Abstract
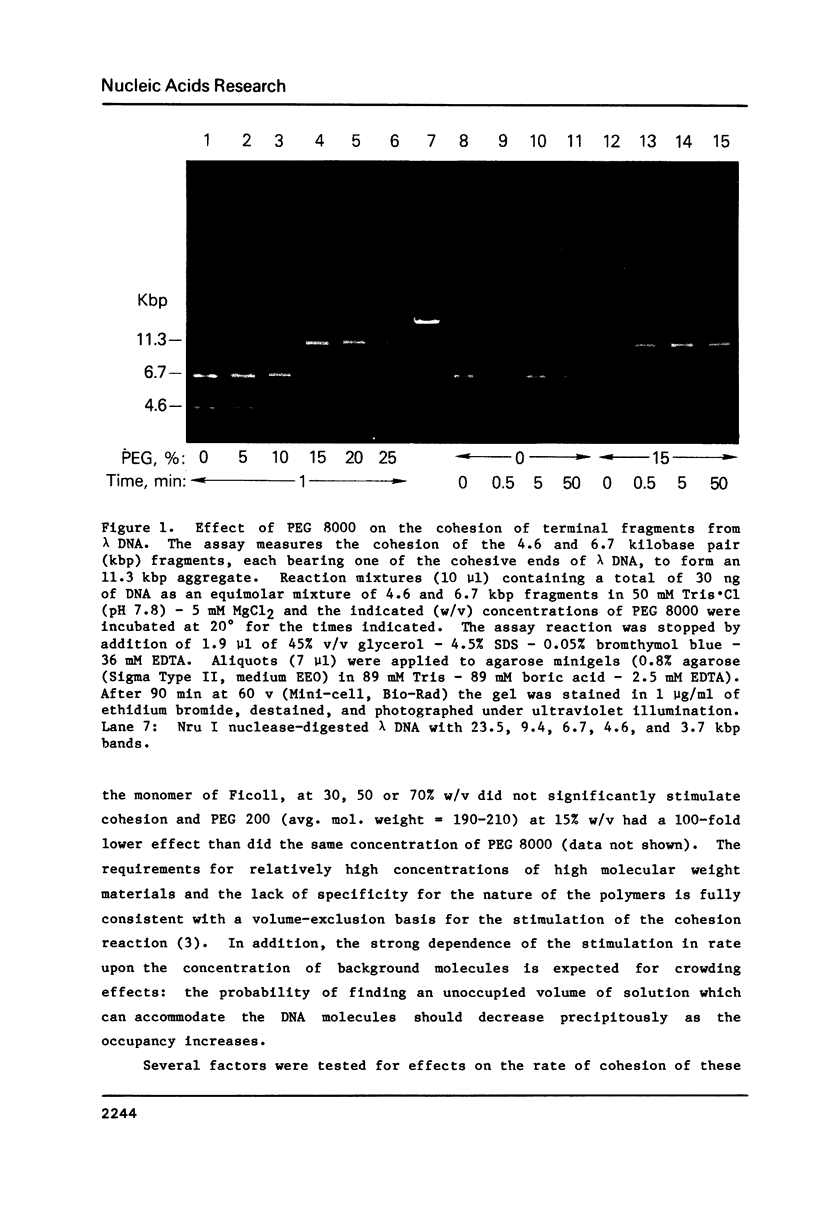
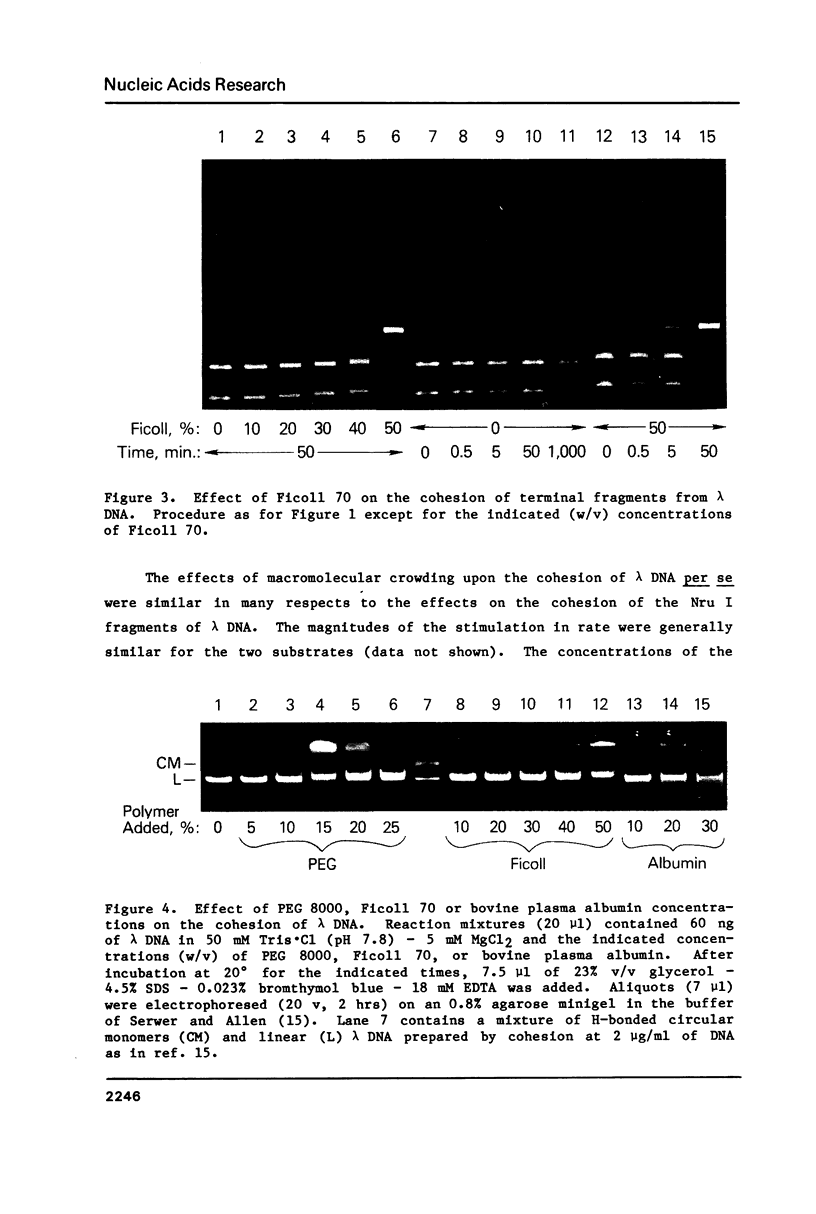
Macromolecular crowding increases the rate of nonenzymatic cohesion of the complementary ends of lambda DNA. Both lambda DNA and DNA fragments bearing the cohesive ends of lambda DNA are similarly affected. High concentrations of plasma albumin or Ficoll 70 increase the rate of cohesion by ca. 100-fold whereas high concentrations of polyethylene glycol 8000 cause greater than 2000-fold stimulation in this rate. These results have implications for the mechanism of polymer-stimulated enzymatic ligation of DNA or RNA. In addition, these crowding effects may help to explain the rapid cohesion of lambda DNA observed in vivo. An improved procedure for the recovery of DNA fragments separated by agarose gel electrophoresis is also described.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bode V. C., Kaiser A. D. Changes in the structure and activity of lambda DNA in a superinfected immune bacterium. J Mol Biol. 1965 Dec;14(2):399–417. doi: 10.1016/s0022-2836(65)80190-5. [DOI] [PubMed] [Google Scholar]
- Chang C. T., Hain T. C., Hutton J. R., Wetmur J. G. Effects of microscopic and macroscopic viscosity on the rate of renaturation of DNA. Biopolymers. 1974;13(9):1847–1858. doi: 10.1002/bip.1974.360130915. [DOI] [PubMed] [Google Scholar]
- Dretzen G., Bellard M., Sassone-Corsi P., Chambon P. A reliable method for the recovery of DNA fragments from agarose and acrylamide gels. Anal Biochem. 1981 Apr;112(2):295–298. doi: 10.1016/0003-2697(81)90296-7. [DOI] [PubMed] [Google Scholar]
- Fulton A. B. How crowded is the cytoplasm? Cell. 1982 Sep;30(2):345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- HERSHEY A. D., BURGI E. COMPLEMENTARY STRUCTURE OF INTERACTING SITES AT THE ENDS OF LAMBDA DNA MOLECULES. Proc Natl Acad Sci U S A. 1965 Feb;53:325–328. doi: 10.1073/pnas.53.2.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison B., Zimmerman S. B. Polymer-stimulated ligation: enhanced ligation of oligo- and polynucleotides by T4 RNA ligase in polymer solutions. Nucleic Acids Res. 1984 Nov 12;12(21):8235–8251. doi: 10.1093/nar/12.21.8235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershey A. D., Burgi E., Ingraham L. COHESION OF DNA MOLECULES ISOLATED FROM PHAGE LAMBDA. Proc Natl Acad Sci U S A. 1963 May;49(5):748–755. doi: 10.1073/pnas.49.5.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerman L. S. A transition to a compact form of DNA in polymer solutions. Proc Natl Acad Sci U S A. 1971 Aug;68(8):1886–1890. doi: 10.1073/pnas.68.8.1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minton A. P. The effect of volume occupancy upon the thermodynamic activity of proteins: some biochemical consequences. Mol Cell Biochem. 1983;55(2):119–140. doi: 10.1007/BF00673707. [DOI] [PubMed] [Google Scholar]
- Pheiffer B. H., Zimmerman S. B. Polymer-stimulated ligation: enhanced blunt- or cohesive-end ligation of DNA or deoxyribooligonucleotides by T4 DNA ligase in polymer solutions. Nucleic Acids Res. 1983 Nov 25;11(22):7853–7871. doi: 10.1093/nar/11.22.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sclafani R. A., Wechsler J. A. High-yield purification of small, single-stranded DNA. Anal Biochem. 1981 Jul 15;115(1):197–202. doi: 10.1016/0003-2697(81)90546-7. [DOI] [PubMed] [Google Scholar]
- Serwer P., Allen J. L. Conformation of double-stranded DNA during agarose gel electrophoresis: fractionation of linear and circular molecules with molecular weights between 3 X 10(6) and 26 X 10(6). Biochemistry. 1984 Feb 28;23(5):922–927. doi: 10.1021/bi00300a020. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Davidson N. Cyclization of phage DNAs. Cold Spring Harb Symp Quant Biol. 1968;33:409–415. doi: 10.1101/sqb.1968.033.01.047. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Davidson N. On the probability of ring closure of lambda DNA. J Mol Biol. 1966 Aug;19(2):469–482. doi: 10.1016/s0022-2836(66)80017-7. [DOI] [PubMed] [Google Scholar]
- Wang J. C., Davidson N. Thermodynamic and kinetic studies on the interconversion between the linear and circular forms of phage lambda DNA. J Mol Biol. 1966 Jan;15(1):111–123. doi: 10.1016/s0022-2836(66)80213-9. [DOI] [PubMed] [Google Scholar]
- Wu R., Taylor E. Nucleotide sequence analysis of DNA. II. Complete nucleotide sequence of the cohesive ends of bacteriophage lambda DNA. J Mol Biol. 1971 May 14;57(3):491–511. doi: 10.1016/0022-2836(71)90105-7. [DOI] [PubMed] [Google Scholar]
- Zimmerman S. B., Pheiffer B. H. Macromolecular crowding allows blunt-end ligation by DNA ligases from rat liver or Escherichia coli. Proc Natl Acad Sci U S A. 1983 Oct;80(19):5852–5856. doi: 10.1073/pnas.80.19.5852. [DOI] [PMC free article] [PubMed] [Google Scholar]






