Abstract
Background
Cyanobacteria possess several cytochrome P450s, but very little is known about their catalytic functions. CYP110 genes unique to cyanaobacteria are widely distributed in heterocyst-forming cyanobacteria including nitrogen-fixing genera Nostoc and Anabaena. We screened the biocatalytic functions of all P450s from three cyanobacterial strains of genus Nostoc or Anabaena using a series of small molecules that contain flavonoids, sesquiterpenes, low-molecular-weight drugs, and other aromatic compounds.
Results
Escherichia coli cells carrying each P450 gene that was inserted into the pRED vector, containing the RhFRed reductase domain sequence from Rhodococcus sp. NCIMB 9784 P450RhF (CYP116B2), were co-cultured with substrates and products were identified when bioconversion reactions proceeded. Consequently, CYP110E1 of Nostoc sp. strain PCC 7120, located in close proximity to the first branch point in the phylogenetic tree of the CYP110 family, was found to be promiscuous for the substrate range mediating the biotransformation of various small molecules. Naringenin and (hydroxyl) flavanones were respectively converted to apigenin and (hydroxyl) flavones, by functioning as a flavone synthase. Such an activity is reported for the first time in prokaryotic P450s. Additionally, CYP110E1 biotransformed the notable sesquiterpene zerumbone, anti-inflammatory drugs ibuprofen and flurbiprofen (methylester forms), and some aryl compounds such as 1-methoxy and 1-ethoxy naphthalene to produce hydroxylated compounds that are difficult to synthesize chemically, including novel compounds.
Conclusion
We elucidated that the CYP110E1 gene, C-terminally fused to the P450RhF RhFRed reductase domain sequence, is functionally expressed in E. coli to synthesize a robust monooxygenase, which shows promiscuous substrate specificity (affinity) for various small molecules, allowing the biosynthesis of not only flavones (from flavanones) but also a variety of hydroxyl-small molecules that may span pharmaceutical and nutraceutical industries.
Keywords: Cyanobacterium, Cytochrome P450, CYP110, Flavone synthase, Zerumbone, Nostoc sp. strain PCC 7120
Background
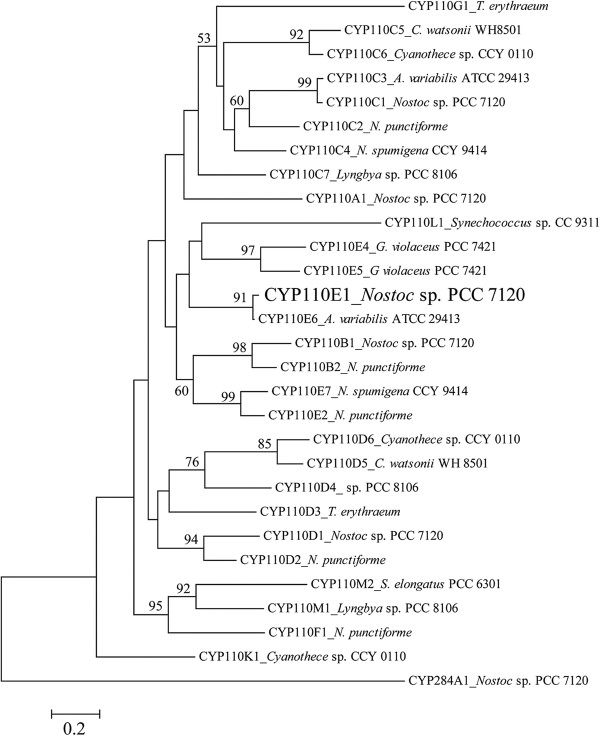
Cyanobacteria possess several cytochrome P450s (P450s), but only a few reports exist regarding their catalytic functions [1-3]. P450 CYP110 is a prominent family found in heterocyst-forming cyanobacteria including nitrogen-fixing genera Nostoc and Anabaena. CYP110 genes are widely distributed in such cyanobacterial strains [1], e.g., Nostoc (also referred to as Anabaena) sp. strain PCC 7120 possesses five CYP110 genes [classified as CYP110A1 (alr1450), CYP110B1 (all3746), CYP110C1 (alr4686), CYP110D1 (alr4766), and CYP110E1 (alr4833)] in addition to one P450 gene of another family (CYP284A1) (Figure 1) [4]. The P450NS gene (CYP110C1) is positioned adjacent to the NS1gene encoding germacrene A synthase (alr4685) [2]. The encoded protein (CYP110C1) was recently confirmed to convert germacrene A to a hydroxylated sesquiterpene (1,2,3,5,6,7,8,8a-octahydro-6-isopropenyl-4,8a-dimethylnaphth-1-ol) [3]. CYP110A1 is the first reported cyanobacterial P450 gene, which was present in a conserved 11.25-kb episomal element (the nifD element), and the encoded protein (CYP110A1) was hypothesized to be a fatty acid ω-hydroxylase, based on its substrate binding profile and amino acid sequence similarities to P450BM3 (CYP102A1) of Bacillus megaterium and the mammalian P450 family 4 fatty acid ω-hydroxylase [1,5].
Figure 1.
Phylogenetic positions of cyanobacterial CYP110 family using 27 CYP110 proteins derived from 11 cyanobacteria, whose amino acid sequences are shown in CyanoBase. The accession numbers in parentheses show those of respective P450 proteins. CYP110E1 is shown in boldface. The phylogenetic tree was constructed using the neighbor-joining method. The number shown next to each node indicates the percentage bootstrap value of 1,000 replicates (only 50% and higher are cited). The scale bar indicates a genetic distance of 0.02 (Knuc).
In order to function as terminal monooxygenases, P450s must be associated with one or two additional proteins (or protein domains) to transfer two electrons from NAD(P)H to the heme domain of the P450 protein [6,7]. The vast majority of bacterial P450s need a FAD-containing ferredoxin reductase to receive electrons from NAD(P)H, and a ferredoxin (small iron-sulfur protein) to receive them from ferredoxin reductase, which subsequently reduces P450 itself [class I system; Additional file 1 Figure S1a[7]. P450RhF (CYP116B2) derived from Rhodococcus sp. NCIMB 9784 was discovered to be a self-sufficient P450 protein, in which the P450 domain is C-terminally fused to a reductase domain (here called RhFRed) [8]. RhFRed contained an FMN-binding ferredoxin reductase subdomain to receive electrons from NAD(P)H and a [2Fe-2S] ferredoxin subdomain [9]. A short linker region of 16 amino acids existed between P450 and RhFRed [8]. The redox chain of P450RhF resembles that of the class I system [Additional file 1 Figure S1c. Thus, vector pRED was constructed for the functional expression of bacterial P450 (class I) genes in Escherichia coli, using the linker sequence and RhFRed domain sequence [( Additional file 1Figure S2[10]. This vector has been demonstrated to be useful for functional expression of the P450cam gene (CYP101A1) [10,11], P450bzo gene (CYP203A) [10], P450balk gene (CYP153A13a) [12,13], other CYP153A genes [14], and P450 PikC gene [15], constituting corresponding self-sufficient P450 monooxygenation enzymes. We elucidate here that the CYP110E1 gene is functionally expressed on pRED in E. coli to synthesize a robust monooxygenase, which shows promiscuous substrate specificity (affinity) for various small molecules.
Results
Screening experiments
Nostoc sp. strain PCC 7120, Nostoc punctiforme PCC 73102, and Anabaena variabilis ATCC 29413 possess six, ten, and four P450s, respectively. We screened the biocatalytic functions of these P450s using 47 small molecules that contain flavonoids, sesquiterpenes, low-molecular-weight drugs, naphthalene derivatives, and other chemicals with benzene rings [ Additional file 1 Figure S3)]. E. coli BL21 (DE3) cells carrying each P450 gene inserted into the pRED vector were co-cultured with the substrates and possible bioconversion products were analyzed by HPLC. Consequently, CYP110E1 of Nostoc PCC 7120 was found to be promiscuous for the substrate range mediating the biotransformation of various small molecules. The CYP110E1 enzyme that is C-terminally fused to RhFRed was confirmed to constitute the active P450 form by CO difference spectral analysis [ Additional file 1Figure S4]. Thus, cells of E. coli BL21 (DE3) carrying plasmid pCYP110E1-Red were used for the following experiments.
Bioconversion of flavanones by E. coli (pCYP110E1-Red)
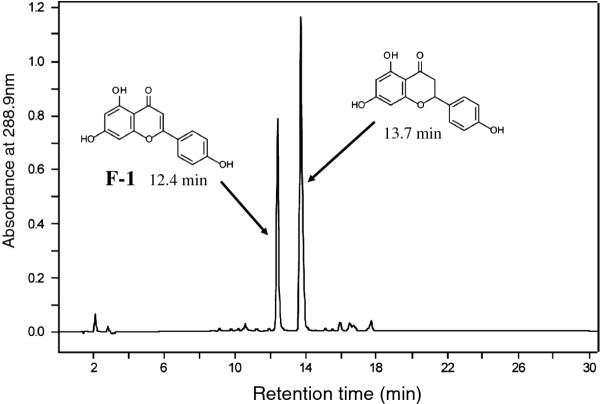
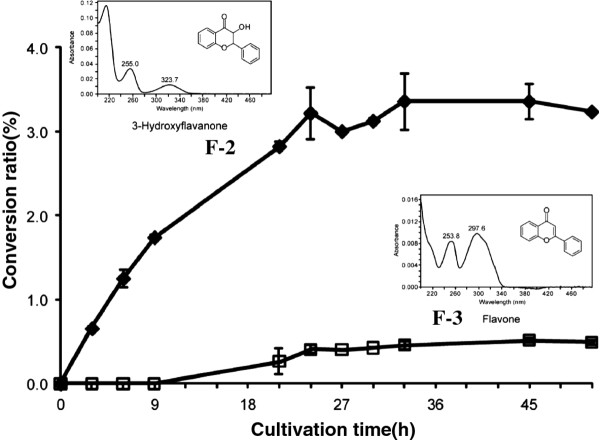
Naringenin was biotransformed to a product (F-1) with a conversion ratio of 31.5% (Figure 2) through co-cultivation with cells of E. coli (pCYP110E1-Red). F-1 was identified as apigenin (4’,5,7-trihydroxyflavone) by its comparison with an authentic sample on HPLC analysis. Flavanone (RT 18.2 min) was converted to products F-2 (RT 15.9 min; 5.7%) and F-3 (RT 17.2 min; 2.1%), which were identified as 3-hydroxyflavanone and flavone, respectively, by their comparison with authentic samples on HPLC analysis. Figure 3 shows their production rate curves. Since this P450 was thought to biotransform various flavanones, we further examined 6-hydroxyflavanone (RT 15.9 min) and 7-hydroxyflavanone (RT 15.4 min). As a result, these hydroxyflavanones were converted to products F-4 (RT 15.1 min; 12.1%) and F-5 (RT 14.5 min; 1.4%), which were identified as 6-hydroxyflavone and 7-hydroxyflavone, respectively, by their comparison with authentic samples on HPLC analysis.
Figure 2.
HPLC analysis of the ethylacetate extract of the co-culture of cells of E. coliBL21 (DE3) carrying plasmid pCYP110E1-Red with naringenin. 31.5% of naringenin was converted to F-1, which was identified as apigenin.
Figure 3.
Conversion rate of flavanone to F-2 [3-hydroxyflavanone (♦)] and F-3 [flavone (□)] byE. coliBL21 (DE3) cells carrying pCYP110E1-Red. Error bars indicate the standard deviations obtained for three independent experiments. The conversion ratio (%) was measured by the ratio of peak area (max plot) in HPLC.
Bioconversion of a sesquiterpene by E. coli (pCYP110E1-Red)
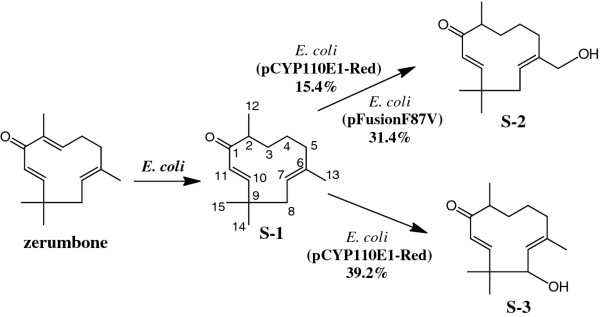
Only zerumbone among the examined terpenes [ Additional file 1: Figure S3] was biotransformed through co-cultivation with cells of E. coli (pCYP110E1-Red). The crude ethyl acetate (EtOAc) extract (152.0 mg) from this bioconversion mixture (200 ml), subjected to silica gel column chromatography (hexane-EtOAc = 2:1), yielded 5.6 mg (12.7%) of S-1 (fr. 10–12), 3.2 mg (6.8%) of S-2 (fr. 19–26), and 1.5 mg (3.2%) of S-3 (fr. 30–40). These spectroscopic data are shown in Additional file 2.
The molecular formula of S-1 was determined to be C15H24O (zerumbone + 2 H) by HREI-MS. Consistent with its molecular formula, S-1 was proposed to be a product obtained by the reduction of a double bond in the substrate. The reduced double bond was determined to be 2,3Δ by the observation of a doublet methyl signal (δH 1.05, H-12), and the 1H-13C long range coupling from this doublet methyl to the ketone carbon (δC 205.1, C-1). The identity of S-1 was thus determined as (6E,10E)-2,6,9,9-tetramethylcycloundeca-2,6-dien-1-one (Figure 4) [16]. Zerumbone was found to be converted to S-1 with nontransformed E. coli BL21 (DE3) cells (data not shown). It was therefore thought that S-2 and S-3 were the genuine products by CYP110E1-Red.
Figure 4.
Bioconversion of sesquiterpene zerumbone with E. coli BL21 (DE3) cells carrying pCYP110E1-Red. % indicates conversion ratio measured by the ratio of peak area (max plot) in HPLC. E. coli BL21 (DE3) cells carrying plasmid pFusionF87V that expressed the P450BM3 (F87V) gene [17] were also examined for the ability to biotransform zerumbone, and the result is shown here.
The molecular formula of S-2 was determined to be C15H24O2 by HREI-MS. Consistent with its molecular formula and 1H-NMR spectrum, the introduction of an alcoholic OH group in S-1 was proposed. The position of the alcoholic OH group was clarified to be C-13 by the observation of an oxymethylene signal (δH 3.92 and δH 4.02, H-13) and the 1H-13C long range coupling from this oxymethylene to C-5 (δC 35.0), C-6 (δC 140.1), and C-7 (δC 126.0). The identity of S-2 was thus determined as (6Z,10E)-6-hydroxymethyl-2,9,9-trimethylcycloundeca-2-ene-1-one (Figure 4). This product (S-2) was a novel compound according to the CAS database.
The molecular formula of S-3 was determined to be C15H24O2 by HREI-MS. Consistent with its molecular formula and 1H-NMR spectrum, the introduction of an alcoholic OH group in S-1 was proposed. The position of the alcoholic OH group was clarified to be C-8 by the observation of an oxymethylene signal (δH 4.24, H-8) and the 1H-13C long range coupling from H-14 (δH 1.12) and H-15 (δH 1.26) to C-8 (δC 75.5). The identity of S-3 was thus determined as (6E,10E)-8-hydroxy-2,6,9,9-tetramethylcycloundeca-2,6-dien-1-one (Figure 4). This product (S-3) was a novel compound according to the CAS database.
Bioconversion of aryl compounds by E. coli (pCYP110E1-Red)
A variety of aryl compounds, which include naphthalene derivatives and low-molecular-weight drugs, were biotransformed through co-cultivation with cells of E. coli (pCYP110E1-Red). Converted compounds were identified by chromatographic and spectroscopic analyses. Spectroscopic data are shown in Additional file 2.
Compounds converted from 1-methoxynaphthalene
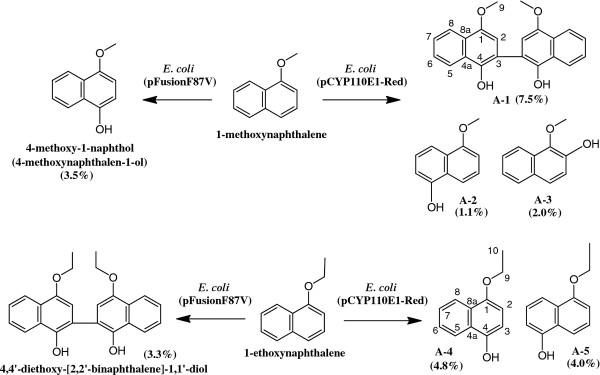
The crude EtOAc extract (156.5 mg) from the bioconversion mixture (200 ml) with 1-methoxynaphthalene and E. coli (pCYP110E1-Red), subjected to silica gel column chromatography (hexane-EtOAc = 6:1), yielded 2.6 mg (7.5%) of A-1 (fr. 13–15), 0.4 mg (1.1%) of A-2 (fr. 19–22), and 0.7 mg (2.0%) of A-3 (fr. 28–35).
The molecular formula of A-1 was determined to be C22H18O4 by HRAPCI-MS. Consistent with its molecular formula and 1H-NMR spectrum, A-1 was proposed to be a secondary product (dimer) obtained by a phenol oxidation coupling reaction of the direct conversion product. The structure of A-1 was determined to be 4,4'-dimethoxy-[2,2'-binaphthalene]-1,1'-diol (Figure 5) by the observation of 1H-13C long range couplings from H-2 (2’) (δ 6.92) to C-1 (1’) (δ 149.3), C-3 (3’) (δ 120.4), and C-4 (4’) (δ 142.1), and 1H vicinal spin network of H-5 (5’) (δ 8.30) – H-6 (6’) (δ 7.54) – H-7 (7’) (δ 7.49) - H-8 (8’) (δ 8.20) [18].
Figure 5.
Bioconversion of 1-methoxynaphthalene and 1-ethoxynaphthalene with E. coli BL21 (DE3) cells carrying pCYP110E1-Red.(%) indicates yield (calculated from the weight of a purified product). The result with E. coli BL21 (DE3) cells carrying plasmid pFusionF87V [17] is also shown for comparison.
The molecular formula of A-2 was determined to be C11H10O2 by HRAPCI-MS. Consistent with its molecular formula and 1H-NMR spectrum, the introduction of one phenolic OH group in the aromatic ring was proposed. The position of this phenolic OH group was determined to be C-5 by the observation of 1H-1H vicinal spin networks of H-2 (δ 6.84) – H-3 (δ 7.39) – H-4 (δ 7.74) and H-6 (δ 6.84) – H-7 (δ 7.30) – H-8 (δ 7.85), and an NOE observed between H-8 and H-9 (δ 3.99). The identity of A-2 was thus determined to be 5-methoxynaphthalen-1-ol (Figure 5) [17].
The molecular formula of A-3 was determined to be C11H10O2 by HRAPCI-MS. Consistent with its molecular formula and 1H-NMR spectrum, the introduction of one phenolic OH group in the aromatic ring was proposed. The position of this phenolic OH group was determined to be C-2 by the observation of 1H-1H vicinal spin coupling of H-3 (δ 7.23, d, J = 8.5 Hz) and H-4 (δ 7.57, d, J = 8.5 Hz) and 1H-13C long range couplings from H-4 to C-2 (δ 145.4) and C-5 (δ 128.3). The identity of A-3 was thus determined to be 1-methoxynaphthalen-2-ol (Figure 5) [19].
Compounds converted from 1-ethoxynaphthalene
The crude EtOAc extract (89.3 mg) from the bioconversion mixture (200 ml) with 1-ethoxynaphthalene and E. coli (pCYP110E1-Red), subjected to silica gel column chromatography (hexane-EtOAc = 6:1), yielded 1.8 mg (4.8%) of A-4 (fr. 10–12) and 1.5 mg (4.0%) of A-5 (fr. 14–16).
The molecular formula of A-4 was determined to be C12H12O2 by HRAPCI-MS. Consistent with its molecular formula and 1H-NMR spectrum, the introduction of one phenolic OH group in the aromatic ring was proposed. The position of this phenolic OH group was determined to be C-4 by the observation of 1H-1H vicinal spin coupling of H-2 (δ 7.55, d, J = 8.7 Hz) and H-3 (δ 7.23, d, J = 8.7 Hz) and the 1H-13C long range couplings from H-5 (δ 7.78) to C-4 (δ 145.8). The identity of A-4 was thus determined to be 4-ethoxynaphthalen-1-ol (Figure 5) [20].
The molecular formula of A-5 was determined to be C12H12O2 by HRAPCI-MS. Consistent with its molecular formula and 1H-NMR spectrum, the introduction of one phenolic OH group in the aromatic ring was proposed. The position of this phenolic OH group was determined to be C-5 by the observation of 1H-1H vicinal spin networks of H-2 (δ 6.83) – H-3 (δ 7.37) – H-4 (δ 7.71), and H-6 (δ 6.85) – H-7 (δ 7.29) – H-8 (δ 7.89), and the 1H-13C long range couplings from H-3 to C-1 (δ 154.8) and C-4a (δ 125.4) and from H-7 to C-5 (δ 151.2) and C-8a (δ 127.3). The identity of A-5 was thus determined to be 5-ethoxynaphthalen-1-ol (Figure 5) [21].
Compounds converted from 2-methylnaphthalene
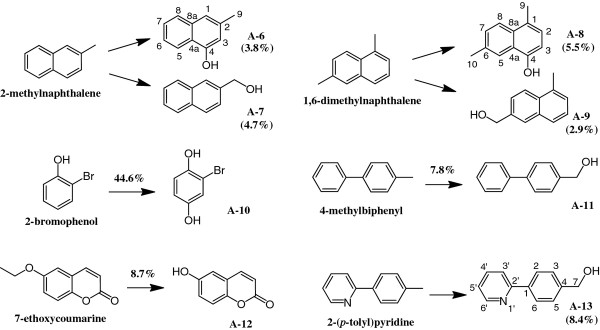
The crude EtOAc extract (175.0 mg) from the bioconversion mixture (200 ml) with 2-methylnaphthalene and E. coli (pCYP110E1-Red), subjected to silica gel column chromatography (hexane-EtOAc = 5:1), yielded 1.2 mg (3.8%) of A-6 (fr. 11–13) and 1.5 mg (4.7%) of A-7 (fr. 20–24).
The molecular formula of A-6 was determined to be C11H10O by HRAPCI-MS. Consistent with its molecular formula and 1H-NMR spectrum, the introduction of one phenolic OH group in the aromatic ring was proposed. The position of this phenolic OH group was determined to be C-4 by the observation of two singlet sp2 methines (δ 6.67 and δ 7.22) and the 1H-1H vicinal spin networks of H-5 (δ 8.10) – H-6 (δ 7.40) – H-7 (δ 7.43) - H-8 (δ 7.71). The identity of A-6 was thus determined to be 3-methylnaphthalen-1-ol (Figure 6). A-7 was identified as naphthalene-2-ylmethanol (Figure 6) with HPLC analysis by its comparison with an authentic sample extracted from co-culture with 2-methylnaphthalene and E. coli BL21 cells carrying plasmid pUCRED-Balk, which expressed the CYP153A13a gene [13].
Figure 6.
Bioconversion of other aryl compounds with E. coli BL21 (DE3) cells carrying pCYP110E1-Red. % and (%) indicate conversion ratio and yield, respectively.
Compounds converted from 1,6-dimethylnaphthalene
The crude EtOAc extract (95.7 mg) from the bioconversion mixture (200 ml) with 1,6-dimethylnaphthalene and E. coli (pCYP110E1-Red), subjected to silica gel column chromatography (hexane-EtOAc = 6:1), yielded 1.9 mg (5.5%) of A-8 (fr. 12–15) and 1.0 mg (2.9%) of A-9 (fr. 21–26).
The molecular formula of A-8 was determined to be C12H12O by HRAPCI-MS. Consistent with its molecular formula and 1H-NMR spectrum, the introduction of one phenolic OH group in the aromatic ring was proposed. The position of this phenolic OH group was determined to be C-4 by the observation of 1H-1H vicinal spin couplings of H-2 (δ 7.05, d, J = 7.9 Hz) and H-3 (δ 6.70, d, J = 7.9 Hz), and H-7 (δ 7.37, d, J = 8.6 Hz) and H-8 (δ 7.84, d, J = 8.6 Hz), and 1H-13C long range couplings from H-9 (δ 2.58) to C-1 (δ 126.6), C-2 (δ 125.1), and C-8a (δ 131.7). The identity of A-8 was thus determined to be 4,7-dimethylnaphthalen-1-ol [17]. A-9 was identified as (5-methylnaphthalen-2-yl)methanol (Figure 6) with HPLC analysis by its comparison with an authentic sample extracted from co-culture with 1,6-dimethylnaphthalene and E. coli BL21 (pUCRED-Balk) [13].
A compound converted from 2-bromophenol
The EtOAc extract from the bioconversion mixture (0.5 ml) with 2-bromophenol and E. coli (pCYP110E1-Red) was subjected to HPLC to yield a product (A-10). A-10 was identified as 2-bromobenzene-1,4-diol (Figure 6) with HPLC by its comparison with an authentic sample extracted from co-culture with 2-bromophenol and E. coli BL21 (pUCRED-Balk) [13].
A compound converted from 4-methylbiphenyl
The EtOAc extract from the bioconversion mixture (0.5 ml) with 4-methylbiphenyl and E. coli (pCYP110E1-Red) was subjected to HPLC to yield a product (A-11). A-11 was identified as [1,1’-biphenyl]-4-ylmethanol (Figure 6) with HPLC by its comparison with an authentic sample extracted from co-culture with 4-methylbiphenyl and E. coli BL21 (pUCRED-Balk) [13].
A compound converted from 7-ethoxycoumarine
The EtOAc extract from the bioconversion mixture (0.5 ml) with 7-ethoxycoumarine and E. coli (pCYP110E1-Red) was subjected to HPLC to yield a product (A-12). A-12 was identified as 6-hydroxy-2 H-chromen-2-one (Figure 6) with HPLC by its comparison with an authentic sample.
A compound converted from 2-(p-tolyl)pyridine
The crude EtOAc extract (103.7 mg) from the bioconversion mixture (200 ml) with 2-(p-tolyl)pyridine and E. coli (pCYP110E1-Red), subjected to silica gel column chromatography (CH2Cl2-MeOH = 20:1), yielded 3.1 mg (8.4%) of A-13 (fr. 23–27). The molecular formula of A-13 was determined to be C12H11NO by HREI-MS. The 1H- and 13C-NMR spectra showed the methyl group in the substrate was oxidized to the corresponding primary alcohol. The identity of A-13 was thus determined to be (4-(pyridin-2-yl)phenyl)methanol (Figure 6), which was also produced through co-culture with 2-(p-tolyl)pyridine and E. coli BL21 (pUCRED-Balk) [13].
A compound converted from ibuprofen methylester
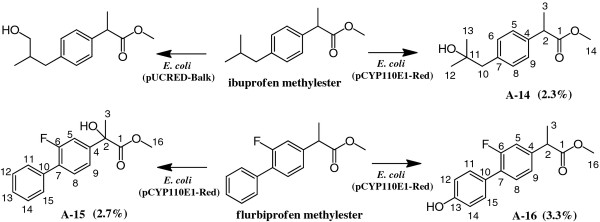
The crude EtOAc extract (225.0 mg) from the bioconversion mixture (200 ml) with ibuprofen methylester and E. coli (pCYP110E1-Red), subjected to silica gel column chromatography (hexane-EtOAc = 4:1), yielded 1.1 mg (2.3%) of A-14. The molecular formula of A-14 was determined to be C14H20O3 by HREI-MS. Consistent with its molecular formula and 1H-NMR, the introduction of one alcoholic OH group in the substrate was proposed. The position of this alcoholic OH group was determined to be C-11 because all signals of H-10, H-12, and H-13 were observed to be singlet. The identity of A-14 was thus determined to be methyl 2-(4-(2-hydroxy-2-methylpropyl)phenyl)propanoate (Figure 7) [22].
Figure 7.
Bioconversion of drugs ibuprofen methylester and flurbiprofen methylester withE. coliBL21 (DE3) cells carrying pCYP110E1-Red. (%) indicates yield. The result with E. coli BL21 cells carrying plasmid pUCRED-Balk that expressed the P450balk gene (CYP153A13a) from an alkane-assimilating marine bacterium Alcanivorax borkumensis[13] is also shown for comparison.
Compounds converted from flurbiprofen methylester
The crude EtOAc extract (254.7 mg) from the bioconversion mixture (200 ml) with flurbiprofen methylester and E. coli (pCYP110E1-Red), subjected to silica gel column chromatography (hexane-EtOAc = 4:1), yielded 1.5 mg (2.7%) of A-15 (fr 10–12) and 1.8 mg (3.3%) of A-16 (fr 23–28).
The molecular formula of A-15 was determined to be C16H15FO3 by HREI-MS. Consistent with its molecular formula and 1H-NMR spectrum, the introduction of one alcoholic OH group in the substrate was proposed. The position of this alcoholic OH group was determined to be C-2 because the signal of H-3 (δ 1.80) was observed to be singlet. The identity of A-15 was thus determined to be methyl 2-(2-fluoro-[1,1'-biphenyl]-4-yl)-2-hydroxypropanoate (Figure 7).
The molecular formula of A-16 was determined to be C16H15FO3 by HRAPCI-MS. Consistent with its molecular formula and 1H-NMR spectrum, the introduction of one phenolic OH group in the aromatic ring was proposed. The position of this phenolic OH group was determined to be C-13 because the signals of H-12 and H-14 were observed to be doublet (J = 8.6 Hz) and at high field (δ 6.89). The identity of A-16 was thus determined to be methyl 2-(2-fluoro-4'-hydroxy-[1,1'-biphenyl]-4-yl)propanoate (Figure 7).
Discussion
The phylogenetic analysis (Figure 1) showed that CYP110E1 of Nostoc sp. strain PCC 7120 was located in close proximity to the first branch point in the phylogenetic tree of the CYP110 family, i.e., among 27 CYP110 proteins derived from 11 cyanobacteria, only four P450s, CYP110K1, CYP110D2, CYP110E6, and CYP110D3, were located closer to the first branch point than CYP110E1. In this study, CYP110E1, whose function had been unknown, was found to function as a substrate-promiscuous monooxygenase when it was C-terminally fused to the RhFRed reductase domain of P450RhF (CYP116B2) by the use of the pRED vector. Naringenin was converted directly to apigenin with a significant conversion ratio (31.5%) (Figure 2). Naringenin and apigenin belong to typical flavonoids that can be biosynthesized in higher plants. Artificial flavonoids, flavanone, 6-hydroxyflavanone and 7-hydroxyflavanone, were also converted to the respective flavones, even if with low conversion ratios, i.e., major parts of the substrates remained without being biotransformed even after 48-h co-culture. These results revealed that CYP110E1 functions as a flavone synthase. Such an activity is reported for the first time in prokaryotic P450s. When using flavanone as the substrate, 3-hydroxyflavanone was additionally generated (Figure 3). It may be possible that 3-hydroxyflavanone is the intermediate from flavanone to flavone. However, E. coli (pCYP110E1-Red) did not biotransform 3-hydroxyflavanone when it was added as the substrate (data not shown). It is therefore likely that flavones and 3-hydroxyflavanone were generated independently from flavanone. The higher-plant CYP93B was characterized as P450-type flavone synthase (FSII) and was proposed to convert flavanones to flavones by way of 2-hydroxyflavanones [23,24]. Such a catalytic route may be the case with CYP110E1.
P450BM3 (also described as P450BM-3 or P450BM3; CYP102A1) derived from Bacillus megaterium, one of the best-characterized prokaryotic P450s, is a natural fusion enzyme composed of a P450 part and a eukaryote-type NADPH-P450 reductase domain Additional file 1: Figure 1d[25-27]. This P450 part is closely related to the CYP110 family [1] and exhibited 24.6% amino acid sequence identity to CYP110E1. P450BM3, whose native substrates are thought to be long-chain fatty acids, has been shown to possess substrate and catalytic promiscuities [28-31]. Specifically, P450BM3 variants incorporating active site mutations that include F87V (or F87A) were found to acquire broader substrate affinity not only for a variety of aryl compounds including substituted naphthalenes [17,32,33], but also for the monoterpene α-pinene and the sesquiterpene amorpha-4,11-diene [30,34]. The P450BM3 variant F87V that was N-terminally fused to an archaeal peptidyl-prolyl cistrans isomerase (PPIase), which was synthesized by the plasmid named pFusionF87V in E. coli cells, was shown to elevate the stability of the P450 protein [17].
Zerumbone, a sesquiterpene contained in the shampoo ginger (Zingiber zerumbet Smith), is a promising chemopreventive agent, since its anti-inflammatory and anti-tumor activities have been investigated [35-38]. E. coli BL21 (DE3) carrying plasmid pCYP110E1-Red was shown to hydroxylate zerumbone to produce two novel compounds (S-2 and S-3) via the endogenous metabolite in E. coli (S-1; Figure 4). On the other hand, E. coli BL21 (DE3) carrying plasmid pFusionF87V was not able to synthesize compound S-3 from zerumbone, although it bioconverted zerumbone to compound S-2 with a higher conversion ratio than that of E. coli (pCYP110E1-Red) (Figure 4). E. coli (pFusionF87V) was able to hydroxylate β-eudesmol (a sesquiterpene contained in edible plants of the Zingiberaceae family) at its C-5 position [17], while E. coli (pCYP110E1-Red) was not able to biotransform β-eudesmol (data not shown). The two prokaryotic P450s, CYP110E1 and P450BM3 (F87V), are likely to have the ability to biotransform some sesquiterpenes of higher-plant origin complementally. CYP109B1 from Bacillus subtilis was also found to possess a wide substrate range for saturated fatty acids, n-alcohol, and some isoprenoids, and convert the sesquiterpene (+)-valencene to (+)-nootkatone, a high added-value compound found in grapefruit juice [39,40].
E. coli (pCYP110E1-Red) was found to biotransform various aryl compounds. 1-Methoxynaphthalene and 1-ethoxynaphthalene were converted to several hydroxylated compounds (Figure 5). When using 1-methoxynaphthalene as the substrate, an aryl coupling reaction was observed to produce compound A-1. This product is likely to be generated via oxidative aryl coupling through a non-enzymatic dimerization process from 4-methoxynaphthalen-1-ol, which is speculated to be the direct enzyme product. Misawa et al. (2011) showed that these two substrates were biotransformed by E. coli (pFusionF87V) to hydroxylated compounds different from those obtained using E. coli (pCYP110E1-Red) (Figure 5) [17]. In this case, oxidative aryl coupling was observed in the reaction, not from 1-methoxynaphthalene but from 1-ethoxynaphthalene to produce 4,4'-diethoxy-[2,2'-binaphthalene]-1,1'-diol [17]. These findings suggest that the two P450s, CYP110E1 and P450BM3 (F87V), are useful for producing a variety of 1-methoxy and 1-ethoxy naphthalene derivatives, whose naphthalene rings acquire one hydroxyl group without eliminating the methyl or ethyl group. Such an elimination is often likely to occur, e.g., CYP153A13a (P450balk) from Alcanivorax borkumensis SK2, which was found to possess a promiscuous substrate range for aryl compounds and aromatic compounds including alkyl groups, converted 1-methoxy naphthalene to 1-naphthol [12,13]. Figure 6 shows other aryl compounds biotransformed with E. coli (pCYP110E1-Red). The reactions towards products A-7A-9A-10A-11, and A-13 were observed also in E. coli BL21 (pUCRED-Balk) that expressed the CYP153A13a gene [13]. On the other hand, E. coli (pFusionF87V) converted 2-methylnaphthalene to compounds 2-methyl-1-naphthol and 6-methy-1-naphthol, which are different from those with E. coli (pCYP110E1-Red), while it converted 1,6-dimethylnaphthalene to the same compound A-8[17].
The present study also showed that E. coli (pCYP110E1-Red) biotransformed the methylester forms of non-steroidal anti-inflammatory drugs ibuprofen and flurbiprofen to produce the respective hydroxyl derivatives that are difficult to synthesize chemically (Figure 7). E. coli (pUCRED-Balk) converted ibuprofen methylester to another hydroxyl substituent [13], while it was not able to biotransform flurbiprofen methylester (data not shown). The four drug metabolites produced by recombinant E. coli cells (Figure 7) or their free carboxylate forms, awaiting the determination of the absolute configuration, could be used as standards in studies on the metabolisms of ibuprofen and flurbiprofen with human P450s [41].
Conclusion
The present study revealed that cyanobacterial cytochrome P450 CYP110E1, C-terminally fused to the P450RhF (CYP116B2) RhFRed reductase domain, is promiscuous for substrate and catalytic ranges and is useful for biosynthesizing not only flavones (from flavanones), but also a variety of hydroxyl- small molecules that are difficult to synthesize chemically, which may span pharmaceutical and nutraceutical industries.
Methods
Bacterial strains and genetic manipulation
Three cyanobacterial strains, Nostoc sp. strain PCC 7120, Nostoc punctiforme PCC 73102 (=ATCC 29133), and Anabaena variabilis ATCC 29413 were obtained from Pasteur Culture collection, Paris, and grown autotrophically in BG 11 medium. For the isolation of genomic DNA, cyanobacteria were harvested from the log phase and were immediately treated with lysozyme (10 mg/ml for 1 h). Genomic DNA was then isolated with the GenElute plant genomic DNA kit (Sigma-Aldrich, St. Louis, MO).
E. coli DH5α (ECOS Competent E. coli DH5α; Nippon Gene, Tokyo, Japan) was utilized as the host for DNA manipulations. E. coli BL21 (DE3) (Nippon Gene) was used for the functional expression of each P450 gene, which was inserted into the pRED vector [10]. PCR amplifications were performed using Prime STAR Max Premix DNA polymerase (Takara Bio, Ohtsu, Japan) and a thermal cycler (Applied Biosystems, Foster City, CA). Restriction enzymes and DNA-modifying enzymes were purchased from New England BioLabs (Beverly, CA) or Takara Bio. A Ligation-Convenience Kit (Nippon Gene) was also used. Plasmid DNA was prepared with a QIAprep Miniprep Kit (Qiagen, Hilden, Germany). All recombinant DNA experiments were carried out according to the suppliers’ manuals or Sambrook and Russell (2001) [42].
Nucleotide sequencing and computer analysis
Nucleotide sequences were confirmed with Bigdye terminator cycle sequencing ready reaction kit version 3.1 (Applied Biosystems) and a model 3730 DNA analyzer (Applied Biosystems). Homologous protein sequences in the protein sequence database were retrieved from CyanoBase of Kazusa DNA Research Institute (http://genome.kazusa.or.jp/cyanobase) with the BLAST program [43], and aligned by Clustal W program in Molecular Evolutionary Genetics Analysis (MEGA) software version 5.0 (http://www.megasoftware.net/). A phylogenetic tree was also constructed according to MEGA 5.0.
Construction of plasmids
Cyanobacterial P450 genes were amplified by PCR from genomic DNA of Nostoc sp. PCC 7120, N. punctiforme PCC 73102, or A. variabilis ATCC 29413. All synthetic oligonucleotides used in this work were listed in Table 1. PCR amplification was performed in a 50 μl reaction mixture containing 25 ng of genomic DNA, 25 μl of 2 × the DNA polymerase, 10 μM of each primer, and 5% dimethyl sulfoxide (DMSO). The PCR conditions used were the following: preincubation at 98 °C for 2 min; a total of 5 cycles at 98 °C for 10 sec, 55 °C for 10 sec, and 75 °C for 15 sec; a total of 30 cycles at 98 °C for 10 sec, 62 °C for 5 sec and 75 °C for 15 sec. An amplified 1.4 kb fragment was digested with NdeI and EcoRI or HindIII, and ligated into the NdeI-EcoRI or NdeI-HindIII site of pRED to construct the desired plasmids. In these plasmids, the stop codons of the respective P450 genes were removed to fuse the N-terminus of RhFRed.
Table 1.
List of primers used in this study
| primer | sequence | species | |
|---|---|---|---|
| CYP284A1 |
Ana1361F |
5'-TAC CAT ATG ATG CTC CAA TAC ATT ACT GCT CTC-3' |
|
| Ana1361R |
5'-TAC GAA TTC ATT TCT CAA CCG AAA GCG CAC T-3' |
|
|
| CYP110A1 |
Ana1450F |
5'-TAC CAT ATG ATG TTG ACT CAA TTA CCA AAT CC-3' |
|
| Ana1450R |
5'-TAC GAA TTC GTT GAA AAT CTT GCT ACT TTG CT-3' |
|
|
| CYP110B1 |
Ana3746F |
5'-TAC CAT ATG ATG CAC CTA CCA AAA GG-3' |
|
| Ana3746R |
5'-CAC GAA TTC ACT TAC AGT AGT TGT TTC TAG-3' |
|
|
| |
|
|
Nostocsp. PCC 7120 |
| CYP110C1 |
Ana4686F |
5'-CAC CAT ATG ATG AAA TAT CAA ATA CAG AGA CC-3' |
|
| Ana4686R |
5'-TAC AAG CTT TGC GTT GAA TGT TGT TGA G-3' |
|
|
| CYP110D1 |
Ana4766F |
5'-TAC CAT ATG ATG ACA GTC ACT CAA AAC C-3' |
|
| Ana4766R |
5'-CAC AAG CTT CGA ATT ACG CAT TCT TTT ATT AG-3' |
|
|
| CYP110E1 |
Ana4833F |
5'-TAC CAT ATG ATG AAA CTT CCA GAT AGT C-3' |
|
| |
Ana4833R |
5'-TAC GAA TTC TAC TTC TAC AGG GTT TTT G-3' |
|
| CYP110C3 |
Ana1981F |
5'-TAC CAT ATG ATG AAG TAT CAA ATA AAG AGA C-3' |
|
| Ana1981R |
5'-TAC GAA TTC TGT TGT GAA TGT TGT TGA G-3' |
|
|
| CYP110E6 |
Ana2103F |
5'-TAC CAT ATG ATG AAA CTT CCA GAT AGT CC-3' |
|
| Ana2103R |
5'-TAC GAA TTC TAC TTC TAC AGG GCT TTT GA-3' |
|
|
| |
|
|
A. variabilisATCC 29413 |
| CYP110A2 |
Ana3921F |
5'-TAC CAT ATG ATG TTG ACT CAA TTA CCA AA-3' |
|
| Ana3921R |
5'-CAC GAA TTC ATT AAA AAT CTT GTT ACT TTG CT-3' |
|
|
| CYP284A3 |
Ana4063F |
5'-TAC CAT ATG ATG CTC CAA TAC GTT ACT GCT C-3' |
|
| |
Ana4063R |
5'-TAC GAA TTC ATT TCT CAA CCG AAA GCG CAC-3' |
|
| CYP110F1 |
Nos0984F |
5'-TAC CAT ATG ATG AAA ATA CTT GAT AGT CTA AC-3' |
|
| Nos0984R |
5'-TAC AAG CTT AGT AGA AAG TAT TGT TTG TCT TT-3' |
|
|
| CYP110B2 |
Nos0985F |
5'-TAC CAT ATG ATG AAA TTA CCA AAA GGC C-3' |
|
| Nos0985R |
5'-TAT GAA TTC AAC AGT GGC TGT CTG-3' |
|
|
| CYP197B1 |
Nos2212F |
5'-TAC CAT ATG ATG GTT GCC GAT GTA TT-3' |
|
| |
5'-CAC GAA TTC TTT AGA AGT GTC TAA TGC AA-3' |
|
|
| CYP284A2 |
Nos2399F |
5'-TAC CAT ATG ATG TTC CAA CAG ATT GCT GC-3' |
|
| Nos2399R |
5'-TAC GAA TTC ACG AGC GAT ATT GTC AGA GT-3' |
|
|
| CYP120B1 |
Nos2686F |
5'-TAC CAT ATG ATG AAA ACT AAT CAA ATT CCT-3' |
|
| Nos2686R |
5'-TAT GAA TTC CCG AGG TTG AAA TCT-3' |
|
|
| |
|
|
N. punctiformePCC 73102 |
| CYP110D2 |
Nos3640F |
5'-CTA C CA TAT GAT GAA AAG TCG TAA CAA TAA AA-3' |
|
| Nos3640R |
5'-TAT GAA TTC AAC TAG GGC TGG C-3' |
|
|
| CYP110C2 |
Nos6291F |
5'-TAC CAT ATG ATG CAA CTA CCT AAT ATT CT-3' |
|
| Nos6291R |
5'-TAT GAA TTC GGA TAG GGG TGT AG-3' |
|
|
| CYP110E2 |
Nos7017F |
5'-GCA GCA TAT GAT GTC TTT ACT TAA ACT G-3' |
|
| Nos7017R |
5'-TCA C GA ATT CAA CTG AAC TAG AGC T-3' |
|
|
| CYP227A1 |
Nos7684F |
5'-ATA C CA TAT GAT GAC ACT TAA AGA TAA AG-3' |
|
| Nos7684R |
5'-TAT GAA TTC CAG TCG TTG AGC AA-3' |
|
|
| CYP120C1 |
Nos8095F |
5'-TAC CAT ATG ATG CAG CAG TTA AAA TCC G-3' |
|
| Nos8095R | 5'-TAC GAA TTC ACT ATC CAA GGG ATG CTT T-3 |
CO difference spectral analysis
CO difference spectral analysis was done as described [12].
Bioconversion experiments
E. coli BL21 (DE3) carrying each plasmid was grown in an LB medium including ampicillin (Ap; 100 μg/ml) at 37°C with rotary shaking for 3–4 h until the absorbance at OD 600 nm reached approximately 0.8. For screening experiments, 1.5 ml of this preculture was inoculated into 125 ml of an LB medium including Ap (100 μg/ml), 5-aminolevulinic acid hydrochloride (5-ALA; 80 μg/ml), ammonium iron (II) sulfate (0.1 mM), and IPTG (0.1 mM) in a baffled Erlenmeyer flask, and cultured at 20°C for 20 h on a rotary shaker (140 rpm; Kuhner Shaker Lab Therm LT-X, Basel, Switzerland). Cells were collected by centrifugation and resuspended in 25 ml of CV-3 buffer [sodium phosphate buffer (50 mM, pH 7.2) containing 5% glycerol] in a baffled Erlenmeyer flask. Five hundred μl of this cell suspension was added into each well of a 96 well sterile plate (PR-Master Block 2ML; Greiner Bio-One, Frickenhausen, Germany), together with 1 mM (final concentration) of substrate dissolved in dimethyl sulfoxide (DMSO). Bioconversion was performed by cultivation at 25°C for 24 h with 300 rpm using the Kuhner Shaker.
For structural determination of products, large scale cultivation was carry out, by inoculation of 5 ml of the preculture into 500 ml of LB medium including Ap (100 μg/ml), 5-ALA (80 μg/ml), ammonium iron (II) sulfate (0.1 mM), and IPTG (0.1 mM) in a baffled Erlenmeyer flask at 20°C for 20 h with 140 rpm on the Kuhner Shaker. Cells were collected by centrifugation, and resuspended in 100 ml of CV-3 buffer in a baffled Erlenmeyer flask. Each substrate dissolved in DMSO was added at a final concentration of 1 mM to the cell suspension and bioconversion was performed by cultivation at 25°C for 48 h with 180 rpm.
Substrates and authentic samples used in this study were purchased from Tokyo Chemical Industry Co. (Tokyo, Japan), Sigma-Aldrich Co. (St. Louis, MO), or Wako Pure Chemical Industries (Osaka, Japan).
Chemical synthesis of ibuprofen methylester and flurbiprofen methylester
Zero point six mol/l of trimethylsilyldiazomethane in hexane (12.5 ml, 7.5 mmol; Tokyo Chemical Industry) was added, drop by drop on ice, to a solution (33 ml, benzene/methanol = 2/1) containing ibuprofen (1.03 g, 5 mmol; Wako Pure Chemical Industries) or flurbiprofen (1.22 g, 5 mmol; Wako Pure Chemical Industries). The reaction mixtures from ibuprofen and flurbiprofen were stirred at room temperature for 5 h. After concentrated to dryness, the respective residues (1.23 g and 1.53 g) were subjected to column chromatography (I.D. 20 mm × 300 mm) with silica gel IR-60-63/210-w (Daiso Co., Osaka Japan), developed in hexane-ethyl acetate (EtOAc) = 2:1 and hexane-EtOAc = 10:1, to yield 0.79 g of ibuprofen methylester as a colorless oil and 1.27 g of flurbiprofen methylester as a white powder.
Extraction and HPLC analysis of products
Five hundred μl of the reaction mixture liquid was added to 100 μl of saturated sodium chloride solution and 500 μl of EtOAc and shaken for 5 min. After centrifugation, the organic phase was analyzed by high pressure liquid chromatography (HPLC; Waters 2695, Waters Corp., Milford, MA) equipped with an on-line photodiode array detector (Waters 2996). An aliquot of the organic phase (20 μl) was applied to HPLC and separated using an XTerra MS C18 column (I.D. 4.6 mm × 100 mm; Waters), and a flow rate of 1 ml/min was used, with solvent A [5% acetonitrile (CH3CN) in 20 mM phosphoric acid] for 3 min, then a linear gradient from solvent A to solvent B (95% CH3CN in 20 mM phosphoric acid) for 25 min, and finally with solvent B for 15 min, with the maximum absorbance being monitored in the range of 200–500 nm (max plot).
Purification and identification of products
The liquid phase containing the reaction mixture (200 ml; 100 ml x 2) was extracted with EtOAc (200 ml x 2 times). The resultant organic layer was concentrated in vacuo and analyzed by thin-layer chromatography (TLC) on silica gel (E. Merck 60 F-254 0.25-mm silica gel plates). Products were purified by column chromatography on Silica Gel 60 [20 mm (diameter) x 250 mm (length); Merck]. To elucidate the structures of these products, high resolution mass spectral data [HREI-MS (Jeol DX505W; Jeol, Tokyo, Japan) or HRAPCI-MS (Jeol JMS-T100LP)], and nuclear magnetic resonance (NMR) spectral data (400 MHz, Bruker AMX400) were applied.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
GS cultured the cyanobacterial strains and prepared genome. TM and HH isolated P450 genes and constructed plasmids for functionally expressing them. TM also carried out screening and bioconversion experiments, and HPLC analyses. TO selected substrates for the screening and participated in the screening and bioconversion experiments and interpretation of the screening results. KS and EK purified the products and did their spectroscopic analyses. NM made substantial contributions to conception and design of experiments and participated in writing the manuscript. All authors read the manuscript and gave final approval of the version to be published.
Supplementary Material
Figure S1. Composition of four distinct P450 monooxygenase systems. Figure S2: Structure of the pRED vector for the functional expression of class I P450 genes inE. coli.Figure S3: List of screened substrates (47 samples). Figure S4: CO difference spectral analysis of CYP110E1 C-terminally fused to RhFRed. Cell extracts from E. coli BL21 (DE3) carrying plasmid pCYP110E1-Red (three samples, S1, S2, and S3) were measured for CO difference spectra. (PPT 424 kb)
Spectroscopic data of the individual converted products. (DOC 48 kb)
Contributor Information
Takuya Makino, Email: takuya1103@topaz.plala.or.jp.
Toshihiko Otomatsu, Email: t_otomatsu@kncweb.co.jp.
Kazutoshi Shindo, Email: kshindo@fc.jwu.ac.jp.
Emi Kitamura, Email: m1122003ke@gr.jwu.ac.jp.
Gerhard Sandmann, Email: sandmann@bio.uni-frankfurt.de.
Hisashi Harada, Email: harada@bio.tottori-u.ac.jp.
Norihiko Misawa, Email: n-misawa@ishikawa-pu.ac.jp.
Acknowledgements
The authors are grateful to the Marine Biotechnology Institute (MBI), Kamaishi-shi, Iwate, Japan (closed on June 30, 2008), since this work was initially in part performed here. T.M. thanks Prof. Takehiko Ogata, School of Fisheries Sciences, Kitasato University, for his valuable guidance.
References
- Torres S, Fjetland CR, Lammers PJ. Alkane-induced expression, substrate binding profile, and immunolocalization of a cytochrome P450 encoded on the nifD excision element of Anabaena 7120. BMC Microbiol. 2005;5:16. doi: 10.1186/1471-2180-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agger SA, Lopez-Gallego F, Hoye TR, Schmidt-Dannert C. Identification of sesquiterpene synthases from Nostoc punctiforme PCC 73102 and Nostoc sp. strain PCC7120. J Bacteriol. 2008;190:6084–6096. doi: 10.1128/JB.00759-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Shindo K, Iki K, Teraoka A, Okamoto S, Yu F, Utsumi R, Hattan J, Misawa N. Efficient functional analysis system for cyanobacterial or plant cytochromes P450 involved in sesquiterpene biosynthesis. Appl Microbiol Biotechnol. 2011;90:467–476. doi: 10.1007/s00253-010-3062-9. [DOI] [PubMed] [Google Scholar]
- Kaneko T, Nakamura Y, Wolk CP, Kuritz T, Sasamoto S, Watanabe A, Iriguchi M, Ishikawa A, Kawashima K, Kimura T, Kishida Y, Kohara M, Matsumoto M, Matsuno A, Muraki A, Nakazaki N, Shimpo S, Sugimoto M, Takazawa M, Yamada M, Yasuda M, Tabata S. Complete genomic sequence of the filamentous nitrogen-fixing cyanobacterium Anabaena sp. strain PCC 7120. DNA Res. 2001;8:205–213. doi: 10.1093/dnares/8.5.205. 227–253. [DOI] [PubMed] [Google Scholar]
- Lammers PJ, McLaughlin S, Papin S, Trujillo-Provencio C, Ryncarz AJ. Developmental rearrangement of cyanobacterial nif genes: nucleotide sequence, open reading frames, and cytochrome P-450 homology of the Anabaena sp. strain PCC 7120 nifD element. J Bacteriol. 1990;172:6981–6990. doi: 10.1128/jb.172.12.6981-6990.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udit AK, Contakes SM, Gray HB. In: Metal ions in life sciences. vol. 3. The ubiquitous roles of cytochrome P450 proteins. Sigel A, Sigel H, Sigel RKO, editor. John Wiley & Sons, Chichester, England; 2007. P450 electron transfer reactions; pp. 157–185. [Google Scholar]
- Hannemann F, Bichet A, Ewen KM, Bernhardt R. Cytochrome P450 systems –biological variations of electron transport chains. Biochim Biophys Acta. 2007;1770:330–344. doi: 10.1016/j.bbagen.2006.07.017. [DOI] [PubMed] [Google Scholar]
- Roberts GA, Grogan G, Greter A, Flitsch SL, Turner NJ. Identification of a new class of cytochrome P450 from a Rhodococcus sp. J Bacteriol. 2002;184:3898–3908. doi: 10.1128/JB.184.14.3898-3908.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunter DJ, Roberts GA, Ost TW, White JH, Muller S, Turner NJ, Flitsch SL, Chapman SK. Analysis of the domain properties of the novel cytochrome P450 RhF. FEBS Lett. 2005;579:2215–2220. doi: 10.1016/j.febslet.2005.03.016. [DOI] [PubMed] [Google Scholar]
- Nodate M, Kubota M, Misawa N. Functional expression system for cytochrome P450 genes using the reductase domain of self-sufficient P450RhF from Rhodococcus sp. NCIMB 9784. Appl Microbiol Biotechnol. 2006;71:455–462. doi: 10.1007/s00253-005-0147-y. [DOI] [PubMed] [Google Scholar]
- Robin A, Roberts GA, Kisch J, Sabbadin F, Grogan G, Bruce N, Turner NJ, Flitsch SL. Engineering and improvement of the efficiency of a chimeric [P450cam-RhFRed reductase domain] enzyme. Chem Commun. 2009;2009:2478–2480. doi: 10.1039/b901716j. [DOI] [PubMed] [Google Scholar]
- Fujita N, Sumisa F, Shindo K, Kabumoto H, Arisawa A, Ikenaga H, Misawa N. Comparison of two vectors for functional expression of a bacterial cytochrome P450 gene in Escherichia coli using CYP153 genes. Biosci Biotechnol Biochem. 2009;73:1825–1830. doi: 10.1271/bbb.90199. [DOI] [PubMed] [Google Scholar]
- Otomatsu T, Bai L, Fujita N, Shindo K, Shimizu K, Misawa N. Bioconversion of aromatic compounds by Escherichia coli that expresses cytochrome P450 CYP153A13a gene isolated from an alkane-assimilating marine bacterium Alcanivorax borkumensis. J Mol Catal B Enzym. 2010;66:234–240. doi: 10.1016/j.molcatb.2010.05.015. [DOI] [Google Scholar]
- Kubota M, Nodate M, Yasumoto-Hirose M, Uchiyama T, Kagami O, Shizuri Y, Misawa N. Isolation and functional analysis of cytochrome P450 CYP153A genes from various environments. Biosci Biotechnol Biochem. 2005;69:2421–2430. doi: 10.1271/bbb.69.2421. [DOI] [PubMed] [Google Scholar]
- Li S, Podust LM, Sherman DH. Engineering and analysis of a self-sufficient biosynthetic cytochrome P450 PikC fused to the RhFRED reductas domain. J Am Chem Soc. 2007;129:12940–12941. doi: 10.1021/ja075842d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao YH, Houghton PJ, Hoult JRS. Novel and known constituents from BuddleJa species and their activity against leukocyte eicosanoid generation. J Nat Prod. 1999;62:1241–1245. doi: 10.1021/np990092+. [DOI] [PubMed] [Google Scholar]
- Misawa N, Nodate M, Otomatsu T, Shimizu K, Kaido C, Kikuta M, Ideno A, Ikenaga H, Ogawa J, Shimizu S, Shindo K. Bioconversion of substituted naphthalenes and β-eudesmol with the cytochrome P450 BM3 variant F87V. Appl Microbiol Biotechnol. 2011;90:147–157. doi: 10.1007/s00253-010-3064-7. [DOI] [PubMed] [Google Scholar]
- Takeya T, Kondo H, Tomita K, Okamoto I, Morita N, Tamura O. One-pot synthesis of dibenzofuran-1,4-diones. Heterocycles. 2007;74:961–968. doi: 10.3987/COM-07-S(W)26. [DOI] [Google Scholar]
- Bell KH, McCaffery LF. Regioselective monomethylation of unsymmetrical naphthalenediols with methanolic hydrogen chloride. Aust J Chem. 1993;46:731–737. doi: 10.1071/CH9930731. [DOI] [Google Scholar]
- Ferreira VF, Schmitz FJ. Reductive-alkylation and aromatic coupling reactions of 1,4-benzoquinone derivatives promoted by ethylaluminum dichloride. J Organomet Chem. 1998;571:1–6. doi: 10.1016/S0022-328X(98)00865-1. [DOI] [Google Scholar]
- Thompson RB, Chenicek JA. Dihydroxynaphthalenes and some derivatives as gasoline antioxidants. J Ind Eng Chem (Washington, D. C.) 1995;47:1431–1432. [Google Scholar]
- Rentmeister A, Arnold FH, Fasan R. Chemo-enzymatic fluorination of unactivated organic compounds. Nat Chem Biol. 2009;5:26–28. doi: 10.1038/nchembio.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akashi T, Fukuchi-Mizutani M, Aoki T, Ueyama Y, Yonekura-Sakakiba K, Tanaka Y, Kusumi T, Ayabe S. Molecular cloning and biochemical characterization of a novel cytochrome P450, flavone synthase II, that catalyzes direct conversion flavanones to flavones. Plant Cell Physiol. 1999;40:1182–1186. doi: 10.1093/oxfordjournals.pcp.a029505. [DOI] [PubMed] [Google Scholar]
- Leonard E, Yan Y, Lim KH, Koffas MAG. Investigation of two distinct flavone synthases for plant-specific flavone biosynthesis in Saccharomyces cerevisiae. Appl Environ Microbiol. 2005;71:8241–8248. doi: 10.1128/AEM.71.12.8241-8248.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narhi LO, Fulco AJ. Characterization of a catalytically self-sufficient 119,000-dalton cytochrome P450 monooxygenase induced by barbiturates in Bacillus megaterium. J Biol Chem. 1986;261:7160–7169. [PubMed] [Google Scholar]
- Ruettinger RT, Wen LP, Fulco AJ. Coding nucleotide, 5’ regulatory, and deduced amino acid sequences of P-450 BM-3, a single peptide cytochrome P-450: NADPH-P-450 reductase from Bacillus megaterium. J Biol Chem. 1989;264:10987–10995. [PubMed] [Google Scholar]
- Li H, Poulos TL. The structure of the cytochrome P450 BM-3 haem domain complexed with the fatty acid structure, palmitoleic acid. Nat Struct Biol. 1997;4:140–146. doi: 10.1038/nsb0297-140. [DOI] [PubMed] [Google Scholar]
- Boddupalli SS, Estabrook RW, Peterson JA. Fatty acid monooxygenation by cytochrome P450 BM-3. J Biol Chem. 1990;265:4233–4239. [PubMed] [Google Scholar]
- Graham-Lorence SE, Truan G, Peterson JA, Falck JR, Wei S, Helvig C, Capdevila JH. An active site substitution, F87V, converts cytochrome P450 BM-3 into a regio- and stereoselectivive (14S,15R)-arachidonic acid epoxygenase. J Biol Chem. 1997;272:1127–1135. doi: 10.1074/jbc.272.2.1127. [DOI] [PubMed] [Google Scholar]
- Dietrich JA, Yoshikuni Y, Fisher KJ, Woolard FX, Ockey D, McPhee DJ, Renninger NS, Chang MCY, Baker D, Keasling JD. A novel semi-biosynthetic route for artemisinin production using engineered substrate-promiscuous P450BM3. ACS Chem Biol. 2009;4:261–267. doi: 10.1021/cb900006h. [DOI] [PubMed] [Google Scholar]
- Misawa N. Pathway engineering for functional isoprenoids. Curr Opin Biotechnol. 2011;22:627–633. doi: 10.1016/j.copbio.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Li QS, Ogawa J, Schmid RD, Shimizu S. Engineering cytochrome P450 for oxidation of polycyclic aromatic hydrocarbons. Appl Environ Microbiol. 2001;67:5735–5739. doi: 10.1128/AEM.67.12.5735-5739.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sulistyaningdyah WT, Ogawa J, Li QS, Maeda C, Yano Y, Schmid RD, Shimizu S. Hydroxylation activity of P450 BM-3 mutant F87V towards aromatic compounds and its application to the synthesis of hydroquinone derivatives from phenolic compounds. Appl Microbiol Biotechnol. 2005;67:556–562. doi: 10.1007/s00253-004-1761-9. [DOI] [PubMed] [Google Scholar]
- Schewe H, Holtmann D, Schrader J. P450BM-3-catalyzed whole-cell biotransformation of α-pinene with recombinant Escherichia coli in an aqueous-organic two-phase system. Appl Microbiol Biotechnol. 2009;83:849–857. doi: 10.1007/s00253-009-1917-8. [DOI] [PubMed] [Google Scholar]
- Murakami A, Tanaka T, Lee JY, Surh YJ, Kim HW, Kawabata K, Nakamura Y, Jiwajinda S, Ohigashi H. Zerumbone suppresses skin tumor initiation and promotion stages in mice: involvement of induction of anti-oxidative and phase II drug metabolizing enzymes and inhibition of NF-kB-mediated cyclooxygenase-2 expression. Int J Cancer. 2004;110:481–490. doi: 10.1002/ijc.20175. [DOI] [PubMed] [Google Scholar]
- Eguchi A, Kaneko Y, Murakami A, Ohigashi H. Zerumbone suppresses phorbol ester-induced expression of multiple scavenger receptor genes in THP-1 human monocytic cells. Biosci Biotechnol Biochem. 2007;71:935–945. doi: 10.1271/bbb.60596. [DOI] [PubMed] [Google Scholar]
- Ohnishi K, Irie K, Murakami A. In vitro covalent binding proteins of zerumbone, a chemopreventive food factor. Biosci Biotechnol Biochem. 2009;73:1905–1907. doi: 10.1271/bbb.90265. [DOI] [PubMed] [Google Scholar]
- Singh CB, Nongalleima K, Brojendrosingh S, Ningomban S, Lokendrajit N, Singh LW. Biological and chemical properties of Zingiber zerumbet Smith: a review. Phytochem Rev. 2012;11:113–125. doi: 10.1007/s11101-011-9222-4. [DOI] [Google Scholar]
- Girhard M, Machida K, Itoh M, Schmid RD, Arisawa A, Urlacher VB. Regioselective biooxidation of (+)-valencene by recombinant E. coli expressing CYP109B1 from Bacillus subtilis in a two-liquid-phase system. Microb Cell Factories. 2009;8:36. doi: 10.1186/1475-2859-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girhard M, Klaus T, Khatri Y, Bernhardt R, Urlacher VB. Characterization of the versatile monooxygenase CYP109B1 from Bacillus subtilis. Appl Microbiol Biotechnol. 2010;87:595–607. doi: 10.1007/s00253-010-2472-z. [DOI] [PubMed] [Google Scholar]
- Chang SY, Li W, Traeger SC, Wang B, Cui D, Zhang H, Wen B, Rodrigues AD. Confirmation that cytochrome P450 2C8 (CYP2C8) plays a minor role in (S)-(+) and (R)-(−)-ibuprofen hydroxylation in vitro. Drug Metab Dispos. 2008;36:2513–2522. doi: 10.1124/dmd.108.022970. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning: a laboratory manual. 3. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York; 2001. [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipmann DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Composition of four distinct P450 monooxygenase systems. Figure S2: Structure of the pRED vector for the functional expression of class I P450 genes inE. coli.Figure S3: List of screened substrates (47 samples). Figure S4: CO difference spectral analysis of CYP110E1 C-terminally fused to RhFRed. Cell extracts from E. coli BL21 (DE3) carrying plasmid pCYP110E1-Red (three samples, S1, S2, and S3) were measured for CO difference spectra. (PPT 424 kb)
Spectroscopic data of the individual converted products. (DOC 48 kb)