Abstract
Background
Alzheimer’s disease (AD) is the most common cause of dementia among the elderly. To clarify pathogenesis of AD, thousands of reports have been accumulating. However, knowledge of signaling pathways in the field of AD has not been compiled as a database before.
Description
Here, we have constructed a publicly available pathway map called “AlzPathway” that comprehensively catalogs signaling pathways in the field of AD. We have collected and manually curated over 100 review articles related to AD, and have built an AD pathway map using CellDesigner. AlzPathway is currently composed of 1347 molecules and 1070 reactions in neuron, brain blood barrier, presynaptic, postsynaptic, astrocyte, and microglial cells and their cellular localizations. AlzPathway is available as both the SBML (Systems Biology Markup Language) map for CellDesigner and the high resolution image map. AlzPathway is also available as a web service (online map) based on Payao system, a community-based, collaborative web service platform for pathway model curation, enabling continuous updates by AD researchers.
Conclusions
AlzPathway is the first comprehensive map of intra, inter and extra cellular AD signaling pathways which can enable mechanistic deciphering of AD pathogenesis. The AlzPathway map is accessible at http://alzpathway.org/.
Background
Alzheimer’s disease (AD) is the most common cause of dementia among the elderly. With the advent of the aging society, 24.3 million people are estimated to suffer dementia worldwide, increasing to 42.3 million people by 2020 and 81.1 million by 2040 [1,2]. The total estimated worldwide costs of dementia are US$604 billion in 2010, and are estimated to increase by 85% to 2030 [3]. The cost for caring for the increasing number of people with dementia will rise dramatically and thus will be disastrous burden to our societies within upcoming 10–20 years. To address this social issue, clarification of the pathogenic mechanism of AD and development of AD drugs are urgently expected.
Genetic association with putative AD susceptibility genes has been studied and collected as a publicly available database called AlzGene (http://www.alzgene.org/) [4]. Pathological signaling has been also studied and two core pathological hallmarks of AD, amyloid plaques and neurofibrillary tangles (NFT) accumulation, have been elucidated intensively. However, wealth of this information has become increasingly difficult to follow, much less interpret, and has not been integrated before. Integration of pathway knowledge bridging amyloid plaques and neurofibrillary tangles has been still missing.
Efforts to construct an AD pathway map have been made before, however, these maps are overviews or partial maps of an AD pathway, and are not comprehensive maps. Manual elaboration is still required to construct a comprehensive and accurate map of a particular signaling pathway [5]. Efforts to manually construct pathway maps of particular signaling, e.g., Toll-like receptor, EGFR, RB/E2F, mTOR signalings, and dendritic cell signaling in response to pathogenes, have been made before [6-10].
In this study, we collected and manually curated over 100 review articles involving in AD, and built an AD pathway map using CellDesigner [11], a modeling editor for biochemical pathways. AlzPathway is the first comprehensive map of intra, inter and extra cellular signaling pathways of particular disease. AlzPathway now contains 34 canonical pathways such as APP, mitochondrial and apoptosis pathways, which are composed of 1347 species (proteins, complexes, simple moecules, genes, RNAs, ions, degraded products, and phenotypes), 1070 reactions, 129 phenotypes in neuron, brain blood barrier, presynaptic, postsynaptic, astrocyte, and microglial cells and their cellular localizations. Our AlzPathway provides a powerful AD pathway map for deciphering pathogenesis of AD, and it serves as a pathway reference database just as AlzGene for risk-gene reference database. AlzPathway allows us to not only evaluate candidate risk genes listed by GWAS studies, but also analyze omics data including DNA microarray data and RNA-seq data to reveal pathogenesis of AD. Our pathway map will be an indispensable basic resource for both clarification of the pathogenic mechanism of AD and development of AD drug to address social issues caused by AD.
Construction and content
Construction of AlzPathway
We collected 123 review articles related to AD accessible from PubMed. We then manually curated these review articles, and have built an AD pathway map by using CellDesigner. In PubMed, the number of research articles involved in Alzheimer’s disease is over 80,000. In fact, we cannot collect and manually curate over 80,000 research articles. To construct AlzPathway, we need to choose articles among over 80,000 research articles. Review articles are attempts to chose important articles and summarize current state of understanding on signaling pathways involved in Alzheimer’s disease. Therefore, we chose to use review articles for the curation. Notations are based on the PD (Process Description) of SBGN (Systems Biology Graphical Notation) [12] and the map is available in standardized SBML (Systems Biology Markup Language) [13] format for file exchange between different tools. Molecules are distinguished by the following types: proteins, complexes, simple molecules, genes, RNAs, ions, degraded products, and phenotypes. Gene symbols are pursuant to the HGNC symbols. Reactions are also distinguished by the following categories: state transition, transcription, translation, heterodimer association, dissociation, transport, unknown transition, and omitted transition. All the reactions have evidences to the references in PubMed ID using the MIRIAM scheme [14]. All the references used for constructing the AlzPathway are listed in the ‘References for AlzPathway’. Cellular types are distinguished by the followings: neuron, astrocyte, and microglial cells. Cellular compartments are also distinguished by the followings: brain blood barrier, presynaptic, postsynaptic, and their inner cellular localizations. AlzPathway is available as both the SBML map for CellDesigner ( Additional file 1; see the section of Additional files) and the high resolution image map ( Additional file 2; see the section of Additional files).
Imprementation of web service of AlzPathway
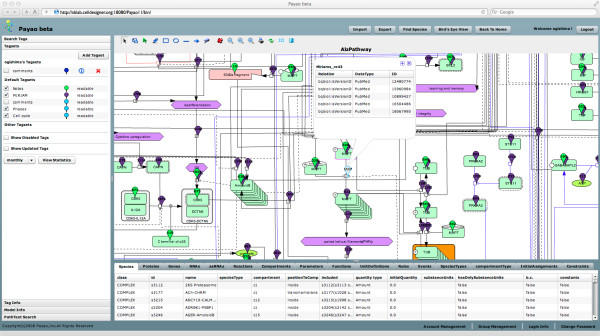
AlzPathway is also available as the web service (online map) implemented by using Payao [15], a community-based, collaborative web service platform for gene-regulatory and biochemical pathway model curation, enabling continuous updates by AD researchers. Payao web service (online map) is accessible from http://alzpathway.org/. Instruction on how to access the AlzPathway web service (online map) is described in the Additional documentation file ( Additional file 3; see the section of Additional files). Using the Payao system would enable AD researchers not only to browse reactions and their references in PubMed ID but also to comment, correct and update AlzPathway in a community-wide collaboration.
Overview of alzheimer’s disease pathway
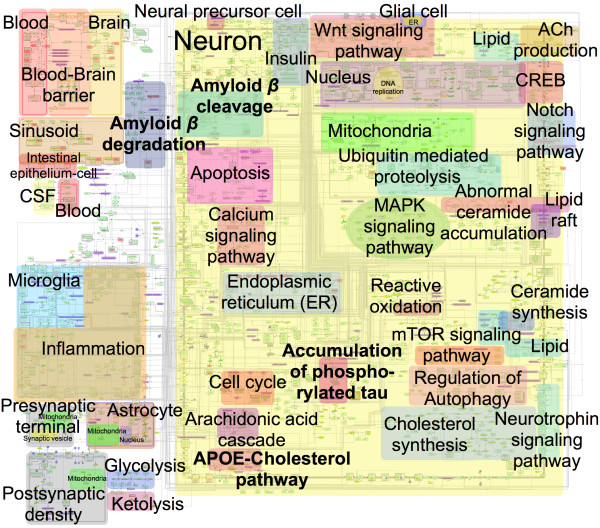
Here, we present a map of Alzheimer’s disease signaling networks (Figure 1). We manually constructed a map of Alzheimer’s disease signaling networks by assembling molecular interactions based on published papers using the modeling software, CellDesigner ver. 4.2 (http://celldesigner.org/).
Figure 1.
Overview of AlzPathway overlaid with canonical pathway annotations for explanation of map. AlzPathway consists of 1347 molecules, 1070 reactions, and 129 phenotypes. Original and high resolution version of AlzPathway is available at http://alzpathway.org/ as both the SBML (Systems Biology Markup Language) map for CellDesigner and the high resolution image map without overlay of canonical pathway annotation. AlzPathway is also available as the web service (online map) implemented by using Payao system.
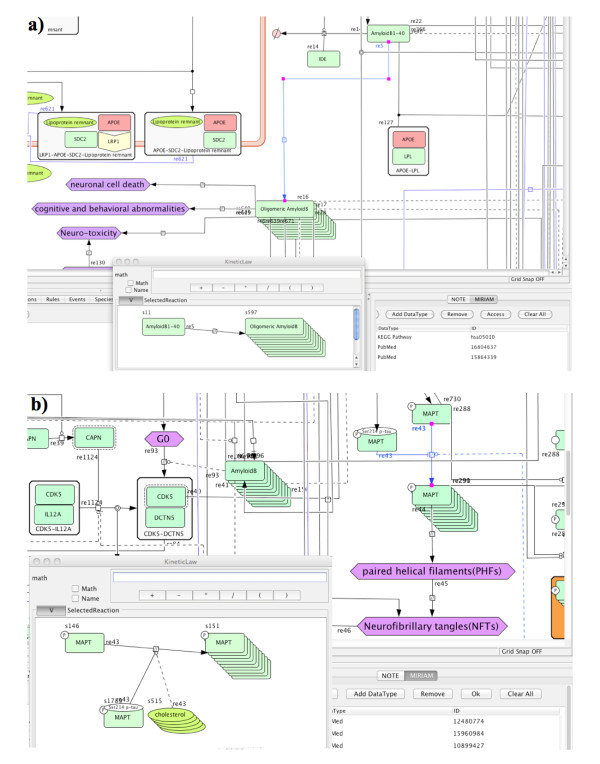
The AlzPathway map consists of 1347 species, 1070 reactions, and 129 phenotypes. The molecues shown on the AlzPathway can be categorized as follows: 650 proteins, 232 complexes, 223 simple molecules, 32 genes, 36 RNAs, 24 ions, and 21 degraded products. The breakdown of reactions is as follows: 401 state transitions, 22 transcriptions, 30 translations, 172 heterodimer associations, 49 dissociations, 87 transports, 20 unknown transitions, and 228 omitted transitions. All the 123 references used for constructing the map are listed in the ‘References for AlzPathway’ [16-138]. The CellDesigner software allows the user to access references that are used as evidences for individual reaction using PubMed ID (Figure 2).
Figure 2.
Detailed view of AlzPathway. (a) oligomeric amyloid β formation, (b) MAPT phosphorylation. Notations are based on SBGN (Systems Biology Graphical Notation). References to the reactions are represented in PubMed ID (PMID) using the MIRIAM scheme.
The map consists of the AD hallmark pathways and canonical pathways. The AD hallmark pathways are amyloid β cleavage, amyloid β degradation, APOE-cholesterol pathway and NFT accumulation, which are major pathological pathways of AD. On the other hand, the canonical pathways are acetylcholine production, cholesterol synthesis, Wnt signaling pathway, Notch signaling pathway, Ubiquitin mediated proteolysis, apoptosis, calcium signaling pathway, ER stress, MAPK signaling pathway, abnormal ceramide accumulation, ceramide synthesis, reactive oxidation process, regulation of Autophagy, neurotrophin signaling pathway, cell cycle, arachidonic acid cascade, mTOR signaling pathway, lipid pathway, lipid raft, inflammation pathway, insulin pathway, and CREB pathway.
AlzPathway is the first comprehensive map of intra, inter and extra cellular signaling pathways of particular disease manually constructed. Manual reconstructions of comprehensive map have been reported before: epidermal growth factor receptor signaling, toll-like receptor signaling network, RB/E2F signaling pathway and mTOR signaling pathways [6-9], which are molecular signaling pathways and are not comprehensive intra, inter and extra cellular signaling pathways of particular disease. That is, AlzPathway comprehensively catalogs not only intra but also inter and extracellular signaling pathways among neuron, glial cell, microglia, presynaptic cell, postsynaptic cell, astrocyte, and blood–brain barrier. The brain and spinal cord are made up of various regions and cells, including neurons and glial cells. To reveal pathogenic mechanism of AD, complicated signaling pathways among neuron, glial cell, microglia, presynaptic cell, postsynaptic cell, astrocyte, and blood–brain barrier should be clarified.
Binary-relation notation and key molecules
As for intuitively understandable notation of AlzPathway, we also constructed AlzPathway not only in SBGN (Systems Biology Graphical Notation) PD (Process Description) notation but also in binary-relation notation (Figure 3(a)). In SBGN PD notation, a reaction consists of reactants, modifiers, and products. We converted this notation to binary-relation notation by decomposing reactions into those between (1) reactants and products, and (2) modifiers and products. Molecules are limited to proteins, complexes, genes, RNAs and phenotype for simplification. SBGN PD notation is precise notation for describing pathways, on the other hand the binary-relation based conventional notation used by molecular biologists in the current literature, is intuitively understandable. To clarifying basic structure of AlzPathway, we constructed AlzPathway in binary-relation notation, and found that amyloid β accumulation and hyperphosphorylated tau accumulation are central pathogenesis signaling pathways in AlzPathway. Binary-relation notation is provided as a SIF (simple interaction format) file ( Additional file 4) which can be opened by using Cytoscape [139].
Figure 3.
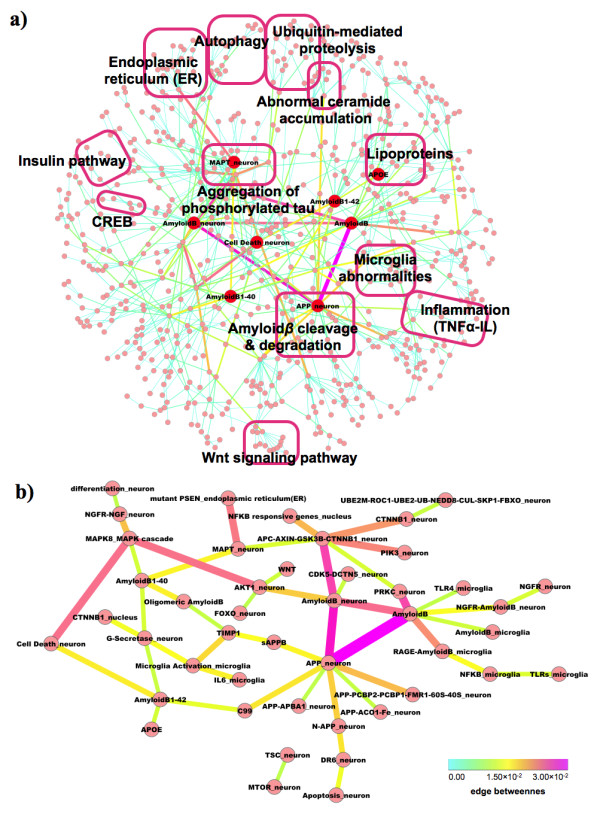
Binary-relation notation and key molecules of AlzPathway. (a) overview of AlzPathway in binary-relation notation, (b) top 50 high centrality relations as highlighted primary pathway of AlzPathway.
According to edge betweenness centrality of each reaction, high centrality relations were highlighted as primary relations. Top 50 high centrality relations are shown in Figure 3(b). Highlighted binary relations were so-called AD hallmark pathways: amyloid plagues (amyloid β accumulation) and NFT accumulation (hyperphosphorylated tau accumulation). The γ-secretase produces amyloid β 1–40, which leads to oligomeric amyloid β (amyloid β accumulation). Amyloid β 1–42 is related to cell death, which is amyloid plagues (amyloid β accumulation) and NFT accumulation crucial for AD progression. On the other hand, APC-AXIN-GSK3B-CTNNB1 complex phosphorylates MAPT and mutant PSEN promotes phosphorylation of MAPT, which lead to MAPT hyperphosphorylation and NFT accumulation.
Utility and discussion
Access to AlzPathway map
The AlzPathway map is accessible at http://alzpathway.org/. As described above, AlzPathway is provided as (1) the SBML map for CellDesigner ( Additional file 1), (2) the web service (online map) implemented by using Payao accessible from http://alzpathway.org/ (see the Additional file 3 for instruction on how to access the AlzPathway online map by Payao), (3) the high resolution image map ( Additional file 2), (4) the binary-relation notation ( Additional file 4), (5) the pure SBML map ( Additional file 5) for compatibility with other SMBL supporting tools, and (6) the BioPAX file for exchanges of pathway data ( Additional file 6). For Notations are based on the PD (Process Description) of SBGN (Systems Biology Graphical Notation). The CellDesigner software and Payao system allow the user to access references that are used as evidences for individual reaction in PubMed ID using the MIRIAM scheme. They also allow the user to access external resources including UniProt and PubChem for individual species using the MIRIAM scheme. AlzGene information is provided for individual species using Notes. Expanded version of AlzPathway map having external links to Gene Ontology is also provided ( Additional file 7). As for usage of CellDesigner software, see the documentations provided at CellDesigner web site. As for usage of Payao system, see the user’s guide provided at Payao web site.
Community driven update of AlzPathway using payao
We constructed AlzPathway to be comprehensive but not necessarily to be complete. While continuous curation of molecular pathways remain a major challenge, the availability of Alzpathway through Payao (http://alzpathway.org/) would enable AD researchers to review, comment and provide feedbacks to the pathway through the web-based interface (Figure 4). Correction and update of AlzPathway can be made through comments and feedback from AD researchers in specific molecules and interactions. By using Payao system, we envision to reach out to the AD community and continuously correct and update AlzPathway. In addition to community-driven correction and update using Payao system, we will keep updating our pathway map by ourselves.
Figure 4.
AlzPathway on Payao system. Payao system provide community-based, collaborative web service (online map) platform for pathway manual curation.
AlzPathway as basic resource for AD study
AlzPathway is comprehensive AD pathway map, and is expected to be a guidance map in the study of AD. Just as AlzGene for risk-gene reference database, AlzPathway serves as a pathway “reference” integrating wealth of pathway information published before. AlzPathway also serves for analysis on omics data including DNA microarray data and RNA-seq data. Availability of AD pathway map in standardized formats (SBML and SBGN) renders the network applicable to systems-biology analyses based on various SBML compliant tools. Therefore, our AD pathway map will be an indispensable basic resource for both clarification of the pathogenic mechanism of AD and development of AD drug.
Conclusions
We constructed a publicly available database called “AlzPathway” that comprehensively catalogs signaling pathways in the field of AD. We have collected and manually curated over 100 review articles involving in AD, and have built an AD pathway map using CellDesigner. AlzPathway is the first comprehensive map of intra, inter and extra cellular signaling pathways of particular disease for deciphering pathogenesis of AD. AlzPathway is currently composed of 1347 molecules, 1070 reactions, and 129 phenotypes in neuron, brain blood barrier, presynaptic, postsynaptic, astrocyte, and microglial cells and their cellular localizations. AlzPathway is available as both the SBML (Systems Biology Markup Language) map for CellDesigner and the high resolution image map. AlzPathway is also available as the web service (online map) implemented on Payao, a community-based, collaborative web service platform for pathway model curation. The molecular level mechanistic interactions captured in AlzPathway, together with a community-driven, web based curation platform, would provide a comprehensive resource to the AD community towards deeper insights into AD pathogenesis and identification of novel therapeutic targets.
Availability and requirements
The AlzPathway map is accessible at http://alzpathway.org/. The Payao system does not work on a 64 bit mode Linux.
Abbreviations
AD, Alzheimer’s disease; SBML, Systems biology markup language; NFT, Neurofibrillary tangles; PD, Process description; SBGN, Systems biology graphical notation.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
SO conceived and designed the project with the help of RK and HT. SM, RI, TM, MK, and SO built the database. AM and RK reviewed the database. YM and SG implemented the web service by using Payao. SO wrote the paper with the help of YM, SG, MK, and TM. All authors read and approved the final manuscript.
Supplementary Material
SBML map file of AlzPathway. The SBML map file alzpathway_sbml_map.xml can be browsed using CellDesigner. Please download CellDesigner at http://www.celldesigner.org/, install it, and open the SBML map file alzpathway_sbml_map.xml to browse AlzPathway map by CellDesigner. As for usage of CellDesigner software, see the documentations provided at CellDesigner web site: http://www.celldesigner.org/documents.html.
High resolution image map of AlzPathway. The PNG file alzpathway_image_map.png contains a high resolution map of AlzPathway. This image map does not contain the reference information used for constructing the AlzPathway. SBML (CellDesigner) map and Online map (Payao) are recommended to browse the AlzPathway map.
Instruction on usage of Payao. The PDF file alzpathway_payao_access_instruction.pdf contains insturction on usage of Payao web service (online map). Payao system requires login. AlzPathway demo user account is prepared for demo use: demo username is “ap_demo” and the corresponding password is “4patients”. As for usage of Payao system, see the user’s guide provided at Payao web site: http://payao.oist.jp:8080/payaologue/doc/PAYAO_Users_GuideE11.pdf.
Binary-relation notation file of AlzPathway. The SIF (simple interaction format) file alzpathway_binary_relation.sif is the binary-relation notation of AlzPathway map which can be opened by using Cytoscape.
Pure SBML map file of AlzPathway. The pure SBML file alzpathway_pure_sbml.xml is for compatibility with other SMBL supporting tools.
BioPAX file of AlzPathway. The BioPAX file alzpathway_biopax.owl is for exchanges of pathway data.
Expanded SBML map file of AlzPathway with Gene Ontology. The SBML map file alzpathway_sbml_map_go.xml is the expanded version of AlzPathway map (alzpathway_sbml_map.xml) having external links to Gene Ontology. The SBML map file with the external links to Gene Ontology requires time to be opened by CellDesigner 4.2, and thus was provided separately as an expanded SBML map.
Contributor Information
Satoshi Mizuno, Email: mizuno@bioinfo.tmd.ac.jp.
Risa Iijima, Email: riijima@bioinfo.tmd.ac.jp.
Soichi Ogishima, Email: ogishima@sysbioevo.org.
Masataka Kikuchi, Email: kikuchi@bioinfo.tmd.ac.jp.
Yukiko Matsuoka, Email: myukiko@sbi.jp.
Samik Ghosh, Email: ghosh@sbi.jp.
Tadashi Miyamoto, Email: miyamoto@bioinfo.tmd.ac.jp.
Akinori Miyashita, Email: miyashi@bri.niigata-u.ac.jp.
Ryozo Kuwano, Email: ryosun@bri.niigata-u.ac.jp.
Hiroshi Tanaka, Email: htanaka@bioinfo.tmd.ac.jp.
Acknowledgements
The authors are grateful for the helpful comments of an editor and anonymous referees. We thank Dr. Takeshi Fukuhara for critical discussion on AlzPathway. Funding: This work was supported by Grant-in-Aid for Scientific Research (22700311) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan. The Payao system is supported by Okinawa Institute of Science and Technology (http://www.oist.jp).
References
- Ballard C, Gauthier S, Corbett A, Brayne C, Aarsland D, Jones E. Alzheimer’s disease. Lancet. 2011;377(9770):1019–1031. doi: 10.1016/S0140-6736(10)61349-9. [DOI] [PubMed] [Google Scholar]
- Ferri CP, Prince M, Brayne C, Brodaty H, Fratiglioni L, Ganguli M, Hall K, Hasegawa K, Hendrie H, Huang Y. et al. Global prevalence of dementia: a Delphi consensus study. Lancet. 2005;366(9503):2112–2117. doi: 10.1016/S0140-6736(05)67889-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alzheimer’s Disease Interanational. World Alzheimer’s Report 2010. http://www.alz.co.uk/research/files/WorldAlzheimerReport2010ExecutiveSummary.pdf.
- Bertram L, McQueen MB, Mullin K, Blacker D, Tanzi RE. Systematic meta-analyses of Alzheimer disease genetic association studies: the AlzGene database. Nat Genet. 2007;39(1):17–23. doi: 10.1038/ng1934. [DOI] [PubMed] [Google Scholar]
- Bauer-Mehren A, Furlong LI, Sanz F. Pathway databases and tools for their exploitation: benefits, current limitations and challenges. Mol Syst Biol. 2009;5:290. doi: 10.1038/msb.2009.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Matsuoka Y, Funahashi A, Kitano H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol Syst Biol. 2005;1:2005.0010. doi: 10.1038/msb4100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K, Kitano H. A comprehensive map of the toll-like receptor signaling network. Mol Syst Biol. 2006;2:2006.0015. doi: 10.1038/msb4100057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calzone L, Gelay A, Zinovyev A, Radvanyi F, Barillot E. A comprehensive modular map of molecular interactions in RB/E2F pathway. Mol Syst Biol. 2008;4:173. doi: 10.1038/msb.2008.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caron E, Ghosh S, Matsuoka Y, Ashton-Beaucage D, Therrien M, Lemieux S, Perreault C, Roux PP, Kitano H. A comprehensive map of the mTOR signaling network. Mol Syst Biol. 2010;6:453. doi: 10.1038/msb.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patil S, Pincas H, Seto J, Nudelman G, Nudelman I, Sealfon SC. Signaling network of dendritic cells in response to pathogens: a community-input supported knowledgebase. BMC Syst Biol. 2010;4:137. doi: 10.1186/1752-0509-4-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funahashi A, Matsuoka Y, Jouraku A, Morohashi M, Kikuchi N, Kitano H. CellDesigner 3.5: a versatile modeling tool for biochemical networks. Proc IEEE. 2008;96(8):1254–1265. [Google Scholar]
- Le Novère N, Hucka M, Mi H, Moodie S, Schreiber F, Sorokin A, Demir E, Wegner K, Aladjem MI, Wimalaratne SM. et al. The Systems Biology Graphical Notation. Nat Biotechnol. 2009;27(8):735–741. doi: 10.1038/nbt.1558. [DOI] [PubMed] [Google Scholar]
- Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A. et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19(4):524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- Laibe C, Le Novère N. MIRIAM Resources: tools to generate and resolve robust cross-references in Systems Biology. BMC Syst Biol. 2007;1:58. doi: 10.1186/1752-0509-1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka Y, Ghosh S, Kikuchi N, Kitano H. Payao: A community platform for SBML pathway model curation. Bioinformatics. 2010;26(10):1381–1383. doi: 10.1093/bioinformatics/btq143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allain H, Bentué-Ferrer D, Tribut O, Gauthier S, Michel BF, Drieu-La Rochelle C. Alzheimer’s disease: the pharmacological pathway. Fundam Clin Pharmacol. 2003;17(4):419–428. doi: 10.1046/j.1472-8206.2003.00153.x. [DOI] [PubMed] [Google Scholar]
- Arendt T, Holzer M, Stöbe A, Gärtner U, Lüth HJ, Brückner MK, Ueberham U. Activated mitogenic signaling induces a process of dedifferentiation in Alzheimer’s disease that eventually results in cell death. Ann N Y Acad Sci. 2000;920:249–255. doi: 10.1111/j.1749-6632.2000.tb06931.x. [DOI] [PubMed] [Google Scholar]
- Atamna H. Heme, iron, and the mitochondrial decay of ageing. Ageing Res Rev. 2004;3(3):303–318. doi: 10.1016/j.arr.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Behl C. Apoptosis and Alzheimer’s disease. J Neural Transm. 2000;107(11):1325–1344. doi: 10.1007/s007020070021. [DOI] [PubMed] [Google Scholar]
- Björkhem I. Crossing the barrier: oxysterols as cholesterol transporters and metabolic modulators in the brain. J Intern Med. 2006;260(6):493–508. doi: 10.1111/j.1365-2796.2006.01725.x. [DOI] [PubMed] [Google Scholar]
- Boonen RA, van Tijn P, Zivkovic D. Wnt signaling in Alzheimer’s disease: up or down, that is the question. Ageing Res Rev. 2009;8(2):71–82. doi: 10.1016/j.arr.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Bosco D, Fava A, Plastino M, Montalcini T, Pujia A. Possible implications of insulin resistance and glucose metabolism in Alzheimer’s disease pathogenesis. J Cell Mol Med. 2011;15(9):1807–1821. doi: 10.1111/j.1582-4934.2011.01318.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinton RD. Estrogen regulation of glucose metabolism and mitochondrial function: therapeutic implications for prevention of Alzheimer’s disease. Adv Drug Deliv Rev. 2008;60(13–14):1504–1511. doi: 10.1016/j.addr.2008.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buhaescu I, Izzedine H. Mevalonate pathway: a review of clinical and therapeutical implications. Clin Biochem. 2007;40(9–10):575–584. doi: 10.1016/j.clinbiochem.2007.03.016. [DOI] [PubMed] [Google Scholar]
- Buoso E, Lanni C, Schettini G, Govoni S, Racchi M. beta-Amyloid precursor protein metabolism: focus on the functions and degradation of its intracellular domain. Pharmacol Res. 2010;62(4):308–317. doi: 10.1016/j.phrs.2010.05.002. [DOI] [PubMed] [Google Scholar]
- Burns M, Duff K. Cholesterol in Alzheimer’s disease and tauopathy. Ann N Y Acad Sci. 2002;977:367–375. doi: 10.1111/j.1749-6632.2002.tb04839.x. [DOI] [PubMed] [Google Scholar]
- Cameron B, Landreth GE. Inflammation, microglia, and Alzheimer’s disease. Neurobiol Dis. 2010;37(3):503–509. doi: 10.1016/j.nbd.2009.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caricasole A, Bakker A, Copani A, Nicoletti F, Gaviraghi G, Terstappen GC. Two sides of the same coin: Wnt signaling in neurodegeneration and neuro-oncology. Biosci Rep. 2005;25(5–6):309–327. doi: 10.1007/s10540-005-2893-6. [DOI] [PubMed] [Google Scholar]
- Caricasole A, Copani A, Caruso A, Caraci F, Iacovelli L, Sortino MA, Terstappen GC, Nicoletti F. The Wnt pathway, cell-cycle activation and beta-amyloid: novel therapeutic strategies in Alzheimer’s disease? Trends Pharmacol Sci. 2003;24(5):233–238. doi: 10.1016/S0165-6147(03)00100-7. [DOI] [PubMed] [Google Scholar]
- Chen YZ. APP induces neuronal apoptosis through APP-BP1-mediated downregulation of beta-catenin. Apoptosis. 2004;9(4):415–422. doi: 10.1023/B:APPT.0000031447.05354.9f. [DOI] [PubMed] [Google Scholar]
- Chong ZZ, Li F, Maiese K. Stress in the brain: novel cellular mechanisms of injury linked to Alzheimer’s disease. Brain Res Brain Res Rev. 2005;49(1):1–21. doi: 10.1016/j.brainresrev.2004.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong ZZ, Maiese K. Targeting WNT, protein kinase B, and mitochondrial membrane integrity to foster cellular survival in the nervous system. Histol Histopathol. 2004;19(2):495–504. doi: 10.14670/hh-19.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chorsky RL, Yaghmai F, Hill WD, Stopa EG. Alzheimer’s disease: a review concerning immune response and microischemia. Med Hypotheses. 2001;56(1):124–127. doi: 10.1054/mehy.2000.1148. [DOI] [PubMed] [Google Scholar]
- Choy YM, Lau KM, Lee CY. Purification and characterization of urinary choriogonadotropin from patients with hydatidiform mole. J Biol Chem. 1979;254(4):1159–1163. [PubMed] [Google Scholar]
- Copani A, Caraci F, Hoozemans JJ, Calafiore M, Sortino MA, Nicoletti F. The nature of the cell cycle in neurons: focus on a “non-canonical” pathway of DNA replication causally related to death. Biochim Biophys Acta. 2007;1772(4):409–412. doi: 10.1016/j.bbadis.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Correia SC, Santos RX, Perry G, Zhu X, Moreira PI, Smith MA. Insulin-resistant brain state: the culprit in sporadic Alzheimer’s disease? Ageing Res Rev. 2011;10(2):264–273. doi: 10.1016/j.arr.2011.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coughlan CM, Breen KC. Factors influencing the processing and function of the amyloid beta precursor protein–a potential therapeutic target in Alzheimer’s disease? Pharmacol Ther. 2000;86(2):111–145. doi: 10.1016/S0163-7258(00)00036-X. [DOI] [PubMed] [Google Scholar]
- Coulson EJ. Does the p75 neurotrophin receptor mediate Abeta-induced toxicity in Alzheimer’s disease? J Neurochem. 2006;98(3):654–660. doi: 10.1111/j.1471-4159.2006.03905.x. [DOI] [PubMed] [Google Scholar]
- Cruts M, Van Broeckhoven C. Loss of progranulin function in frontotemporal lobar degeneration. Trends Genet. 2008;24(4):186–194. doi: 10.1016/j.tig.2008.01.004. [DOI] [PubMed] [Google Scholar]
- Cuello AC, Bruno MA. The failure in NGF maturation and its increased degradation as the probable cause for the vulnerability of cholinergic neurons in Alzheimer’s disease. Neurochem Res. 2007;32(6):1041–1045. doi: 10.1007/s11064-006-9270-0. [DOI] [PubMed] [Google Scholar]
- Cuenda A, Rousseau S. p38 MAP-kinases pathway regulation, function and role in human diseases. Biochim Biophys Acta. 2007;1773(8):1358–1375. doi: 10.1016/j.bbamcr.2007.03.010. [DOI] [PubMed] [Google Scholar]
- Das UN. Acetylcholinesterase and butyrylcholinesterase as possible markers of low-grade systemic inflammation. Med Sci Monit. 2007;13(12):RA214–RA221. [PubMed] [Google Scholar]
- Deutsch SI, Rosse RB, Deutsch LH. Faulty regulation of tau phosphorylation by the reelin signal transduction pathway is a potential mechanism of pathogenesis and therapeutic target in Alzheimer’s disease. Eur Neuropsychopharmacol. 2006;16(8):547–551. doi: 10.1016/j.euroneuro.2006.01.006. [DOI] [PubMed] [Google Scholar]
- Dmitriev LF. Shortage of lipid-radical cycles in membranes as a possible prime cause of energetic failure in aging and Alzheimer disease. Neurochem Res. 2007;32(8):1278–1291. doi: 10.1007/s11064-007-9322-0. [DOI] [PubMed] [Google Scholar]
- Eckert A, Keil U, Marques CA, Bonert A, Frey C, Schüssel K, Müller WE. Mitochondrial dysfunction, apoptotic cell death, and Alzheimer’s disease. Biochem Pharmacol. 2003;66(8):1627–1634. doi: 10.1016/S0006-2952(03)00534-3. [DOI] [PubMed] [Google Scholar]
- Eckert A, Marques CA, Keil U, Schüssel K, Müller WE. Increased apoptotic cell death in sporadic and genetic Alzheimer’s disease. Ann N Y Acad Sci. 2003;1010:604–609. doi: 10.1196/annals.1299.113. [DOI] [PubMed] [Google Scholar]
- Farooqui AA, Ong WY, Farooqui T. Lipid mediators in the nucleus: Their potential contribution to Alzheimer’s disease. Biochim Biophys Acta. 2010;1801(8):906–916. doi: 10.1016/j.bbalip.2010.02.002. [DOI] [PubMed] [Google Scholar]
- Filiz G, Price KA, Caragounis A, Du T, Crouch PJ, White AR. The role of metals in modulating metalloprotease activity in the AD brain. Eur Biophys J. 2008;37(3):315–321. doi: 10.1007/s00249-007-0244-1. [DOI] [PubMed] [Google Scholar]
- Florent-Béchard S, Desbène C, Garcia P, Allouche A, Youssef I, Escanyé MC, Koziel V, Hanse M, Malaplate-Armand C, Stenger C. et al. The essential role of lipids in Alzheimer’s disease. Biochimie. 2009;91(6):804–809. doi: 10.1016/j.biochi.2009.03.004. [DOI] [PubMed] [Google Scholar]
- Fraser PE, Yang DS, Yu G, Lévesque L, Nishimura M, Arawaka S, Serpell LC, Rogaeva E, St George-Hyslop P. Presenilin structure, function and role in Alzheimer disease. Biochim Biophys Acta. 2000;1502(1):1–15. doi: 10.1016/S0925-4439(00)00028-4. [DOI] [PubMed] [Google Scholar]
- Fuentealba RA, Farias G, Scheu J, Bronfman M, Marzolo MP, Inestrosa NC. Signal transduction during amyloid-beta-peptide neurotoxicity: role in Alzheimer disease. Brain Res Brain Res Rev. 2004;47(1–3):275–289. doi: 10.1016/j.brainresrev.2004.07.018. [DOI] [PubMed] [Google Scholar]
- Gandy S. The role of cerebral amyloid beta accumulation in common forms of Alzheimer disease. J Clin Invest. 2005;115(5):1121–1129. doi: 10.1172/JCI25100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner U, Holzer M, Arendt T. Elevated expression of p21ras is an early event in Alzheimer’s disease and precedes neurofibrillary degeneration. Neuroscience. 1999;91(1):1–5. doi: 10.1016/S0306-4522(99)00059-7. [DOI] [PubMed] [Google Scholar]
- Gasparini L, Xu H. Potential roles of insulin and IGF-1 in Alzheimer’s disease. Trends Neurosci. 2003;26(8):404–406. doi: 10.1016/S0166-2236(03)00163-2. [DOI] [PubMed] [Google Scholar]
- Ghosh AK, Gemma S, Tang J. beta-Secretase as a therapeutic target for Alzheimer’s disease. Neurotherapeutics. 2008;5(3):399–408. doi: 10.1016/j.nurt.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Saijo K, Winner B, Marchetto MC, Gage FH. Mechanisms underlying inflammation in neurodegeneration. Cell. 2010;140(6):918–934. doi: 10.1016/j.cell.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodenough S, Schäfer M, Behl C. Estrogen-induced cell signalling in a cellular model of Alzheimer’s disease. J Steroid Biochem Mol Biol. 2003;84(2–3):301–305. doi: 10.1016/s0960-0760(03)00043-8. [DOI] [PubMed] [Google Scholar]
- Halliday G, Robinson SR, Shepherd C, Kril J. Alzheimer’s disease and inflammation: a review of cellular and therapeutic mechanisms. Clin Exp Pharmacol Physiol. 2000;27(1–2):1–8. doi: 10.1046/j.1440-1681.2000.03200.x. [DOI] [PubMed] [Google Scholar]
- Hamaguchi T, Ono K, Yamada M. Anti-amyloidogenic therapies: strategies for prevention and treatment of Alzheimer’s disease. Cell Mol Life Sci. 2006;63(13):1538–1552. doi: 10.1007/s00018-005-5599-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdane M, Delobel P, Sambo AV, Smet C, Bégard S, Violleau A, Landrieu I, Delacourte A, Lippens G, Flament S. et al. Neurofibrillary degeneration of the Alzheimer-type: an alternate pathway to neuronal apoptosis? Biochem Pharmacol. 2003;66(8):1619–1625. doi: 10.1016/S0006-2952(03)00533-1. [DOI] [PubMed] [Google Scholar]
- Harold D, Abraham R, Hollingworth P, Sims R, Gerrish A, Hamshere ML, Pahwa JS, Moskvina V, Dowzell K, Williams A. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat Genet. 2009;41(10):1088–1093. doi: 10.1038/ng.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann D, Tournoy J, Saftig P, Annaert W, De Strooper B. Implication of APP secretases in notch signaling. J Mol Neurosci. 2001;17(2):171–181. doi: 10.1385/JMN:17:2:171. [DOI] [PubMed] [Google Scholar]
- Hartmann T. Intracellular biology of Alzheimer’s disease amyloid beta peptide. Eur Arch Psychiatry Clin Neurosci. 1999;249(6):291–298. doi: 10.1007/s004060050102. [DOI] [PubMed] [Google Scholar]
- Hegde AN, Upadhya SC. The ubiquitin-proteasome pathway in health and disease of the nervous system. Trends Neurosci. 2007;30(11):587–595. doi: 10.1016/j.tins.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Hicks D, John D, Makova NZ, Henderson Z, Nalivaeva NN, Turner AJ. Membrane targeting, shedding and protein interactions of brain acetylcholinesterase. J Neurochem. 2011;116(5):742–746. doi: 10.1111/j.1471-4159.2010.07032.x. [DOI] [PubMed] [Google Scholar]
- Hölscher C, Li L. New roles for insulin-like hormones in neuronal signalling and protection: new hopes for novel treatments of Alzheimer’s disease? Neurobiol Aging. 2010;31(9):1495–1502. doi: 10.1016/j.neurobiolaging.2008.08.023. [DOI] [PubMed] [Google Scholar]
- Hooper NM. Roles of proteolysis and lipid rafts in the processing of the amyloid precursor protein and prion protein. Biochem Soc Trans. 2005;33(Pt 2):335–338. doi: 10.1042/BST0330335. [DOI] [PubMed] [Google Scholar]
- Inestrosa NC, Varela-Nallar L, Grabowski CP, Colombres M. Synaptotoxicity in Alzheimer’s disease: the Wnt signaling pathway as a molecular target. IUBMB Life. 2007;59(4–5):316–321. doi: 10.1080/15216540701242490. [DOI] [PubMed] [Google Scholar]
- Issad T, Masson E, Pagesy P. O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab. 2010;36(6 Pt 1):423–435. doi: 10.1016/j.diabet.2010.09.001. [DOI] [PubMed] [Google Scholar]
- Iwata N, Saido TC. Amyloid-beta peptide metabolism and Alzheimer’s disease. Nihon Yakurigaku Zasshi. 2003;122(1):5–14. doi: 10.1254/fpj.122.5. [DOI] [PubMed] [Google Scholar]
- Johnson GV, Bailey CD. The p38 MAP kinase signaling pathway in Alzheimer’s disease. Exp Neurol. 2003;183(2):263–268. doi: 10.1016/S0014-4886(03)00268-1. [DOI] [PubMed] [Google Scholar]
- Kamoun P. Endogenous production of hydrogen sulfide in mammals. Amino Acids. 2004;26(3):243–254. doi: 10.1007/s00726-004-0072-x. [DOI] [PubMed] [Google Scholar]
- Katayama T, Imaizumi K, Manabe T, Hitomi J, Kudo T, Tohyama M. Induction of neuronal death by ER stress in Alzheimer’s disease. J Chem Neuroanat. 2004;28(1–2):67–78. doi: 10.1016/j.jchemneu.2003.12.004. [DOI] [PubMed] [Google Scholar]
- Kim D, Tsai LH. Bridging physiology and pathology in AD. Cell. 2009;137(6):997–1000. doi: 10.1016/j.cell.2009.05.042. [DOI] [PubMed] [Google Scholar]
- Klysik J, Theroux SJ, Sedivy JM, Moffit JS, Boekelheide K. Signaling crossroads: the function of Raf kinase inhibitory protein in cancer, the central nervous system and reproduction. Cell Signal. 2008;20(1):1–9. doi: 10.1016/j.cellsig.2007.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kontush A. Amyloid-beta: an antioxidant that becomes a pro-oxidant and critically contributes to Alzheimer’s disease. Free Radic Biol Med. 2001;31(9):1120–1131. doi: 10.1016/S0891-5849(01)00688-8. [DOI] [PubMed] [Google Scholar]
- Kopan R, Goate A. Aph-2/Nicastrin: an essential component of gamma-secretase and regulator of Notch signaling and Presenilin localization. Neuron. 2002;33(3):321–324. doi: 10.1016/S0896-6273(02)00585-8. [DOI] [PubMed] [Google Scholar]
- Layfield R, Alban A, Mayer RJ, Lowe J. The ubiquitin protein catabolic disorders. Neuropathol Appl Neurobiol. 2001;27(3):171–179. doi: 10.1046/j.1365-2990.2001.00335.x. [DOI] [PubMed] [Google Scholar]
- Lee CY, Landreth GE. The role of microglia in amyloid clearance from the AD brain. J Neural Transm. 2010;117(8):949–960. doi: 10.1007/s00702-010-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre T, Dehennaut V, Guinez C, Olivier S, Drougat L, Mir AM, Mortuaire M, Vercoutter-Edouart AS, Michalski JC. Dysregulation of the nutrient/stress sensor O-GlcNAcylation is involved in the etiology of cardiovascular disorders, type-2 diabetes and Alzheimer’s disease. Biochim Biophys Acta. 2010;1800(2):67–79. doi: 10.1016/j.bbagen.2009.08.008. [DOI] [PubMed] [Google Scholar]
- Li H, Wolfe MS, Selkoe DJ. Toward structural elucidation of the gamma-secretase complex. Structure. 2009;17(3):326–334. doi: 10.1016/j.str.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahley RW, Ji ZS. Remnant lipoprotein metabolism: key pathways involving cell-surface heparan sulfate proteoglycans and apolipoprotein E. J Lipid Res. 1999;40(1):1–16. [PubMed] [Google Scholar]
- Mariño G, López-Otín C. Autophagy: molecular mechanisms, physiological functions and relevance in human pathology. Cell Mol Life Sci. 2004;61(12):1439–1454. doi: 10.1007/s00018-004-4012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters CL, Beyreuther K. Alzheimer’s centennial legacy: prospects for rational therapeutic intervention targeting the Abeta amyloid pathway. Brain. 2006;129(Pt 11):2823–2839. doi: 10.1093/brain/awl251. [DOI] [PubMed] [Google Scholar]
- Mattson MP. Cellular actions of beta-amyloid precursor protein and its soluble and fibrillogenic derivatives. Physiol Rev. 1997;77(4):1081–1132. doi: 10.1152/physrev.1997.77.4.1081. [DOI] [PubMed] [Google Scholar]
- Mattson MP, Chan SL, Camandola S. Presenilin mutations and calcium signaling defects in the nervous and immune systems. Bioessays. 2001;23(8):733–744. doi: 10.1002/bies.1103. [DOI] [PubMed] [Google Scholar]
- McGeer PL, McGeer EG. Anti-inflammatory drugs in the fight against Alzheimer’s disease. Ann N Y Acad Sci. 1996;777:213–220. doi: 10.1111/j.1749-6632.1996.tb34421.x. [DOI] [PubMed] [Google Scholar]
- McLoughlin DM, Miller CC. The FE65 proteins and Alzheimer’s disease. J Neurosci Res. 2008;86(4):744–754. doi: 10.1002/jnr.21532. [DOI] [PubMed] [Google Scholar]
- McTaggart SJ. Isoprenylated proteins. Cell Mol Life Sci. 2006;63(3):255–267. doi: 10.1007/s00018-005-5298-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mischoulon D, Fava M. Role of S-adenosyl-L-methionine in the treatment of depression: a review of the evidence. Am J Clin Nutr. 2002;76(5):1158S–1161S. doi: 10.1093/ajcn/76/5.1158S. [DOI] [PubMed] [Google Scholar]
- Moley KH, Mueckler MM. Glucose transport and apoptosis. Apoptosis. 2000;5(2):99–105. doi: 10.1023/A:1009697908332. [DOI] [PubMed] [Google Scholar]
- Morgan C, Colombres M, Nuñez MT, Inestrosa NC. Structure and function of amyloid in Alzheimer’s disease. Prog Neurobiol. 2004;74(6):323–349. doi: 10.1016/j.pneurobio.2004.10.004. [DOI] [PubMed] [Google Scholar]
- Nagai Y, Ogasawara A, Heese K. Possible mechanisms of A beta(1–40)- or A beta(1–42)-induced cell death and their rescue factors. Nihon Yakurigaku Zasshi. 2004;124(3):135–143. doi: 10.1254/fpj.124.135. [DOI] [PubMed] [Google Scholar]
- Nelson TJ, Alkon DL. Insulin and cholesterol pathways in neuronal function, memory and neurodegeneration. Biochem Soc Trans. 2005;33(Pt 5):1033–1036. doi: 10.1042/BST20051033. [DOI] [PubMed] [Google Scholar]
- Nixon RA. A “protease activation cascade” in the pathogenesis of Alzheimer’s disease. Ann N Y Acad Sci. 2000;924:117–131. [PubMed] [Google Scholar]
- Pascale A, Amadio M, Govoni S, Battaini F. The aging brain, a key target for the future: the protein kinase C involvement. Pharmacol Res. 2007;55(6):560–569. doi: 10.1016/j.phrs.2007.04.013. [DOI] [PubMed] [Google Scholar]
- Poirier J. Apolipoprotein E and Alzheimer’s disease. A role in amyloid catabolism. Ann N Y Acad Sci. 2000;924:81–90. doi: 10.1111/j.1749-6632.2000.tb05564.x. [DOI] [PubMed] [Google Scholar]
- Poirier J. Apolipoprotein E, cholesterol transport and synthesis in sporadic Alzheimer’s disease. Neurobiol Aging. 2005;26(3):355–361. doi: 10.1016/j.neurobiolaging.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Puglielli L. Aging of the brain, neurotrophin signaling, and Alzheimer’s disease: is IGF1-R the common culprit? Neurobiol Aging. 2008;29(6):795–811. doi: 10.1016/j.neurobiolaging.2007.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid PC, Urano Y, Kodama T, Hamakubo T. Alzheimer’s disease: cholesterol, membrane rafts, isoprenoids and statins. J Cell Mol Med. 2007;11(3):383–392. doi: 10.1111/j.1582-4934.2007.00054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers JT, Bush AI, Cho HH, Smith DH, Thomson AM, Friedlich AL, Lahiri DK, Leedman PJ, Huang X, Cahill CM. Iron and the translation of the amyloid precursor protein (APP) and ferritin mRNAs: riboregulation against neural oxidative damage in Alzheimer’s disease. Biochem Soc Trans. 2008;36(Pt 6):1282–1287. doi: 10.1042/BST0361282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo LE, Fernández JA, Maccioni AA, Jimenez JM, Maccioni RB. Neuroinflammation: implications for the pathogenesis and molecular diagnosis of Alzheimer’s disease. Arch Med Res. 2008;39(1):1–16. doi: 10.1016/j.arcmed.2007.10.001. [DOI] [PubMed] [Google Scholar]
- Rosner M, Hanneder M, Siegel N, Valli A, Fuchs C, Hengstschläger M. The mTOR pathway and its role in human genetic diseases. Mutat Res. 2008;659(3):284–292. doi: 10.1016/j.mrrev.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Rossner S. New players in old amyloid precursor protein-processing pathways. Int J Dev Neurosci. 2004;22(7):467–474. doi: 10.1016/j.ijdevneu.2004.07.004. [DOI] [PubMed] [Google Scholar]
- Rossner S, Ueberham U, Schliebs R, Perez-Polo JR, Bigl V. The regulation of amyloid precursor protein metabolism by cholinergic mechanisms and neurotrophin receptor signaling. Prog Neurobiol. 1998;56(5):541–569. doi: 10.1016/S0301-0082(98)00044-6. [DOI] [PubMed] [Google Scholar]
- Rushworth JV, Hooper NM. Lipid Rafts: Linking Alzheimer’s Amyloid-β Production, Aggregation, and Toxicity at Neuronal Membranes. Int J Alzheimers Dis. 2010;2011:603052. doi: 10.4061/2011/603052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen A, Ojala J, Kauppinen A, Kaarniranta K, Suuronen T. Inflammation in Alzheimer’s disease: amyloid-beta oligomers trigger innate immunity defence via pattern recognition receptors. Prog Neurobiol. 2009;87(3):181–194. doi: 10.1016/j.pneurobio.2009.01.001. [DOI] [PubMed] [Google Scholar]
- Schöneich C. Methionine oxidation by reactive oxygen species: reaction mechanisms and relevance to Alzheimer’s disease. Biochim Biophys Acta. 2005;1703(2):111–119. doi: 10.1016/j.bbapap.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Selkoe DJ. Notch and presenilins in vertebrates and invertebrates: implications for neuronal development and degeneration. Curr Opin Neurobiol. 2000;10(1):50–57. doi: 10.1016/S0959-4388(99)00054-9. [DOI] [PubMed] [Google Scholar]
- Selkoe D, Kopan R. Notch and Presenilin: regulated intramembrane proteolysis links development and degeneration. Annu Rev Neurosci. 2003;26:565–597. doi: 10.1146/annurev.neuro.26.041002.131334. [DOI] [PubMed] [Google Scholar]
- Sinha S, Lieberburg I. Cellular mechanisms of beta-amyloid production and secretion. Proc Natl Acad Sci U S A. 1999;96(20):11049–11053. doi: 10.1073/pnas.96.20.11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SA. Retromer sorting: a pathogenic pathway in late-onset Alzheimer disease. Arch Neurol. 2008;65(3):323–328. doi: 10.1001/archneurol.2007.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small SA, Duff K. Linking Abeta and tau in late-onset Alzheimer’s disease: a dual pathway hypothesis. Neuron. 2008;60(4):534–542. doi: 10.1016/j.neuron.2008.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreghan B, Thomas SN, Yang AJ. Aberrant sphingomyelin/ceramide metabolic-induced neuronal endosomal/lysosomal dysfunction: potential pathological consequences in age-related neurodegeneration. Adv Drug Deliv Rev. 2003;55(11):1515–1524. doi: 10.1016/j.addr.2003.07.007. [DOI] [PubMed] [Google Scholar]
- Stark DT, Bazan NG. Neuroprotectin D1 induces neuronal survival and downregulation of amyloidogenic processing in Alzheimer’s disease cellular models. Mol Neurobiol. 2011;43(2):131–138. doi: 10.1007/s12035-011-8174-4. [DOI] [PubMed] [Google Scholar]
- Tafti M, Ghyselinck NB. Functional implication of the vitamin A signaling pathway in the brain. Arch Neurol. 2007;64(12):1706–1711. doi: 10.1001/archneur.64.12.1706. [DOI] [PubMed] [Google Scholar]
- Takashima A. Drug development targeting the glycogen synthase kinase-3beta (GSK-3beta)-mediated signal transduction pathway: role of GSK-3beta in adult brain. J Pharmacol Sci. 2009;109(2):174–178. doi: 10.1254/jphs.08R29FM. [DOI] [PubMed] [Google Scholar]
- Takatori Y. Mechanisms of neuroprotective effects of therapeutic acetylcholinesterase inhibitors used in treatment of Alzheimer’s disease. Yakugaku Zasshi. 2006;126(8):607–616. doi: 10.1248/yakushi.126.607. [DOI] [PubMed] [Google Scholar]
- Toledo EM, Colombres M, Inestrosa NC. Wnt signaling in neuroprotection and stem cell differentiation. Prog Neurobiol. 2008;86(3):281–296. doi: 10.1016/j.pneurobio.2008.08.001. [DOI] [PubMed] [Google Scholar]
- Townsend KP, Obregon D, Quadros A, Patel N, Volmar C, Paris D, Mullan M. Proinflammatory and vasoactive effects of Abeta in the cerebrovasculature. Ann N Y Acad Sci. 2002;977:65–76. doi: 10.1111/j.1749-6632.2002.tb04799.x. [DOI] [PubMed] [Google Scholar]
- Turner PR, O’Connor K, Tate WP, Abraham WC. Roles of amyloid precursor protein and its fragments in regulating neural activity, plasticity and memory. Prog Neurobiol. 2003;70(1):1–32. doi: 10.1016/S0301-0082(03)00089-3. [DOI] [PubMed] [Google Scholar]
- Uthayathas S, Karuppagounder SS, Thrash BM, Parameshwaran K, Suppiramaniam V, Dhanasekaran M. Versatile effects of sildenafil: recent pharmacological applications. Pharmacol Rep. 2007;59(2):150–163. [PubMed] [Google Scholar]
- Van Nostrand WE, Melchor J, Wagner M, Davis J. Cerebrovascular smooth muscle cell surface fibrillar A beta. Alteration of the proteolytic environment in the cerebral vessel wall. Ann N Y Acad Sci. 2000;903:89–96. doi: 10.1111/j.1749-6632.2000.tb06354.x. [DOI] [PubMed] [Google Scholar]
- Vasto S, Candore G, Listì F, Balistreri CR, Colonna-Romano G, Malavolta M, Lio D, Nuzzo D, Mocchegiani E, Di Bona D. et al. Inflammation, genes and zinc in Alzheimer’s disease. Brain Res Rev. 2008;58(1):96–105. doi: 10.1016/j.brainresrev.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Vetrivel KS, Thinakaran G. Membrane rafts in Alzheimer’s disease beta-amyloid production. Biochim Biophys Acta. 2010;1801(8):860–867. doi: 10.1016/j.bbalip.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LH, Besirli CG, Johnson EM. Mixed-lineage kinases: a target for the prevention of neurodegeneration. Annu Rev Pharmacol Toxicol. 2004;44:451–474. doi: 10.1146/annurev.pharmtox.44.101802.121840. [DOI] [PubMed] [Google Scholar]
- Weggen S, Rogers M, Eriksen J. NSAIDs: small molecules for prevention of Alzheimer’s disease or precursors for future drug development? Trends Pharmacol Sci. 2007;28(10):536–543. doi: 10.1016/j.tips.2007.09.004. [DOI] [PubMed] [Google Scholar]
- Wei J, Bhattacharyya S, Varga J. Peroxisome proliferator-activated receptor γ: innate protection from excessive fibrogenesis and potential therapeutic target in systemic sclerosis. Curr Opin Rheumatol. 2010;22(6):671–676. doi: 10.1097/BOR.0b013e32833de1a7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellington CL, Hayden MR. Caspases and neurodegeneration: on the cutting edge of new therapeutic approaches. Clin Genet. 2000;57(1):1–10. doi: 10.1034/j.1399-0004.2000.570101.x. [DOI] [PubMed] [Google Scholar]
- Woodgett JR. Judging a protein by more than its name: GSK-3. Sci STKE. 2001;2001(100):re12. doi: 10.1126/stke.2001.100.re12. [DOI] [PubMed] [Google Scholar]
- Wu HY, Tomizawa K, Matsui H. Calpain-calcineurin signaling in the pathogenesis of calcium-dependent disorder. Acta Med Okayama. 2007;61(3):123–137. doi: 10.18926/AMO/32905. [DOI] [PubMed] [Google Scholar]
- Wyss-Coray T, Rogers J. Inflammation in Alzheimer Disease−−A Brief Review of the Basic Science and Clinical Literature. Cold Spring Harbor Perspect Med. 2012;2(1):a006346. doi: 10.1101/cshperspect.a006346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y, Gaynor RB. Therapeutic potential of inhibition of the NF-kappaB pathway in the treatment of inflammation and cancer. J Clin Invest. 2001;107(2):135–142. doi: 10.1172/JCI11914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang SY, He XY, Miller D. HSD17B10: a gene involved in cognitive function through metabolism of isoleucine and neuroactive steroids. Mol Genet Metab. 2007;92(1–2):36–42. doi: 10.1016/j.ymgme.2007.06.001. [DOI] [PubMed] [Google Scholar]
- Yoshimura T, Arimura N, Kaibuchi K. Molecular mechanisms of axon specification and neuronal disorders. Ann N Y Acad Sci. 2006;1086:116–125. doi: 10.1196/annals.1377.013. [DOI] [PubMed] [Google Scholar]
- Zhao WQ, Townsend M. Insulin resistance and amyloidogenesis as common molecular foundation for type 2 diabetes and Alzheimer’s disease. Biochim Biophys Acta. 2009;1792(5):482–496. doi: 10.1016/j.bbadis.2008.10.014. [DOI] [PubMed] [Google Scholar]
- Zipp F, Waiczies S, Aktas O, Neuhaus O, Hemmer B, Schraven B, Nitsch R, Hartung HP. Impact of HMG-CoA reductase inhibition on brain pathology. Trends Pharmacol Sci. 2007;28(7):342–349. doi: 10.1016/j.tips.2007.05.001. [DOI] [PubMed] [Google Scholar]
- Zlokovic BV. New therapeutic targets in the neurovascular pathway in Alzheimer’s disease. Neurotherapeutics. 2008;5(3):409–414. doi: 10.1016/j.nurt.2008.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoot ME, Ono K, Ruscheinski J, Wang PL, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27(3):431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
SBML map file of AlzPathway. The SBML map file alzpathway_sbml_map.xml can be browsed using CellDesigner. Please download CellDesigner at http://www.celldesigner.org/, install it, and open the SBML map file alzpathway_sbml_map.xml to browse AlzPathway map by CellDesigner. As for usage of CellDesigner software, see the documentations provided at CellDesigner web site: http://www.celldesigner.org/documents.html.
High resolution image map of AlzPathway. The PNG file alzpathway_image_map.png contains a high resolution map of AlzPathway. This image map does not contain the reference information used for constructing the AlzPathway. SBML (CellDesigner) map and Online map (Payao) are recommended to browse the AlzPathway map.
Instruction on usage of Payao. The PDF file alzpathway_payao_access_instruction.pdf contains insturction on usage of Payao web service (online map). Payao system requires login. AlzPathway demo user account is prepared for demo use: demo username is “ap_demo” and the corresponding password is “4patients”. As for usage of Payao system, see the user’s guide provided at Payao web site: http://payao.oist.jp:8080/payaologue/doc/PAYAO_Users_GuideE11.pdf.
Binary-relation notation file of AlzPathway. The SIF (simple interaction format) file alzpathway_binary_relation.sif is the binary-relation notation of AlzPathway map which can be opened by using Cytoscape.
Pure SBML map file of AlzPathway. The pure SBML file alzpathway_pure_sbml.xml is for compatibility with other SMBL supporting tools.
BioPAX file of AlzPathway. The BioPAX file alzpathway_biopax.owl is for exchanges of pathway data.
Expanded SBML map file of AlzPathway with Gene Ontology. The SBML map file alzpathway_sbml_map_go.xml is the expanded version of AlzPathway map (alzpathway_sbml_map.xml) having external links to Gene Ontology. The SBML map file with the external links to Gene Ontology requires time to be opened by CellDesigner 4.2, and thus was provided separately as an expanded SBML map.