Abstract
Glycogen, a branched polymer of glucose, acts as an intracellular carbon and energy reserve in many tissues and cell types. An important pathway for its degradation is by transport to lysosomes in an autophagy-like process. It has been proposed that starch-binding domain-containing protein 1 (Stbd1) may participate in this mechanism by anchoring glycogen to intracellular membranes. In addition, Stbd1 has been reported to interact with a known autophagy protein, GABARAPL1, a member of the Atg8 family. Here, we confirm this interaction and identify an Atg8 interacting motif (AIM) in Stbd1 necessary for GABARAPL1 binding as judged by co-immunoprecipitation from cell extracts and co-localization in cells as evidenced by immunofluorescence microscopy. The AIM sequence of Stbd1 200HEEWEMV206 lies within a predicted disordered region of the molecule and fits the consensus of other AIM sequences in cargo-specifying proteins such as p62 and Nix. Mutation of the AIM, including single point mutations of either W203 or V206, eliminated the co-localization of Stbd1 with both over-expressed and endogenous GABARAPL1. Stbd1 may therefore function as a novel cargo binding protein that delivers glycogen to lysosomes in an autophagic pathway that could be termed “glycophagy”.
Keywords: Stbd1, GABARAPL1, GABARAP, Atg8 family, Selective autophagy, AIM/LIR, Glycogen
1. Introduction
Glycogen, a branched glucose polymer, acts as an intracellular carbon and energy reserve in many cell types [1,2]. Many studies of glycogen metabolism have focused on its cytosolic synthesis and degradation. Although glycogen is usually considered cytosolic, glycogen particles have been found by electronic microscopy close to intracellular membranes such as the endoplasmic reticulum (ER) in liver [3] and the sarcoplasmic reticulum (SR) in muscle [4]. Glycogen is also present in the lysosomes of mammalian cells where it is directly hydrolyzed by lysosomal acid α-glucosidase (acid maltase, GAA) [5]. The significance of the lysosomal hydrolysis pathway is underscored by the fact that defects in the glucosidase cause a severe glycogen storage disease, Pompe disease, which eventually destroys tissues by over-accumulating glycogen in lysosomes [6].
The molecular mechanism for glycogen trafficking to the lysosome is not well understood although autophagy or an autophagy-like process is likely to be involved [7,8]. Initial clues for a mechanism came from a recent study of starch-binding domain-containing protein 1 (Stbd1), also known as genethonin 1 [9]. Stbd1 contains a conserved N-terminal hydrophobic sequence and a conserved C-terminal carbohydrate binding module of the CBM20 family (see Supplemental Fig. 2 of Jiang et al. [7]). The intervening region within Stbd1, which is overall much less conserved [7], is predicted to be disordered by analysis of the Stbd1 sequence using the “predictors of natural disordered regions” (PONDR) algorithm [10]. Stbd1 binds glycogen in vitro and is associated with glycogen in cells [7]. Jiang et al. [7] proposed that Stbd1 functions to anchor glycogen to membranes via its N-terminus. A further linkage to vesicular transport of glycogen is suggested by a yeast two-hybrid screen using human Stbd1 lacking the first 171 residues as bait. Among the targets identified were two autophagy related proteins, GABARAP and GABARAPL1, which are members of the Atg8 family [11]. Because GABARAPL1 more strictly co-distributed with Stbd1 in cells, it may be the preferred physiological binding partner. GABARAPL1 was originally cloned as an estrogen-regulated message in guinea pig endometrial glandular epithelial cells (GEC) and given the name Gec1 [12] which is still in use. Several functions have been proposed for GABARAPL1, including a role in autophagy, but much remains to be learned about its physiological function [13].
In mammalian cells, macroautophagy was initially thought to be an essentially random process for recycling cellular materials in response to nutritional deprivation [14–18]. However, there is emerging evidence that autophagic pathways can be more selective [17,18]. The autophagic disposal of several organelles has been described by processes named pexophagy (peroxisomes), mitophagy (mitochondria), ribophagy (ribsosomes) and reticulophagy (surplus edoplasmic reticulum). Aggrephagy describes removal of protein inclusions called aggrosomes and lipophagy refers to the disposal of oxidized lipids. Xenophagy is a process to eliminate intracellular pathogens like bacteria and viruses [16–18]. One idea to explain such selectivity is that cargo-specific receptors are coupled to individual selective autophagic pathways. Several cargo adaptor proteins have been identified, including p62, NBR1 and Nix. The ubiquitin-binding protein p62, also called sequestosome protein-1 (SQSTM1), is proposed to function as a cargo receptor for autophagic degradation of ubiquitinated protein substrates [17,19,20]. NBR1 has been suggested to function as an SQSTM1/p62 partner in disposal of misfolded proteins [21]. Nix has been proposed to selectively target the clearance of mitochondria in a ubiquitin-independent manner [22]. These receptors are all proposed to act by binding to Atg8 family members via short, specific and conserved sequences in the cargo receptors, termed Atg8-family interacting motifs (AIM) [23] or LC3 interacting regions (LIR) [20]. We propose that Stbd1 acts as a cargo receptor for glycogen and report the identification of the AIM in Stbd1 responsible for its interaction with GABARAPL1.
2. Materials and methods
2.1. Plasmid construction
Mammalian expression vectors containing HA-tagged hStbd1 (hStbd1-HA) and different truncation mutants thereof (ΔN24-HA and ΔC96-HA) for mammalian expression were made by PCR amplification of a human cDNA with addition of an HA tag at the C-terminus. The products were subcloned into BamHI/EcoRI sites of the pcDNA3 vector. Plasmids with double or single point mutations as well as deletion of the potential AIM regions ((W203A, V206A)-HA, (W212A, V215A)-HA, W203A-HA, V206L-HA and Δ198–222–HA) were constructed by site-directed mutation using pcDNA3-hStbd1-HA as template. The primers containing the mutated sequences were designed using the on-line service of PrimerX and were synthesized by Invitrogen. The Pfu Turbo DNA polymerase (Stratagene, 600250) was used for the PCR. The PCR product was digested with DpnI (New England Biolabs, R0176S) at 37 °C for 2–3 h to remove the parental DNA. The PCR product was transformed into competent cells to generate pcDNA3-(W203A, V206A)-HA, pcDNA3-(W212A, V215A)-HA, pcDNA3-W203A-HA, pcDNA3-V206L-HA and pcDNA3-(Δ198–222)-HA. cDNA encoding GABARAPL1 were respectively subcloned into Eco-RI/XbaI sites of pFLAGCMV-2 vectors. Sequences of all constructs were verified by the DNA Sequencing Core Facility, Indiana University School of Medicine.
2.2. Antibodies
Rabbit polyclonal anti-HA epitope tag antibodies were from Rockland (600-401-384). Mouse monoclonal anti-HA epitope tag antibodies were from Covance (MMS-101P). Rabbit polyclonal GABARAPL1 antibodies were from Protein Tech Group (11010-1-AP). Mouse monoclonal anti-FLAG M2 antibodies were from Sigma–Aldrich (F3165).
2.3. Cell culture and transfections
COSM9 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Media Tech, MT10013CV) with 25 mM glucose and 10% fetal bovine serum (FBS) (Atalanta Biologicals, S11550). Transfections were performed with FuGENE 6 (Roche Applied Science, 11814443001) following the manufacturer’s instructions. All cells were incubated at 37 °C with 5% CO2.
2.4. Preparation of cell lysate and immunoblotting
Cultured cells were lysed in buffer containing 50 mM Tris–HCl, 100 mM NaCl, pH 7.5, 0.5% Triton X-100, 1 mM PMSF, 0.1 mM TLCK, 1 mM benzamidine, 1 µg/ml aprotinin, pepstatin, and leupeptin. The cell lysates were centrifuged at 10,000g for 15 min at 4 °C to pellet insoluble materials. Protein concentration was determined by the Bradford method using BSA as standard [24]. Samples were subjected to 10% SDS–PAGE. Proteins were transferred to nitrocellulose membranes and incubated with antibodies, followed by horseradish peroxidase (HRP)-conjugated secondary antibodies and ECL (Thermo Scientific, PI32106).
2.5. Co-immunoprecipitation of Stbd1 and GABARAPL1
COSM9 cells co-transfected with pcDNA3-hStbd1-HA and pCMVFLAG-GABARAPL1, or related vectors expressing mutated hStbd1, were lysed and centrifuged at 8000 g for 15 min at 4 °C. EZ-view red anti-FLAG gel (Sigma–Aldrich, F2426) was equilibrated with lysis buffer before use. The supernatant from cell lysates was mixed with affinity gel and incubated at 4 °C overnight. The samples were centrifuged at 8000g for 30 s to pellet the agarose. The agarose was washed by lysis buffer three times, and loading buffer was added for SDS–PAGE and immunoblotting.
2.6. Immunofluorescence staining and microscopy
Cells were grown on glass coverslips for 24 or 48 h before fixation. Cells were fixed in PBS with 4% paraformaldehyde. Cells were then quenched and permeabilized in PBS with 100 mM glycine and 0.2% Triton X-100. Nonspecific binding sites were blocked with 5% bovine serum albumin (BSA) (Sigma–Aldrich, A7906) in PBS before addition of primary antibodies diluted in PBS with 2% BSA. Antibody dilutions were as follows: anti-HA, 1:1000; anti-FLAG, 1:1000; anti-GABARAPL1, 1:100. Cells were then washed in PBS with 2% BSA and developed with secondary antibodies (1:400, Invitrogen), conjugated with either Texas Red or Alexa488 fluorophores. Nuclei were visualized by staining with 1 µg/ml Hoechst (Invitrogen, H1398). The specimens were imaged using a Zeiss Axio Observer Z1 microscope with a Plan Apochromat X63 oil immersion objective (Zeiss) as structured light acquired via an Apotome (Zeiss). Images were processed with Zeiss Axiovision 4.7.
3. Results
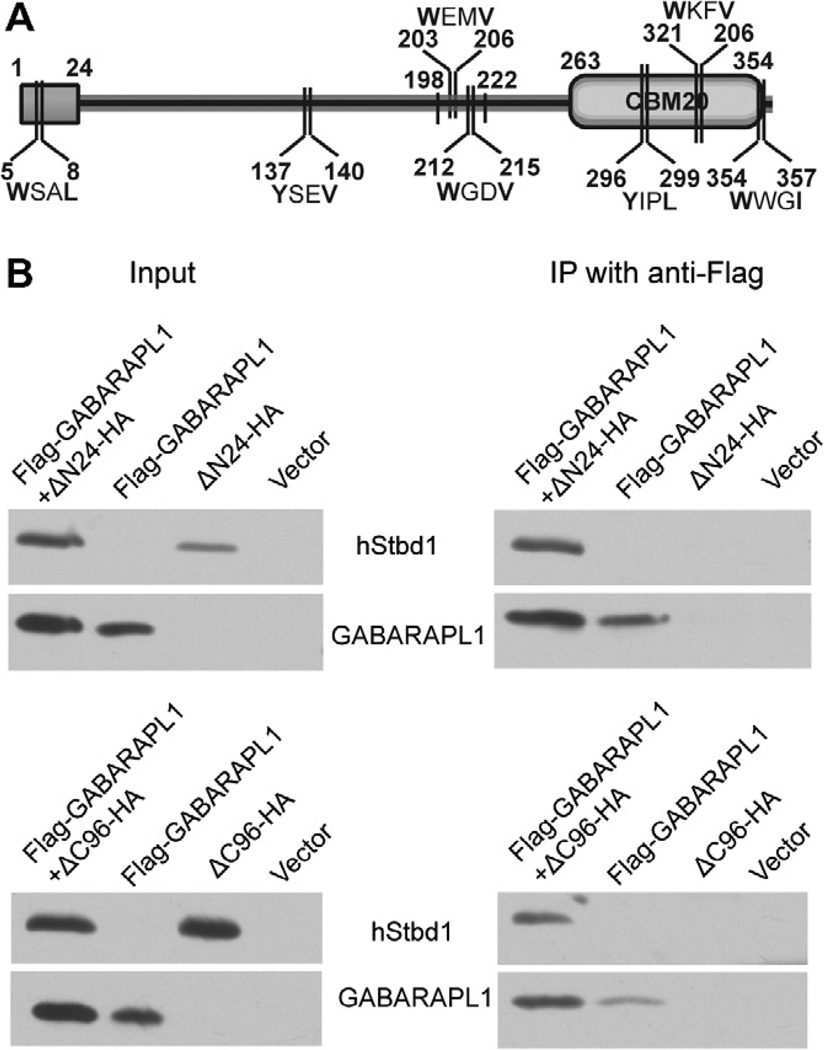
The goal of the current study was to identify whether Stbd1 contained a functional AIM motif, which would both define its interacting region with GABARAPL1 and strengthen the argument that Stbd1 shares the basic mechanism for autophagic targeting defined for other Atg8-family interacting cargo receptors. AIM sequences were initially defined by the simple consensus motif WxxL [25] that was later refined to x−3x−2x−1W/F/Yx1x2L/I/V, where x1 and/or x2 and at least one of x−3–x−2–x−1 would be acidic residues [23]. Seven occurrences of the more inclusive motif W/F/YxxL/I/V were present in Stbd1 (Fig. 1), one in the N-terminal region and three in the C-terminus. Because Flag-tagged GABARAPL1 co-immunoprecipitated both N-terminal (ΔN24-HA) and C-terminal (ΔC96-HA) truncation mutants of Stbd1 from extracts of COSM9 cells (Fig. 1), none of these four motifs likely mediate the interaction of Stbd1 with GABARAPL1. The data with the N-terminal truncation mutant are consistent with the fact that the original identification of GABARAPL1 by yeast two-hybrid screening had used as bait Stbd1 that was N-terminally truncated [7].
Fig. 1.
Interaction of GABARAPL1 with truncated mutants of Stbd1. Truncated hStbd1 lacking 1–24 residues or 262–358 residues with a C-terminal HA-tag (ΔN24-HA or Δ96-HA) and N-terminal Flag-tagged GABARAPL1 were expressed alone, or in the indicated combination in COS M9 cells. Control cells were transfected with empty pcDNA3 vector (Vector). (A) Schematic of potential AIM sites. (B) Immunoblotting of the cell lysates with the indicated antibody. Immunoprecipitation of GABARAPL1 with Anti-Flag antibodies covalently bound to agarose followed by immunoblotting with the indicated antibody.
The other three putative AIMs, SRSYSEV (Y137 and V140), HEEWEMV (W203 and V206) and HSSWGDV (W212 and V215) all reside in the disordered central region of the Stbd1 protein sequence. The SRSYSEV sequence was excluded because it is poorly conserved in different mammalian species [7]. The remaining two putative AIMs were located in the most conserved portion of the otherwise non-conserved disordered region. Specifically, W203, V206 and W212 are invariant in all 16 mammalian Stbd1 protein sequences that were analyzed [7]. V215 is conserved in 12 out of 16 Stbd1 sequences, and is replaced by Ile, another large hydrophobic residue, in the other four proteins. We therefore engineered several mutations in this region: (1) a deletion Δ198–222 that removes both putative AIMs of Stbd1, (2) two double point mutations, W203A, V206A and W212A, V215A, and (3) four single point mutations, W203A, V206A, W212A and V215A. All the mutants had an HA tag at their C-termini like the full length protein that we had studied. We previously reported that overexpression of wild type Stbd1 localized in distinctive perinuclear structures that sequestered glycogen as well as either endogenous or ectopically expressed GABARAPL1. When each Stbd1 mutant was individually overexpressed in COSM9 cells, the familiar perinuclear enlarged vesicle-like structure was observed in all cases (Fig. S1) indicating that none of the mutations per se affected the subcellular distribution of Stbd1.
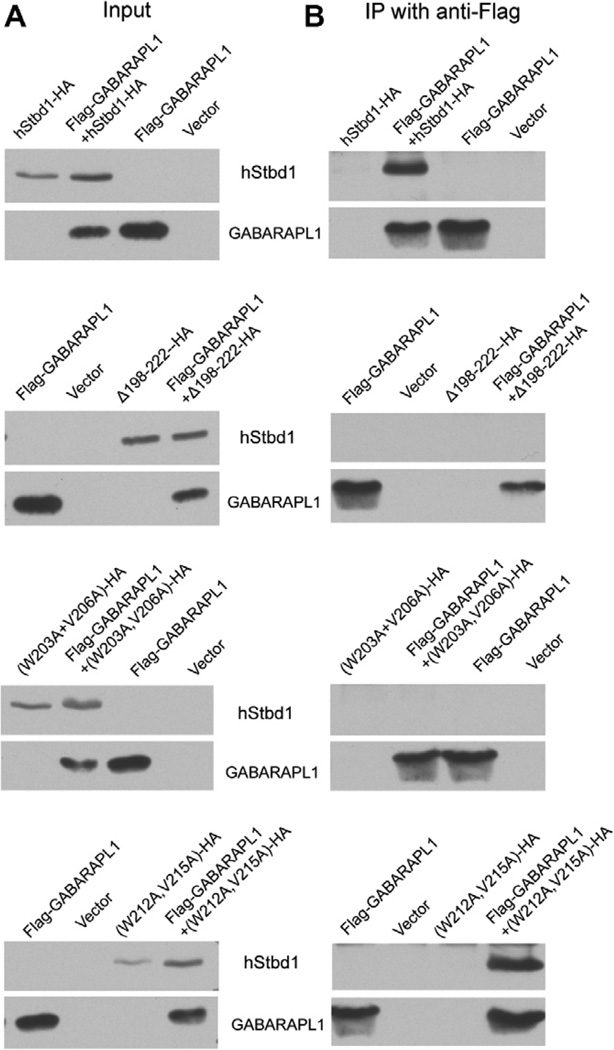
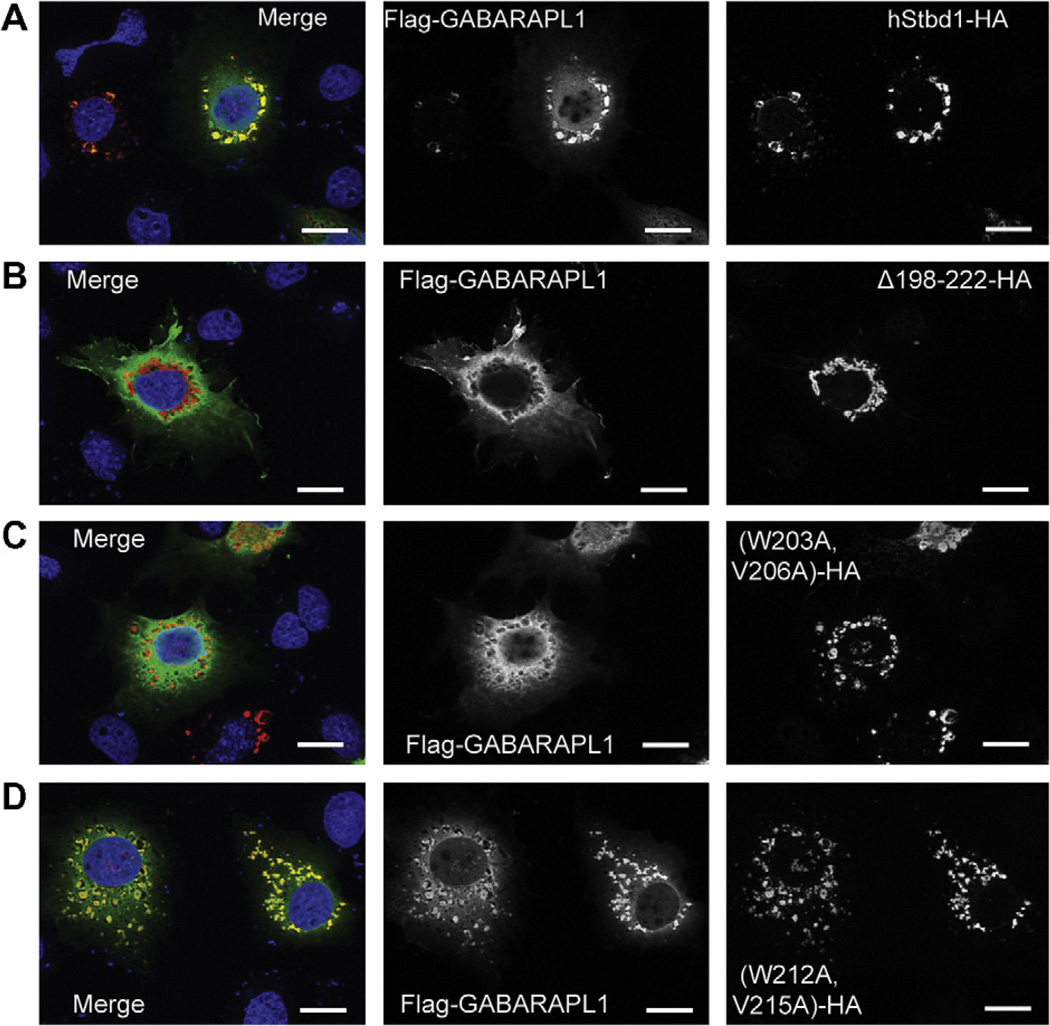
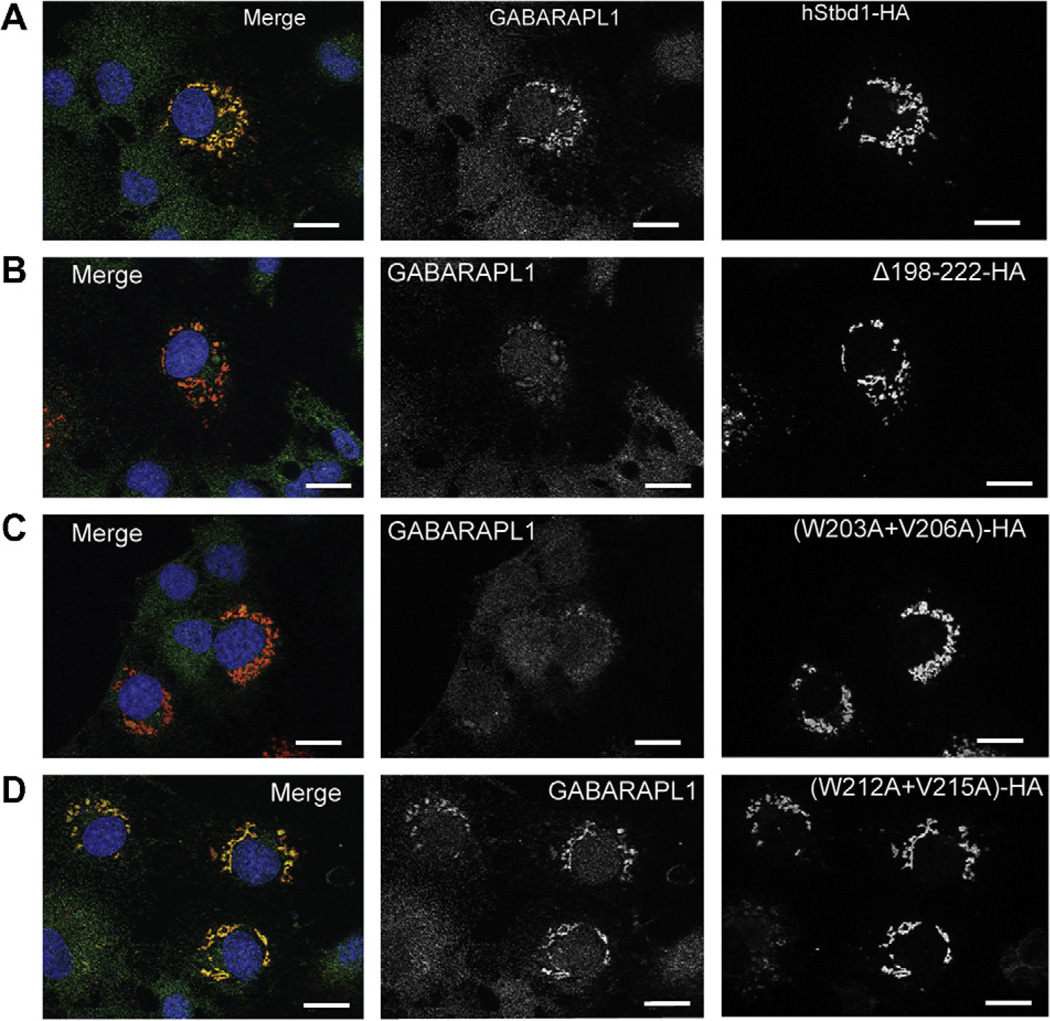
In other experiments, Flag-GABARAPL1 was co-expressed with each mutant. The deletion mutant, Δ198–222–HA, failed to immunoprecipitate Flag-GABARAPL1 (Fig. 2) and did not co-localize with GABARAPL1 as visualized by immunfluorescent staining (Fig. 3B), even though Δ198–222–HA appeared in the perinuclear structures. Similar analyses of the two double mutations indicated that only (W203A, V206A)-HA reproduced what was observed with Δ198–222–HA (Fig. 3C). (W212A, V215A)-HA acted like wild type full length Stbd1-HA (Fig. 3D). Mutation of either W203 or V206 in Stbd1 was sufficient to eliminate association with GABARAPL1 (Fig. S2A and B). In contrast, the other single mutants, W212A-HA and V215A-HA, behaved like wild-type Stbd1, in terms of subcellular co-localization with Flag-GABARAPL1 (Fig. S2C and D). We had previously shown that mutation of W293 in the CBM domain eliminated co-localization of glycogen with Stbd1 in cells, while largely retaining the characteristic perinuclear localization of Stbd1 [7]. We analyzed cells expressing a W293G-HA mutant, which does not bind to glycogen, and observed that co-localization with Flag-GABARAPL1 was unaffected (Fig. S2E). Thus, the ability to recruit glycogen is not required for Stbd1 interaction with GABARAPL1. To further validate the results with overexpressed GABARAPL1, we also analyzed the co-localization of Stbd1 mutants with endogenous GABARAPL1. The results were essentially identical to those observed with ectopically expressed GABARAPL1 (Fig. 4). There was no co-localization with the Stbd1 Δ198–222 HA and (W203A, V206A)-HA mutants whereas the (W212A, V215A)-HA mutant behaved like wild type Stbd1. Therefore, we conclude that the AIM defined by W203 and V206, in the sequence HEEWEMV, is necessary for interaction with GABARAPL1.
Fig. 2.
Interaction of GABARAPL1 with Stbd1 and potential Atg8 family interacting motif (AIM) mutants of Stbd1. hStbd1, full-length or mutants lacking 198–222 residues or double point mutations at potential AIMs with a C-terminal HA-tag (Δ198–222–HA (W203A, V206A)-HA, or (W212A, V215A)-HA) and N-terminal Flag-tagged GABARAPL1 were expressed alone, or in the indicated combination in COS M9 cells. Control cells were transfected with empty pcDNA3 vector (Vector). (A) Immunoblotting of the cell lysates with the indicated antibody (left panels). (B) Immunoprecipitation of GABARAPL1 with Anti-Flag antibodies covalently bound to agarose followed by immunoblotting with the indicated antibody (right panels).
Fig. 3.
Subcellular localization of GABARAPL1 and Atg8 family interacting motif (AIM) mutants of Stbd1 co-expressed in COS M9 cells. Mutated hStbd1 with a C-terminal HA-tag was co-expressed in COS M9 cells with N-terminal Flag-tagged GABARAPL1 and immunostained with anti-HA antibodies (red) or anti-Flag antibodies (green). (A) Co-localization of hStbd1 and GABARAPL1 (merged in left panel) in cells co-expressing C-terminal HA-tagged full length hStbd1 (right panel) and N-terminal Flag-tagged GABARAPL1 (middle panel). (B) Loss of co-localization (merged in left panel) of Flag-tagged GABARAPL1 (middle panel) with potential AIM deletion mutant of hStbd1, Δ198–222–HA (right panel). (C) Impaired co-localization (merged in left panel) of Flag-tagged GABARAPL1 (middle panel) with double mutation in a potential AIM on hStbd1, (W203A, V206A)-HA (right panel). (D) Unaffected co-localization (merged in left panel) of Flag-tagged GABARAPL1 (middle panel) with double mutation in another potential AIM on hStbd1, (W212A, V215A)-HA (right panel). Nuclei were stained with Hoechst (blue). The scale bar is 20 µm.
Fig. 4.
Subcellular localization of endogenous GABARAPL1 and overexpressed Stbd1 with Atg8 family interacting motif (AIM) mutations in COS M9 cells. Overexpressed full length or mutated hStbd1 with a C-terminal HA-tag and endogenous GABARAPL1 in COS M9 cells was immunostained with anti-HA antibodies (red) or anti-GABARAPL1 antibodies (green). (A) Co-localization of hStbd1 and GABARAPL1 (merged in left panel) in cells expressing C-terminal HA-tagged full length hStbd1 (right panel) and endogenous GABARAPL1 (middle panel). (B) Loss of co-localization (merged in left panel) of endogenous GABARAPL1 (middle panel) with potential AIM deletion mutant of hStbd1, Δ198–222–HA (right panel). (C) Impaired co-localization (merged in left panel) of endogenous GABARAPL1 (middle panel) with double mutation in potential AIM on hStbd1, (W203A, V206A)-HA (right panel). (D) Unaffected co-localization (merged in left panel) of endogenous GABARAPL1 (middle panel) with double mutation in another potential AIM on hStbd1, (W212A, V215A)-HA (right panel). Nuclei were stained with Hoechst (blue). The scale bar is 20 µm.
4. Discussion
A relationship between glycogen and autophagy or an autophagy-like process has long been inferred even though the mechanism is not well-defined. The fact that Pompe disease is associated with massive over-accumulation of glycogen had implicated a lysosomal degradative pathway [8] and the more recent work of Raben and Plotz has explicitly connected autophagy and glycogen [26]. Physiologic contexts for this process have been described in the liver, muscle and heart of newborn animals [27,28] as a mechanism to meet the extensive energy requirements during the period after birth until the start of suckling [29]. In the liver of newborns, glycogen autophagy is proposed to act in addition to the phosphorolytic degradation of glycogen to oppose hypoglycemia since the gluconeogenic pathway is not well developed at birth [30]. Further links between glycogen and autophagy came from genetic studies of the yeast Saccharomyces cerevisiae [31].
Initial mechanistic insights into the mechanism of glycogen transfer to the lysosome have come from study of the glycogen binding protein Stbd1 [7]. We proposed that Stbd1 targets glycogen to a membrane compartment in the cell. Based on the observation that Stbd1 interacts in cells with GABARAPL1, and to a lesser extent GABARAP, it could represent a new cargo-specifying receptor. Stbd1 was also identified by Behrends et al. [32] as interacting with GABARAP, GABARAPL1 and GABARAPL2. In recent years, it has become apparent that not all autophagy is random and, depending on the cargo, there are multiple selective autophagy pathways including aggrephagy, pexophagy, mitophagy, reticulophagy, ribophagy, lipophagy and xenophagy [16–18], as discussed in the Introduction. The three best studied mammalian cargo receptors are SQSTM1/p62, NBR1 and Nix although new candidates are rapidly emerging [17]. These, and other Atg8-family-interacting proteins, share a specific conserved interacting motif or AIM (Table 1). From the work described herein, Stbd1 contains a single AIM, 200HEEWEMV206, that matches well the canonical sequence, including acidic residues in the x−1 x−2 and x1 positions [23]. This sequence is highly conserved in mammals and is located in a segment of the molecule predicted to be disordered, a property that is associated with increased likelihood of participation in protein–protein interactions [33].
Table 1.
Comparison of AIM sequences in selective autophagy receptors.
| Receptor | AIM sequence |
|---|---|
| SQSTM1/p62 | DDDWTHLSS |
| NBR1 | SEDYIIILP |
| Nix | NSFWVELPM |
| Stbd1 | HEEWEMVPR |
Our proposal is that Stbd1 is a cargo receptor for glycogen and, like SQSTM1/p62, NBR1 and Nix, delivers its cargo to an autophagic pathway resulting in the transport of glycogen to lysosomes. In parallel to the naming of other selective autophagy pathways mentioned above, the process could be designated “glycophagy”. Stbd1 binds more tightly to glycogen that has fewer than normal branch points [7]. Overaccumulation of such poorly branched polysaccharide can have serious consequences and is associated with several genetic glycogen storage diseases, including Andersen disease, [34] Tarui disease [35] and Lafora disease [36–39]. Therefore, poorly branched glycogen is to be avoided and Stbd1 could have a housekeeping function to preferentially dispose of abnormal glycogen via lysosomes. Alternatively, Stbd1-dependent glycophagy could be regulated by as yet undefined mechanisms. Nor can we exclude the possibility that multiple pathways exist for the trafficking of glycogen to the lysosome.
Supplementary Material
Acknowledgment
Supported in part by NIH grants R37 DK27221 and R01 NS056454.
Abbreviations
- Stbd1
starch-binding domain-containing protein 1
- GABA
γ-aminobutyric acid
- GABARAP
GABAA receptor-associated protein
- GABARAPL1
GABARAP-like 1
- AIM
Atg8 family interacting motif
- LIR
LC3 interacting region
- TLCK
N-p-tosyl-l-lysine chloromethyl ketone.
Footnotes
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.bbrc.2011.08.106.
References
- 1.Roach PJ. Glycogen and its metabolism. Curr. Mol. Med. 2002;2:101–120. doi: 10.2174/1566524024605761. [DOI] [PubMed] [Google Scholar]
- 2.Greenberg CC, Jurczak MJ, Danos AM, Brady MJ. Glycogen branches out: new perspectives on the role of glycogen metabolism in the integration of metabolic pathways. Am. J. Physiol. Endocrinol. Metab. 2006;291:E1–E8. doi: 10.1152/ajpendo.00652.2005. [DOI] [PubMed] [Google Scholar]
- 3.Cardell RR, Jr, Michaels JE, Hung JT, Cardell EL. SERGE, the subcellular site of initial hepatic glycogen deposition in the rat: a radioautographic and cytochemical study. J. Cell Biol. 1985;101:201–206. doi: 10.1083/jcb.101.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shearer J, Graham TE. Novel aspects of skeletal muscle glycogen and its regulation during rest and exercise. Exerc. Sport Sci. Rev. 2004;32:120–126. doi: 10.1097/00003677-200407000-00008. [DOI] [PubMed] [Google Scholar]
- 5.Rosenfeld EL. Alpha-glucosidases (gamma-amylases) in human and animal organisms. Pathol. Biol. (Paris) 1975;23:71–84. [PubMed] [Google Scholar]
- 6.Hirschhorn R, Reuser AJ. Glycogen Storage Disease Type II: Acid alpha-Glucosidase (Acid Maltase) Deficiency. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic & molecular bases of inherited disease. New York: McGraw-Hill; 2000. pp. 3389–3420. [Google Scholar]
- 7.Jiang S, Heller B, Tagliabracci VS, Zhai L, Irimia JM, DePaoli-Roach AA, Wells CD, Skurat AV, Roach PJ. Starch binding domain-containing protein 1/genethonin 1 is a novel participant in glycogen metabolism. J. Biol. Chem. 2010;285:34960–34971. doi: 10.1074/jbc.M110.150839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fukuda T, Roberts A, Ahearn M, Zaal K, Ralston E, Plotz PH, Raben N. Autophagy and lysosomes in Pompe disease. Autophagy. 2006;2:318–320. doi: 10.4161/auto.2984. [DOI] [PubMed] [Google Scholar]
- 9.Janecek S. A motif of a microbial starch-binding domain found in human genethonin. Bioinformatics. 2002;18:1534–1537. doi: 10.1093/bioinformatics/18.11.1534. [DOI] [PubMed] [Google Scholar]
- 10.Romero P, Obradovic Z, Dunker AK. Natively disordered proteins: functions and predictions. Appl. Bioinformatics. 2004;3:105–113. doi: 10.2165/00822942-200403020-00005. [DOI] [PubMed] [Google Scholar]
- 11.Weidberg H, Shvets E, Shpilka T, Shimron F, Shinder V, Elazar Z. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 2010;29:1792–1802. doi: 10.1038/emboj.2010.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pellerin I, Vuillermoz C, Jouvenot M, Ordener C, Royez M, Adessi GL. Identification and characterization of an early estrogen-regulated RNA in cultured guinea-pig endometrial cells. Mol. Cell Endocrinol. 1993;90:R17–R21. doi: 10.1016/0303-7207(93)90161-c. [DOI] [PubMed] [Google Scholar]
- 13.Le Grand JN, Chakrama FZ, Seguin-Py S, Fraichard A, Delage-Mourroux R, Jouvenot M, Boyer-Guittaut M. GABARAPL1 (GEC1): Original or copycat? Autophagy. 2011;7 doi: 10.4161/auto.7.10.15904. [DOI] [PubMed] [Google Scholar]
- 14.Yang Z, Klionsky DJ. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 2010;22:124–131. doi: 10.1016/j.ceb.2009.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang Z, Klionsky DJ. An overview of the molecular mechanism of autophagy. Curr. Top. Microbiol. Immunol. 2009;335:1–32. doi: 10.1007/978-3-642-00302-8_1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rabinowitz JD, White E. Autophagy and metabolism. Science. 2010;330:1344–1348. doi: 10.1126/science.1193497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johansen T, Lamark T. Selective autophagy mediated by autophagic adapter proteins. Autophagy. 2011;7:1–18. doi: 10.4161/auto.7.3.14487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Komatsu M, Ichimura Y. Selective autophagy regulates various cellular functions. Genes Cells. 2010;15:923–933. doi: 10.1111/j.1365-2443.2010.01433.x. [DOI] [PubMed] [Google Scholar]
- 19.Vadlamudi RK, Joung I, Strominger JL, Shin J. P62, a phosphotyrosine-independent ligand of the SH2 domain of p56lck, Belongs to a new class of ubiquitin-binding proteins. J. Biol. Chem. 1996;271:20235–20237. doi: 10.1074/jbc.271.34.20235. [DOI] [PubMed] [Google Scholar]
- 20.Pankiv S, Clausen TH, Lamark T, Brech A, Bruun JA, Outzen H, Overvatn A, Bjorkoy G, Johansen T. P62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007;282:24131–24145. doi: 10.1074/jbc.M702824200. [DOI] [PubMed] [Google Scholar]
- 21.Kirkin V, Lamark T, Sou YS, Bjorkoy G, Nunn JL, Bruun JA, Shvets E, McEwan DG, Clausen TH, Wild P, Bilusic I, Theurillat JP, Overvatn A, Ishii T, Elazar Z, Komatsu M, Dikic I, Johansen T. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 2009;33:505–516. doi: 10.1016/j.molcel.2009.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Novak I, Kirkin V, McEwan DG, Zhang J, Wild P, Rozenknop A, Rogov V, Lohr F, Popovic D, Occhipinti A, Reichert AS, Terzic J, Dotsch V, Ney PA, Dikic I. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2010;11:45–51. doi: 10.1038/embor.2009.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noda NN, Ohsumi Y, Inagaki F. Atg8-family interacting motif crucial for selective autophagy. FEBS Lett. 2010;584:1379–1385. doi: 10.1016/j.febslet.2010.01.018. [DOI] [PubMed] [Google Scholar]
- 24.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 25.Noda NN, Kumeta H, Nakatogawa H, Satoo K, Adachi W, Ishii J, Fujioka Y, Ohsumi Y, Inagaki F. Structural basis of target recognition by Atg8/LC3 during selective autophagy. Genes Cells. 2008;13:1211–1218. doi: 10.1111/j.1365-2443.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 26.Raben N, Roberts A, Plotz PH. Role of autophagy in the pathogenesis of Pompe disease. Acta Myol. 2007;26:45–48. [PMC free article] [PubMed] [Google Scholar]
- 27.Schiaffino S, Hanzlikova V. Autophagic degradation of glycogen in skeletal muscles of the newborn rat. J. Cell Biol. 1972;52:41–51. doi: 10.1083/jcb.52.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kondomerkos DJ, Kalamidas SA, Kotoulas OB, Hann AC. Glycogen autophagy in the liver and heart of newborn rats. The effects of glucagon, adrenalin or rapamycin. Histol. Histopathol. 2005;20:689–696. doi: 10.14670/HH-20.689. [DOI] [PubMed] [Google Scholar]
- 29.Kotoulas OB, Kalamidas SA, Kondomerkos DJ. Glycogen autophagy in glucose homeostasis. Pathol. Res. Pract. 2006;202:631–638. doi: 10.1016/j.prp.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Dawes GS, Shelley HJ. Physiological aspects of carbohydrate metabolism in the foetus and newborn. In: Dickens F, Randle PJ, Whelan WJ, editors. Carbohydrate metabolism and its disorders. London; New York: Academic Press; 1968. pp. 87–116. [Google Scholar]
- 31.Wang Z, Wilson WA, Fujino MA, Roach PJ. Antagonistic controls of autophagy and glycogen accumulation by Snf1p, the yeast homolog of AMP-activated protein kinase, and the cyclin-dependent kinase Pho85p. Mol. Cell Biol. 2001;21:5742–5752. doi: 10.1128/MCB.21.17.5742-5752.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr. Opin. Struct. Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 34.Chen Y-T, Burchell A. Glycogen storage diseases. In: Scriver CR, Beaudet AL, Sly WS, Valle D, editors. The metabolic and molecular bases of inherited disease. New York: McGraw-Hill, Heath Professions Division; 1995. pp. 935–965. [Google Scholar]
- 35.Nakajima H, Hamaguchi T, Yamasaki T, Tarui S. Phosphofructokinase deficiency: recent advances in molecular biology. Muscle Nerve. 1995;3:S28–S34. doi: 10.1002/mus.880181408. [DOI] [PubMed] [Google Scholar]
- 36.Andrade DM, Turnbull J, Minassian BA. Lafora disease, Seizures and sugars. Acta Myol. 2007;26:83–86. [PMC free article] [PubMed] [Google Scholar]
- 37.Delgado-Escueta AV. Advances in lafora progressive myoclonus epilepsy. Curr. Neurol. Neurosci. Rep. 2007;7:428–433. doi: 10.1007/s11910-007-0066-7. [DOI] [PubMed] [Google Scholar]
- 38.Gentry MS, Dixon JE, Worby CA. Lafora disease: insights into neurodegeneration from plant metabolism. Trends Biochem. Sci. 2009;34:628–639. doi: 10.1016/j.tibs.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ganesh S, Puri R, Singh S, Mittal S, Dubey D. Recent advances in the molecular basis of Lafora’s progressive myoclonus epilepsy. J. Hum. Genet. 2006;51:1–8. doi: 10.1007/s10038-005-0321-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.