Abstract
Direct reprogramming of somatic cells to induced pluripotent stem cells (iPSCs) provides an invaluable resource for regenerative medicine. Because of some ethical and logistical barriers, human iPSCs cannot be used to generate a chimera, which is one of markers representing pluripotency. As the most attractive model for preclinical studies, pigs offer another path to improve clinical medicine. In this study, porcine adult stem cells (pASCs), including adipose mesenchymal stem cells (AMSCs) and bone marrow mesenchymal stem cells (BMSCs), were collected and cultured under the same conditions in vitro. Real-time PCR, immunocytochemical staining, apoptosis analysis, and induced differentiation and reprogramming techniques were used to investigate the proliferative capacity and pluripotent characteristics of pASCs. Our results showed that both AMSCs and BMSCs displayed a similar immunophenotype, and their proliferative capacity appeared as a downward trend as the cell passage number increased. The cell proliferative capacity of AMSCs was significantly lower than that of BMSCs (p<0.05). Moreover, each type of pASCs went through 20 passages without undergoing alterations in the expression of reprogramming transcriptional factors (Oct4, Sox2, c-Myc, and Nanog). All pASCs had adipogenic and osteogenic differentiation potential. In addition, they also could be reprogrammed to pig induced pluripotent stem cells (piPSCs) with similar time and efficiency. In conclusion, porcine BMSCs had a higher proliferative capacity than AMSCs, and the pluripotency of pASCs was stable in long-term culture.
Introduction
Stem cells mainly include embryonic stem cells (ESCs) and adult stem cells (ASCs). Because stem cells are derived from different sources, they possess distinct characteristics. ESCs are totipotent cells that are derived from blastulas, whereas ASCs are pluripotent and are derived from adult tissues. ASCs are found in many tissues, including the adult central nervous system, epidermis, intestine, bone marrow, liver, and adipose tissue (Gnecchi and Melo, 2009; Zuk et al., 2001). An important property of ASCs is that they generate various types of cells, which may enhance the development of regenerative medicine (Deans and Moseley, 2000). In 2006, the generation of induced pluripotent stem cells (iPSCs) provided a unique tool for dissecting the molecular events that permit the conversion of one cell type to another. The initial conversion of iPSCs was first achieved by ectopic expression of a select group of transcription factors to directly reprogram somatic cells to induced pluripotency (Takahashi and Yamanaka, 2006). Since the success of fibroblast reprogramming, a multitude types of cells, including stomach (Aoi et al., 2008), liver (Stadtfeld et al., 2008), neural progenitor cells (Eminli et al., 2008), and some ASCs, including mouse adult neural stem cells (Kim et al., 2009b) and porcine primary bone marrow cells (BMCs) (Wu et al., 2009), had been reprogrammed.
It is well known that porcine physiology is remarkably similar to that of humans, and porcine skin has been used for human transplantation and also as a biomedical model for skin replacement therapy, including engineered skin substitutes (Kim et al., 2009a). In light of their compatibility with human physiology, pigs are the most attractive animal model for preclinical studies. Recently, it was reported that adult human adipose stem cells could be reprogrammed to iPSCs (Sun et al., 2009). However, the same research was not done on porcine adipose mesenchymal stem cells (AMSCs). Therefore, this study was conducted to investigate the difference between porcine AMSCs and bone marrow mesenchymal stem cells (BMSCs) in the context of cell proliferative capacity and pluripotent characteristics to determine whether porcine AMSCs could be induced to iPSCs or not. We also investigated whether or not the two types of porcine (p) ASCs would retain their pluripotency with long-term culture in vitro.
Materials and Methods
Ethics Statement
All animal studies were conducted according to the experimental practices and standards approved by the Animal Welfare and Research Ethics Committee at Jilin University (Approval ID: 20101008-2).
Chemicals
Chemicals and media were purchased from Sigma (St. Louis, MO, USA) unless otherwise stated.
Isolation and culture of pASCs
Porcine BMSCs were isolated from the bone marrow of a 1-week-old female pig as previously described (Fang et al., 2003). Subcutaneous adipose tissue was obtained from the same animal, and cells were isolated from adipose tissue as previously described. In brief, adipose tissue was harvested and enzymatically dissociated using 1.0 mg/mL collagenase type I. Digestion was carried out under continuous agitation for 90 min at 37°C, and was terminated by Dulbecco's modified Eagle's medium (DMEM) with 10% fetal bovine serum (FBS) and followed by a 10-min centrifugation (1000×g). The supernatant was removed, and the sediment was resuspended in complete medium (consisting of DMEM with 10% FBS and 1% penicillin-streptomycin). The cells were then recovered and plated on to 10-cm culture plates; these initial plates represented the first passage (P0). Cells were washed with phosphate-buffered saline (PBS) at 24-h intervals to remove contaminating erythrocytes and other unattached cells. Cells were maintained at 37°C with 5% CO2 in tissue culture dishes or flasks, and medium was changed twice each week. Cells reached confluence within 10–14 days of P0. Once 80% confluence was reached, the adherent cells were detached with 0.25% trypsin, replated at 1×104 cells/cm2, and passaged every 3–5 days until ready for analysis. In this study, pASCs were collected at P0, P5, P10, P15, and P20 to obtain samples for testing.
Reprogramming transcriptional factors analysis by quantitative RT-PCR
Total RNA was extracted from cells using Invitrogen Trizol reagent according to the manufacturer's instructions. Total RNA (1 μg) from each sample was reverse-transcribed using the BioRT cDNA First Synthesis kit (BioRT, China) following the manufacturer's instructions with a downscaled reaction volume of 20 μL. To determine the expression levels of Oct4, Sox2, Klf4, c-Myc, and Nanog in different passages of pASCs, total RNA extracted from each passage of pASCs was subjected to quantitative real-time PCR analysis. Single-stranded cDNA was synthesized from total RNA by using the Revertaid™ First Strand cDNA Synthesis kit and diluted 10 times. Filtered distilled water was used as the negative control. Template without reverse transcriptase was used to check for contaminating genomic DNA. All pairs of gene-specific primers were designed to amplify Oct4, Nanog, Sox2, Klf4, and c-Myc transcript products of 157 bp, 613 bp, 85 bp, 119 bp, and 184 bp, respectively. The PCR products were sequenced to verify the specificity of the PCR primers. Primers for glyceraldehyde 3-phosphate dehydrogenase (GAPDH) were used to amplify a 272-bp fragment as a reference. A SYBR Green RT-PCR assay was conducted to determine Oct4, Nanog, Sox2, Klf4, c-Myc, and GAPDH mRNA expression. The PCR temperature profile and reaction conditions were specified by the manufacturer of the SYBR Premix Ex TaqTM (TaKaRa, China) on an ABI StepTwo Real-Time PCR system (Applied Biosystems 7500, USA). The reaction was performed at 95°C for 30 sec followed by 40 cycles of 95°C for 5 sec, 57°C, 59°C, 59°C, 59°C, 55.1° C, and 60.3°C (for Oct4, Nanog, Sox2, Klf4, c-Myc, and GAPDH, respectively) for 31 sec to obtain the dissociation curves. The standard curves tested derived from a series of diluted cDNA (10-fold serial dilution with 10 levels: 1010×, 109×, 108×, 107×, 106×, 105×, 104×, 103×, 102×, 10×, for Oct4, Nanog, Sox2, Klf4, c-Myc, and GAPDH). The R2 values of the standard curves were 0.99954, 0.999096, 0.999124, 0.999656, 0.999434, and 0.999708, respectively, and the dissociation curves showed a single peak. The relative ratios of Oct4, Nanog, Sox2, Klf4, and c-Myc to GAPDH mRNA were calculated using the standard curves to confirm the reliability of the real-time PCR data. Primers are listed in Table 1.
Table 1.
All Pairs of Pluripotency-Specific Marker Gene and GAPDH-Specific Primers
| Primer names | Primer (5′ to 3′) | Annealing temperature (°C)×cycle number | Product size (bp) |
|---|---|---|---|
| Oct4 | FP: GTCGCCAGAAGGGCAAAC RP: CAGGGTGGTGAAGTGAGGG |
57×40 | 157 |
| Sox2 | FP: CCCTGCAGTACAACTCCATGAC RP: GGTGCCCTGCTGCGAGTA |
59×40 | 85 |
| Klf4 | FP: CGGCAAAACCTACACGAAGAGT RP: AGTTCATCTGAGCGGGCAAAT |
59×40 | 119 |
| Nanog | FP: CTTATTCAGGACAGCCCTGATTCTTC RP: AAGACGGCCTCCAAATCACTG |
59×40 | 613 |
| c-Myc | FP: GGATTCCGCCTCGTT RP: TCTCCAAGCATCACTCG |
55.1×40 | 184 |
| GAPDH | FP: ACCTGCCGCCTGGAGAAACC RP: GACCATGAGGTCCACCACCCTG |
60.3×40 | 272 |
GAPDH, glyceraldehyde 3-phosphate dehydrogenase.
Immunocytochemical staining of the reprogramming transcriptional factors
For the analysis of reprogramming transcriptional factors, the pASCs were cultured in a 24-well tissue culture plate for 2 days with daily changes of fresh culture medium. Transcription factors of pASCs were analyzed using rabbit polyclonal immunoglobulin G (IgG) of Oct4, Nanog (Abcam, USA), Sox2, Klf4, and c-Myc antibodies (Sigma, USA). After removal of the medium and two rinses with PBS, cells were fixed with 4% paraformaldehyde for 20 min, followed by three 5-min rinses with 2 mL of Dulbecco's PBS (D-PBS). Cells were permeabilized with PBS-Tween (PBST) solution at 4°C for 10 min, rinsed with PBS for 5 min at room temperature, and then blocked for nonspecific interactions with PBS-BSA (PBS-B) solution at 37°C for 30 min. Samples were then incubated at 4°C with Oct4 (1:100, Abcam, ab18976), Nanog (1:100, Abcam, ab80892), Sox2 (1:100 AV38232), Klf4 (1:100 AV35721), and c-Myc (1:100 A7470) antibodies for overnight. After removal of the primary antibody solution, cells were washed with PBS three times, for 5 min each time, and then incubated with goat anti-rabbit immunofluorescent secondary antibody (1:100, Millipore, 12-507 and AP307R) at room temperature for 1 h. After washing cells with D-PBS three times, for 5 min each time, the cells were counterstained with 1 μg/mL 4′,6-diamidino-2-phenylindole (DAPI) solution for 10 min and observed with a confocal microscope (Olympus, Japan). The green and red color staining indicated the positive reaction of transcript factors and the blue color indicated the nucleus.
Survival rate of pASCs by Trypan Blue exclusion test
The passaged cells were detached with 0.25% trypsin, and then stained with a 1:1 dilution of 0.4% Trypan Blue solution. A drop (20 μL) of solution containing stained pASCs was transferred into a hemocytometer, covered with cover glass, and observed by light microscopy to examine the cellular content. Cellular viability was determined by the presence or absence of blue color. The survival rate of pASCs was calculated using the formula: number of blue positive cells/total cell number×100.
Colony-forming unit fibroblast assay of pASCs
The cells were seeded in each well of a six-well tissue culture plate at a concentration of 5×105 and cultured for 2 weeks with regular change of a fresh culture medium at 3-day intervals. After removing the culture medium on the 14th day, the wells of the tissue culture plate were air dried and fixed with pure methanol for 5 min. The cells in the tissue culture plate wells were stained with 4% Giemsa solution for 5 min and washed with distilled water to remove the excess stain. The plates were then observed under a light microscope for colony-forming unit fibroblast (CFU-F) colonies ranging from 1 mm to 8 mm in diameter. This assay was conducted with six replicates.
Cell apoptosis determination by flow cytometry
The cells were detached with 0.25% trypsin. Suspended cells were collected by centrifugation (2000×g) for 5 min, and the supernatant was aspirated (treating the adherent cells with the trypsin too long could cause a false positive). The cells were washed twice with cold PBS, and collected by centrifugation (2000×g) for 5 min. Cell sediment was suspended with 400 μL of Annexin V solution, and the cell numbers were controlled at approximately 1×106 cells/mL. Then 5 μL of Annexin V-fluorescein isothiocyanate (FITC) staining solution was added to the suspensions, followed by incubation in the dark at 4°C for 15 min. Ten microliters of propidium iodide (PI) staining solution was added to the suspensions, which were then incubated in the dark at 4°C for 5 min. Cells were analyzed by flow cytometry within 1 h.
Analysis of apoptosis-related genes
Following the extraction of total RNA from pASCs using Invitrogen TRIzol reagent (Invitrogen, USA), cDNAs were reverse-transcribed using the BioRT cDNA First Synthesis kit according to the manufacturer's instructions. The specific primers used in this study are listed in Table 2. PCR amplification was carried out in an automated thermal cycler. Maxime PCR Premix Kit in 25 μL was used with each cycle, consisting of: an initial denaturation step at 94°C for 2 min, followed by 35 cycles at 94°C for 30 s, 55°C and 59°C for 30 s; an elongation step at 72°C for 90 s; and a 10-min final extension at 72°C. The PCR product was mixed with gel loading dye (Bromophenol Blue and xylene cyanol) in a 5:1 ratio and loaded on to 1.5 % agarose gel containing 1 μg/mL ethidium bromide and electrophoresed at 120 V for 25 min. Gel images were analyzed using band scan 5.0 Software.
Table 2.
Apoptosis Relative Gene Primers
| Primer names | Primer (5 ‘to 3’) | Annealing temperature (°C)×cycle number | Product size (bp) |
|---|---|---|---|
| Bax | FP: TCTACCAAGAAGTTGAGCGAGTGT RP: CATCCTCTGCAGCTCCATGTTA |
55×35 | 80 |
| Bcl2 | FP: GAAACCCCTAGTGCCATCAA RP: GGGACGTCAGGTCACTGAAT |
59×35 | 86 |
Differentiation of pASCs
pASCs were evaluated for developmental potential using in vitro assays described previously (Zuk et al., 2002; Stenderup et al., 2003). Briefly, adipogenic differentiation was induced by incubating cells on P0, P10, and P20 in low-glucose DMEM including 10% FBS, 1 mM dexamethasone, 500 mM isobutylmethylxanthine, 200 mM indomethacin, and 10 mM insulin for 21 days. Osteogenic differentiation was accomplished by incubating cells in low-glucose DMEM including 10% FBS supplement, 0.1 mM dexamethasone, 10 mM β-glycerophosphate, and 50 mg/mL ascorbic acid for 21 days. Differentiation along the adipogenic and osteogenic lineages was assessed qualitatively based on cell morphology and cytochemistry (Oil Red O and Alizarin Bordeaux stain, respectively), as previously described (Zuk et al., 2002).
Retroviral transduction
pMX plasmids containing mouse Sox2, Klf4, Oct4, and c-Myc have been described previously (Takahashi and Yamanaka, 2006). HEK293T cells were transfected with Lipofectamine 2000 (Invitrogen) following the instructions of the manufacturer. Two rounds (24 h each) of supernatants were collected after stopping the transfection, filtered (0.45-μm pore size), and added onto sparse porcine embryo fibroblasts (PEFs), AMSCs, and BMSCs split the day before. Polybrene (8 μg/mL) was added to increase infection efficiency.
Analysis of piPSCs by alkaline phosphatase staining
Alkaline phosphatase (AP) staining was performed as follows. piPSCs were cultured at low to medium density for 5 days prior to analyzing AP activity. (Note: This time period is critical if the activity level of AP needs to be observed. According to our finding, 5 days of culture are optimal for good AP stain visualization). On day 5, medium was aspirated and piPSCs were fixed with a fixative (e.g., 4% paraformaldehyde in PBS) for 1–2 min. (Note: Do not overfix. Fixing cells longer than 2 min will result in the inactivation of alkaline phosphatase). Fixative was aspirated and cells were rinsed with Rinse Buffer. (Note: Do not allow the wells to dry). Reagents were prepared for AP staining as described above. Enough stain solution was added to cover each well (e.g., 0.5 mL for each well of a 24-well plate). Cells were incubated in the dark at room temperature for 15 min. Staining solution was aspirated and the wells were rinsed with rinse buffer. Cells were covered with PBS to prevent drying, and the number of colonies expressing AP (red stem cell colonies) versus the number of differentiated colonies (colorless) was counted. AP staining criteria were: greater than 90% of colonies should remain undifferentiated and express AP in the well containing 103 units of leukemia inhibitory factor (lif). The p value was ≥0.05.
Analysis of piPSCs by immunostaining
Immunostaining was carried out similarly as previously described (Xiao et al., 2006). The primary antibodies used were anti-Nanog (1:100, Abcam, ab80892), anti-SSEA4 (1:100, Abcam, ab16278), anti-Rex1 (1:100, Abcam, ab50828), and anti-Oct4 (1:100, Abcam, ab18976).
Analysis of exogenous transgenes and endogenous genes by RT-PCR
Total RNA of piPSCs was extracted as previously described. Exogenous transgenes and endogenous genes were detected by PCR. Exogenous transgenes primers are listed in Table 3. The endogenous gene primers were given in Table 1.
Table 3.
Exogenous Transgene Primers
| Primer names | Primer (5′ to 3′) | Annealing temperature (°C)×cycle number |
|---|---|---|
| pMX vector | FP: GCCGACACCAGACTAAGAACCTAGAACCTC | |
| ExoSox2 | RP:GCTTCAGCTCCGTCTCCATCATGTTATACAT | 65×35 |
| ExoOct4 | RP:AGTATGCCATCCCTCCGCAGAACTCGTATG | 66×35 |
| ExoKlf4 | RP:AGGATAAAGTCTAGGTCCAGGAGGTCGTTG | 66×35 |
| Exoc-Myc | RP:AGTCGTAGTCGAGGTCATAGTTCCTGTTGG | 60×35 |
In vitro differentiation of piPSCs
Cells were treated by 0.25% trypsinization and transferred to bacterial culture dishes in ES0 medium [knockout DMEM with 15% knockout serum replacement (KSR), Invitrogen]. After 3 days, embryoid bodies (EBs) were generated. Total RNA of the EBs was extracted as previously described. Germ layer–relevant genes were detected by PCR. The primers are listed in Table 4.
Table 4.
Germ Layer–Relevant Gene Primers
| Primer names | Primer (5′ to 3′) | Annealing temperature (°C)×cycle number | Product size (bp) |
|---|---|---|---|
| GFAP | FP: CTCGCCGCTCCTATGTCT RP: CGCCTTGTTCTGCTGCTC |
57×35 | 248 |
| AFP | FP: TATTGGAGAAATGTTCGCAGTC RP: CCAGGGTTTATGGGCATC |
57×35 | 349 |
| BMP4 | FP: ATTATGCCAAGTCCTGCTA RP: GTGGGTCTGCTCTTCCTC |
51×35 | 268 |
Statistical analysis
Replicate trials were conducted for each treatment, and the mean percentage±standard error of the mean (SEM)] was calculated for each experimental group. Data were analyzed by one-way analysis of variance (ANOVA) using SPSS software (Statistics Production for Service Solution, Version 12.0, USA) after being transformed via least significant difference (LSD). The difference was considered significant when p<0.05.
Results
Analysis of reprogramming transcriptional factors Oct4, Nanog, Sox2, Klf4, and c-Myc by qRT-PCR
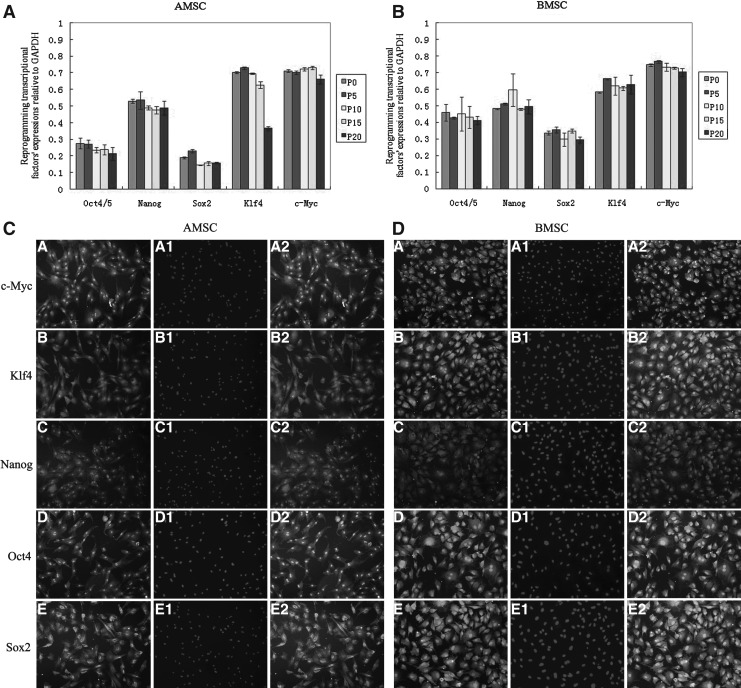
Porcine BMSCs and AMSCs were cultured in vitro for 20 passages, and samples were collected at P0, P5, P10, P15, and P20. The results of qRT-PCR analysis determined that as the cells went through several passages both pASCs showed that the reprogramming transcriptional factors content became lower, albeit not significantly. Compared with AMSCs, the expression of Oct4 and Sox2 in all the BMSCs passages was higher, but Klf4 was expressed at lower levels in BMSCs than in AMSCs (Fig. 1A and B).
FIG. 1.
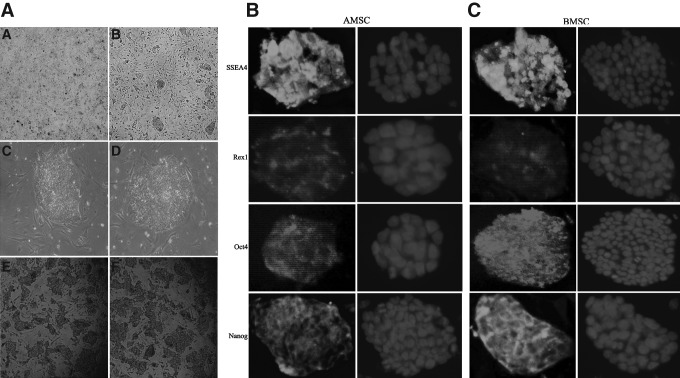
(A and B) The expression of reprogramming transcriptional factors. Quantitative analysis was performed on reprogramming transcriptional factors expression relative to GAPDH in the following cell passages: P0, P5, P10, P15, and P20 of AMSCs and BMSCs. Expression data for each passage were analyzed three times. Vertical bars represent the mean±standard error. (C) Morphological observations and expression of reprogramming transcriptional factors in AMSCs. A–E represent c-Myc, Klf4, Nanog, Oct4, and Sox-2, respectively; A1–E1 are stained with DAPI; A2–E2 are confocal pictures. (D) Morphological observations and expression of reprogramming transcriptional factors in BMSCs. A–E represent c-Myc, Klf4, Nanog, Oct4, and Sox-2, respectively; A1–E1 are stained with DAPI; A2–E2 are confocal pictures. (The fluorescence intensity of coal maceral did not represent protein level).
Morphological observation and expression of reprogramming transcriptional factors
The morphological appearance of pASCs attaching to the culture dish was similar to that of fibroblasts. The reprogramming transcriptional factors Oct4, Nanog, Sox2, Klf4, and c-Myc were expressed in both the nuclei and cytoplasm of individual cells (Fig. 1C and D).
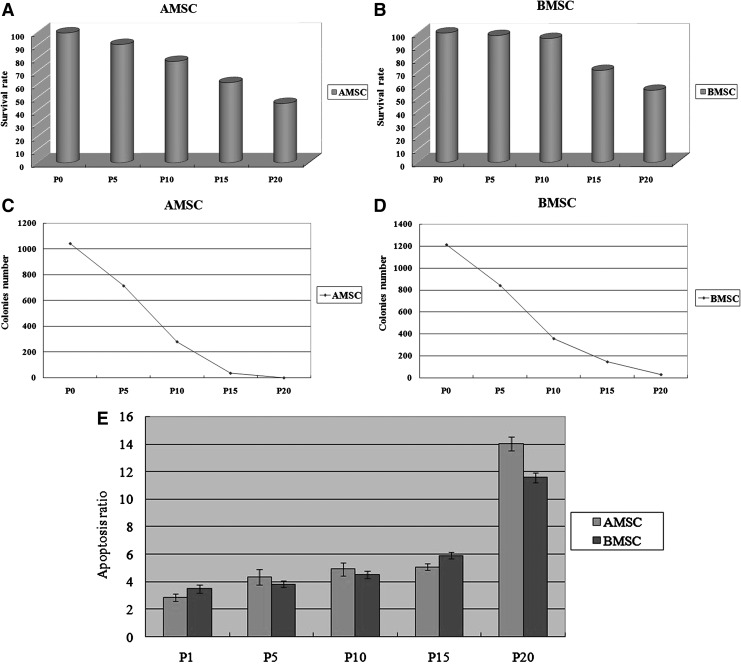
Survival rate of pASCs
The percentage of survival of pASCs in long-term culture is shown in Figure 2A and B. The survival rates of AMSCs at P0, P5, P10, P15, and P20 were 99.5±0.3%, 90.7±1.1%, 77.7±1.5%, 61.5±0.6%, and 45.5±2.4%, respectively. The survival rates of BMSCs at P0, P5, P10, P15, and P20 were 100%, 98.1±0.5%, 85.8±0.7%, 71.1±1.3%, and 55.7±1.9%, respectively. No obvious decreases were observed in P0 and P5 for two types of cells, whereas the survival rate was significantly lower in P15 and P20 groups (p<0.05).
FIG. 2.
(A and B) Survival rate of pASCs. No obvious decreases were observed in P0 and P5 for two types of cells, whereas the survival rate was significantly lower in P15 and P20 groups (p<0.05). (C and D) CFU-F assay result of pASCs. There was a significant decline in the numbers of colonies in higher passages of both cell types (p<0.05), and significant difference was observed between the two pASCs. (E) The cell apoptosis determination result of pASCs. Before P10 of pASCs, apoptosis was not significant, but after P10, the results were significantly higher (p<0.05). And overall, the percentage of apoptotic cells in AMSCs tended to be higher than BMSCs.
CFU-F assay of pASCs
The results of the CFU-F assay with long-term culture of pASCs in vitro are shown in Figure 2C and D. The number of colonies ranging from 1 mm to 8 mm in diameter was observed in all groups of pASCs. The numbers of BMSCs colonies at P0, P5, P10, P15, and P20 were 1210±42.1, 838±20.2, 355±16.1, 143±15.6, and 27.5±3.5, respectively. For AMSCs, the numbers of colonies at each passage were 1041±19.3, 713±22.1, 280±16.7, 36±2.9, and 0, respectively. There was a significant decline in the numbers of colonies in higher passages of both cell types (p<0.05), and significant difference was observed between the two pASCs.
Cell apoptosis determination
To determine the growth kinetics of pASCs, cell apoptosis levels were evaluated from P1 to P20. Percentages of apoptosis cells were increased in pASCs as they were passaged. Overall, the percentage of apoptosis cells in AMSCs tended to be higher than that of BMSCs (Fig. 2E, p<0.05).
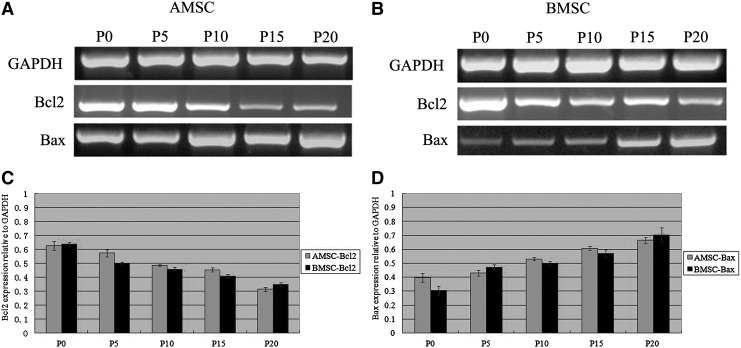
Expression of Bax and Bcl2 genes in pASCs
Expression patterns of Bax and Bcl2 genes in pASCs with long-term culture in vitro are presented in Figure 3A and B. After long-term culturing, all of the pASCs at lower passages expressed markedly higher levels of Bcl2 than those in higher passages (mainly P15 and P20); in contrast, there were slight differences in gene expression levels among the lower passages (P0, P5, and P10). However, P15 and P20 pASCs expressed Bax significantly more than P0, P5, and P10 cells (p<0.05), although nearly no change was observed before P10. GAPDH was an internal control to compare the gene expression levels in pASCs at different passages (Fig. 3C and D).
FIG. 3.
(A and B) Expression of the apoptosis-related genes Bax and Bcl2 in pASCs with different cell passages using RT-PCR. GAPDH was an internal control. (C and D) RT-PCR result of Bax and Bcl2 in pASCs. Light-filled histogram represents the AMSCs and the dark-filled histogram represents the BMSCs.
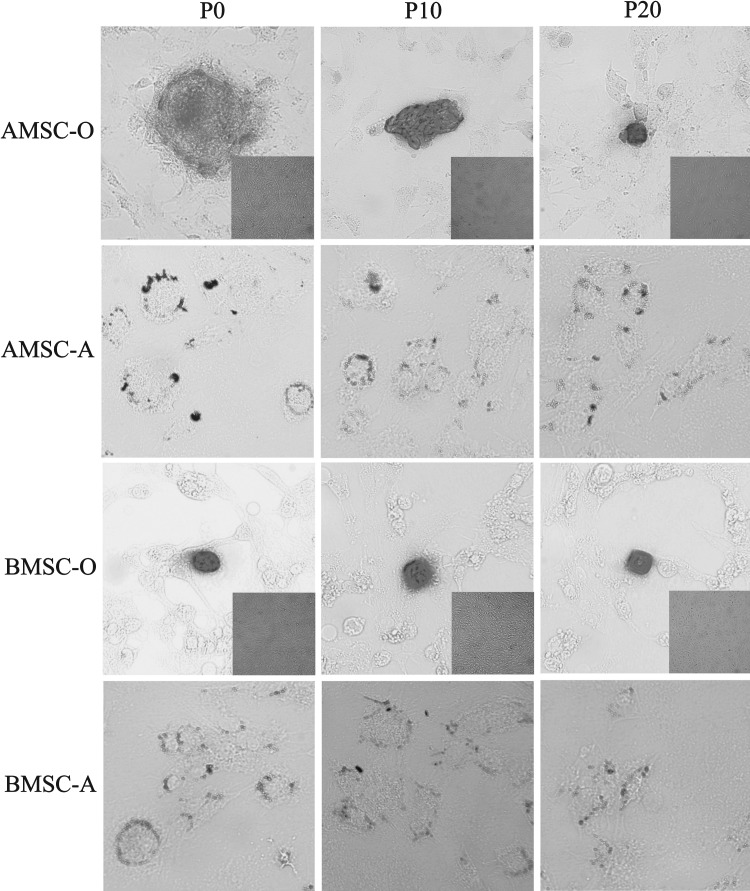
Differentiation of pASCs
To compare the differentiation potential of AMSCs and BMSCs toward the adipogenic and osteogenic lineages, cells at P0, P10, and P20 were cultured in lineage-specific inductive media. After adipogenic induction, both pASCs showed accumulation of lipid vesicles, thus confirming the adipogenic differentiation potential of pASCs. The potential for osteogenic differentiation was also verified in both pASCs (Fig. 4).
FIG. 4.
Adipogenic and osteogenic differentiation of pASCs. Oil Red O staining of pASCs after 3 weeks of treatment with adipogenic media. Dark staining denotes lipid droplets [AMSC-A(P0), AMSC-A(P10), AMSC-A(P20), BMSC-A(P0), BMSC-A(P10), and BMSC-A(P20)]. Alizarin Bordeaux staining of pASCs after 3 weeks of treatment with osteogenic media. Dark staining denotes calcium nodules [AMSC-O(P0), AMSC-O(P10), AMSC-O(P20), BMSC-O(P0), BMSC-O(P10), and BMSC-O(P20)]. The small insets show matched ASC cultures with nondifferentiating media as controls. Magnification, 200×.
The generation of piPSCs
We first focused our characterization efforts on reprogramming the pASCs to piPSCs. The pASCs that expressed Oct4, Sox2, Klf4, and c-Myc genes endogenously were reprogrammed into piPSC colonies within 8 days. The reprogramming efficiency of AMSCs, 291±11.2 AP+ colonies per 5×105 cells, and of BMSCs was 371±24.1; both pASCs were significantly higher than porcine embryo fibroblasts (p<0.05), which were only 150±33.2 per 5×105 cells. It is interesting to note that the time of reprogramming pASCs (8 days) was significantly shorter than that of induced PEFs (12 days) (Table 5). In addition, the reprogramming efficiency of BMSCs was little higher than AMSCs, but not significantly. Cells morphology and AP staining are presented in Figure 5A.
Table 5.
The Generation Efficiency and Speed of piPSCs from pASCs and PEFs
| Cell type | Cells number | AP+number | Efficiency (%) | Time (day) |
|---|---|---|---|---|
| AMSCs | 5×105 | 292 | 0.050 | 8 |
| AMSCs | 5×105 | 311 | 0.062 | 8 |
| AMSCs | 5×105 | 272 | 0.054 | 8 |
| BMSCs | 5×105 | 381 | 0.076 | 8 |
| BMSCs | 5×105 | 407 | 0.081 | 8 |
| BMSCs | 5×105 | 325 | 0.065 | 8 |
| PEFs | 5×105 | 170 | 0.034 | 12 |
| PEFs | 5×105 | 195 | 0.039 | 12 |
| PEFs | 5×105 | 85 | 0.017 | 12 |
piPSCs, porcine induced pluripotential stem cells; pASCs, porcine adult stem cells; PEFs, porcine embryonic fibroblasts; AP, alkaline phosphatase; AMSCs, adipose mesenchymal stem cells; BMSCs, bone marrow mesenchymal stem cells.
FIG. 5.
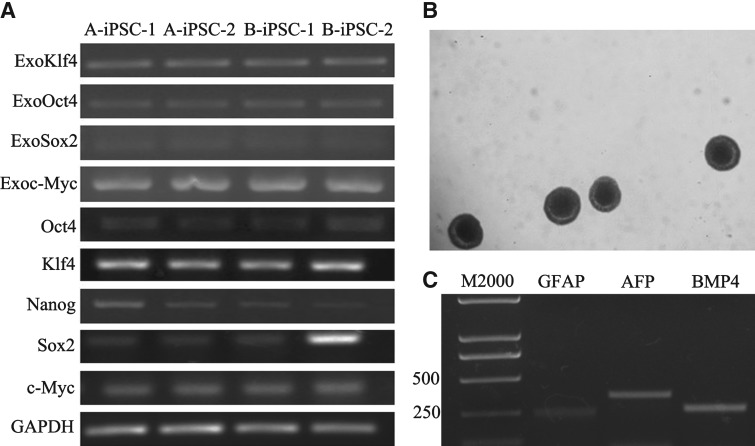
(A) The generation of piPSCs and AP results. (A) Cell morphology after transfection 5 days. (B) Clone morphology. (C) AMSCs induced piPSCs morphology. (D) BMSCs induced piPSCs morphology. (E) AP staining of AMSC induced piPSCs. (F) AP staining of BMSC induced piPSCs. (B and C) Immunostaining results of pASC induced iPSC clones. These iPSCs express pluripotency markers.
Immunostaining of piPSCs
Immunostaining of piPSCs was positive for the human ESCs specific glycoprotein Rex1, mouse ESCs specific glycoprotein SSEA4, and the transcription factors Nanog and Oct4 (Fig. 5B and C).
Analysis of exogenous transgenes and endogenous genes by RT-PCR
The analysis of integrated transgenes by RT-PCR with primers that specifically amplify the mRNA product showed no silencing in selected iPSC lines. And the piPSCs still expressed the endogenous genes (Fig. 6A).
FIG. 6.
(A) Analysis of exogenous transgenes and endogenous genes by RT-PCR. (B) EB morphology. (C) Analysis of germ layer–relevant genes. GFAP, ectoderm; AFP, endoderm; BMP4, mesoderm.
In vitro differentiation of iPS Cells
Porcine iPSCs cultured in the ES0 medium without LIF in bacterial culture dishes formed EBs (Fig. 6B), and they expressed the germ layers' relevant genes (Fig. 6C).
Discussion
In general, stem cells obtained from adult sources are pluripotent and could be induced to differentiate, whereas pASCs were essentially similar to fibroblasts in terms of morphology. ASCs expressed reprogramming transcriptional factors to a greater degree than fibroblasts, but at reduced levels when compared to ESCs (Kim et al., 2009a; Shi et al., 2008). Cells that express reprogramming transcriptional factors (Carlin et al., 2006) could therefore be used to determine the presence of adult stem cells (Kuleshova et al., 2009). In the present study, the reprogramming transcriptional factors were expressed mainly in both the nucleus and cytoplasm of adult stem cells, which is similar to observations made in earlier reports (Kues et al., 2005). The pASCs isolated from porcine adult sources exhibited the typical pASC characteristics of fibroblast-like structures and colony formation when cultured in vitro. These results are in accordance with previous reports (Ringe et al., 2002; Vacanti et al., 2005). These findings clearly demonstrated the successful isolation and expansion of pASCs derived from porcine adult tissues.
In previous studies, the fibroblasts could be cultured for approximately 10 passages in vitro, whereas mouse ESCs can be cultured in vitro for as long as 250 passages without losing totipotency (Zalzman et al., 2010). Unlike ESCs and fibroblast cells, pASCs are pluripotent, and their differentiation ability is restricted to the cell types of a particular tissue responsible for organ regeneration and longevity. In 2006, Takahashi converted adult mouse fibroblasts to induced iPSCs by ectopically expressing a select group of transcription factors to accomplish the direct reprogramming of somatic cells to achieve pluripotency (Takahashi and Yamanaka, 2006). They ultimately determined that four transcription factors (Oct4, Sox2, c-Myc, and Klf4) could mediate reprogramming. In 2007, direct reprogramming was achieved in human cells (Takahashi et al., 2007). This study also used four transcription factors to reprogram, although they were different from those used in mouse fibroblasts and included Nanog and Lin28 instead of c-Myc and Klf4. Wu et al. successfully converted porcine primary BMCs to iPSCs in 2009 (Wu et al., 2009). In the same year, it was shown that iPSCs also could be generated from adult human adipose stem cells (hASCs). As more and more adult stem cells have been reprogrammed, we found that induction time and efficiency became reduced and increased respectively when compared with adult fibroblasts (Kim et al., 2009b). Recently, many studies have demonstrated that iPSCs transcription factors could be replaced or removed (Blelloch et al., 2007; Nakagawa et al., 2008; Montserrat et al., 2011) and iPSCs generated successfully. Distinct from fibroblasts, pASCs could be cultured for longer periods in vitro, and they expressed stem cell reprogramming transcriptional factors (Oct4, Sox2, and c-Myc), although at a lower level than ESCs. However, fibroblasts barely express these genes, so these characteristics made them more amenable to reprogramming. Up to now, some groups reported the generation of porcine iPSCs (Esteban et al., 2009; Ezashi et al., 2009; Wu et al., 2009); however, the exogenous transgenes were not adequately silenced in piPSCs from these studies.
In the present study, we have demonstrated that porcine adult stem cells could be cultured in vitro for 20 passages. As the passage number increased, both types of pASCs proliferative capacity displayed a downward trend. At under 10 passages, the survival rate of pASCs did not have an obvious change; however, after 10 passages, there was significant decline in the survival of AMSCs (p<0.01). In both pASCs, as the cell passage increased, the results of the CFU-F assay declined significantly for all samples (p<0.01). Furthermore, pASCs differentially expressed apoptosis-related genes Bax and Bcl2 at different passages. While the antiapoptotic gene Bcl2 promotes cell survival, Bax is a proapoptotic gene that accelerates cell death under various internal and external conditions. We also used flow cytometry to test the level of cell apoptosis. The results suggested that the percentage of apoptosis cells in BMSCs tended to be lower than in AMSCs. In addition, we successfully induced the P0, P10, and P20 of pASCs into adipocyte and osteogenic cells, illustrating that pASCs have differentiation potential.
Although the efficiency of iPSCs reprogramming has undergone enormous developments, an indispensable requirement has remained in all pertinent studies: reprogrammed cells must be low-passage cells (usually lower than P3), which is a limitation for induced cells. To determine whether higher passages of pASCs that are cultured in vitro retain pluripotency, we detected the reprogramming transcriptional factors' mRNA expression levels at different passages of porcine BMSCs and AMSCs in vitro, relative to GAPDH as an internal control, using real-time RT-PCR. The results demonstrated that the increased passage of cells did not significantly reduce the expression of the aforementioned genes. In P5 of BMSCs, the expression of Sox2, Klf4, and c-Myc was highest; however, the expression of Oct4 was highest in P0, and Nanog was in P10 (p>0.05). Unlike BMSCs, the highest expression of c-Myc was in P15 of AMSCs; however, the expression of the other four genes was highest in P5 (p>0.05). Relative to AMSCs, the expression of Oct4, Sox2, and c-Myc in different BMSCs passages was higher, although Nanog and Klf4 were expressed at lower levels in BMSCs than AMSCs (P0, P5, and P15). With the exception of the Oct4 and Sox2 genes, all of the above differences were not statistically significant. Every BMSC sample expressed Oct4 and Sox2 at a higher level than AMSCs (p<0.01). These findings suggest that culturing AMSCs and BMSCs in vitro could permit retention of their pluripotency and stable expression of reprogramming transcriptional factors (except the expression of Klf4 in P20 AMSCs was lower than other passages). BMSCs come from bone marrow, whereas AMSCs derive from adipose tissue. Although these two adult tissues originate from mesoderm, their developmental mechanisms have some differences, which may be the reason why they differentially express the genes studied here; this will require further investigation. To compare the reprogramming efficiency and speed between the AMSCs and BMSCs, we induced the P1 cells of pASCs into piPSCs. The results demonstrated that two types of pASCs had the advantage over PEF on reprogramming efficiency and time, and there was no difference between them. The staining of piPSCs was positive for the ESCs maker genes, and their genomes were integrated with the four transgenes. All of the piPSC lines acquired generated EBs, but were not fully reprogrammed. Our results indicated that porcine AMSCs and BMSCs were easy to be reprogrammed into iPSCs when compared with fibroblasts. Because porcine BMSCs have advantages over AMSCs in proliferative capacity in higher passages, BMSCs were the better choice when induced cells aged.
In conclusion, porcine AMSCs and BMSCs had some similar biological characteristics, but had different proliferative capacities. Under P5, there was no difference between AMSCs and BMSCs regarding the proliferative capacity. However, when they were cultured over P5, proliferative capacity of BMSCs was better than that of AMSCs. Porcine AMSCs and BMSCs could be reprogrammed into piPSCs with similar reprogramming efficiency, but they were not completely reprogrammed.
Acknowledgments
This work was supported by grants from the National Basic Research Program (No. 2009CB941001 and No. 2011CBA01003) and the National Natural Science Foundation (No. 31072027) in China.
Author Disclosure Statement
The authors declare that no competing financial interests exist.
References
- Aoi T. Yae K. Nakagawa M., et al. Generation of pluripotent stem cells from adultmouse live and stomach cells. Science. 2008;321:699–702. doi: 10.1126/science.1154884. [DOI] [PubMed] [Google Scholar]
- Blelloch R. Venere M. Yen J., et al. Generation of induced pluripotent stem cells in the absence of drug selection. Cell Stem Cell. 2007;1:245–247. doi: 10.1016/j.stem.2007.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlin R. Davis D. Weiss M., et al. Expression of early transcription factors Oct-4, Sox-2 and Nanog by porcine umbilical cord (PUC) matrix cells. Reprod. Biol. Endocrinol. 2006;4:8. doi: 10.1186/1477-7827-4-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deans R.J. Moseley A.B. Mesenchymal stem cells: Biology and potential clinical uses. Exp. Hematol. 2000;28:875–884. doi: 10.1016/s0301-472x(00)00482-3. [DOI] [PubMed] [Google Scholar]
- Eminli S. Utikal J. Arnold K., et al. Reprogramming of neural progenitor cells into iPS cells in the absence of exogenous Sox2 expression. Stem Cells. 2008;26:2467–2474. doi: 10.1634/stemcells.2008-0317. [DOI] [PubMed] [Google Scholar]
- Esteban M. A. Xu J. Yang J., et al. Generation of induced pluripotent stem cell lines from Tibetan miniature pig. J. Biol. Chem. 2009;284:17634–17640. doi: 10.1074/jbc.M109.008938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezashi T. Telugu B. Alexenko A.P., et al. Derivation of induced pluripotent stem cells from pig somatic cells. Proc. Natl. Acad. Sci. USA. 2009;106:10993–10998. doi: 10.1073/pnas.0905284106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang L.J. Fu X.B. Sun T.Z., et al. Isolation and culture of multipotent mesenchymal stem cells from porcine bone marrow. Zhongguo. Wei. Zhong. Bing. Ji. Jiu. Yi. 2003;15:606–608. [PubMed] [Google Scholar]
- Gnecchi M. Melo L.G. Bone marrow-derived mesenchymal stem cells: isolation, expansion, characterization, viral transduction, and production of conditioned medium. Methods Mol. Biol. 2009;482:281–294. doi: 10.1007/978-1-59745-060-7_18. [DOI] [PubMed] [Google Scholar]
- Kim H.I. Yu J.E. Lee S.Y., et al. The effect of composite porcine islet-human endothelial cell grafts on the instant blood-mediated inflammatory reaction. Cell. Transplant. 2009a;18:31–37. doi: 10.3727/096368909788237113. [DOI] [PubMed] [Google Scholar]
- Kim J.B. Sebastiano V. Wu G., et al. Oct4-induced pluripotency in adult neural stem cells. Cell. 2009b;136:411–419. doi: 10.1016/j.cell.2009.01.023. [DOI] [PubMed] [Google Scholar]
- Kues W.A. Petersen B. Mysegades W., et al. Isolation of murine and porcine fetal stem cells from somatic tissue. Biol. Reprod. 2005;72:1020–1028. doi: 10.1095/biolreprod.104.031229. [DOI] [PubMed] [Google Scholar]
- Kuleshova L.L. Tan F.C. Magalhães R., et al. Effective cryopreservation of neural stem or progenitor cells without serum or proteins by vitrification. Cell. Transplant. 2009;18:135–144. [PubMed] [Google Scholar]
- Montserrat N. de Oñate L. Garreta E., et al. Generation of feeder free porcine induced pluripotent stem cells without Pou5f1. Cell. Transplant. Sep. 2011;23 doi: 10.3727/096368911X601019. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- Nakagawa M. Koyanagi M. Tanabe K., et al. Generation of induced pluripotent stem cells without Myc from mouse and human fibroblasts. Nat. Biotechnol. 2008;26:101–106. doi: 10.1038/nbt1374. [DOI] [PubMed] [Google Scholar]
- Ringe J. Kaps C. Schmitt B., et al. Porcine mesenchymal stem cells. Induction of distinct mesenchymal cell lineages. Cell. Tissue. Res. 2002;307:321–327. doi: 10.1007/s00441-002-0525-z. [DOI] [PubMed] [Google Scholar]
- Shi Y. Desponts C. Do J.T., et al. Induction of pluripotent stem cells from mouse embryonic fibroblasts by Oct4 and Klf4 with small-molecule compounds. Cell. Stem. Cell. 2008;3:568–574. doi: 10.1016/j.stem.2008.10.004. [DOI] [PubMed] [Google Scholar]
- Stadtfeld M. Nagaya M. Utikal J., et al. Induced pluripotent stem cells generated without viral integration. Science. 2008;322:945–949. doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenderup K. Justesen J. Clausen C., et al. Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone. 2003;33:919–926. doi: 10.1016/j.bone.2003.07.005. [DOI] [PubMed] [Google Scholar]
- Sun N. Panetta N.J. Gupta D.M., et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc. Natl. Acad. Sci. USA. 2009;106:15720–15725. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi K. Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Takahashi K. Tanabe K. Ohnuki M., et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- Vacanti V. Kong E. Suzuki G., et al. Phenotypic changes of adult porcine mesenchymal stem cells induced by prolonged passaging in culture. J. Cell. Physiol. 2005;205:194–201. doi: 10.1002/jcp.20376. [DOI] [PubMed] [Google Scholar]
- Wu Z. Chen J. Ren J., et al. Generation of pig induced pluripotent stem cells with a drug-inducible system. J. Mol. Cell Biol. 2009;1:46–54. doi: 10.1093/jmcb/mjp003. [DOI] [PubMed] [Google Scholar]
- Xiao L. Yuan X. Sharkis SJ. Activin A maintains self-renewal and regulates fibroblast growth factor, Wnt, and bone morphogenic protein pathways in human embryonic stem cells. Stem Cells. 2006;24:1476–1486. doi: 10.1634/stemcells.2005-0299. [DOI] [PubMed] [Google Scholar]
- Zalzman M. Falco G. Sharova L.V., et al. Zscan4 regulates telomere elongation and genomic stability in ES cells. Nature. 2010;464:858–863. doi: 10.1038/nature08882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk P.A. Zhu M. Mizuno H., et al. Multineage cells from human adipose tissue: implications for cell-based therapies. Tissue. Eng. 2001;7:211–228. doi: 10.1089/107632701300062859. [DOI] [PubMed] [Google Scholar]
- Zuk PA. Zhu M. Ashjian P, et al. Human adipose tissue is a source of multipotent stem cells. Mol Biol Cell. 2002;13:4279–4295. doi: 10.1091/mbc.E02-02-0105. [DOI] [PMC free article] [PubMed] [Google Scholar]