Abstract
The tryptophan isomers of the cyclic tetrapeptide CJ-15,208, reported to be a kappa opioid receptor (KOR) antagonist [Saito, T.; Hirai, H.; Kim, Y. J.; Kojima, Y.; Matsunaga, Y.; Nishida, H.; Sakakibara, T.; Suga, O.; Sujaku, T.; Kojima, N. J. Antibiot. (Tokyo) 2002, 55, 847–854.], were synthesized to determine the tryptophan stereochemistry in the natural product. A strategy was developed to select linear precursor peptides that favor cyclization using molecular modeling, and optimized cyclization conditions are reported. The optical rotation of the l-Trp isomer is consistent with that of the natural product. Unexpectedly both isomers exhibit similar nanomolar affinity for KOR.
Keywords: Cyclic tetrapeptide, Cyclization, Molecular modeling
Kappa opioid receptor (KOR) antagonists have demonstrated promising activity in animal models as antidepressants, anxiolytics, and therapeutics for cocaine and opiate abuse.1 Thus, there is considerable interest in developing new KOR antagonists as potential therapeutic agents. Several nonpeptide selective KOR antagonists (i.e., norbinaltorphimine (norBNI), 5′-guanidinonaltrindole (GNTI), and (3R)-7-hydroxy-N-[(1S)-1-[[(3R,4R)-4-(3-hydroxyphenyl)-3,4-dimethyl-1-piperidinyl]methyl]-2-methylpropyl]-1,2,3,4-tetrahydro-3-isoquinolinecarboxamide (JDTic)) have been identified and used as pharmacological tools to study KOR.1a,2 However, all these compounds exhibit exceptionally long antagonist activity, lasting for weeks to more than a month after a single injection,1a a profile which could potentially limit their use as therapeutic agents. In contrast, peptide antagonists are expected to exhibit shorter durations of activity in vivo as a result of metabolism by proteases.3 While linear peptides are often metabolized too rapidly for use in vivo, cyclization can increase metabolic stability and improve pharmacokinetic properties.3,4
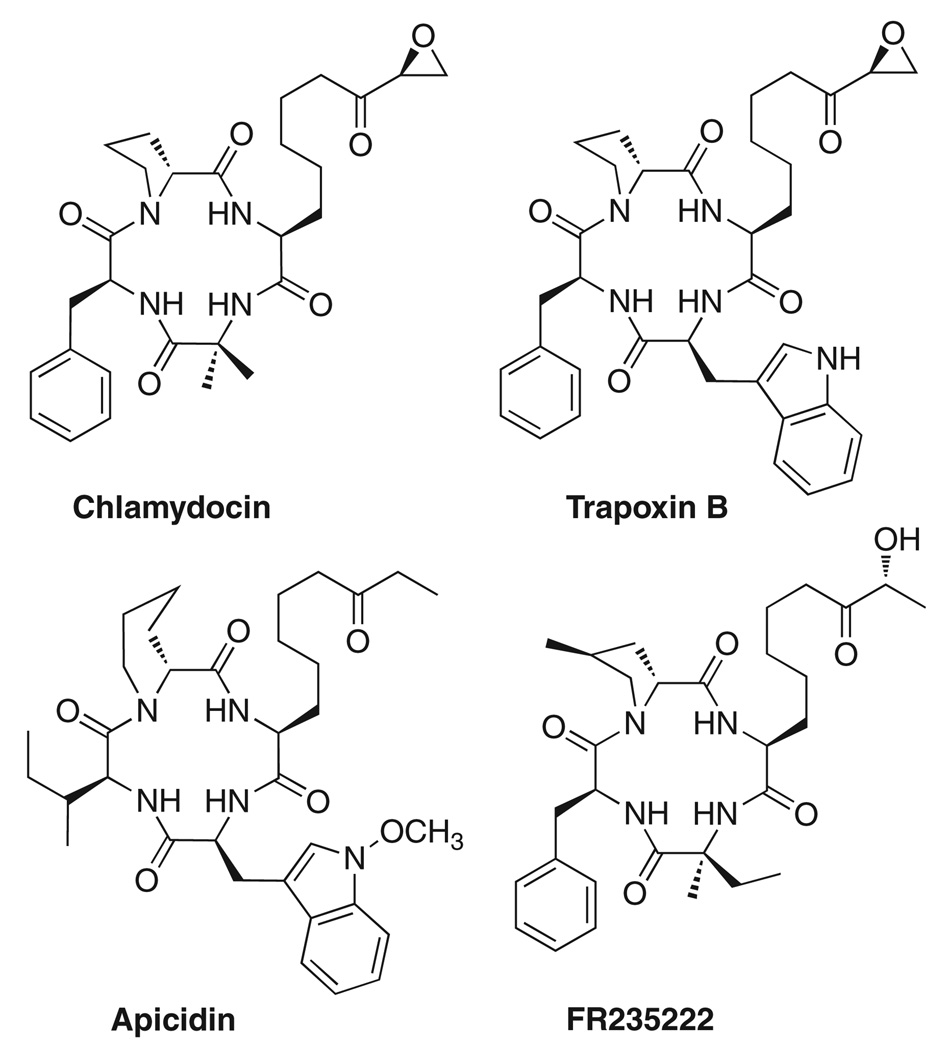
Head-to-tail cyclic peptides represent a unique class of peptides that exhibit biological activity at a broad range of targets. A number of cyclic tetrapeptides with diverse biological activities have been obtained from different natural sources, including the cytotoxic chlamydocin and immunosuppressant FR 235222 (Fig. 1).5 Constrained cyclic peptides have substantially fewer degrees of freedom than linear peptides and therefore may provide valuable information about the spatial requirements necessary for the interaction of ligand functional groups with the target binding site.
Figure 1.
Cyclic tetrapeptide natural products.
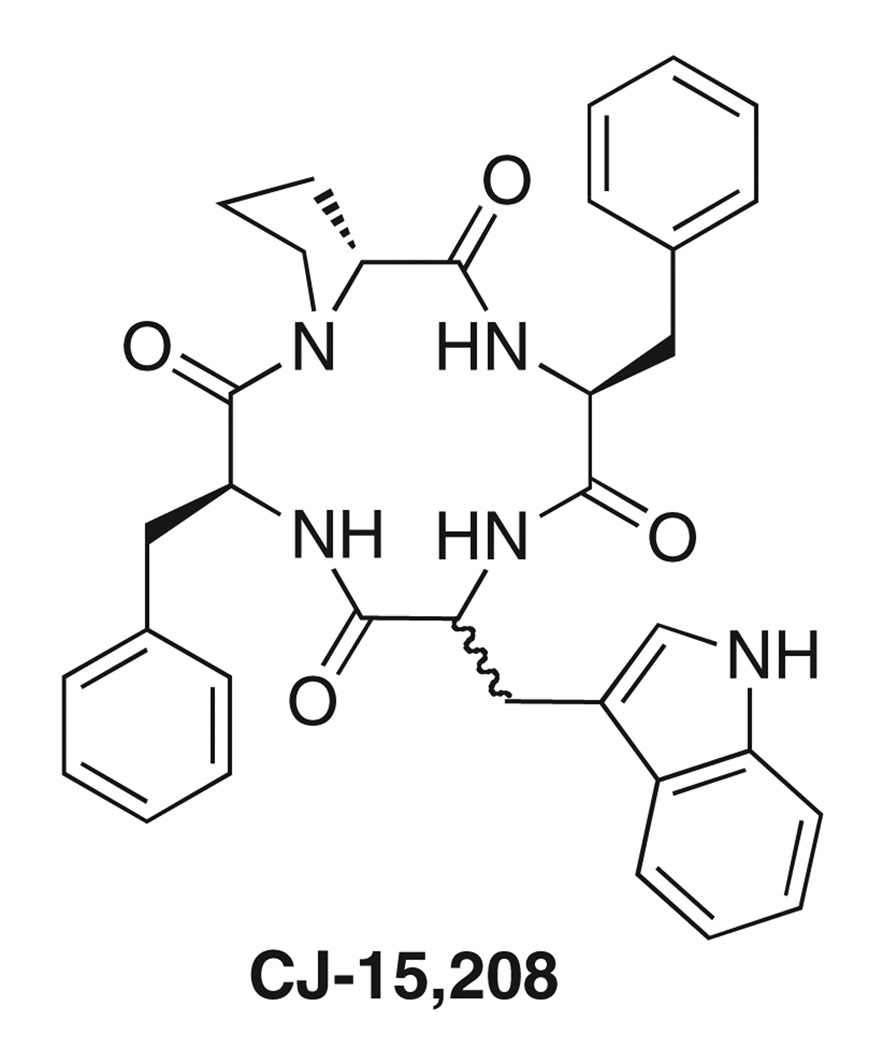
Saito et al.6 reported the isolation and characterization of the cyclic tetrapeptide CJ-15,208 (Fig. 2) from the fermentation broth of the fungus Ctenomyces serratus ATCC 15502. In binding assays this peptide was reported to preferentially bind to KOR with an IC50 value of 47 nM and antagonized the KOR agonist asimadoline in the electrically-stimulated rabbit vas deferens.6 Thus, this peptide provides a novel lead for the development of new KOR antagonists.
Figure 2.
Cyclic tetrapeptide natural product CJ-15,208.
Saito et al. elucidated the structure of CJ-15,208 using high resolution fast atom bombardment mass spectrometry and NMR to establish the sequence and cyclic structure of this peptide.6 This cyclic tetrapeptide consists of a proline, a tryptophan, and two phenylalanine residues (Fig. 2). While amino acid analysis and HPLC of chiral derivatives established the identity and stereochemistry of the Pro and Phe residues, the stereochemistry of the Trp residue was not determined due to destruction during acid hydrolysis.6 While there is a brief reference to the l-Trp isomer in a conference proceeding,7 no details on the synthesis were provided nor was evidence presented to establish whether this isomer was the natural product. Therefore, we undertook the synthesis of both isomers of CJ-15,208 varying in the stereochemistry of the Trp residue (Fig. 2) to conclusively identify the natural product and to develop synthetic schemes that may be applied to the synthesis of future analogs.8
Difficulties have often been encountered during the synthesis of cyclic tetrapeptides due to the constrained 12-membered ring structure of the product. Most cyclic tetrapeptide natural products contain at least one turn-inducing residue such as Pro, an N-substituted amino acid, Gly, or Aib (α-aminoisobutyric acid) that favors a folded conformation and is essential for cyclization.9 The cyclization reaction still often leads to the formation of the cyclic octapeptide, which in some cases is the predominant product.10 The amino acid sequence of the linear precursor and the method of activation of the carboxy terminus also have significant influence on the results of the cyclization reaction.10a,11
Initially the syntheses of the isomers of CJ-15,208 were attempted using a similar strategy to that described for trapoxin B12 based on the similarity in backbone structure. The cyclization reaction was performed using the linear sequences Phe-Trp-Phe-d-Pro and Phe-d-Trp-Phe-d-Pro with d-proline at the C-terminus. Despite a variety of attempts using different reaction conditions and coupling reagents, including the pentafluorophenyl ester methodology reported by Schmidt et al.,9a,11b the desired cyclic tetrapeptides were not observed.
The sequence of the precursor linear peptide can have a marked influence on the success of the cyclization reaction.11a In cyclic tetrapeptides such as CJ-15,208 there are four different linear sequences that can be used to attempt cyclization. However, not all of the linear sequences may give the desired cyclic tetrapeptide in high yield, with some sequences yielding little or no desired product.11a,b Also which linear sequences will successfully cyclize differs for different cyclic tetrapeptides.11a,b
Molecular modeling and conformational analysis can aid in identifying linear precursor peptides that may adopt conformations that can undergo cyclization.13 Therefore we utilized molecular modeling to choose linear sequences for cyclization to increase the probability of cyclic tetrapeptide formation. Thus, we initiated a molecular modeling study with the four possible linear precursors for both tryptophan isomers of CJ-15,208 (Table 1).14
Table 1.
Analysis of the conformations of the linear peptides by molecular modeling
| Linear sequence | Hydrogen bonds | Conformationa | Distanceb (Å) |
|---|---|---|---|
| l-Trp isomer | |||
| Phe-Trp-Phe-d-Pro | — | Extended | 11.5 |
| Trp-Phe-d-Pro-Phe | 2 | γ-Turn | 6.5 |
| Phe-d-Pro-Phe-Trp | 1 | β-Turn | 3.4 |
| d-Pro-Phe-Trp-Phe | — | Extended | 11.9 |
| d-Trp isomer | |||
| Phe-d-Trp-Phe-d-Pro | — | Extended | 10.4 |
| d-Trp-Phe-d-Pro-Phe | 2 | γ-Turn | 2.9 |
| Phe-d-Pro-Phe-d-Trp | 2 | γ-Turn | 3.1 |
| d-Pro-Phe-d-Trp-Phe | — | Extended | 11.6 |
Preliminary assignment based on hydrogen bonding patterns.
The distance was measured from the carboxy terminal carbonyl carbon to the amino terminal nitrogen.
The results of the molecular modeling suggested which linear sequences were likely to be compatible with cyclization. The conformation of the linear precursor peptide plays a very important role in successful cyclizations, and intramolecular hydrogen bonds can stabilize a folded conformation that is favorable for cyclization.4 Analysis of the linear sequence Phe-Trp-Phe-d-Pro with d-proline at the C-terminus suggested that it preferentially existed in an extended conformation with a distance of about 11 Å between the amino and carboxy termini (Table 1), which is consistent with our inability to obtain the cyclic tetrapeptide from this linear precursor. In contrast, analysis of the linear peptides with proline at the Ci+1 or Ci+2 position resulted in folded conformations with the amino and carboxy termini in closer proximity. A similar analysis of the possible linear sequences containing d-Trp was also performed. Based on this analysis cyclization of the linear sequences Phe-d-Pro-Phe-Trp for the l-Trp peptide and d-Trp-Phe-d-Pro-Phe or Phe-d-Pro-Phe-d-Trp for the d-Trp isomer was expected to yield the required cyclic tetrapeptides.
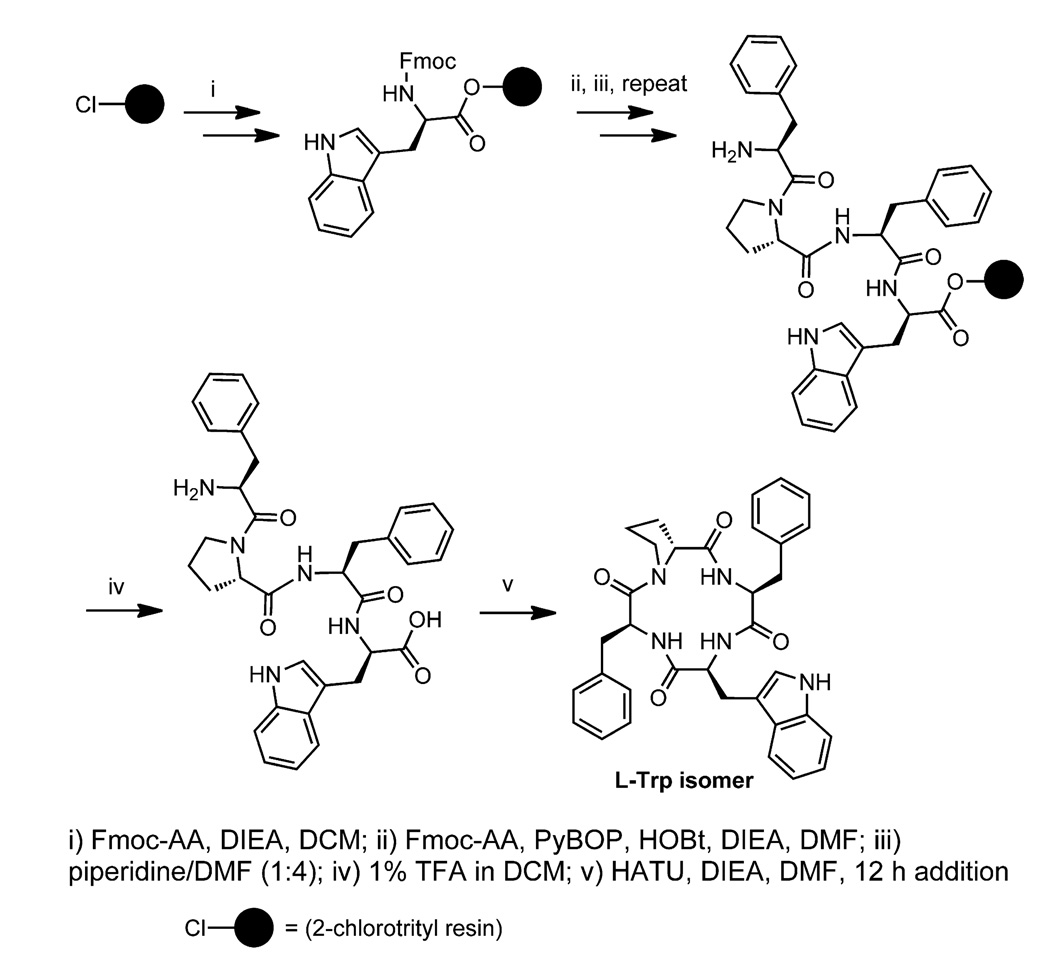
The linear peptide precursors were synthesized on the 2-chlorotrityl resin (Scheme 1). Attachment of the first amino acid to the resin was performed using a twofold excess of amino acid and DIEA (N,N-diisopropylethylamine) (Scheme 1).15 The synthesis of the linear peptides was performed using Fmoc-protected amino acids and standard coupling and deprotection protocols. Previously Mou and Singh reported that the protection of the indole nitrogen of Trp in the precursor of apicidin was necessary for the successful cyclization using the pentafluorophenyl ester method.11c Therefore the indole nitrogen of the Trp residue in our linear precursor peptides was initially protected with the Boc-protecting group.8 This proved unnecessary using the cyclization conditions described below, however, and the cyclic peptides were subsequently synthesized using Fmoc-Trp without side chain protection (Scheme 1). The crude linear peptides were ≥98% pure by analytical HLPC following cleavage from the resin with 1% trifluoroacetic acid (TFA) in dichloromethane (DCM).15
Scheme 1.
Synthesis of the cyclic tetrapeptide containing l-Trp.
The cyclization of the linear peptide Phe-d-Pro-Phe-Trp was performed in solution. The conditions used for the cyclization had a marked influence on the results of this reaction, in terms of the formation of the dimeric cyclic octapeptide versus the desired cyclic tetrapeptide. Highly dilute conditions minimize the formation of the cyclic octapeptide.12 Also the use of the azabenzotriazole-derived uronium reagent HATU (2-(1H-7-azabenzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate) has been reported to facilitate a difficult cyclization and decrease cyclodimerization compared to benzotriazole-derived coupling agents,10a most likely due to neighboring group assistance by the ring nitrogen. When the cyclization of Phe-d-Pro-Phe-Trp was performed in DCM/MeCN, however, even in dilute solution with slow addition of the linear peptide, the cyclic octapeptide was obtained in addition to the desired cyclic tetrapeptide. The rate of addition of Phe-d-Pro-Phe-Trp also affected the yields of the cyclic tetrapeptide versus cyclic octapeptide; even in dilute solution the rapid addition of the linear peptide resulted in the cyclic octapeptide predominating, even when DMF (N,N-dimethylformamide) was used as the solvent.16 Based on these results and procedures reported previously for the other cyclic tetrapeptides,10a,12 optimized conditions for the cyclization were developed which involved the slow addition of the linear peptide in DMF over 12 h to a dilute solution of HATU and DIEA in DMF.17 Mass spectrometry and HPLC analysis of the isolated product indicated the formation of the desired cyclic tetrapeptide cyclo[Phe-d-Pro-Phe-Trp]18 with minimum formation of the cyclic octapeptide (<5%) under these conditions.
The same optimized conditions were also used to cyclize d-Trp-Phe-d-Pro-Phe to give the d-Trp isomer of CJ-15,208, which was analyzed by mass spectrometry and HPLC.19 In contrast to the l-Trp peptide, cyclization of d-Trp-Phe-d-Pro-Phe yielded minimal cyclic octapeptide even under conditions where the linear l-Trp precursor yielded almost exclusively the cyclic dimer. Thus the second d-amino acid in the d-Trp peptide facilitated the folding of the linear precursor into a conformation that favors cyclization to give the cyclic tetrapeptide, making the synthesis of this cyclic peptide more straight forward than the l-Trp isomer.
One concern during head-to-tail cyclizations is epimerization of the C-terminal residue. Azabenzotriazole reagents such as HATU minimize epimerization of the C-terminal residue of peptides during activation and coupling.20 The l-Trp and d-Trp cyclic tetrapeptides had very different retention times on HPLC (tR = 26.0 and 35.5 min, respectively), and the d-Trp peptide was not detected during cyclization of Phe-d-Pro-Phe-Trp, suggesting minimum epimerization of the C-terminal residue that occurred during cyclization under the conditions described above.
The optical rotation of the l-Trp cyclic tetrapeptide closely matched the reported value for the natural product (αD = −63.6° (c 0.05, DMSO), lit.6 (αD = −64.0° (c 0.05, DMSO)), while the optical rotation of the isomer containing d-Trp had the opposite sign (αD = +19.8° (c 0.05, DMSO)). Surprisingly both cyclic tetrapeptides exhibited nanomolar affinity for KOR (Ki (± SEM) = 21.2 ± 5.6 and 46.9 ± 16.1 nM for the l-Trp and d-Trp isomers, respectively), in competition radioligand-binding assays using mouse brain membranes and [3H]U69,593 as the radioligand). These results are similar to those recently reported for the d-Trp isomer of CJ-15,208 by Dolle et al.21
In conclusion, we successfully synthesized the two isomers of the cyclic tetrapeptide CJ-15,208 containing l- and d-tryptophan. Using the molecular modeling approach described here appropriate linear sequences were chosen for cyclization. The synthesis of the l-Trp isomer still proved challenging, however, because of the tendency of the linear precursor to dimerize during cyclization. Optimized cyclization conditions were identified that minimized the formation of the cyclic octapeptide and maximized the yield of the desired cyclic tetrapeptide. The optical rotation of the l-Trp isomer agrees with the optical rotation reported for the natural product CJ-15,208.6 Unexpectedly, both compounds exhibit nanomolar affinity for KOR and thus can serve as excellent leads for further structural modification. Such studies are currently underway in our laboratory. Cyclization using a linear peptide that is preselected based on molecular modeling as described here will assist in the synthesis of analogues of these and other cyclic tetrapeptides.
Supplementary Material
Acknowledgments
This research was supported by NIDA grant R01 DA018832. The authors thank Robert Drake of the KU Mass Spectrometry Laboratory for his assistance with the ESI spectra and Wendy Hartsock for her assistance with HPLC and for preparation of the manuscript.
Footnotes
Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.tetlet.2010.07.086.
References and notes
- 1.(a) Metcalf MD, Coop A. AAPS J. 2005;7:E704–E722. doi: 10.1208/aapsj070371. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Aldrich JV, McLaughlin JP. AAPS J. 2009;11:312–322. doi: 10.1208/s12248-009-9105-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aldrich JV, Vigil-Cruz SC. In: Burger’s Medicinal Chemistry & Drug Discovery. Abraham DJ, editor. Vol. 6. New York: John Wiley & Sons; 2003. pp. 329–481. [Google Scholar]
- 3.Aldrich JV, Patkar KA, McLaughlin JP. Proc. Natl. Acad. Sci. U.S.A. 2009;106:18396–18401. doi: 10.1073/pnas.0910180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gilon C, Halle D, Chorev M, Selinger Z, Byk G. Biopolymers. 1991;31:745–750. doi: 10.1002/bip.360310619. [DOI] [PubMed] [Google Scholar]
- 5.(a) Closse A, Huguenin R. Helv. Chim. Acta. 1974;57:533–545. doi: 10.1002/hlca.19740570306. [DOI] [PubMed] [Google Scholar]; (b) Itazaki H, Nagashima K, Sugita K, Yoshida H, Kawamura Y, Yasuda Y, Matsumoto K, Ishii K, Uotani N, Nakai H, Terui A, Yoshimatsu S, Ikenishi Y, Nakagawa Y. J. Antibiot. (Tokyo) 1990;43:1524–1532. doi: 10.7164/antibiotics.43.1524. [DOI] [PubMed] [Google Scholar]; (c) Singh SB, Zink DL, Polishook JD, Dombrowski AW, Darkin-Rattray SJ, Schmatz DM, Goetz MA. Tetrahedron Lett. 1996;37:8077–8080. [Google Scholar]; (d) Mori H, Urano Y, Kinoshita T, Yoshimura S, Takase S, Hino M. J. Antibiot. (Tokyo) 2003;56:181–185. doi: 10.7164/antibiotics.56.181. [DOI] [PubMed] [Google Scholar]
- 6.Saito T, Hirai H, Kim YJ, Kojima Y, Matsunaga Y, Nishida H, Sakakibara T, Suga O, Sujaku T, Kojima N. J. Antibiot. (Tokyo) 2002;55:847–854. doi: 10.7164/antibiotics.55.847. [DOI] [PubMed] [Google Scholar]
- 7.Seale PW, Stead P, Jaxa-Chamiec A. In: Peptides 2000. Martinez J, Fehrentz JA, editors. Paris, France: EDK; 2001. pp. 271–272. [Google Scholar]
- 8.Kulkarni SS, Ross NC, McLaughlin JP, Aldrich JV. Adv. Exp. Med. Biol; Preliminary results were presented at the 20th American Peptide Symposium; Montreal, Canada. 2009. pp. 269–270. [Google Scholar]
- 9.(a) Schmidt U, Langner J. J. Pept. Res. 1997;49:67–73. doi: 10.1111/j.1399-3011.1997.tb01122.x. [DOI] [PubMed] [Google Scholar]; (b) Davies JS. J. Pept. Sci. 2003;9:471–501. doi: 10.1002/psc.491. [DOI] [PubMed] [Google Scholar]
- 10.(a) El Haddadi M, Cavelier F, Vives E, Azmani A, Verducci J, Martinez J. J. Pept. Sci. 2000;6:560–570. doi: 10.1002/1099-1387(200011)6:11<560::AID-PSC275>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]; (b) Berst F, Ladlow M, Holmes A. B. Chem. Commun. 2002:508–509. doi: 10.1039/b110068h. [DOI] [PubMed] [Google Scholar]
- 11.(a) Pastuszak J, Gardner JH, Singh J, Rich DH. J. Org. Chem. 1982;47:2982–2987. [Google Scholar]; (b) Schmidt U, Beutler U, Lieberknecht A. Angew. Chem., Int. Ed. Engl. 1989;28:333–334. [Google Scholar]; (c) Mou L, Singh G. Tetrahedron Lett. 2001;42:6603–6606. [Google Scholar]; (d) Meutermans WD, Bourne GT, Golding SW, Horton DA, Campitelli MR, Craik D, Scanlon M, Smythe ML. Org. Lett. 2003;5:2711–2714. doi: 10.1021/ol034907o. [DOI] [PubMed] [Google Scholar]
- 12.Taunton J, Collins JL, Schreiber SL. J. Am. Chem. Soc. 1996;118:10412–10422. [Google Scholar]
- 13.(a) Cavelier-Frontin F, Pepe G, Verducci J, Siri D, Jacquier R. J. Am. Chem. Soc. 1992;114:8885–8890. [Google Scholar]; (b) Besser D, Olender R, Rosenfeld R, Arad O, Reissmann S. J. Pept. Res. 2000;56:337–345. doi: 10.1034/j.1399-3011.2000.00735.x. [DOI] [PubMed] [Google Scholar]
- 14.The linear precursors were constructed and minimized using the Tripos force field in SYBYL7.0 (Tripos, Inc., St. Louis, MO). Electrostatic charges were generated using the Gasteiger and Marsili method within SYBYL; an 8Å non-bonded cut-off was employed and a dielectric constant of 4 was applied for minimizations. A three-step minimization protocol, consisting of steepest decent gradient (100 iterations), conjugate gradient (10,000 iterations) and then a BFGS gradient minimization with convergence criterion of 0.001 kcal/mol, was utilized to obtain convergence.
- 15.Chan WC, White PD. Fmoc Solid Phase Peptide Synthesis: A Practical Approach. New York: Oxford University Press; 2000. pp. 41–76. [Google Scholar]
- 16.The yields of cyclic tetrapeptide and cyclic octapeptide measured by HPLC analysis following rapid addition were 28% and 44%, respectively.
- 17.The crude linear peptide (50 mg, 0.072 mmol) in DMF (10 mL) was added dropwise at a rate of 1.6 mL/h (using a KD Scientific single infusion syringe pump) to a solution of HATU (46 mg, 0.12 mmol) and DIEA (0.10 mL, 0.58 mmol) in DMF (150 mL) over 6 h. After 6 h a second portion of HATU (46 mg, 0.12 mmol) was added to the reaction in one portion, and additional linear peptide (50 mg, 0.072 mmol) in DMF (10 mL) was added dropwise at a rate of 1.6 mL/h as described above. The reaction was then allowed to stir at room temperature for an additional 12–24 h, and the solvent evaporated. Following dissolution in EtOAc or DCM, the solution was washed (with 2 N citric acid, sat bicarbonate and brine), dried (MgSO4), and the solvent was removed to give the crude cyclic peptide.
- 18.ESI-MS (M+Na)+ = 600.2566 (calcd 600.2587); HPLC: tR = 26.0 min (30–70% aqueous MeOH over 40 min starting after 1 min, flow rate 0.8 mL/min).
- 19.ESI-MS (M+Na)+ = 600.2345 (calcd 600.2587); HPLC: tR = 35.5 min.
- 20.Albericio F, Bofill JM, El-Faham A, Kates SA. J. Org. Chem. 1998;63:9678–9683. [Google Scholar]
- 21.Dolle RE, Michaut M, Martinez-Teipel B, Seida PR, Ajello CW, Muller AL, DeHaven RN, Carroll PJ. Bioorg. Med. Chem. Lett. 2009;19:3647–3650. doi: 10.1016/j.bmcl.2009.04.105. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.