Abstract
Background:
As more scientific work is published, it is important to improve access to the biomedical literature. Since 2000, when Medical Subject Headings (MeSH) Concepts were introduced, the MeSH Thesaurus has been concept based. Nevertheless, information retrieval is still performed at the MeSH Descriptor or Supplementary Concept level.
Objective:
The study assesses the benefit of using MeSH Concepts for indexing and information retrieval.
Methods:
Three sets of queries were built for thirty-two rare diseases and twenty-two chronic diseases: (1) using PubMed Automatic Term Mapping (ATM), (2) using Catalog and Index of French-language Health Internet (CISMeF) ATM, and (3) extrapolating the MEDLINE citations that should be indexed with a MeSH Concept.
Results:
Type 3 queries retrieve significantly fewer results than type 1 or type 2 queries (about 18,000 citations versus 200,000 for rare diseases; about 300,000 citations versus 2,000,000 for chronic diseases). CISMeF ATM also provides better precision than PubMed ATM for both disease categories.
Discussion:
Using MeSH Concept indexing instead of ATM is theoretically possible to improve retrieval performance with the current indexing policy. However, using MeSH Concept information retrieval and indexing rules would be a fundamentally better approach. These modifications have already been implemented in the CISMeF search engine.
Highlights.
The introduction of Medical Subject Headings (MeSH) Concepts (creating subgroups of entry terms within MeSH Descriptors) has not changed overall indexing or retrieval practices in MEDLINE.
The use of MeSH Concepts could significantly improve the precision of retrieval for PubMed searches related to rare and chronic diseases.
In-depth knowledge of MeSH is not required for users to benefit from improved search performance using MeSH Concepts.
Implications.
Information professionals can use their advanced knowledge of the MeSH thesaurus to make changes to indexing and retrieval practices that are transparent to users and enhance their search experience.
Information professionals can use MeSH Concepts to conduct more precise searches in some cases, for example, rare and chronic diseases.
INTRODUCTION
The coverage of MEDLINE and the volume of the literature in biomedicine and health are increasing rapidly, and the National Library of Medicine (NLM) consistently undertakes projects to improve access to biomedical information through PubMed. For instance, recent research efforts have addressed the evaluation of ranking and querying strategies for the PubMed search engine [1–3] and the development of a disease sensor for facilitating access to trustworthy disease-related information through PubMed [4, 5]. In addition, a recent review found that another twenty-eight institutes worldwide are devoting efforts to the development of web tools designed to assist users in quickly and efficiently searching and retrieving relevant publications from MEDLINE [6]. The Catalog and Index of French-language Health Internet (CISMeF) team has made significant contributions to these efforts by providing access to MEDLINE using queries in French [7, 8] and proposing a method for improving the precision of PubMed Automatic Term Mapping (ATM) [9]. All of this work shows that there is a need for and an interest from the research community for continued improvement in access to the biomedical literature.
Since 2000, the underlying structure of the Medical Subject Headings (MeSH) Thesaurus has changed from a term-based system to a concept-oriented system to make it more compatible with the Unified Medical Language System (UMLS) [10]. In its 2011 version, the MeSH Thesaurus contains 26,142 Descriptors, 83 Qualifiers, 25,801 Entry Terms, 200,676 Supplementary Concepts, and 317,554 Concepts. A MeSH Descriptor is now viewed as a class of MeSH Concepts and a MeSH Concept as a class of entry terms [10]. Specifically, in this concept-oriented system, MeSH Concepts consist of subgroups of entry terms created within MeSH Descriptors. Each MeSH Concept (or group of entry terms) thus formed provides a finer-grained definition of the relationship between the MeSH Descriptors and their MeSH Entry Terms.
A MeSH Descriptor class consists of one or more MeSH Concepts closely related to each other in meaning. Several relationships may exist between MeSH Concept and MeSH Descriptor or MeSH Supplementary Concept (substance name and, since 2011, names of some rare diseases): “preferred term,” “related,” “narrower,” and “broader.” Currently, multiple concepts are still combined in one class in the MEDLINE bibliographic database, and descriptors, rather than MeSH Concepts, continue to be used for the purposes of indexing, retrieval, and organization of the literature [10].
For MeSH Concepts, the same MeSH Descriptor or Supplementary Concept is to be used both for indexing and for searching the MEDLINE bibliographic database via the PubMed interface. Therefore, a query referring to a MeSH Concept is currently performed on the MeSH Descriptor or on the MeSH Supplementary Concept, but not on the MeSH Concept itself. For example, a query referring to “Drooling,” which is a MeSH Concept, is currently performed on the MeSH Descriptor “Sialorrhea.” The study reported here assesses the benefits of using MeSH Concepts for searching.
Each MeSH Descriptor (or Supplementary Concept) is linked to one unique preferred MeSH Concept in the MeSH Thesaurus. Among 317,554 MeSH Concepts, 226,818 are preferred Concepts and 90,736 are non-preferred or subordinate Concepts. The MeSH subordinate Concepts have one specific relationship with one MeSH Descriptor or one MeSH Supplementary Concept: 78,036 are related with the relationship “narrower than,” 6,627 with the relationship “broader than,” and 6,073 with the relationship “related.” In contrast, preferred MeSH Concepts are identical to their MeSH Descriptor or Supplementary Concept; it is a reflexive relationship. Table 1 shows the number of MeSH Descriptors and Supplementary Concepts that have a relationship with MeSH Concepts.
Table 1.
Type and frequency of relationships between Medical Subject Headings (MeSH) descriptors and MeSH Concepts, and between MeSH Supplementary Concept and MeSH Concepts (in August 2011)
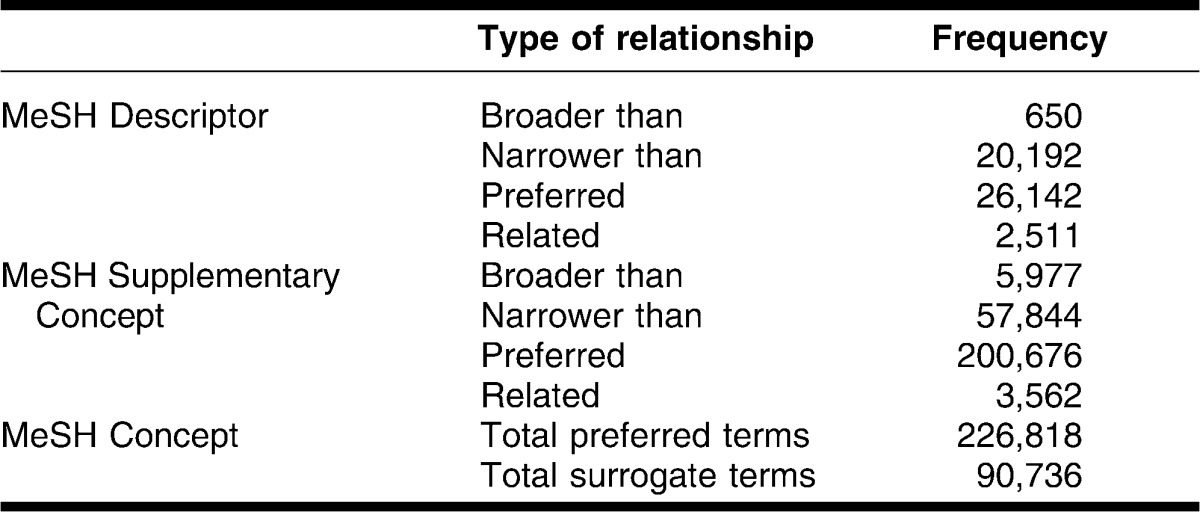
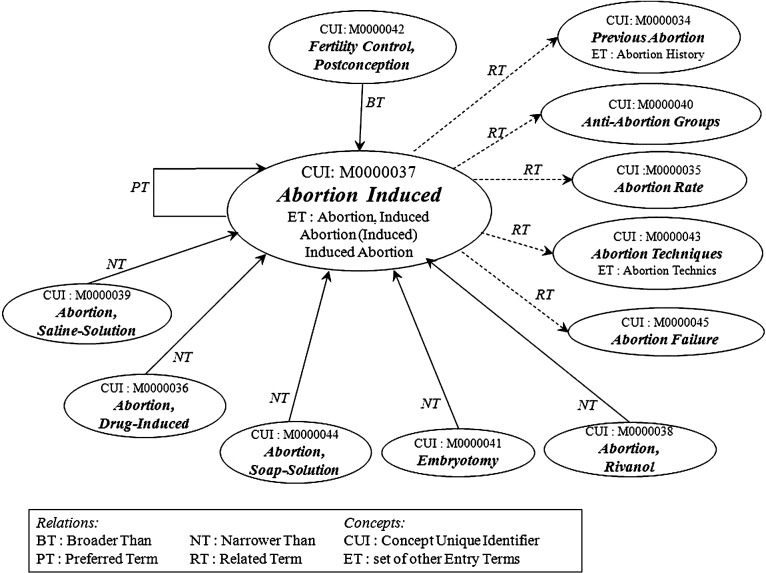
Figure 1 presents a simplified illustration of the standard concept view accessible from the MeSH browser for a sample descriptor.† It shows the relationships between the twelve concepts and fifteen entry terms grouped under the MeSH Descriptor “Abortion, Induced.” This descriptor has four types of relationships with the MeSH Concepts grouped under it: (i) a reflexive relationship to the preferred MeSH Concept “Abortion Induced,” which has two entry terms; (ii) a “narrower than” relationship with the subordinate concepts “Abortion, Drug-Induced,” “Abortion, Rivanol” “Abortion, Saline-Solution,” and “Abortion, Soap-Solution”; (iii) a “broader than” relationship with the subordinate concept, “Fertility Control, Postconception”; and (iv) a “related” relationship with the subordinate concepts “Abortion Failure,” “Abortion Rate,” “Abortion Techniques,” “Anti-Abortion Groups,” and “Previous Abortion.” Each MeSH Concept class could be given its own definition if desired.
Figure 1.
A simplified illustration of the standard concept view, accessible from the Medical Subject Headings (MeSH) browser
The objective of the study was to test the hypothesis that using subordinate (“non-preferred”) MeSH Concepts to index MEDLINE citations, assuming that they are more precise than their related MeSH Descriptors or MeSH Supplementary Concepts, will yield two benefits: (1) provide more precise indexing for citations and (2) improve the quality of information retrieval.
METHODS
To test this hypothesis, the field of experiment was restricted to 2 different subjects: (a) rare diseases and (b) chronic diseases. Rare diseases are mainly defined by their prevalence, with criteria that may vary from country to country. For instance, in the United States, a rare disease is defined as a condition that affects less than 1 person in 1,500 (i.e., fewer than 200,000 patients in the United States); in Europe, the cut-off is set at 1 in 2,000 (e.g., fewer than 30,000 patients in France). Rare diseases were chosen as a focus for this study because of their relative frequency (>7,000) in the MeSH Thesaurus. Chronic diseases were chosen because they are a known public health problem. Some rare and chronic diseases are grouped in 1 MeSH Descriptor related to several MeSH Concepts.
Choice of Medical Subject Headings Concepts
Non-preferred or subordinate MeSH Concepts describing rare or chronic diseases that had the relationship “narrower than” with one MeSH Descriptor or one MeSH Supplementary Concept were used to test the hypothesis. MeSH Concepts that have the relationships “broader than” or “related” were excluded for 2 reasons: (1) the relationship “narrower than” is the most common one in MeSH (78,036/90,736; 86.0% of non-preferred or subordinate MeSH Concepts), without taking into account “preferred term,” and (2) the 2 other relationships “broader than” and “related” are not adequate to test the hypothesis. “Broader than” should test the opposite hypothesis, as it should provide more citations than the related MeSH Descriptor or MeSH Supplementary Concept, whereas “related” would be difficult to analyze.
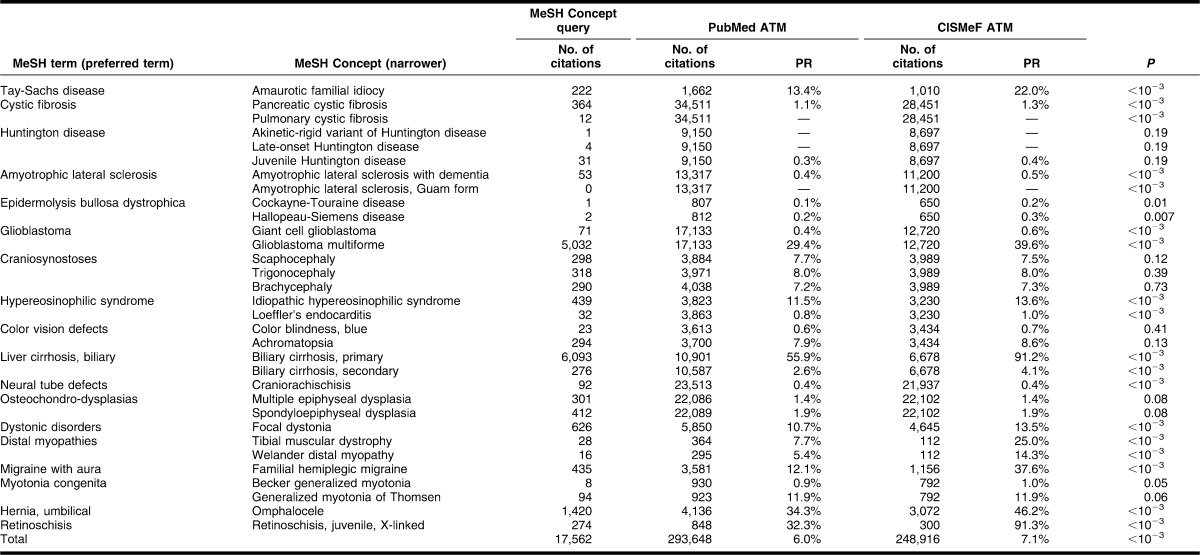
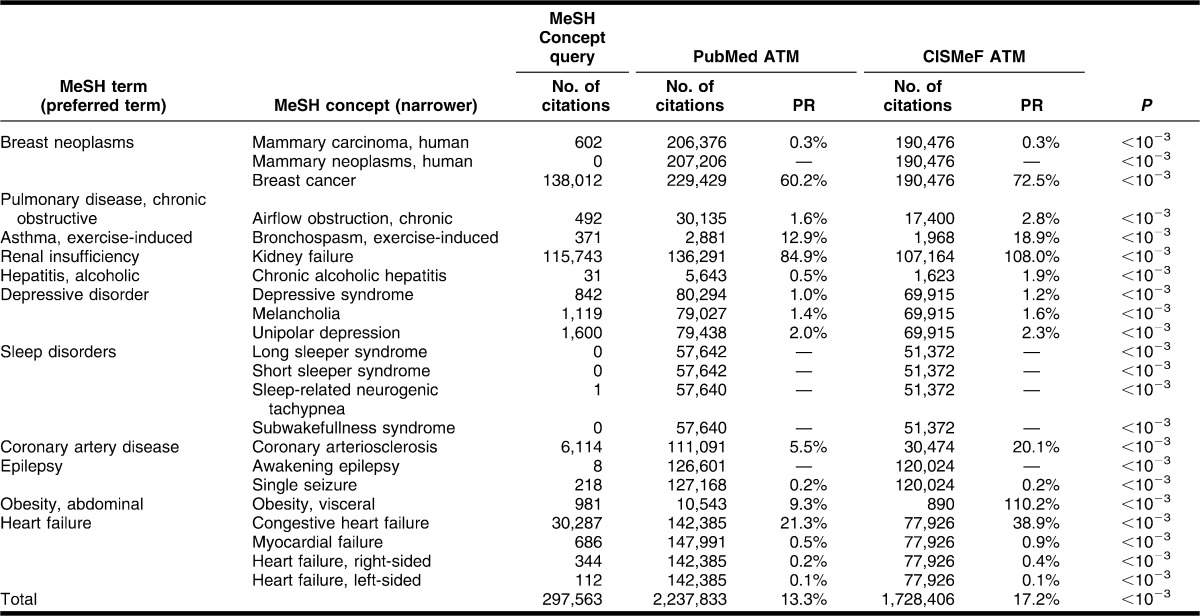
The most frequent rare and chronic diseases were used for the study. Rare diseases were selected based on a recent literature review of rare disease prevalence published by the Orphanet information website for rare diseases [11]. MEDLINE frequency counts, according to the 2011 MEDLINE baseline repository data [12], were used to identify the most common chronic diseases. The list of rare diseases is displayed in Table 2, and the list of chronic diseases is displayed in Table 3.
Table 2.
Comparison of PubMed and Catalog and Index of French-language Health Internet (CISMeF) Automatic Term Mapping (ATM) for rare diseases, number of citations and precision ratio (PR) (in August 2011)
Table 3.
Comparison of PubMed and CISMeF ATM for chronic diseases, number of citations and PR (in August 2011)
Three different PubMed queries
The MEDLINE bibliographic database was searched using the PubMed interface for each of the MeSH Concepts shown in Tables 2 and 3. Three different queries were used: (1) the default PubMed ATM query, (2) the corresponding CISMeF ATM query [9], and (3) a specific query to extrapolate the MEDLINE citations that should be indexed with a MeSH Concept.
The first two queries provided the current results of a PubMed search on the selected MeSH Concepts (the second query being a more precise variant of the first one [9]), which currently pools together all relevant MeSH Concepts at the descriptor level. The third query aimed to model the retrieval of documents for the sole MeSH Concept of interest, which would become the default search with MEDLINE indexing at the MeSH Concept level, and therefore retrieval at the MeSH Concept level, instead of MeSH Descriptor level, as is currently the case. Comparing the results of the third query to the other two provided an indication of the benefits of MeSH Concept indexing for retrieval in MEDLINE. Specifically, it was assumed that the citations retrieved by the third query were the ones that should have been indexed with the MeSH Concept of interest and, therefore, the only relevant ones for the search. Based on this assumption, two precision scores were computed:
 |
 |
This extrapolation slightly underestimates the true number of citations that should be indexed with MeSH Concepts, as some papers without any mention of the MeSH Concept in the title or in the abstract could still need to be indexed with that MeSH Concept.
The subordinate MeSH Concept “Amaurotic familial idiocy” related to the MeSH Descriptor “Tay-Sachs disease” provides an illustration of these three types of queries (Figures 2 and 3). The main differences are:
Figure 2.

Default PubMed ATM query for “Tay-Sachs Disease” in PubMed syntax
Figure 3.

CISMeF ATM query for “Tay-Sachs Disease” in PubMed syntax ([TIAB] = Title or Abstract, [SB] = subset)
The CISMeF ATM constructs the same query whether the end-user query contains MeSH preferred terms or MeSH entry terms. This is not the case for PubMed ATM.
The CISMeF ATM employs semantic expansion, using all the entry terms associated with a MeSH Descriptor or MeSH Supplementary Concept, without taking into account their relationships. The goal of this semantic expansion is to improve recall, while limiting the loss of precision by applying it only to the retrieval of citations that have not yet been manually indexed.
The CISMeF query was shown to be more precise than the default PubMed ATM query in a 2008 study [9].
For the third query, the following format was used to locate the MEDLINE citations that should be indexed with a MeSH Concept x (called MeSH Concept query): x [TW] OR synonyms (x)[TW]. In the example of the subordinate MeSH Concept “Amaurotic familial idiocy,” the query is: “Amaurotic familial idiocy”[TW] OR “Familial Amaurotic Idiocy”[TW].
The format of the MeSH Concept query was constructed by the two librarians (Letord and Thirion) on the assumption that all the articles where a MeSH Concept x appears in the title or in the abstract should be indexed with the concept. As noted above, this likely underestimates the total number of articles that would actually be indexed with any MeSH Concept, because articles where a concept x appears neither in the title or abstract may require indexing with the concept.
The evaluation was performed on 32 MeSH Concepts for rare diseases and 22 MeSH Concepts for chronic diseases. A statistical analysis was performed comparing the 2 precision ratios (PubMed ATM versus CISMeF ATM) using the χ2 test (significance level: 0.05) for each of 54 MeSH Concepts (32 for rare diseases and 22 for chronic diseases).
RESULTS
Main results are displayed in Table 2 for rare diseases and Table 3 for chronic diseases.
For rare diseases, the average precision of the default PubMed ATM query when searching a narrower MeSH Concept was quite low (5.98%); the precision was a bit better when using the CISMeF PubMed query (7.06%). The PubMed ATM provided more results than the CISMeF ATM in 21 out of the 32 rare diseases (Table 2 shows the P values). No statistical difference was found in the other 11 rare diseases.
For chronic diseases, the average precision of the default PubMed ATM query when searching a narrower MeSH Concept was low (13.30%), whereas the precision was once again slightly better when using the CISMeF PubMed query (17.22%). The PubMed ATM provided more results than the CISMeF ATM for all 22 chronic diseases (Table 3 shows the P values). Paradoxically, for 2 MeSH Concepts (“Obesity” and “Kidney Failure”), the MeSH Concept query provided more results than the CISMeF query. The MeSH Concept query never provided more results than the PubMed query.
These results were considered by the CISMeF team to be sufficient grounds to implement the following rules in the CISMeF catalogue [13]:
When manually indexing, index with the subordinate MeSH Concept (if it exists) AND the MeSH Descriptor OR the MeSH Supplementary Concept related to it. This rule is very similar to the one already used in MEDLINE, which instructs the curator to index with both a MeSH Supplementary Concept and with the MeSH Descriptor related to it. This addition introduces a fourth item for indexing, MeSH Concepts after MeSH Descriptors, MeSH Supplementary Concepts, and MeSH Qualifiers. For preferred MeSH Concepts, no modification is required.
In the case of information retrieval, no modification is needed for preferred MeSH Concepts either. For subordinate or non-preferred MeSH Concepts, the query will differ according to the relationship: for MeSH Concepts related to a MeSH Descriptor or MeSH Supplementary Concept with the relationship “narrower than” or “related,” the query on the MeSH Concept is limited to the single MeSH Concept, as is the case for MeSH Supplementary Concepts. There is no semantic expansion with the related MeSH Descriptor, because the query in that case would introduce too many irrelevant results. For MeSH Concepts related to MeSH Descriptors or Supplementary Concepts with the relationship “broader than,” semantic expansion is employed only in the case of manual indexing. The query on the MeSH Concept is transformed into the following: MeSH Concept OR MeSH Descriptor (or MeSH Supplementary Concept). If the MeSH Concept is related to a MeSH Descriptor, this implies the explosion of the descriptor, which is valid in this case (and not in the previous one with relationships “narrower than” or “related”).
An example of MeSH Concept indexing is available in the CISMeF search engine, with the MeSH Concept “Belatacept” linked to the MeSH Supplementary Concept “Abatacept” with the “Narrower than” relationship. The MeSH Concept–based query in CISMeF‡ retrieves only one citation, while the Supplementary Concept–based query§ in CISMeF retrieves 14 citations, including the unique citation retrieved by the MeSH Concept-based query and 13 additional citations addressing aspects of the Supplementary Concept “Abatacept” that are not at all relevant to the concept “Belatacept.”** This example illustrates that MeSH Concept indexing provides more precise results. Citations manually indexed by CISMeF librarians with the MeSH Concept “Belatacept” were also indexed, applying the above rules, with the MeSH Supplementary Concept “Abatacept.” Therefore, all the citations indexed with MeSH Concept and retrieved by the MeSH Concept query are also retrieved by both PubMed and CISMeF ATM queries.
DISCUSSION
The authors agree with the NLM MeSH Section that MeSH Concepts have a fundamental role in the underlying structure of the MeSH Thesaurus [13]. In addition, previous research [14] has shown that the method used by search engines to map users' queries to MeSH has a direct impact on the specificity and effectiveness of retrieved results. Therefore, it can be expected that users' search experiences in MEDLINE will be enhanced by techniques whereby both database and search engine developers make full use of the MeSH structure. This paper shows the potential benefits of using MeSH Concepts for indexing and retrieval in MEDLINE, with an illustration of the CISMeF search tool.
Nonetheless, this study has several limitations. It has focused on precision and was not intended to measure recall. To measure the precision of this new approach, the authors assumed that all the articles where the MeSH Concept appears in the title or in the abstract should be indexed in the citation. This is not necessarily a safe assumption, especially with regard to words in the abstract. In the example of the CISMeF search tool, medical librarians manually indexed articles using the MeSH Concepts. Therefore, this limitation of the study could be overcome if MeSH Concept indexing were used in the future, in particular for the MEDLINE database. Some entry terms (e.g., “amaurotic familial idiocy”) could also be also outdated. In that case, performing information retrieval with the MeSH Concept could lead to very old citations.
CONCLUSION
This experiment on fifty-four rare and chronic disease MeSH Concepts shows that higher retrieval precision can be obtained with queries based on MeSH Concepts rather than MeSH Descriptors, which is the current default. This illustrates the conclusion of Lipscomb in her historical overview of MeSH after the introduction of MeSH Concepts in 2000: “an important role remains for MeSH in organizing information in a way that provides precision and power in retrieval” [15].
In practice, the specific querying strategy that was used in this experiment (type 3 query) could be applied for modifying the PubMed ATM query for relevant concepts (i.e., non-preferred MeSH Concepts that are narrower than the preferred concept in the relevant MeSH Descriptor). While this strategy offers the advantage of not requiring any changes to the current indexing policy, using concept indexing combined with some indexing rules applied to MeSH Supplementary Concepts (chemical substances and rare disease terms that are not MeSH terms) likely would be a fundamentally better approach. This improvement could be easily integrated into the PubMed interface to increase precision when querying the MEDLINE bibliographic database, in particular for rare diseases where there are multiple MeSH Concepts for one MeSH Descriptor. To do so, the authors strongly suggest creating 1 MeSH Supplementary Concept for each subordinate MeSH Concept that is not a preferred concept (n = 90, 736) (Table 1) and using these for indexing and for information retrieval, thereby extending the addition of some rare diseases to the Supplementary Concepts list introduced in MeSH in 2011. This change could be transparent to users. A simple query automatically mapped to the relevant MeSH Concept would yield improved results without requiring any advanced knowledge of MeSH, which has been shown to be a challenge for many nonprofessional searchers [16].
Footnotes
This author was supported by the Intramural Research Program of the National Institutes of Health, National Library of Medicine.
2011 Medical Subject Headings (MeSH) Descriptor data for “Abortion, Induced” <http://www.nlm.nih.gov/cgi/mesh/2011/MB_cgi?mode=&index=28&view=concept>.
These two drugs, abatacept and belatacept, have two different World Health Organization (WHO)-Anatomical Therapeutic Chemical (ATC) codes.
Catalog and Index of French-language Health Internet (CISMeF) Concept-based query <http://doccismef.chu-rouen.fr/servlets/Simple?Mot=belatacept.co&aff=4&tri=20&datt=1&msh=msh&debut=0>.
CISMeF Supplementary Concept–based query <http://doccismef.chu-rouen.fr/servlets/Simple?Mot=abatacept.mr&aff=4&tri=20&datt=1&cis=cis&pha=pha&msh=msh&debut=0>.
REFERENCES
- 1.Lu Z, Kim W, Wilbur W.J. Evaluation of query expansion using MeSH in PubMed. Inf Retr Boston. 2009;12(1):69–80. doi: 10.1007/s10791-008-9074-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lu Z, Kim W, Wilbur W.J. Evaluating relevance ranking strategies for MEDLINE retrieval. J Am Med Inform Assoc. 2009 Jan–Feb;16(1):32–6. doi: 10.1197/jamia.M2935. Epub 2008 Oct 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Névéol A, Kim W, Lu Z. Scenario-specific information retrieval in the biomedical domain. Proc AMIA Annu Symp. 2010. p. 1192.
- 4.Névéol A, Kim W, Wilbur W.J, Lu Z. Exploring two biomedical text genres for disease recognition. BioNLP '09. 4–5 Jun 2009. pp. 144–52. Proceedings of the Workshop on Current Trends in Biomedical Natural Language Processing; Boulder, CO; p.
- 5.Névéol A, Jiang G, Lu Z. Integrated access to disease information: the PubMed disease sensor. Proc AMIA Annu Symp. 2011. p. 1901.
- 6.Lu Z. PubMed and beyond: a survey of web tools for searching biomedical literature. Database (Oxford) 2011 Jan 18;2011:baq036. doi: 10.1093/database/baq036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Névéol A, Pereira S, Soualmia L.F, Thirion B, Darmoni S.J. A method of cross-lingual consumer health information retrieval. Stud Health Technol Inform. 2006;124:601–8. [PubMed] [Google Scholar]
- 8.Thirion B, Pereira S, Névéol A, Dahamna B, Darmoni S. French MeSH browser: a cross-language tool to access MEDLINE/PubMed. AMIA Annu Symp Proc. 2007 Oct 11. p. 1132. [PubMed]
- 9.Thirion B, Robu I, Darmoni S.J. Optimization of the PubMed Automatic Term Mapping. Stud Health Technol Inform. 2009;150:238–42. [PubMed] [Google Scholar]
- 10.Savage A. Changes in MeSH data structure. NLM Tech Bull [Internet] 2000 Mar–Apr. p. e2. [cited 24 Aug 2011]. < http://www.nlm.nih.gov/pubs/techbull/ma00/ma00_mesh.html>.
- 11.Prévalence des maladies rares: données bibliographiques [Internet] Les Cahiers d'Orphanet. serie Maladies Rares. 2011 Nov(1). [cited 31 Dec 2011]. < http://www.orpha.net/orphacom/cahiers/docs/FR/Prevalence_des_maladies_rares_par_ordre_alphabetique.pdf>.
- 12.National Library of Medicine. MEDLINE baseline repository data [Internet] The Library [cited 24 Aug 2011]. < http://mbr.nlm.nih.gov/Download/index.shtml#MeSH>.
- 13.Darmoni S.J, Leroy J.P, Baudic F, Douyère M, Piot J, Thirion B. CISMeF: a structured health resource guide. Methods Inf Med. 2000 Mar;39(1):30–5. [PubMed] [Google Scholar]
- 14.Gault L.V, Shultz M, Davies K.J. Variations in Medical Subject Headings (MeSH) mapping: from the natural language of patron terms to the controlled vocabulary of mapped lists. J Med Lib Assoc. 2002 Apr;90(2):173–80. [PMC free article] [PubMed] [Google Scholar]
- 15.Lipscomb C.E. Medical Subject Headings (MeSH) [historical notes] Bull Med Lib Assoc. 2000 Jul;88(3):265–6. [PMC free article] [PubMed] [Google Scholar]
- 16.Delozier E.P, Lingle V.A. MEDLINE and MeSH: challenges for end users [review] Med Ref Serv Q. 1992 Fall;11(3):29–46. doi: 10.1300/J115V11N03_03. [DOI] [PubMed] [Google Scholar]