsp90 functions, nascent chain folding, protein refolding | DNAJA2 (DNAJ [Hsp40]) |
| HSP60 | ↑1.6 | Mitochondrial chaperonin, nascent chain folding, protein refolding | HSPD1 |
| HSP10 | ↑1.5 | Hsp60 co-chaperonin, protein refolding | HSPE1 |
| MPD1 | ↓1.9 | Endoplasmic reticulum chaperone for glycoproteins | |
| SBA1 | ↓1.5 | Co-chaperone, binds to and regulates Hsp90 family, regulates telomerase activity | p23 |
Essential genes are underlined.
The Rpn4 transcription factor that stimulates expression of proteasomal genes, positively regulates DNA repair, and physically interacts with Blm10 (SGD Project 2010), was upregulated nearly twofold in mutant cells in the current studies (Table S1). However, the expression of genes encoding components of the 19S and 20S proteasome subunits with which Blm10 physically associates (e.g. Pre1-Pre10, Pup2, Pup3, Rpt6, and Scl12; SGD Project 2010) were not significantly altered in cells lacking Blm10, with the exception of PRE6 and PUP2, which were downregulated 31% and 23%, respectively, and PUP3 which was upregulated 28% (Table S1).
By analogy to the downregulated genes, all of the upregulated genes in these groups may not lead to upregulated protein expression following posttranscriptional and posttranslational modification, or result directly from deleting BLM10. Nevertheless, the concurrent upregulated expression of this specific subset of genes produces a chaperone-mediated, protein-folding signature.
In Figure 2, UBP3/BLM3 is tightly connected and geometrically proximal to SSA1 and SSA2 with high confidence (1.0 and 0.9, respectively). Also proximal to these chaperone proteins is SNF2 (0.65 confidence), encoding the catalytic subunit of the SWI/SNF chromatin remodeling complex involved in nucleosome modification, transcriptional regulation, and DNA double-strand break repair. NGG1 is also proximal (0.62 confidence), and is involved in chromatin modification as a constituent of histone acetyltransferase complexes.
Proteolysis
Having discovered UBP3/BLM3 is linked with the highest confidence to both highly upregulated genes most central in the network of upregulated genes in blm10Δ/blm10Δ cells (Figure 2), we compared the degradation of β-galactosidase in blm3-1 cells with that in wild-type and blm10Δ cells. Cells were transformed with pUB23, a plasmid containing an ubiquitin-lacZ gene fusion under the control of a GAL promoter that allows gene expression to be controlled by the amount of galactose supplied in media (Bachmair et al. 1986). After induction of the fusion protein, ubiquitinated β-galactosidase is produced, and the ubiquitin tag automatically targets β-galactosidase for degradation by proteasomes (Bachmair et al. 1986).
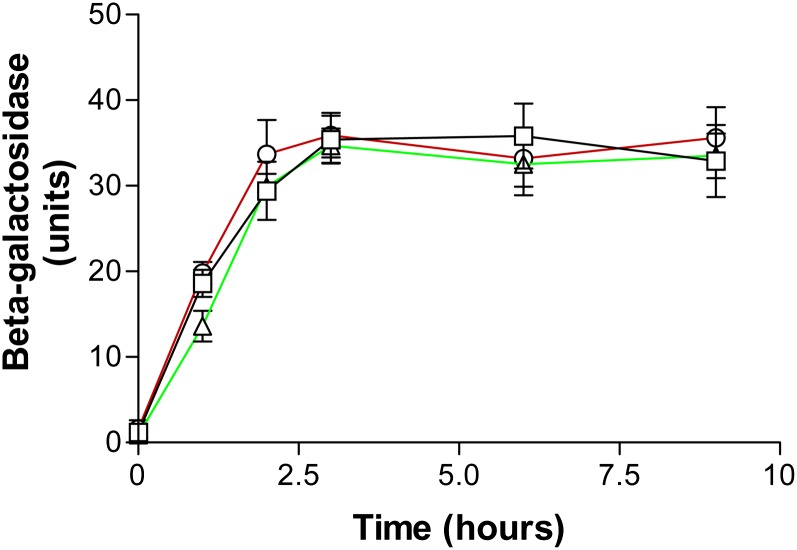
Time-dependent β-galactosidase activities in the three strains are shown in Figure 3. Activities were barely detectable at time zero but increased after the first hour and for two additional hours before reaching a plateau. Comparable activities of the three genotypes indicated Ubp3/Blm3 and Blm10 do not stimulate hydrolysis of this model and intact, folded protein. Our Blm10 results are consistent with structural restraints of the small dome-like opening of Blm10/PA200-proteasome complexes through which small peptides, but not proteins, might pass (recently reviewed in Savulescu and Glickman 2011; Stadtmueller and Hill 2011; Dange et al. 2011). They also are consistent with previous reports that Blm10 appears not to contribute significantly to the degradation of other ubiquitinated proteasome substrates (Schmidt et al. 2005a), and that Blm10/PA200 binding to the core particle activates it for hydrolysis of some peptides (Ustrell et al. 2002; Schmidt et al. 2005a; Iwanczyk et al. 2006; Lehmann et al. 2008; Lopez et al. 2011; Dange et al. 2011). The specific peptide substrate affects the amount of hydrolysis by Blm10/PA200 in vitro (Ustrell et al. 2002; Fehlker et al. 2003; Lopez et al. 2011; Dange et al. 2011).
Figure 3 .
Degradation of β-galactosidase by proteasomes in normal (□), blm3-1 (Ο), and blm10Δ (Δ) strains. Enzymatic activities in cells were determined spectrophotometrically at the indicated time points.
Global DNA damage is conferred by the blm10Δ and blm3-1 mutations
As a logical follow-up to the comparative genome analyses, we evaluated the genomic integrity in diploid cells without Blm10 and included blm3-1/ blm3-1 diploid cells in these studies. For these chromosomal studies, we used pulsed-field gel electrophoresis, during which individual chromosomes separate into distinct bands according to molecular weight and electric field interaction, and double-strand breaks in DNAs cause bands to lessen in intensity or disappear in a dose-dependent manner (Moore et al. 2000). Degraded chromosomes either leave the gel or accumulate at the bottom as a diffuse smear. The bleomycin-phleomycin family of chemical congeners is used as a tool in DNA damage studies (e.g. Moore 1990, 1999; Moore et al. 2000), and we used phleomycin to examine the quality of chromosomal DNAs in wild-type diploid cells (BLM10/BLM10, BLM3/BLM3) and mutant diploid cells in which either BLM10 was deleted (blm10Δ/blm10Δ, BLM3/BLM3) or UBP3/BLM3 was truncated (BLM10/BLM10, blm3-1lblm3-1) on homologous chromosomes.
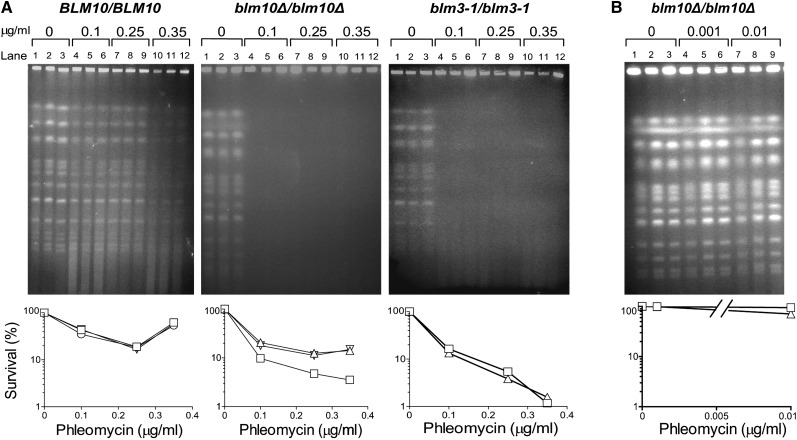
Without treatment, chromosomes from all three diploids consistently produced strong bands (Figure 4, A and B, lanes 1−3). In wild-type cells, chromosomal bands remained relatively strong after 0.1 μg/mL (lanes 4−6; Figure 4A) and 0.25 μg/mL (lanes 7−9; Figure 4A) phleomycin treatments, and underwent moderate degradation after 0.35 μg/mL treatments (lanes 10−12; Figure 4A). However, chromosomes in blm10Δ/blm10Δ and blm3-1/blm3-1 cells degraded following the same three treatments (lanes 4−12 vs. lanes 1−3; Figure 4A), indicating marked deficiencies. Exposure to 10- to 100-fold lower doses left chromosomes in blm10Δ/blm10Δ cells intact (Figure 4B).
Figure 4 .
Pulsed-field gel electrophoretic analyses comparing chromosomal damage and killing after no treatments and after 30-min phleomycin treatments. Diploid genotypes with respect to BLM10 and BLM3 were BLM10/BLM10, BLM3/BLM3; blm10Δ/blm10Δ, BLM3/BLM3; and BLM10/BLM10, blm3-1/blm3-1. Treated populations were divided and incubated under nongrowing conditions for 24 or 48 hr, during which competent strains can reconstruct their chromosomes (Moore et al. 2000). Routine microscopic examination and visual counting of cells before and after these LH periods confirmed cell populations did not bud or grow, and cell lysis was never observed. (A) Lanes 1, 4, 7, and 10, no LH. Lanes 2, 5, 8, and 11, 24-hr LH. Lanes 3, 6, 9 and 12, 48-hr LH. (B) Lanes 1, 4, 7, no LH. Lanes 2, 5, 8, 24-hr LH. Lanes 3, 6, 9, 48-hr LH. Corresponding survival data: squares, 0 LH; inverted triangles, 24-hr LH; triangles, 48-hr LH. Pulsed-field gel electrophoreses and survival analyses are representative of three independent experiments and of multiple diploid constructions of the same genotypes.
Cell death, measured in parallel with DNA damage in wild-type, blm10Δ/blm10Δ, and blm3-1/blm3-1 cells in each experiment, was low in wild-type cells and increased after 0.1 and 0.25 µg/mL phleomycin treatments (Figure 4A). Cell death was greater in mutant cells than in wild-type cells (Figure 4, A and B), indicating protective functions of Blm10 and Ubp3/Blm3. The order of decreasing resistance among the diploids was BLM10/BLM10, BLM3/BLM3 > blm10Δ/blm10Δ, BLM3/BLM3 > BLM10/BLM10, blm3-1lblm3-1 > blm10Δ/blm10Δ, blm3-1lblm3-1.
Consistently, we observed lower cell death in wild-type cells after 0.35 µg/mL treatments than after 0.25 µg/mL treatments, but not in mutant cells (Figure 4A). The reduced cell death in wild-type cells is most likely attributable to induced DNA repair and cellular recovery observed after this drug family causes extensive chromosomal damage (Moore et al. 2000).
Susceptibilities to lethal effects of agents with different mechanisms of action
The comparative genome analyses together with finding elevated DNA damage and killing after phleomycin treatments led us to examine whether the absence of Blm10 heightened susceptibilities to agents with different mechanisms of action. For these studies, we selected agents we previously had not investigated.
As shown in Figure 5, Blm10 loss reduced resistance to low concentrations of methyl methanesulfonate and strikingly compromised defenses against low-dose treatments of hydrogen peroxide, 5-fluorouracil, hydroxyurea, and doxorubicin. Considered oxidant mutagens, the bleomycin-phleomycin family and hydrogen peroxide cause similar DNA damage (Demple and Harrison 1994). Some of the gene functions important for resistance to these agents (e.g. Moore 1978, 1982; Moore et al. 1992, 2000; Aouida et al. 2004; Cloos et al. 2006) overlap with those required for resistance to ionizing radiation (Bennett et al. 2001) and some other chemicals (Parsons et al. 2004). In contrast, anticancer 5-fluorouracil and hydroxyurea are antimetabolites (Brunton et al. 2006; Friedberg et al. 2006; Nitiss and Heitman 2007). 5-fluorouracil is a pyrimidine antagonist that is synthesized into 5-fluoro-2-deoxyuridine, a nucleotide that inhibits the 2-deoxythymidine synthesis by thymidine synthetase. Incorporation of 5-fluorouracil into DNA interferes with DNA synthesis and inhibits RNA production. Hydroxyurea inhibits DNA and RNA synthesis by blocking ribonucleotide reductase and causes site-specific DNA damage through the formation of hydrogen peroxide and nitric oxide (Sakano et al. 2001). The anthracycline doxorubicin intercalates between DNA bases, causing DNA breaks by blocking topoisomerase type II (Brunton et al. 2006). Moreover, DNA can be damaged by oxidative free radicals generated during the metabolism of doxorubicin (Turner et al. 1990).
Figure 5 .
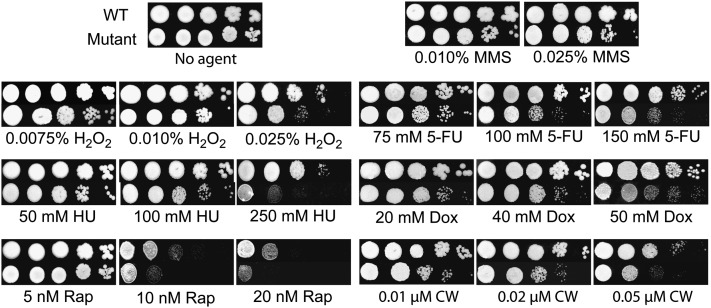
Dose-dependent susceptibilities of normal and mutant diploids. From left to right are fivefold serial dilutions of each genotype. MMS indicates methyl methanesulfonate; H2O2, hydrogen peroxide; 5-FU, 5-fluorouracil; HU, hydroxyurea; Dox, doxorubicin; Rap, rapamycin; CW, calcofluor white. Results are representative of two to four independent experiments.
Mutant cells also lost protection against rapamycin (Figure 5), a novel anticancer antibiotic first identified as antifungal (Brunton et al. 2006; Nitiss and Heitman 2007). Immunosuppressant rapamycin arrests cells at G1/S and inhibits highly conserved nutrient sensing pathways. The elevated sensitivity of the mutant cells to rapamycin perhaps could be explained by the knowledge that this drug strongly induces BLM10 (Hardwick et al. 1999) and increases BLM10 mRNA levels (Lopez et al. 2011), and these events would not be possible in blm10-deleted cells.
Finally, bleomycin damages fungal cell walls (Moore et al. 1992; Beaudouin et al. 1993; Moore et al. 2003), as does cell-wall perturbing calcofluor which binds chitin, thereby inhibiting chitin synthase and proper cell wall biosynthesis (Nitiss and Heitman 2007). As Figure 5 shows, cells lacking Blm10 were hypersusceptible to calcofluor. It is tempting to suggest that this sensitivity could be attributable to the downregulation of MEC3, cell-cycle checkpoints, transcription factors, and cytokinesis in these cells (Table 4). As a whole, the results shown in Figure 5 suggest inhibition by Blm10/PA200 could dually target cancer and fungal cells.
In similar experiments, normal and mutant strains grew equally well after mitomycin C, ultraviolet light irradiation, and ethidium bromide treatments (data not shown). This could reveal discernment of the type of DNA damage because the modes of action of these agents differ from those in Figure 5. Mitomycin C is a potent DNA cross-linker (Friedberg et al. 2006; Deans and West 2011). Ultraviolet radiation produces cyclobutane pyrimidine dimers in DNAs, along with pyrimidine-pyrimidone (6-4) and other photoproducts (Friedberg et al. 2006). Ethidium bromide intercalates in DNA (Reinhardt and Krugh 1978), but differs from doxorubicin in its action.
In a previous study, BLM10 and blm10Δ strains were reported to grow equally after treatments with gamma irradiation, methyl methanesulfonate, ultraviolet irradiation, and camptothecin (Iwanczyk et al. 2006). In the same study these strains also grew equally after treatments with bleomycin, phleomycin, and hydroxyurea (Iwanczyk et al. 2006), in contrast to the hypersusceptibilities blm10Δ cells exhibited to these drugs in the current studies (Figures 4 and 5) and to the bleomycin-phleomycin family in previous studies (Febres et al. 2001; Doherty et al. 2004; Schmidt et al. 2005a).
Growth of BLM10/BLM10 and blm10Δ/blm10Δ strains at 37°
Previously, a temperature-sensitive phenotype was reported for blm10Δ cells growing at 37° in one strain background (Fehlker et al. 2003) but not in others (Schmidt et al. 2005a; Iwanczyk et al. 2006). We compared the growth of BLM10/BLM10 and blm10Δ/blm10Δ strains in several experiments in the current studies and found that they grew equally well at 37°. This finding was confirmed in quantitative growth measurements by hemacytometer counts of cells from 0 to 72 hr in three independent experiments (K. Doherty and J. Lukose, unpublished data).
Respiratory deficiency
The functional state of mitochondria affects resistance to killing by the bleomycin-phleomycin family of DNA-cleaving drugs, as respiratory-deficient cells lacking mitochondrial DNA (ρ0) are up to 100-fold more resistant than isogenic respiratory-proficient (ρ+) cells (Davermann et al. 2002). During the current studies, we found that blm10Δ and blm10Δ/blm10Δ strains growing in the absence of bleomycin or phleomycin produced up to tenfold higher frequencies of respiratory-deficient petite colonies than BLM10 and BLM10/BLM10 strains (L. Pride, K. Doherty, and C.W. Moore, unpublished data). Other blm10Δ haploid strain backgrounds also produced increased yields of petite colonies (Sadre-Bazzaz et al. 2010).
Blm10 localization during the cell cycle
The cellular localization of the YFP-Blm10 fusion protein was determined in live cells by fluorescence microscopy. YFP-Blm10 was the sole copy of Blm10 in the cell and gene expression was driven from its endogenous promoter in its native chromosomal context. The protein is completely functional in totally relieving phleomycin hypersensitivity in blm10Δ and blm10Δ/blm10Δ mutant cells.
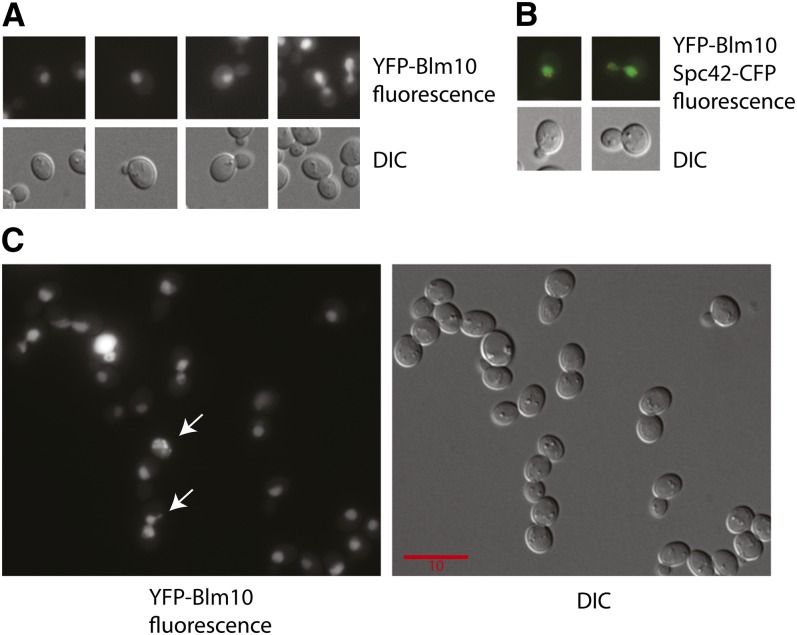
YFP-Blm10 localized to nuclei in all budded and unbudded cells (Figure 6A), consistent with our findings that Blm10 protects against DNA damage, and that the yeast 26S proteasome complex is predominantly nuclear (80%; Russell et al. 1999). The identification of the nucleus was confirmed by coexpressing YFP-Blm10 with CFP-tagged Spc42 (Figure 6B), a component of the spindle pole body that is embedded in the nuclear membrane (Muller et al. 2005).
Figure 6 .
Nuclear localization and colocalization of Blm10. Shown are representative YFP and differential interference contrast (DIC) images of living cells. The scale bar applies for all images in the panels as all images are scaled equally. Representative cells from populations of 4000 to 10,000 cells are shown to illustrate the following: (A) Nuclear localization, showing progression from unbudded to large-budded cells. (B) Colocalization of nuclear Spc42-CFP and YFP-Blm10. (C) Nuclear localization after growth on medium supplemented with 20 μg/mL phleomycin D1. Arrows indicate cells with evidence for nuclear membrane fragmentation.
The levels of Blm10 in the nucleus were measured quantitatively as described in Materials and Methods and found not to be constant during the cell cycle. The fluorescence intensity of YFP-Blm10 in budded cells, before nuclear migration to the neck, was approximately 40% greater than in unbudded G1 cells. Thus, after DNA replication Blm10 becomes enriched in the nucleus during late S and early M phases, before anaphase and cell division. The enrichment at the S/M transition of the cell cycle is when the state of the DNA is monitored by checkpoint controls.
Blm10 localization signal and pattern after DNA damage
We compared the localization signals and patterns after DNA damage to those before DNA damage. Cells were treated with 20 µg/mL phleomycin D1 for 4 hr. After this treatment, fluorescence was relatively uniform throughout the nucleus. A heterogeneous response was observed, with some cells showing a large increase in the level of YFP-Blm10 in the nucleus and some cells showing evidence of nuclear fragmentation (Figure 6C).
Identification of conserved regions in different parts of the Blm10 protein
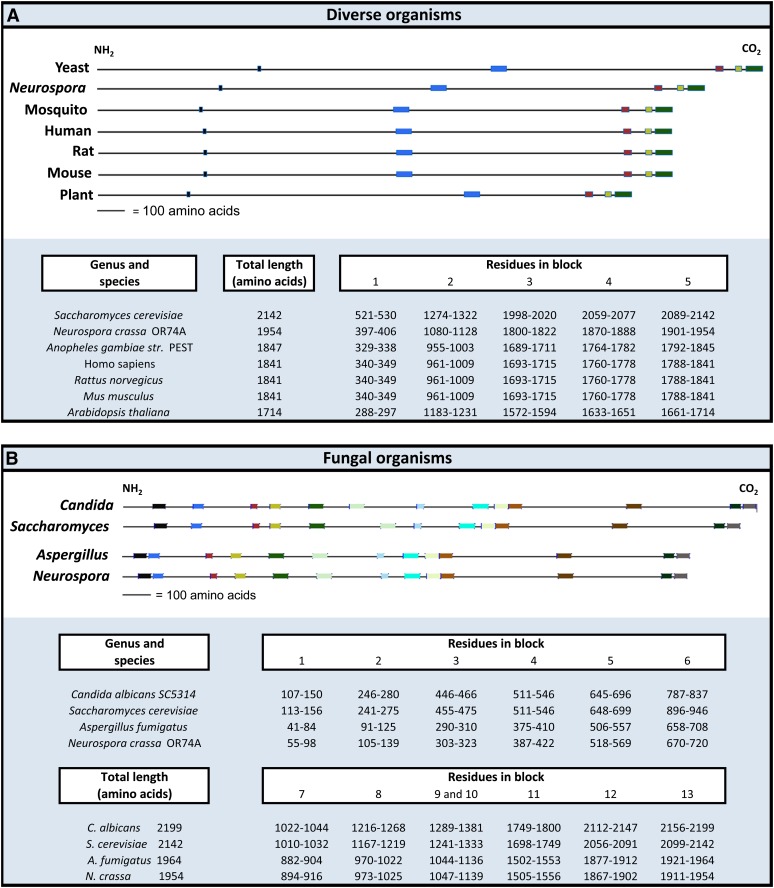
The Blm10 protein is highly conserved among diverse organisms, from yeast to humans. Blm10/PA200 is the most conserved proteasome activator (Finley 2009). In addition, all homologs are quite large, with 240-kD Blm10 being the largest (http://blast.ncbi.nlm.nih.gov/Blast.cgi).
We arbitrarily selected seven of these homologs and used the Blocks database (Henikoff et al. 2000) to multiply align ungapped Blm10 regions to them. This identified five conserved regions (Figure 7A). Comparing the yeast sequence to the other sequences, we found that the positions of the first two conserved regions, and the distances between these two conserved regions, varied most (Figure 7A). In contrast, the positions and spacing of the three carboxyl-terminal regions were more conserved among all seven sequences. The sizes of the human, rat, and mouse proteins and the positions of all five conserved sequences in those proteins were identical (Figure 7A). The mosquito sequence was most similar to these three in size and locations of conservation.
Figure 7 .
Evolutionarily conserved sequences among Blm10 homologs. The sequences are kept to scale to show the relative location and spatial arrangements of the conserved regions. Homologs were aligned by the global multiple sequence alignment program, CLUSTALW (http://www.ddbj.nig.ac.jp/search/clustalw-j.html). Proteins are arranged in order of decreasing size in (A) and (B). (A) Dissimilar distances among some of the homologs between the N-terminus and first conserved block, first and second block, and second and third block contrast with the relatively similar distances between carboxyl-conserved regions in all homologs. (B) Similarities among conserved regions as described in the text.
The Blm10 sequences of two molds and two fungi that grow as yeasts also were arbitrarily selected and compared (Figure 7B). Interestingly, the sizes and locations of the conserved regions of the Neurospora crassa homolog were most similar to those in the other mold, Aspergillus fumigatus. By comparison, the sizes and locations of conserved regions of Blm10 in S. cerevisiae were most similar to those in Candida albicans.
Protection does not require the largest conserved region at the carboxyl-terminus
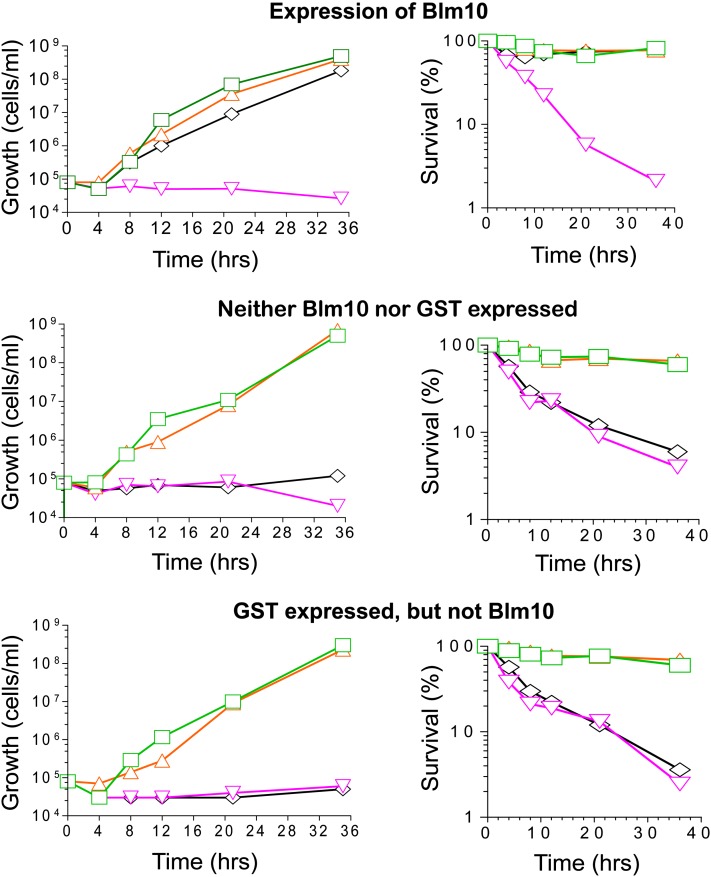
We investigated activities of a truncated Blm10 protein (Figure 1) lacking the three carboxyl-terminal conserved regions (blocks 3, 4, and 5 in Figure 7A). Without drug, mutant cells grew normally whether or not Blm10(-339aa) was induced (Figure 8). However, Blm10(-339aa) rescued growth-inhibition and cell death in the presence of 0.5, 1, 3, 5, 7, and 9 µg/mL phleomycin. For example, as shown in Figure 8, blm10Δ conferred complete inhibition of growth in the presence of 5 µg/mL phleomycin, and survival of the treated cells decreased to 2% by 35 hr unless Blm10(-339aa) was produced (Figure 8, top row). These experiments demonstrated Blm10(-339aa) was functional when produced as a GST-fusion, and its expression rescued growth inhibition and cell death. Similarly, the largest conserved region was not required for relief of phleomycin hypersusceptibility in blm3-1 mutant strains (Febres et al. 2001; Doherty et al. 2004). In experiments not shown here, overexpression of Blm10(-339aa) did not produce additional phenotypes.
Figure 8 .
Resistance conferred by truncated Blm10. Expression of Blm10(-339aa)-GST from an inducible pCUP1 promoter was controlled by adjusting the amounts of copper added to media (Mascorro-Gallardo et al. 1996). Top: blm10Δ cells expressing BLM10. Middle: blm10Δ cells not expressing BLM10 or GST. Bottom: blm10Δ cells expressing GST but not BLM10. Left: cell densities during growth. Right: survival. Green rectangles indicate no copper or phleomycin; orange triangles, 50 μM copper, no phleomycin; inverted pink triangles, 5 μg/mL phleomycin, no copper; black diamonds, 50 μM copper, 5 μg/mL phleomycin.
Control experiments confirmed Blm10(-339aa) expression was required to rescue growth inhibition and cell death because mutant cells without plasmid-expressed Blm10(-339aa) were growth inhibited and survived poorly (Figure 8, middle and bottom rows). Copper had no effect on growth or killing because each was comparable with and without copper.
Of note, haploid and homozygous diploid strains encoding Blm10(-339aa) under the control of its endogenous promoter exhibit higher sensitivities to killing by phleomycin than strains of the same ploidy with the entire gene deleted, and their sensitivities were nearly as high as blm3-1 strains (Febres et al. 2001; Doherty et al. 2004). In a different strain background, a truncated deletion also caused higher bleomycin sensitivity than the full deletion (Schmidt et al. 2005a). Heterozygous strains bearing the truncated BLM10 allele exhibited intermediate resistance (Febres et al. 2001; D. Febres and K. Doherty, unpublished data).
Localization of Blm10(-339aa)
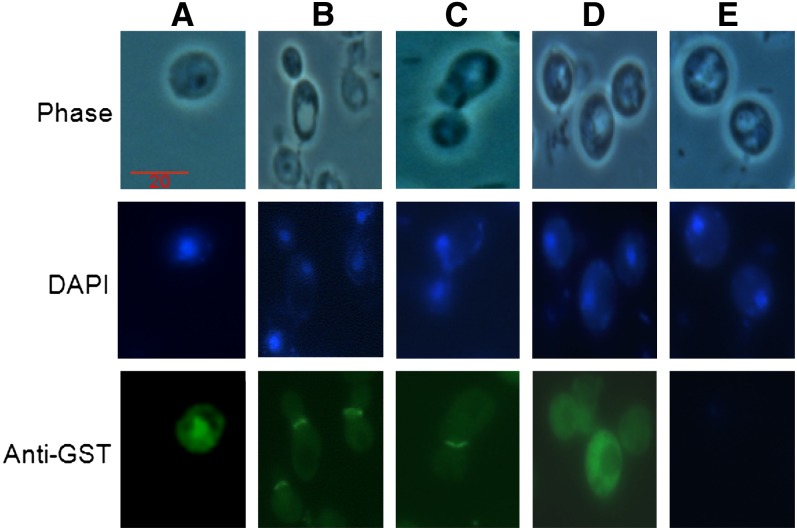
Using the same Blm10(-339aa)-GST, we investigated its localization and whether the conserved C-terminus was required for nuclear localization. Immunofluorescence microscopy localized Blm10(-339aa)-GST with GST antibodies, and various copper concentrations controlled protein amounts. Like full-length Blm10, Blm10(-339aa) localized in nuclei of stationary phase (G0) cells, as confirmed by DAPI staining of DNA (e.g., Figure 9A). During G1, S, G2, and M phases, the fluorescence formed a disk at the bud neck or septum between mother and daughter cells (e.g. Figure 9, B and C). It appeared to arch and follow the contour of the mother cell rather than the daughter (e.g., Figure 9C). Expressing GST alone revealed diffuse cytoplasmic staining (Figure 9D), eliminating the GST as the cause for the localization patterns. Without staining, no significant autofluorescence was observed (Figure 9E).
Figure 9 .
Blm10(-339aa) localization. The truncated Blm10 protein was induced with 50 μM copper because this concentration produced a stable protein and functionally relieved drug hypersensitivity. The representative wild-type cells illustrate two populations of cells, one unbudded (A) and the other budded (B and C). The nuclear localization is maintained in unbudded cells, in contrast to the bud-neck localization in budded cells. Top row: phase contrast. Middle row: DAPI staining of DNA. Bottom row: cells after GST antibody staining. Column A: unbudded cells. Columns B and C: different populations of budded cells. Column D: cells expressing GST but not Blm10. Column E: no antibody treatment. Because of the truncated protein, these cells are somewhat distended. In addition, before treatments with DAPI and anti-GST antibody, cells are converted to spheroplasts, which causes distortion of cells that have lost their cell wall integrity.
The localization we observed in budded cells and the cytoplasmic distribution of Blm10 observed previously (Schmidt et al. 2005a) suggest that a nuclear localization sequence could be encoded in the truncated region. Database searches identified a strong, although not canonical, nuclear localization sequence in that region (Schmidt et al. 2005a).
Discussion
Cells have multiple protein complexes to keep the genome intact and functional. Based on the current studies, the relationship among the components of the complexes may well differ between cells with and without the Blm10-20S proteasome activator. Loss of the activator downregulated numerous genes crucial for maintaining genomic stability, heightened DNA damage, and selectively sensitized cells to agents with different mechanisms of action. Although the exact or direct cause of the elevated DNA damage in Blm10 cells was not investigated for the current studies, the simultaneous downregulated expression appears important and suggests ways that Blm10 loss predisposes cells to lethal effects of agents with diverse mechanisms of action. We propose that modulated chromatin structure could compromise DNA integrity in blm10Δ/blm10Δ mutant cells, and conclude that the hypersusceptibilities establish Blm10 as a guardian against cellular stresses.
Even without external stimuli, cells were stressed without Blm10 because components of the chaperone and co-chaperone Hsp90 complex, Hsp70 subfamily, and Hsp100 family were selectively upregulated. This upregulation took place despite the fact that the chemostat regime guaranteed the growth rate remained the same in wild-type and mutant cultures. Consistent with its protection of chromosomes, Blm10 remained in nuclei throughout the cell cycle. The findings contribute knowledge that provides a strong foundation for future studies of the role(s) of Blm10 in relationship to functional groups of proteins, and of the mechanisms of the downregulated and upregulated genes, pathways and signaling networks.
The results also have implications for translational studies of ubiquitin-independent targeting in anticancer therapy. PA200/Blm10 inhibition could be a novel approach to cancer treatment, either alone or in combination with targeted inhibition of one or more of the upregulated chaperones. In fact, cytokinesis was the most enriched process among genes downregulated 25% or more in the current study, suggesting that inhibiting Blm10/PA200 could cause cancer cells to fail to divide. That proteasome inhibitors upregulate molecular chaperones in mammalian cells (Wyttenbach et al. 2000; Mitsiades et al. 2002; Pritts et al. 2002; Awasthi and Wagner 2005; Zaarur et al. 2006) supports our predictions. However, control experiments such as the steady-state conditions in the current report were not possible in mammalian cells to determine whether the general mammalian stress response was induced.
Blm10 physically associates with proteins that can assist in mediating protection and maintaining the integrity of chromosomal DNA (SGD Project 2010). Three of these proteins, Sir4, Zds2, and Dun1, are involved in maintaining chromosomal integrity and promoting DNA repair. Genetic interaction between Blm10 and the Dun1 DNA damage checkpoint kinase causes a synthetic growth defect (Pan et al. 2006). Without Blm10, DUN1 was downregulated 50% in the current studies, ZDS2 was upregulated 10%, and the expression of SIR4 was not significantly changed (Table S1). Sir4 and Zds2 are important for efficient DNA repair by nonhomologous end-joining (Lewis and Resnick 2000), the major DNA repair pathway in human cells. Sir4 leaves telomeres after DNA damage and relocates to double-strand breaks, where it binds the well-established component of double-strand break repair, yKu70 (Martin et al. 1999). Zds2 confers resistance to the anticancer DNA damaging drug cisplatin (Burger et al. 2000), suppresses defects resulting from histone mutations (Ma et al. 1996), and suppresses mutations in the CDC20 gene required for chromosome segregation (Yu et al. 1996).
Interacting proteins also include the molecular chaperones, Hsp104, Hsc82, Ssz1, Ump1, and Zuo1. When Blm10 was absent, HSP104 and HSC82 were upregulated fourfold and 2.3-fold, respectively (Table 6). SSZ1 and UMP1 were upregulated 30% and 10%, respectively (Table S1).
Blm10 and Ubp3/Blm3 physically associate or exhibit genetic interactions with four of the same proteins, Hsc82, Sir4, Rpn4, and Ump1, a short-lived chaperone involved in ubiquitin-mediated proteolysis and protein folding (SGD Project 2010). These shared interactions may have facilitated compensation for the blm3-1 mutant defect by Blm10 overexpression (Febres et al. 2001; Doherty et al. 2004) and could link the two proteins. It is unknown why the blm3-1mutant phenotype was not suppressed by Blm10 overexpression from a different construct in a later study (McCullock et al. 2006).
Based upon all of the results presented in the current report, and strengthened by the functional knowledge of proteins with which Blm10 physically and genetically interact, we propose that Blm10 acts to mediate DNA damage and other stresses. For example, 20S proteasomes effectively degrade and remove oxidatively damaged histones and other proteins. Chromatin reorganization would then allow for efficient DNA repair. We further propose that the activated Blm10-CP complex may be involved in removing toxic agents or detoxifying them. In future studies, it would be informative to determine which of the downregulated genes contribute to the vulnerability of blm10Δ cells to agents with different mechanisms of action. It is possible that at least some of the downregulated genes may cause the slower growth of blm10Δ/blm10Δ mutant diploids than wild-type diploids that we observe in and on limited or synthetic media outside of chemostats.
We also propose a role for Ubp3/Blm3 in protection based on its central position and connection to SSA1 and SSA2 in the upregulated protein network (Figure 2) and the excessive chromosomal damage and lethality in blm3-1/blm3-1 cells (Figure 4). UBP3 was isolated as a multicopy suppressor of the temperature sensitivity of S. cerevisiae cells doubly mutant for the SSA1 and SSA2 molecular chaperone genes (Baxter and Craig 1998). Interestingly, these two chaperone-encoding genes were the most highly upregulated in blm10Δ/blm10Δ cells in the current studies (Table 6). In addition, it is known that Ubp3 binds Sir4 and regulates chromosomal silencing, possibly by controlling the activity or assembly of the Sir complex (Moazed and Johnson 1996). It has been proposed that by deubiquitinating misfolded proteins, Ubp3 permits protein refolding, stability and function (Brew and Huffaker 2002). The S. cerevisiae Rad4 protein binds ultraviolet light-damaged DNA and promotes nucleotide excision repair (Mao and Smerdon 2010). Recently, it was suggested that Ubp3 physically interacts with the 26S proteasome and the Rad4 protein to help degrade Rad4 and suppress DNA repair (Mao and Smerdon 2010).
Results in this report indicate Blm10 protects cells from genomic instability and cell death. Consistent with our findings, PA200-knockdown cells showed genomic instability and reduced survival after ionizing irradiation (Blickwedehl et al. 2008). We reason that protein interactions and signaling cascades respond to DNA damage to arrest the cell cycle and repair DNA, recruit chromatin remodeling and DNA repair proteins, and recruit chaperones to assist with ridding cells of dysfunctional proteins or toxic agents. Molecular chaperones can repair nonfunctional or misfolded proteins or proteins can be ubiquitinated and targeted to the 26S proteasome for degradation. Neither PA200 nor Blm10 activates the proteasomal 20S catalytic chamber in response to ubiquitinated proteins or requires the activity of ATPase for proteasomal cleavage (Ustrell et al. 2002; Schmidt et al. 2005a), unlike the ubiquitin-dependent, ATPase-dependent 19S regulatory particle.
Although the protective function of Blm10 does not require its carboxyl-terminal region (Figure 8), it does require the two new conserved regions identified in these studies (Figure 7A). The truncated protein is missing the last three residues (TyrTyrAla) that structural analyses show make very close contact with the 20S proteasome (Sadre-Bazzaz et al. 2010). In addition, manipulation of these residues by mutagenesis alters Blm10 function (Lopez et al. 2011; Dange et al. 2011). Thus, the protective function appears not to require that Blm10 associate with the 20S proteasome, or the truncated Blm10 can associate with the proteasome without this region.
The truncated protein may be missing a domain that disallows its association or interaction with one or more other proteins. The carboxyl-terminus is essential for retention in the nucleus at all stages of growth, since the truncated protein mislocalized in a prior study (Schmidt et al. 2005a) and in budded cells in the current studies. It is not known if the septin localization relates to the physical association of Blm10 with a protein involved in bud site selection, Bud20 (Ho et al. 2002), which could pull the complex to the septin. This could underlie the increased calcofluor sensitivity of blm10Δ/blm10Δ cells (Figure 5).
Finally, results presented here suggest a 20s proteasome activator could be a target for proteasome inhibition in combination anticancer therapies. Multiple myeloma cells, for example, have increased proteasome levels and activity (Wada et al. 1993; Edwards et al. 2009) and circulating proteasomes (Jakob et al. 2007). Proteasome inhibition in these cells inhibits proliferation, induces apoptosis, and overcomes drug resistance (Hideshima et al. 2001; Edwards et al. 2009). In addition to targeting PA200/Blm10 and one or more of the upregulated proteins, targeting it with a conventional treatment could be effective.
Supplementary Material
Acknowledgments
We thank Agnes Baudin, Eric Phizicky, Philip James, Susan Henry, and Arthur Goldberg for strains, plasmids, and assistance and Martin Rechsteiner and Tim Formosa for primers. For invaluable assistance and suggestions, we thank Trisha Davis, David Gresham, Alexandra Ward, Louis Mazzella, Avanella James, Vanessa Lee, Donna Febres, Sanford Silverman, Jessica Buckles, Donna Storton, Matt Hibbs, Curtis Huttenmeyer, Maitreya Dunham, Chad Myers, and Miho Murashima. This work was supported by National Institutes of Health grants GM08168 and RR/AIO3060 (C.W.M.), P41RR11823 (E.G.M.), and P50GM071508 and R01GM046406 (D.B.), and MCB-9874878 and HRD-9628726 grants (C.W.M.) from the National Science Foundation.
Footnotes
Communicating editor: B. J. Andrews
Literature Cited
- Aouida M., Page N., Leduc A., Peter M., Ramotar D., 2004. A genome-wide screen in Saccharomyces cerevisiae reveals altered transport as a mechanism of resistance to the anticancer drug bleomycin. Cancer Res. 64: 1102–1109 [DOI] [PubMed] [Google Scholar]
- Awasthi N., Wagner B. J., 2005. Upregulation of heat shock protein expression by proteasome inhibition: an antiapoptotic mechanism in the lens. Invest. Ophthalmol. Vis. Sci. 46: 2082–2091 [DOI] [PubMed] [Google Scholar]
- Bachmair A., Finley D., Varshavsky A., 1986. In vivo half-life of a protein is a function of its amino-terminal residue. Science 234: 179–186 [DOI] [PubMed] [Google Scholar]
- Baudin A., Ozier-Kalogeropoulos O., Denouel A., Lacroute F., Cullin C., 1993. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 21: 3329–3330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter B. K., Craig E. A., 1998. Isolation of UBP3, encoding a de-ubiquitinating enzyme, as a multicopy suppressor of a heat-shock mutant strain of S. cerevisiae. Curr. Genet. 33: 412–419 [DOI] [PubMed] [Google Scholar]
- Beaudouin R., Lim S. T., Steide J. A., Powell M., McKoy J., et al. , 1993. Bleomycin affects cell wall anchorage of mannoproteins in Saccharomyces cerevisiae. Antimicrob. Agents Chemother. 37: 1264–1269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett C. B., Lewis L. K., Karthikeyan G., Lobachev K. S., Jin Y. H., et al. , 2001. Genes required for ionizing radiation resistance in yeast. Nat. Genet. 29: 426–434 [DOI] [PubMed] [Google Scholar]
- Blickwedehl J., McEvoy S., Wong I., Kousis P., Clements J., et al. , 2007. Proteasomes and proteasome activator 200 kDa (PA200) accumulate on chromatin in response to ionizing radiation. Radiat. Res. 167: 663–674 [DOI] [PubMed] [Google Scholar]
- Blickwedehl J., Agarwal M., Seong C., Pandita R. K., Melendy T., et al. , 2008. Role for proteasome activator PA200 and postglutamyl proteasome activity in genomic stability. Proc. Natl. Acad. Sci. USA 105: 16165–16170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M. J., Saldanha A. J., Dolinski K., Botstein D., 2005. Homeostatic adjustment and metabolic remodeling in glucose-limited yeast cultures. Mol. Biol. Cell 16: 2503–2517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer M. J., Huttenhower C., Airoldi E. M., Rosenstein R., Matese J. C., et al. , 2008. Coordination of growth rate, cell cycle, stress response, and metabolic activity in yeast. Mol. Biol. Cell 19: 352–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brew C. T., Huffaker T. C., 2002. The yeast ubiquitin protease, Ubp3p, promotes protein stability. Genetics 162: 1079–1089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunton L. L., Lazo J. S., Parker K. L., editors, 2006. Goodman and Gilman's The Pharmacological Basis of Therapeutics. McGraw-Hill, New York
- Burger H., Capelloa A., Schenka P. W., Stotera G., Brouwerb J., et al. , 2000. A genome-wide screening in Saccharomyces cerevisiae for genes that confer resistance to the anticancer agent cisplatin. Biochem. Biophys. Res. Commun. 269: 767–774 [DOI] [PubMed] [Google Scholar]
- Cloos J., De Boer W. P., Snel M. H., Van Den Ijssel P., Ylstra B., et al. , 2006. Microarray analysis of bleomycin-exposed lymphoblastoid cells for identifying cancer susceptibility genes. Mol. Cancer Res. 4: 71–77 [DOI] [PubMed] [Google Scholar]
- Coughlan C., Brodsky J., 2003. Yeast as a model system to investigate protein conformational diseases, pp. 77–90 in Protein Misfolding and Disease: Principles and Protocols, edited by P. Bross, and N. Gregersen. Humana Press Inc., Totowa, NJ
- Dange T., Smith D., Noy T., Rommel P. C., Jurzitza L., et al. , 2011. Blm10 protein promotes proteasomal substrate turnover by an active gating mechanism. J. Biol. Chem. 286: 42830–42839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davermann D., Martinez M., McKoy J., Patel N., Averbeck D., et al. , 2002. Impaired mitochondrial function protects against free radical-mediated cell death. Free Radic. Biol. Med. 33: 1209–1220 [DOI] [PubMed] [Google Scholar]
- Deans A. J., West S. C., 2011. Mitomycin C DNA interstrand crosslink repair and cancer. Nat. Rev. Cancer 11: 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demartino G. N., Gillette T. G., 2007. Proteasomes: machines for all reasons. Cell 129: 659–662 [DOI] [PubMed] [Google Scholar]
- Demple B., Harrison L., 1994. Repair of oxidative damage to DNA: enzymology and biology. Annu. Rev. Biochem. 63: 915–948 [DOI] [PubMed] [Google Scholar]
- Dobson C. M., 2003. Protein folding and misfolding. Nature 426: 884–890 [DOI] [PubMed] [Google Scholar]
- Doherty K., Pramanik A., Pride L., Lukose J., Moore C. W., 2004. Expression of the expanded YFL007w ORF and assignment of the gene name BLM10. Yeast 21: 1021–1023 [DOI] [PubMed] [Google Scholar]
- Edwards C. M., Lwin S. T., Fowler J. A., Oyajobi B. O., Zhuang J., et al. , 2009. Myeloma cells exhibit an increase in proteasome activity and an enhanced response to proteasome inhibition in the bone marrow microenvironment in vivo. Am. J. Hematol. 84: 268–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Febres D. E., Pramanik A., Caton M., Doherty K., McKoy J., et al. , 2001. The novel BLM3 gene encodes a protein that protects against lethal effects of oxidative damage. Cell Mol Biol (Noisy-le-grand) 47: 1149–1162 [PubMed] [Google Scholar]
- Fehlker M., Wendler P., Lehmann A., Enenkel C., 2003. Blm3 is part of nascent proteasomes and is involved in a late stage of nuclear proteasome assembly. EMBO Rep. 4: 959–963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finley D., 2009. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 78: 477–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Főrster A., Hill C. P., 2006. Proteasome activators, pp. 89–110 in Protein Degradation: The Ubiquitin-Proteasome System, edited by R. J. Mayer, A. Ciechanover, and M. Rechsteiner. Wiley-VCH Verlag GmbH & Co., KGaA, Weinheim, Germany
- Friedberg E. C., Walker G. C., Siede W., Wood R. D., Schultz R. A., et al. , 2006. DNA Repair and Mutagenesis, Ed. 2. American Society for Microbiology Press, Washington, DC
- Gietz R. D., Woods R. A., 2002. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method, 87–96 Methods in Enzymology, edited by Guthrie C., Fink G. R. Academic Press, Inc., San Diego, California: [DOI] [PubMed] [Google Scholar]
- Gilbert C. S., Van Den Bosch M., Green C. M., Vialard J. E., Grenon M., et al. , 2003. The budding yeast Rad9 checkpoint complex: chaperone proteins are required for its function. EMBO Rep. 4: 953–958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glickman M. H., Raveh D., 2005. Proteasome plasticity. FEBS Lett. 579: 3214–3223 [DOI] [PubMed] [Google Scholar]
- Goldberg A. L., 2003. Protein degradation and protection against misfolded or damaged proteins. Nature 426: 895–899 [DOI] [PubMed] [Google Scholar]
- Gollub J., Ball C. A., Binkley G., Demeter J., Finkelstein D. B., et al. , 2003. The Stanford Microarray Database: data access and quality assessment tools. Nucleic Acids Res. 31: 94–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardwick J. S., Kuruvilla F. G., Tong J. K., Shamji A. F., Schreiber S. L., 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. USA 96: 14866–14870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff J. G., Greene E. A., Pietrokovski S., Henikoff S., 2000. Increased coverage of protein families with the blocks database servers. Nucleic Acids Res. 28: 228–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hideshima T., Richardson P., Chauhan D., Palombella V. J., Elliott P. J., et al. , 2001. The proteasome inhibitor PS-341 inhibits growth, induces apoptosis, and overcomes drug resistance in human multiple myeloma cells. Cancer Res. 61: 3071–3076 [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., et al. , 2002. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature 415: 180–183 [DOI] [PubMed] [Google Scholar]
- Huo Q., 2010. Protein complexes/aggregates as potential cancer biomarkers revealed by a nanoparticle aggregation immunoassay. Colloids Surf. B Biointerfaces 78: 259–265 [DOI] [PubMed] [Google Scholar]
- Iwanczyk J., Sadre-Bazzaz K., Ferrell K., Kondrashkina E., Formosa T., et al. , 2006. Structure of the Blm10–20 S proteasome complex by cryo-electron microscopy. Insights into the mechanism of activation of mature yeast proteasomes. J. Mol. Biol. 363: 648–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakob C., Egerer K., Liebisch P., Turkmen S., Zavrski I., et al. , 2007. Circulating proteasome levels are an independent prognostic factor for survival in multiple myeloma. Blood 109: 2100–2105 [DOI] [PubMed] [Google Scholar]
- Kajava A. V., Gorbea C., Ortega J., Rechsteiner M., Steven A. C., 2004. New HEAT-like repeat motifs in proteins regulating proteasome structure and function. J. Struct. Biol. 146: 425–430 [DOI] [PubMed] [Google Scholar]
- Khor B., Bredemeyer A. L., Huang C. Y., Turnbull I. R., Evans R., et al. , 2006. Proteasome activator PA200 is required for normal spermatogenesis. Mol. Cell. Biol. 26: 2999–3007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan N. J., Lam M. H., Fillingham J., Keogh M. C., Gebbia M., et al. , 2004. Proteasome involvement in the repair of DNA double-strand breaks. Mol. Cell 16: 1027–1034 [DOI] [PubMed] [Google Scholar]
- Kubitschek H. E., 1970. Introduction to Research With Continuous Cultures, Prentice Hall, Englewood Cliffs, NJ [DOI] [PubMed]
- Lehmann A., Jechow K., Enenkel C., 2008. Blm10 binds to pre-activated proteasome core particles with open gate conformation. EMBO Rep. 9: 1237–1243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis L. K., Resnick M. A., 2000. Tying up loose ends: nonhomologous end-joining in Saccharomyces cerevisiae. Mutat. Res. 451: 71–89 [DOI] [PubMed] [Google Scholar]
- Li X., Kusmierczyk A. R., Wong P., Emili A., Hochstrasser M., 2007. beta-Subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 26: 2339–2349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez A. D., Tar K., Krügel U., Dange T., Ros I. G., et al. , 2011. Proteasomal degradation of Sfp1 contributes to the repression of ribosome biogenesis during starvation and is mediated by the proteasome activator Blm10. Mol. Biol. Cell 22: 528–540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X. J., Lu Q., Grunstein M., 1996. A search for proteins that interact genetically with histone H3 and H4 amino termini uncovers novel regulators of the Swe1 kinase in Saccharomyces cerevisiae. Genes Dev. 10: 1327–1340 [DOI] [PubMed] [Google Scholar]
- Mao P., Smerdon M. J., 2010. Yeast deubiquitinase Ubp3 interacts with the 26 S proteasome to facilitate Rad4 degradation. J. Biol. Chem. 285: 37542–37550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marques A. J., Glanemann C., Ramos P. C., Dohmen R. J., 2007. The C-terminal extension of the beta7 subunit and activator complexes stabilize nascent 20 S proteasomes and promote their maturation. J. Biol. Chem. 282: 34869–34876 [DOI] [PubMed] [Google Scholar]
- Martin S. G., Laroche T., Suka N., Grunstein M., Gasser S. M., 1999. Relocalization of telomeric Ku and SIR proteins in response to DNA strand breaks in yeast. Cell 97: 621–633 [DOI] [PubMed] [Google Scholar]
- Martzen M. R., McCraith S. M., Spinelli S. L., Torres F. M., Fields S., et al. , 1999. A biochemical genomics approach for identifying genes by the activity of their products. Science 286: 1153–1155 [DOI] [PubMed] [Google Scholar]
- Mascorro-Gallardo J. O., Covarrubias A. A., Gaxiola R., 1996. Construction of a CUP1 promoter-based vector to modulate gene expression in Saccharomyces cerevisiae. Gene 172: 169–170 [DOI] [PubMed] [Google Scholar]
- McCullock S., Kinard T., McCullough L., Formosa T., 2006. blm3–1 is an allele of UBP3, a ubiquitin protease that appears to act during transcription of damaged DNA. J. Mol. Biol. 363: 660–672 [DOI] [PubMed] [Google Scholar]
- Mitsiades N., Mitsiades C. S., Poulaki V., Chauhan D., Fanourakis G., et al. , 2002. Molecular sequelae of proteasome inhibition in human multiple myeloma cells. Proc. Natl. Acad. Sci. USA 99: 14374–14379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moazed D., Johnson D., 1996. A deubiquitinating enzyme interacts with SIR4 and regulates silencing in S. cerevisiae. Cell 86: 667–677 [DOI] [PubMed] [Google Scholar]
- Moore C. W., 1978. Responses of radiation-sensitive mutants of Saccharomyces cerevisiae to lethal effects of bleomycin. Mutat. Res. 51: 165–180 [DOI] [PubMed] [Google Scholar]
- Moore C. W., 1982. Control of in vivo (cellular) phleomycin sensitivity by nuclear genotype, growth phase, and metal ions. Cancer Res. 42: 929–933 [PubMed] [Google Scholar]
- Moore C. W., 1988. Internucleosomal cleavage and chromosomal degradation by bleomycin and phleomycin in yeast. Cancer Res. 48: 6837–6843 [PubMed] [Google Scholar]
- Moore C. W., 1990. Degradation of DNA and structure-activity relationship between bleomycins A2 and B2 in the absence of DNA repair. Biochemistry 29: 1342–1347 [DOI] [PubMed] [Google Scholar]
- Moore C. W., 1991. Further characterizations of bleomycin-sensitive (blm) mutants of Saccharomyces cerevisiae with implications for a radiomimetic model. J. Bacteriol. 173: 3605–3608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. W., 1999. Bleomycin, pp. 292–297 in Encyclopedia of Molecular Biology, edited by T. E. Creighton. John Wiley and Sons, Inc., New York
- Moore C. W., Del Valle R., McKoy J., Pramanik A., Gordon R. E., 1992. Lesions and preferential initial localization of [S-methyl-3H]bleomycin A2 on Saccharomyces cerevisiae cell walls and membranes. Antimicrob. Agents Chemother. 36: 2497–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. W., McKoy J., Dardalhon M., Davermann D., Martinez M., et al. , 2000. DNA damage-inducible and RAD52-independent repair of DNA double-strand breaks in Saccharomyces cerevisiae. Genetics 154: 1085–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore C. W., McKoy J., Del Valle R., Armstrong D., Bernard E. M., et al. , 2003. Fungal cell wall septation and cytokinesis are inhibited by bleomycins. Antimicrob. Agents Chemother. 47: 3281–3289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller E. G., Snydsman B. E., Novik I., Hailey D. W., Gestaut D. R., et al. , 2005. The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell 16: 3341–3352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers C. L., Robson D., Wible A., Hibbs M. A., Chiriac C., et al. , 2005. Discovery of biological networks from diverse functional genomic data. Genome Biol. 6: R114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaraj N. S., Singh O. V., Merchant N. B., 2010. Proteomics: a strategy to understand the novel targets in protein misfolding and cancer therapy. Expert Rev. Proteomics 7: 613–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitiss J. L., Heitman J., editors, 2007. Yeast as a Tool in Cancer Research. Springer, Dordrecht, The Netherlands [Google Scholar]
- Orlowski R. Z., Kuhn D. J., 2008. Proteasome inhibitors in cancer therapy: lessons from the first decade. Clin. Cancer Res. 14: 1649–1657 [DOI] [PubMed] [Google Scholar]
- Ortega J., Heymann J. B., Kajava A. V., Ustrell V., Rechsteiner M., et al. , 2005. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J. Mol. Biol. 346: 1221–1227 [DOI] [PubMed] [Google Scholar]
- Pan X., Ye P., Yuan D. S., Wang X., Bader J. S., et al. , 2006. A DNA integrity network in the yeast Saccharomyces cerevisiae. Cell 124: 1069–1081 [DOI] [PubMed] [Google Scholar]
- Parsons A. B., Brost R. L., Ding H., Li Z., Zhang C., et al. , 2004. Integration of chemical-genetic and genetic interaction data links bioactive compounds to cellular target pathways. Nat. Biotechnol. 22: 62–69 [DOI] [PubMed] [Google Scholar]
- Prein B., Natter K., Kohlwein S. D., 2000. A novel strategy for constructing N-terminal chromosomal fusions to green fluorescent protein in the yeast Saccharomyces cerevisiae. FEBS Lett. 485: 29–34 [DOI] [PubMed] [Google Scholar]
- Pritts T. A., Hungness E. S., Hershko D. D., Robb B. W., Sun X., et al. , 2002. Proteasome inhibitors induce heat shock response and increase IL-6 expression in human intestinal epithelial cells. Am. J. Physiol. Regul. Integr. Comp. Physiol. 282: R1016–R1026 [DOI] [PubMed] [Google Scholar]
- Reinhardt C. G., Krugh T. R., 1978. A comparative study of ethidium bromide complexes with dinucleotides and DNA: direct evidence for intercalation and nucleic acid sequence preferences. Biochemistry 17: 4845–4854 [DOI] [PubMed] [Google Scholar]
- Rose M., Botstein D., 1983. Construction and use of gene fusions to lacZ (beta-galactosidase) that are expressed in yeast. Methods Enzymol. 101: 167–180 [DOI] [PubMed] [Google Scholar]
- Rose M. D., Broach J. R., 1991. Cloning genes by complementation in yeast. Methods Enzymol. 194: 195–230 [DOI] [PubMed] [Google Scholar]
- Rosenzweig R., Glickman M. H., 2008. Chaperone-driven proteasome assembly. Biochem. Soc. Trans. 36: 807–812 [DOI] [PubMed] [Google Scholar]
- Russell S. J., Steger K. A., Johnston S. A., 1999. Subcellular localization, stoichiometry, and protein levels of 26 S proteasome subunits in yeast. J. Biol. Chem. 274: 21943–21952 [DOI] [PubMed] [Google Scholar]
- Sadre-Bazzaz K., Whitby F. G., Robinson H., Formosa T., Hill C. P., 2010. Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening. Mol. Cell 37: 728–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano K., Oikawa S., Hasegawa K., Kawanishi S., 2001. Hydroxyurea induces site-specific DNA damage via formation of hydrogen peroxide and nitric oxide. Jpn. J. Cancer Res. 92: 1166–1174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savulescu A. F., Glickman M. H., 2011. Proteasome ativator 200: the HEAT is on…. Mol. Cellular Proteomics 10: R110.006890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M., Haas W., Crosas B., Santamaria P. G., Gygi S. P., et al. , 2005a The HEAT repeat protein Blm10 regulates the yeast proteasome by capping the core particle. Nat. Struct. Mol. Biol. 12: 294–303 [DOI] [PubMed] [Google Scholar]
- Schmidt M., Hanna J., Elsasser S., Finley D., 2005b Proteasome-associated proteins: regulation of a proteolytic machine. Biol. Chem. 386: 725–737 [DOI] [PubMed] [Google Scholar]
- Scott M., Frydman J., 2003. Aberrant protein folding as the molecular basis of cancer, 67–76 Protein Misfolding and Disease: Principles and Protocols, edited by Bross P., Gregersen N. Humana Press Inc., Totowa, NJ: [DOI] [PubMed] [Google Scholar]
- Sealfon R. S., Hibbs M. A., Huttenhower C., Myers C. L., Troyanskaya O. G., 2006. GOLEM: an interactive graph-based gene-ontology navigation and analysis tool. BMC Bioinformatics 7: 443–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- SGD Project, 2010 Saccharomyces Genome Database. Available at: http://www.yeastgenome.org/July 14,
- Shimogawa M. M., Wargacki M. M., Muller E. G., Davis T. N., 2010. Laterally attached kinetochores recruit the checkpoint protein Bub1, but satisfy the spindle checkpoint. Cell Cycle 9: 3619–3628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtmueller B. M., Hill C. P., 2011. Proteasome activators. Mol. Cell 41: 8–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner M., Everman D., Ellington S., Fields C., 1990. Detection of free radicals during the cellular metabolism of adriamycin. Free Radic. Biol. Med. 9: 415–421 [DOI] [PubMed] [Google Scholar]
- Ustrell V., Hoffman L., Pratt G., Rechsteiner M., 2002. PA200, a nuclear proteasome activator involved in DNA repair. EMBO J. 21: 3516–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Den Bosch M., Lowndes N. F., 2004. Remodelling the Rad9 checkpoint complex: preparing Rad53 for action. Cell Cycle 3: 119–122 [PubMed] [Google Scholar]
- Voorhees P. M., Orlowski R. Z., 2006. The proteasome and proteasome inhibitors in cancer therapy. Annu. Rev. Pharmacol. Toxicol. 46: 189–213 [DOI] [PubMed] [Google Scholar]
- Wada M., Kosaka M., Saito S., Sano T., Tanaka K., et al. , 1993. Serum concentration and localization in tumor cells of proteasomes in patients with hematologic malignancy and their pathophysiologic significance. J. Lab. Clin. Med. 121: 215–223 [PubMed] [Google Scholar]
- Wyttenbach A., Carmichael J., Swartz J., Furlong R. A., Narain Y., et al. , 2000. Effects of heat shock, heat shock protein 40 (HDJ-2), and proteasome inhibition on protein aggregation in cellular models of Huntington's disease. Proc. Natl. Acad. Sci. USA 97: 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y., Kitagaki J., Wang H., Hou D. X., Perantoni A. O., 2009. Targeting the ubiquitin-proteasome system for cancer therapy. Cancer Sci. 100: 24–28 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y., Jiang Y. W., Wellinger R. J., Carlson K., Roberts J. M., et al. , 1996. Mutations in the homologous ZDS1 and ZDS2 genes affect cell cycle progression. Mol. Cell. Biol. 16: 5254–5263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaarur N., Gabai V. L., Porco J. A., Jr, Calderwood S., Sherman M. Y., 2006. Targeting heat shock response to sensitize cancer cells to proteasome and Hsp90 inhibitors. Cancer Res. 66: 1783–1791 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.