Abstract
Chimera formation is a standard test for pluripotency of stem cells in vivo. Interspecific chimera formation between distantly related organisms offers also an attractive approach for propagating endangered species. Parameters influencing interspecies chimera formation have remained poorly elucidated. Here, we report interordinal chimera formation between medaka and zebrafish, which separated ∼320 million years ago and exhibit a more than 2-fold difference in developmental speed. We show that, on transplantation into zebrafish blastulae, both noncultivated blastomeres and long-term cultivated embryonic stem (ES) cells of medaka adopted the zebrafish developmental program and differentiated into physiologically functional cell types including pigment cells, blood cells, and cardiomyocytes. We also show that medaka ES cells express differentiation gene markers during chimeric embryogenesis. Therefore, the evolutionary distance and different embryogenesis speeds do not produce donor-host incompatibility to compromise chimera formation between medaka and zebrafish, and molecular markers are valuable for analyzing lineage commitment and cell differentiation in interspecific chimeric embryos.
Introduction
An increasing number of stem cell lines have been derived from developing embryos of mouse [1,2], human [3], and lower vertebrates including fish [4,5]. Chimera formation by cell transplantation into early developing embryo hosts represents the gold standard assay to test the pluripotency of putative stem cells from several model organisms including mouse [6–8], chicken [9], and fish [10–13]. Early embryos undergo cell proliferation, lineage commitment, cell fate decision, and differentiation. On transplantation into blastocyst/blastula embryos, blastomeres or embryonic stem (ES) cell cultures can participate in normal embryogenesis and contribute to various somatic lineages and the germline leading to the production of fertile animals in diverse vertebrate species such as mammals [14,15], birds [16], and fish [11,12,17–22]. In developmental biology, chimera formation provides direct evidence for cell-autonomous or nonautonomous control [23]. Such experiments usually use intraspecies chimera formation, because donor cells are transplanted into host embryos of the same species.
Since the report of chimera formation between sheep and goats in 1949 [24], interspecific chimera formation (ISCF) has attracted considerable attention in developmental biology [25,26]. ISCF remains the only option for testing the pluripotency of stem cells in vivo in certain cases, where host embryos are rarely available as in nonhuman primates [27] or even not allowed for chimera experiments as in humans [28,29]. Recently, ISCF has become an emerging reproductive biotechnology for animal production in conservation biology [16,22]. ISCF exploits cell transplantation between closely related species to propagate endangered organisms. In this approach, the host and donor species are chosen on their phylogenetic relationship, the closer the better, because it has been unclear whether evolutionary distance would compromise and even abolish chimera formation. There are cases where a distantly related species has to be used as the host, because of the lack of sister species. However, distantly related species often show considerable differences in embryology and developmental speed. It remains to be determined whether a difference in developmental speed prevents chimera formation.
In chimeric experiments, a donor-specific marker is required for tracing the distribution and behavior of donor cells. Pigmentation is one of the easiest visible markers and has been widely used in chimera production [11,19,20]. Genetically labeling by using fluorescent proteins green fluorescent protein (GFP) and red fluorescent protein (RFP) has become a routine tool to visualize donor cells in chimera [18,19]. The investigation in live embryos of differentiation of donor stem cells into specialized cells usually relies on the peculiar phenotypes and location of a limited number of cell types, such as pigment cells, skin epithelia, cardiomyocytes, blood cells, and primordial germ cells [11,13,18–20]. Although these visible markers and cellular phenotypes are instrumental in chimeric assays, numerous molecular markers are required for tracing multiple lineage differentiation of donor stem cells into many specialized cell types. ISCF between distantly related species will allow for the use of species-specific molecular markers to unambiguously trace the behaviors and differentiation of donor cells in various lineages and cell types.
We choose medaka and zebrafish as test organisms for ISCF. Medaka and zebrafish belong to Cypriniformes and Beloniformes, which separated ∼320 million years ago [30]. The genome has been sequenced in both fish species (www.ensembl.org/index.html). The remarkable phylogenetic distance coincides with molecular divergence in gene sequence. The twin fish models exhibit a more than 2-fold difference in developmental speed. The genetic distance and different developmental speeds make the combination of medaka and zebrafish an excellent model system to study the biology of ISCF. More importantly, sequence divergence provides species-specific molecular markers for unambiguously tracing donor cells during lineage commitment and differentiation by RNA detection.
Medaka and zebrafish are the excellent twin model organisms for analyzing vertebrate development [31] and stem cell biology [32–34]. Both species have ideal features for chimera experiments, including daily embryo supply, external embryology, and transparency for live imaging throughout embryogenesis. Chimera formation by blastula cell transplantation has been well established in both medaka [13,19] and zebrafish [11]. In medaka, we have derived diploid and haploid ES cells [4,5]. Specifically, one of the medaka diploid ES cell lines, MES1, has been characterized as being pluripotent in vitro and in vivo. In vitro, MES1 is capable of spontaneous and directed differentiation [4,35]. MES1 can activate the totipotency-specific mouse Oct4 promoter and can be maintained free of spontaneous differentiation [10]. MES1 retains the developmental pluripotency after stable gene transfer and long-term drug selection [36,37]. In vivo on transplantation into medaka blastula hosts, MES1 can proliferate and differentiate into many functional cell types that contribute to various organs [18]. Pluripotency has also proved medaka haploid ES cell lines including HX1 being capable of whole animal production via semi-cloning [5].
This work aimed at the development of ISCF between medaka and zebrafish to analyze the fate and behaviors of noncultivated blastomeres and ES cell donors in a heterologous host and to test whether lineage-specific molecular markers can be explored to study lineage restriction and differentiation of donor cells.
Materials and Methods
Fish
Work with fish followed the guidelines on the Care and Use of Animals for Scientific Purposes of the National Advisory Committee for Laboratory Animal Research in Singapore (permit number 27/09). Medaka strain HB32C and zebrafish strain AB were maintained as described [4,5,18,19,23].
Cell culture
The medaka diploid ES cell line MES1 and haploid ES cell line HX1 were maintained in ESM4 medium on gelatin-coated plasticware as described [4,5].
Cell transfection
Cell transfection with pCVpf and pCVpr was performed in 12-well plates by using the GeneJuice reagent (Novagen) as previously described [37,38]. Drug selection was performed by using puromycin at 1 μg/mL to cells 18–48 h post-transfection and terminated by medium change [37]. Pure populations of stable transgenic cells expressing GFP (HX1) and RFP (MES1) were obtained by clonal growth and expansion as described [37].
Cell transplantation
Medaka blastomeres were mechanically dissociated from embryos at the midblastula stage as described [39]. Single MES1 cells were obtained by trypsinization [19]. Blastomeres or MES1 cells were suspended in cell transplantation medium (100 mM NaCl, 5 mM KCl, 5 mM Hepes, pH 7.1). Zebrafish embryos were collected in zebrafish egg water (ZEW; 0.3‰ Instant Ocean Salt containing 2 ppm of methylene blue), treated at the sphere stage for 2 min with pronase E (1 mg/mL; Sigma) at 26°C for dechorionation, rinsed 5 times, and were arranged in a single row on V-shaped 1.5%-agarose ramps on 6-cm dishes covered with ZEW containing antibiotics at 22°C until use. Approximately 20 blastomeres or 200 ES cells were injected into the deep cell layer of each midblastula recipient [11,19]. The injected embryos were incubated in ZEW at 28°C and regularly monitored.
Microscopy
Microscopy was done as described [5,40]. Briefly, live embryos and fry were visualized using a Leica MZFLIII stereo microscope equipped with a Fluo III UV-light system and a GFP2 filter and photographed by using a Nikon E4500 digital camera (Nikon Corp.). For documentation at larger magnification, live embryos and fry were observed and photographed on a Zeiss Axiovert2 invert microscope equipped with a Zeiss AxioCam MRc digital camera and AxioVision 4 software.
Reverse transcription–polymerase chain reaction
Total RNA was isolated from cell cultures and embryos and subjected to reverse transcription–polymerase chain reaction (RT-PCR) analyses by using primers as described [5,41].
Results
Embryo–embryo transplantation
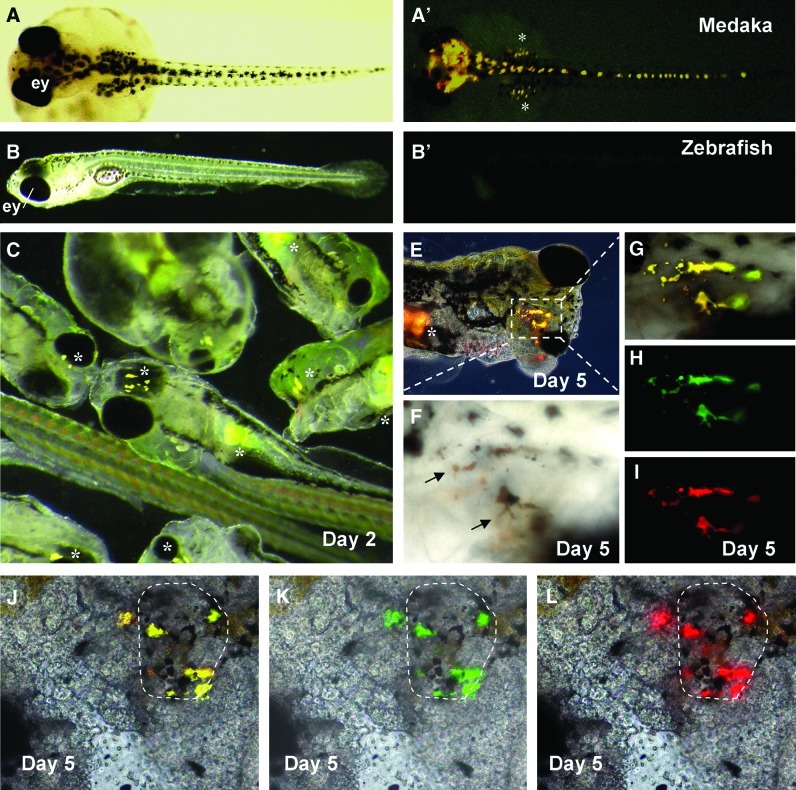
Medaka and zebrafish differ considerably in developmental speed. For example, a cleavage needs 35 and 15 min in medaka and zebrafish, respectively. Embryogenesis until hatching requires 10 days in medaka compared with only 2–3 days in zebrafish. This difference may raise a potential barrier for chimera formation between the 2 species. To test the possibility for ISCF between medaka and zebrafish, we first performed embryo-embryo chimera formation by transplanting noncultivated medaka blastomeres into zebrafish midblastula hosts. We utilized pigmentation as a marker to monitor the survival and differentiation of medaka donor blastomeres in the zebrafish host. In medaka, the pigment lineage comprises of melanin-producing black melanocytes and other autofluorescent pigment cells such as guanophores [18–20]. These pigment cells became visible after 3 days postfertilization (dpf), as illustrated in Fig. 1A and A′. The zebrafish embryo has melanocytes until 2 dpf and lacks autofluorescent pigment cells (Fig. 1B, B′).
FIG. 1.
Chimera formation between medaka blastomeres and zebrafish embryos. (A and A') Micrograph of a 3-day-old embryo of medaka strain HB32C, showing black-pigmented melanophores (A) and autofluorescent guanophores (asterisks; A'). (B and B') Micrograph of a 3-day-old zebrafish embryo, showing black pigmentation in the eye (B) and the absence of autofluorescent cells (B'). (C) Micrograph of 2-day-old chimeric embryos, showing medaka blastomere-derived autofluorescing guanophores (asterisks) in zebrafish hosts. (E–I) Micrographs of a chimeric fry at 5 days postfertilization (dpf), showing the presence of 2 clusters of guanophores in the trunk (asterisk) and dorsal head surface (frame) of the chimera (E). The medaka guanophores (arrows) are yellow and brown in color under bright field optics (F), and are positive for yellow (G), green (H), and red fluorescence (I) under fluorescent optics. (J–L) Merged micrographs of a posthatching chimeric fry at 5 dpf, showing guanophores in the eye (circle). Color images available online at www.liebertonline.com/scd
By transplanting ∼20 medaka blastomeres into each of the 82 zebrafish blastula embryo hosts, we obtained 54 survivors at 1 dpf, giving rise to a 66% survival rate, which is slightly lower than ∼80% for noninjected, dechorionated zebrafish control embryos. We found donor-derived autofluorescing pigment cells in 43 out of the 54 survivors (Fig. 1C), producing an 80% efficiency for pigmented chimera formation. Certain autofluorescent cells turned out to be guanophores, because they became brown colored on the body surface of chimeric fry at 5 dpf (Fig. 1E–I). Autofluorescent guanophores were also seen in the eye of chimeric host embryo (Fig. 1J–L). Notably, the appearance of donor-derived medaka guanophores in zebrafish embryos occurred at 2 dpf when endogenous pigmentary cells of the zebrafish became visible, which is earlier than 3 dpf required for the appearance of donor-derived medaka pigmentary cells in the medaka embryo host [18,20]. Taken together, ISCF by embryo-embryo transplantation is efficient between medaka and zebrafish, and medaka donor blastomeres appear to adopt the developmental program of the zebrafish host for pigmentary cell differentiation.
ES cell-embryo transplantation
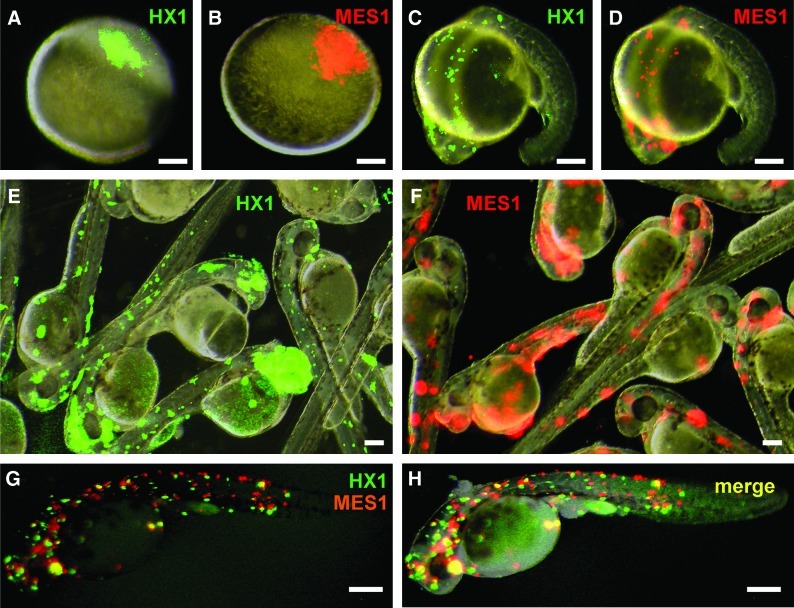
We furthered our experiments to ES cell-embryo transplantation. As the donors, we used the haploid ES cell line HX1 [5] and diploid ES cell line MES1 [4], which were labeled with GFP and RFP, respectively, and co-transplanted at a 1:1 ratio into zebrafish blastula hosts (Fig. 2A, B). In total, we transplanted 61 zebrafish blastulae and obtained 43 survivors at 1 dpf, producing a 70% survival rate. All the survivors had both HX1 and MES1 cells that were distributed widely over the surface of entire embryos at 1 dpf (Fig. 2C, D), 2 dpf (Fig. 2E, F) and 3 dpf (Fig. 2G, H). Therefore, transplanted medaka ES cells do not compromise the survival rate and development of zebrafish host embryos, and are able to participate in the chimeric embryogenesis.
FIG. 2.
Chimera formation between medaka embryonic stem (ES) cells and zebrafish embryos. (A and B) Micrograph of a zebrafish embryo host, showing transplanted medaka haploid ES cell line HX1 (green; A) and diploid ES cell line MES1 (red; B). (C and D) Micrograph of 1-day-old chimera, showing similar distribution of HX1 (C) and MES1 (D). (E and F) Micrograph of 2-day-old chimeras, showing wide distribution of HX1 (E) and MES1 (F). (G and H) Merged micrograph of a 3-day-old chimera, showing a similarly wide distribution of HX1 and MES1 donor cells. Scale bars, 100 μm. Color images available online at www.liebertonline.com/scd
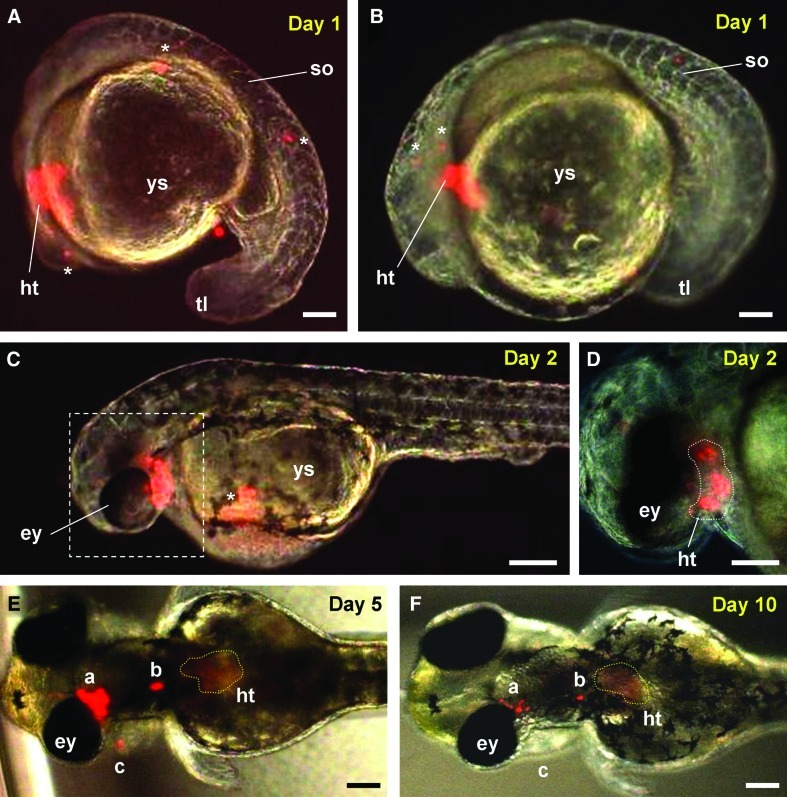
We wanted to examine the ability and time course of medaka ES cells' differentiation into a terminally differentiated cell type in the zebrafish host. To this end, we focused our attention on cardiomyocytes of the heart because of the ease with which physiologically functional cells can unambiguously be identified by rhythmic contraction. On transplantation into the zebrafish host at the blastula stage, RFP-labeled MES1 cells were found in the heart when they formed at 1 dpf (Fig. 3A, B). Evidence for differentiation into functional cardiomyocytes came from the observation that these RFP-labeled MES1 derivatives were indistinguishable from the host cardiomyocytes in cellular phenotype and rhythmic contraction of the pumping heart in chimeric embryos during different stages ranging from 1 dpf (Fig. 3A, B) over 2 dpf (Fig. 3C, D) and 5 dpf (Fig. 3E) to 10 dpf (Fig. 3F). We also observed MES1 derivatives outside the heart (Fig. 3A–F). In addition, MES1 was able to produce blood cells in circulation at 1 dpf when it commenced (data not shown). In medaka, the heart contraction and blood circulation commence until 2 dpf [36]. Therefore, medaka ES cells are able to adopt the developmental program of the zebrafish host for differentiation into functional heart cells and blood cells.
FIG. 3.
Medaka ES cell donors adopt the zebrafish developmental program. Red fluorescent protein (RFP)-labeled MES1 cells were transplanted into zebrafish blastulae and monitored for differentiation into cardiomyocytes in the heart. (A–D) Micrographs of chimeras at the lateral view at 1 dpf (A and B) and 2 dpf (C, D). (D) Larger magnification of the area framed in (C), highlighting MES1-derived cardiomyocytes in the heart. (E) Micrographs of chimeras at the dorsal view, highlighting a reduction in the relative MES1 signal in 3 areas (a, b and c) and the heart (circled) from 5 dpf (E) to 10 dpf (F). One and the same chimera at different days of development is shown in (B–F). The anterior is to the left. Asterisks depict MES1 derivatives outside the heart. ey, eye; ht, heart; so, somites; tl, tail; ys, yolk sac. Micrographs are merges between red fluorescent optics and bright-field optics. Scale bars, 100 μm. Color images available online at www.liebertonline.com/scd
Survival and persistence of medaka ES cell derivatives in zebrafish host
Due to differences in speed and time period of embryonic development, we were interested in determining the behavior of medaka ES cell derivatives in developing zebrafish embryos and larvae. We noticed that the relative MES1 contribution was essentially unchanged from 1 dpf (Fig. 3B) to 2 dpf (Fig. 3D). However, we observed a reduction in the relative signal of MES1 derivatives inside and outside the heart of chimeric larvae from 5 dpf (Fig. 3E) to 10 dpf (Fig. 3F). In addition, 1 area of MES1 derivative, area c, disappeared when development proceeded from 5 dpf (Fig. 3E) to 10 dpf (Fig. 3F). In zebrafish, hatching occurs at 2–3 dpf. Thus, 5 and 10 dpf are equivalent to 2–3 and 7–8 days posthatching, respectively. These results suggest that medaka ES cell derivatives become diluted during late embryogenesis and posthatching development.
Molecular analyses of ES cell differentiation in zebrafish host
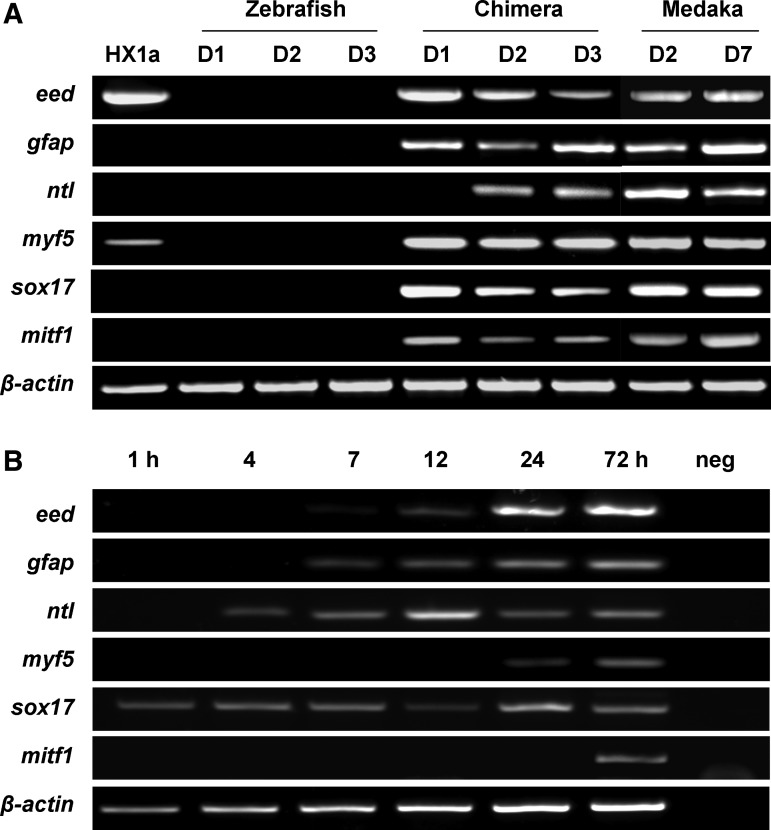
Finally, we were interested in the use of molecular markers to investigate ES cell differentiation in the heterologous host. To this end, zebrafish and medaka control embryos and chimeras produced between MES1 cells, and zebrafish hosts were subjected to an RT-PCR analysis of the expression of lineage markers by using medaka cDNA-specific primers (Fig. 4). Of 6 genes analyzed, the ectodermal marker eed and mesodermal marker myf5 are expressed at a moderate and low level in ES cells, respectively; both of them became dramatically up-regulated in chimeric embryos; whereas the 4 remaining genes, namely the ectodermal marker gfap [33], the mesodermal marker ntl (no tail or brachyury [42]), the endodermal marker sox17 [43], and the neural crest marker mitf [35], were absent in MES1 cells and zebrafish controls but easily detectable in medaka controls and, more importantly, the interspecific chimeras at 1–3 dpf (Fig. 4A).
FIG. 4.
Molecular analyses of medaka ES cell differentiation in zebrafish host. (A) After transplantation at the blastula stage with medaka haploid ES cell line HX1, zebrafish, and medaka embryos were collected at day 1–7 (D1–D7) postfertilization and subjected to reverse transcription–polymerase chain reaction (RT-PCR) analysis of gene expression profiles by using primers specific to medaka cDNAs, except for β-actin primers that amplify the β-actin cDNA of both medaka and zebrafish. The haploid ES cell line HX1 clone HX1a and nontransplanted embryos of medaka and zebrafish were used for comparisons. For β-actin, PCR was run for 28 cycles with 25 ng of cDNA. For other genes, PCR was run for 38 cycles with 25 ng of cDNA (HX1 cells, zebrafish, and medaka embryos) and for 40 cycles with 100 ng of cDNA (chimeras). (B) Medaka embryos. Time course of gene expression was examined in developing embryos at indicated intervals in hours postfertilization. neg, negative control PCR without cDNA. For β-actin, PCR was run for 28 cycles with 25 ng of cDNA. For other genes, PCR was run for 35 cycles with 25 ng of cDNA except for mitf1, which was run for 38 cycles with 50 ng of cDNA.
RT-PCR analysis revealed that the 6 genes examined did exhibit considerable differences in temporal expression patterns during medaka embryogenesis (Fig. 4B). Specifically, sox17 is maternally supplied, whereas the transcripts of ntl, gfap, eed, myf5, and mitf1 were detectable until 4, 7, 12, 24, and 72 hpf, respectively, when embryogenesis reaches the early morula (stage 8), midblastula (between stages 10 and 11), early gastrula (stage 13), late gastrula (stage 16), and somitogenesis (32 somites between stages 28 and 29). These data suggest that the onset of marker expression may far precede the actual commitment of germ layers and cell lineages and the appearance of differentiated cell types in medaka. Specifically, mitf expression is detectable at 2 dpf in medaka embryos but at early as 1 dpf in chimeric embryos, indicating that medaka ES cell donors are capable of adopting the zebrafish host program at the molecular level.
Discussion
In this study, we have established ISCF between medaka and zebrafish, the most popular lower vertebrate models for the experimental analysis of vertebrate development and stem cell biology. We show that a distant evolutionary distance of 320 million years and a salient difference in development speed do not compromise chimera formation and proper differentiation of donor cells in a heterologous host environment. Strikingly, by using pigmentary cells, blood cells, and cardiomyocytes as easily identifiable cell types in phenotype, we reveal that medaka stem cells, either noncultivated blastomeres or ES cell cultures, are capable of adopting the developmental program of the heterologous host for differentiation into physiologically functional cell types that are properly integrated into the host organ systems.
A critical parameter for ISCF is the donor-host compatibility. Factors that determine this compatibility have remained poorly understood. Chimeras have successfully been produced in several combinations of organisms, including birds of different genera [16,25,26], fish of different genera [22], and even between mouse and chicken [44]. However, ISCF in mammals appears to be more complicated. On the one hand, human ES cells can engraft into mouse blastocysts, where they proliferate and differentiate in vitro and persist in chimeric embryos that implant and develop in the uterus of pseudo-pregnant foster mice [29]. On the other hand, in interspecific chimeras similarly produced between monkey ES cells and mouse embryos, the donor ES cells engraft into mouse preimplantation embryos but not postimplantation fetuses [27]; and production of intergeneric chimeras between mice and voles did not succeed [45]. These studies suggest that the phylogenetic distance does not prevent ISCF in general. The medaka-zebrafish combination used in this study represents the most distant organisms thus far tested for ISCF. Our success in ISCF by using either medaka blastomeres or ES cells in zebrafish hosts clearly demonstrates that a great evolutionary distance does not necessarily produce donor-host incompatibility to prevent ISCF in fish. In this regard, medaka and zebrafish may provide a versatile host ideal for testing the pluripotency of putative stem cells from various fish species by ISCF. In support of this notion is the report that ES-like cells from the sea perch (Lateolabrax japonicus) are able to engraft into zebrafish embryos [46].
Several other factors may also be responsible for donor-host compatibility. A difference in cell cycle length, for instance, has been proposed to preclude primate ES cells from participation in development after implantation [27]. In this case, the development speed also differs significantly between rodents and primates: Blastocysts develop within 3.5–4 days in mice but 5–6 days in humans and a week or more in monkeys. It is likely that differences in the cell cycle affect the relative proliferation of monkey ES cells within the mouse embryo. It is also worth mentioning that mouse gestation is around 3 weeks, whereas monkeys require over a half-year. Similar differences in cell cycle, developmental speed, and duration of embryogenesis also exist between medaka and zebrafish. Therefore, our finding that medaka ES cells are able to adopt the zebrafish developmental speed demonstrates that these differences do not abolish the donor-host compatibility. In this regard, the medaka-zebrafish combination is similar to the primate-mouse situation in transplanting slowly dividing/developing cells into a fast developing host. In fact, we have previously shown that a cell cycle difference between donor cells and host embryos does not prevent intra-species chimera formation [20].
Indeed, we have observed the reduction in, and even loss of, medaka ES cell derivatives in posthatching larval chimeras. The doubling time for MES1 cells in culture is 44–48 h [4,5]. The cleavage cell cycle in the zebrafish host embryo is 15 min until the midblastula stage when cell transplantation is performed. Our finding that medaka ES cell donors can adopt the zebrafish developmental program for differentiation strongly suggests that the cell cycle difference has little influence on the differentiation of donor ES cells during early embryogenesis. Our observation that the medaka ES cell derivatives are diluted and even lost during posthatching development suggests a potential role for the cell cycle difference in the proportional donor contribution in chimeras at advanced stages of development. The cell cycle length in subsequent stages of zebrafish development remains unknown, but apparently must be much quicker than for cultured ES cells, as evidenced by a rapid increase in cell number during gastrulation and organogenesis. The fact that medaka ES cell derivatives persist throughout zebrafish embryogenesis points to the usefulness of medaka-zebrafish chimeras for analyzing differentiation of stem cells in vivo.
The chromosome ploidy level plays a role for ES cells' contribution in mammalian chimeras. In mouse aggregation chimeras, for example, tetraploid embryonic cells are confined to extraembryonic compartments, whereas diploid ES cell donors are limited to the inner cell mass [8]. Previously, we have shown that medaka haploid ES cells are indistinguishable from their diploid counterparts in growth and wide embryonic contribution in the medaka host [5]. In this study, we have also observed a similarly wide contribution for medaka haploid and diploid ES cells in the heterologous zebrafish host. Therefore, a difference in chromosome ploidy does not affect donor-host compatibility and the ability of ES cells to adopt the host developmental program for proper differentiation.
ISCF as an emerging reproductive biotechnology for animal production in conservation biology has so far been tested between closely related species, for example, between the pheasant and chicken [16] and between the rainbow trout and salmon [22]. A critical issue for this approach in ISCF between distantly related species is the ability of donor cells to persist throughout embryonic and postembryonic development. The zebrafish embryo hatches at 2–3 dpf. In this study, we have made our observation up to 2–3 days posthatching (namely 5 dpf) for embryo→embryo chimeras and up to 7–8 days posthatching (namely 10 dpf) for ES cell→embryo chimeras. We reveal the survival and functionality (eg, beating muscles in the heart) of medaka cells in zebrafish embryos and posthatched fry. Future work is needed to examine whether medaka cells can persist throughout development into adulthood.
ISCF between medaka and zebrafish provides an obvious advantage in practice. The medaka embryo has a tough chorion that is a barrier for cell transplantation. Dechorionation requires the use of a hatching enzyme preparation from a large number of hatching embryos [18–20], which represents a tedious step for chimera formation in medaka. The chorion of zebrafish embryos is thin and can easily be removed by using the commercially available pronase E, as we have demonstrated in this study. Therefore, chimera formation in zebrafish is robust. In addition, the speed of zebrafish embryogenesis is faster, allowing for observation throughout embryonic development within 3 days compared with 10 days in medaka. Our observation that medaka blastomeres and ES cells can cope with the zebrafish program of embryonic development suggests the usefulness of this ISCF system in studying the pluripotency of heterologous stem cells in zebrafish.
Although chimera formation provides a unique opportunity to identify as many different types of differentiated cells as possible and to analyze the time course of their differentiation during chimeric embryogenesis, the identification and analysis remain an elusive challenge in stem cell biology. An ideal approach will be the use of many species-specific molecular markers characteristic of different germ layers, lineages, and cell types. In this study, we show that medaka donor ES cells are indeed able to express several marker genes of germ layers and cell types. Importantly, we show that the expression of medaka mitf1 as a marker of melanocytes of the neural crest origin becomes already detectable in 1-day-old chimeras comparable to 3 dpf in medaka. Divergent sequences and proper expression of donor genes associated with differentiation make medaka and zebrafish an excellent system for studying chimera biology and analyzing cell fate decision and differentiation of pluripotent stem cells by ISCF.
Conclusion
We have demonstrated the successful interordinal chimera formation between distantly related medaka and zebrafish. We reveal that medaka blastomeres and ES cells adopt the developmental program of the host for differentiation into physiologically functional cells. Most importantly, we show that lineage restriction and cell differentiation can be easily analyzed by detecting the expression of donor-specific molecular markers. We anticipate that ISCF between medaka and zebrafish provides a useful tool for analyzing lineage commitment and cell differentiation of stem cells in vivo.
Acknowledgments
The authors thank J. Deng for fish breeding, and CM. Foong for laboratory management. This work was supported by the Sun Yat-Sen University (Grant No. 42000-3181303), the Ministry of Education of Singapore (R-154-000-388-112), the Biomedical Research Council (R154-000-427-305), and the National Research Foundation of Singapore (NRF2010NRF-CRP002-019).
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Evans MJ. Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 2.Bernemann C. Greber B. Ko K. Sterneckert J. Han DW. Arauzo-Bravo MJ. Scholer HR. Distinct developmental ground states of epiblast stem cell lines determine different pluripotency features. Stem Cells. 2011;29:1496–1503. doi: 10.1002/stem.709. [DOI] [PubMed] [Google Scholar]
- 3.Reubinoff BE. Pera MF. Fong CY. Trounson A. Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 4.Hong Y. Winkler C. Schartl M. Pluripotency and differentiation of embryonic stem cell lines from the medakafish (Oryzias latipes) Mech Dev. 1996;60:33–44. doi: 10.1016/s0925-4773(96)00596-5. [DOI] [PubMed] [Google Scholar]
- 5.Yi M. Hong N. Hong Y. Generation of medaka fish haploid embryonic stem cells. Science. 2009;326:430–433. doi: 10.1126/science.1175151. [DOI] [PubMed] [Google Scholar]
- 6.Dewey MJ. Martin DW., Jr. Martin GR. Mintz B. Mosaic mice with teratocarcinoma-derived mutant cells deficient in hypoxanthine phosphoribosyltransferase. Proc Natl Acad Sci U S A. 1977;74:5564–5568. doi: 10.1073/pnas.74.12.5564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradley A. Evans M. Kaufman MH. Robertson E. Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 8.Nagy A. Gocza E. Diaz EM. Prideaux VR. Ivanyi E. Markkula M. Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 9.van de Lavoir MC. Mather-Love C. Leighton P. Diamond JH. Heyer BS. Roberts R. Zhu L. Winters-Digiacinto P. Kerchner A, et al. High-grade transgenic somatic chimeras from chicken embryonic stem cells. Mech Dev. 2006;123:31–41. doi: 10.1016/j.mod.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 10.Hong Y. Winkler C. Liu T. Chai G. Schartl M. Activation of the mouse Oct4 promoter in medaka embryonic stem cells and its use for ablation of spontaneous differentiation. Mech Dev. 2004;121:933–943. doi: 10.1016/j.mod.2004.03.028. [DOI] [PubMed] [Google Scholar]
- 11.Lin S. Long W. Chen J. Hopkins N. Production of germ-line chimeras in zebrafish by cell transplants from genetically pigmented to albino embryos. Proc Natl Acad Sci U S A. 1992;89:4519–4523. doi: 10.1073/pnas.89.10.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma C. Fan L. Ganassin R. Bols N. Collodi P. Production of zebrafish germ-line chimeras from embryo cell cultures. Proc Natl Acad Sci U S A. 2001;98:2461–2466. doi: 10.1073/pnas.041449398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wakamatsu Y. Ozato K. Hashimoto H. Kinoshita M. Sakaguchi M. Iwamatsu T. Hyodo-Taguchi Y. Tomita H. Generation of germ-line chimeras in medaka (Oryzias latipes) Mol Marine Biol Biotechnol. 1993;2:325–332. [Google Scholar]
- 14.Tanaka M. Hadjantonakis AK. Vintersten K. Nagy A. Aggregation chimeras: combining ES cells, diploid, and tetraploid embryos. Methods Mol Biol. 2009;530:287–309. doi: 10.1007/978-1-59745-471-1_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wood SA. Allen ND. Rossant J. Auerbach A. Nagy A. Non-injection methods for the production of embryonic stem cell-embryo chimaeras. Nature. 1993;365:87–89. doi: 10.1038/365087a0. [DOI] [PubMed] [Google Scholar]
- 16.Kang SJ. Choi JW. Kim SY. Park KJ. Kim TM. Lee YM. Kim H. Lim JM. Han JY. Reproduction of wild birds via interspecies germ cell transplantation. Biol Reprod. 2008;79:931–937. doi: 10.1095/biolreprod.108.069989. [DOI] [PubMed] [Google Scholar]
- 17.Wong TT. Saito T. Crodian J. Collodi P. Zebrafish germline chimeras produced by transplantation of ovarian germ cells into sterile host larvae. Biol Reprod. 2011;84:1190–1197. doi: 10.1095/biolreprod.110.088427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hong N. Li M. Zeng Z. Yi M. Deng J. Gui J. Winkler C. Schartl M. Hong Y. Accessibility of host cell lineages to medaka stem cells depends on genetic background and irradiation of recipient embryos. Cell Mol Life Sci. 2010;67:1189–1202. doi: 10.1007/s00018-009-0247-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hong Y. Winkler C. Schartl M. Production of medakafish chimeras from a stable embryonic stem cell line. Proc Natl Acad Sci U S A. 1998;95:3679–3684. doi: 10.1073/pnas.95.7.3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong Y. Winkler C. Schartl M. Efficiency of cell culture derivation from blastula embryos and of chimera formation in the medaka (Oryzias latipes) depends on donor genotype and passage number. Dev Genes and Evolution. 1998;208:595–602. doi: 10.1007/s004270050220. [DOI] [PubMed] [Google Scholar]
- 21.Joly JS. Kress C. Vandeputte M. Bourrat F. Chourrout D. Irradiation of fish embryos prior to blastomere transfer boosts the colonisation of their gonads by donor-derived gametes. Mol Reprod Dev. 1999;53:394–397. doi: 10.1002/(SICI)1098-2795(199908)53:4<394::AID-MRD4>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 22.Takeuchi Y. Yoshizaki G. Takeuchi T. Biotechnology: surrogate broodstock produces salmonids. Nature. 2004;430:629–630. doi: 10.1038/430629a. [DOI] [PubMed] [Google Scholar]
- 23.Li M. Hong N. Xu H. Yi M. Li C. Gui J. Hong Y. Medaka vasa is required for migration but not survival of primordial germ cells. Mech Dev. 2009;126:366–381. doi: 10.1016/j.mod.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 24.Warwick BL. Berry RO. Inter-generic and intra-specific embryo transfers in sheep and goats. J Hered. 1949;40:297–303. doi: 10.1093/oxfordjournals.jhered.a105963. illustration. [DOI] [PubMed] [Google Scholar]
- 25.Le Douarin NM. Developmental patterning deciphered in avian chimeras. Dev Growth Differentiation. 2008;50(Suppl. 1):S11–S28. doi: 10.1111/j.1440-169X.2008.00989.x. [DOI] [PubMed] [Google Scholar]
- 26.Le Lievre CS. Le Douarin NM. Mesenchymal derivatives of the neural crest: analysis of chimaeric quail and chick embryos. J Embryol Exp Morphol. 1975;34:125–154. [PubMed] [Google Scholar]
- 27.Simerly C. McFarland D. Castro C. Lin CC. Redinger C. Jacoby E. Mich-Basso J. Orwig K. Mills P. Ahrens E. Navara C. Schatten G. Interspecies chimera between primate embryonic stem cells and mouse embryos: monkey ESCs engraft into mouse embryos, but not post-implantation fetuses. Stem Cell Res. 2011;7:28–40. doi: 10.1016/j.scr.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glover JC. Boulland JL. Halasi G. Kasumacic N. Chimeric animal models in human stem cell biology. ILAR J. 2009;51:62–73. doi: 10.1093/ilar.51.1.62. [DOI] [PubMed] [Google Scholar]
- 29.James D. Noggle SA. Swigut T. Brivanlou AH. Contribution of human embryonic stem cells to mouse blastocysts. Dev Biol. 2006;295:90–102. doi: 10.1016/j.ydbio.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 30.Yamanoue Y. Miya M. Inoue JG. Matsuura K. Nishida M. The mitochondrial genome of spotted green pufferfish Tetraodon nigroviridis (Teleostei: Tetraodontiformes) and divergence time estimation among model organisms in fishes. Genes Genet Syst. 2006;81:29–39. doi: 10.1266/ggs.81.29. [DOI] [PubMed] [Google Scholar]
- 31.Furutani-Seiki M. Wittbrodt J. Medaka and zebrafish, an evolutionary twin study. Mech Dev. 2004;121:629–637. doi: 10.1016/j.mod.2004.05.010. [DOI] [PubMed] [Google Scholar]
- 32.Hong N. Li Z. Hong Y. Fish stem cell cultures. Int J Biol Sci. 2011;7:392–402. doi: 10.7150/ijbs.7.392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Alunni A. Hermel JM. Heuze A. Bourrat F. Jamen F. Joly JS. Evidence for neural stem cells in the medaka optic tectum proliferation zones. Dev Neurobiol. 2010;70:693–713. doi: 10.1002/dneu.20799. [DOI] [PubMed] [Google Scholar]
- 34.Yi M. Hong N. Li Z. Yan Y. Wang D. Zhao H. Hong Y. Medaka fish stem cells and their applications. Sci China Life Sci. 2010;53:426–434. doi: 10.1007/s11427-010-0079-3. [DOI] [PubMed] [Google Scholar]
- 35.Bejar J. Hong Y. Schartl M. Mitf expression is sufficient to direct differentiation of medaka blastula derived stem cells to melanocytes. Development. 2003;130:6545–6553. doi: 10.1242/dev.00872. [DOI] [PubMed] [Google Scholar]
- 36.Iwamatsu T. Stages of normal development in the medaka Oryzias latipes. Mech Dev. 2004;121:605–618. doi: 10.1016/j.mod.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 37.Hong Y. Chen S. Gui J. Schartl M. Retention of the developmental pluripotency in medaka embryonic stem cells after gene transfer and long-term drug selection for gene targeting in fish. Transgenic Res. 2004;13:41–50. doi: 10.1023/b:trag.0000017172.71391.fa. [DOI] [PubMed] [Google Scholar]
- 38.Hong Y. Liu T. Zhao H. Xu H. Wang W. Liu R. Chen T. Deng J. Gui J. Establishment of a normal medakafish spermatogonial cell line capable of sperm production in vitro. Proc Natl Acad Sci U S A. 2004;101:8011–8016. doi: 10.1073/pnas.0308668101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong Y. Schartl M. Isolation and differentiation of medaka embryonic stem cells. Methods Mol Biol. 2006;329:3–16. doi: 10.1385/1-59745-037-5:3. [DOI] [PubMed] [Google Scholar]
- 40.Xu H. Li Z. Li M. Wang L. Hong Y. Boule is present in fish and bisexually expressed in adult and embryonic germ cells of medaka. PLoS One. 2009;4:e6097. doi: 10.1371/journal.pone.0006097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang D. Manali D. Wang T. Bhat N. Hong N. Li Z. Wang L. Yan Y. Liu R. Hong Y. Identification of pluripotency genes in the fish medaka. Int J Biol Sci. 2011;7:440–451. doi: 10.7150/ijbs.7.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Araki K. Okamoto H. Graveson AC. Nakayama I. Nagoya H. Analysis of haploid development based on expression patterns of developmental genes in the medaka Oryzias latipes. Dev Growth Differentiation. 2001;43:591–599. doi: 10.1046/j.1440-169x.2001.00601.x. [DOI] [PubMed] [Google Scholar]
- 43.Kobayashi D. Jindo T. Naruse K. Takeda H. Development of the endoderm and gut in medaka, Oryzias latipes. Dev Growth Differentiation. 2006;48:283–295. doi: 10.1111/j.1440-169X.2006.00870.x. [DOI] [PubMed] [Google Scholar]
- 44.Fontaine-Perus J. Cheraud Y. Mouse-chick neural chimeras. Int J Dev Biol. 2005;49:349–353. doi: 10.1387/ijdb.041943jf. [DOI] [PubMed] [Google Scholar]
- 45.Mystkowska ET. Development of mouse-bank vole interspecific chimaeric embryos. J Embryol Exp Morphol. 1975;33:731–744. [PubMed] [Google Scholar]
- 46.Chen SL. Sha ZX. Ye HQ. Liu Y. Tian YS. Hong Y. Tang QS. Pluripotency and chimera competence of an embryonic stem cell line from the sea perch (Lateolabrax japonicus) Mar Biotechnol. 2007;9:82–91. doi: 10.1007/s10126-006-6050-1. [DOI] [PubMed] [Google Scholar]