Abstract
Purpose: The aim of this study was to quantify the effect of four image registration methods on lung texture features extracted from serial computed tomography (CT) scans obtained from healthy human subjects.
Methods: Two chest CT scans acquired at different time points were collected retrospectively for each of 27 patients. Following automated lung segmentation, each follow-up CT scan was registered to the baseline scan using four algorithms: (1) rigid, (2) affine, (3) B-splines deformable, and (4) demons deformable. The registration accuracy for each scan pair was evaluated by measuring the Euclidean distance between 150 identified landmarks. On average, 1432 spatially matched 32 × 32-pixel region-of-interest (ROI) pairs were automatically extracted from each scan pair. First-order, fractal, Fourier, Laws’ filter, and gray-level co-occurrence matrix texture features were calculated in each ROI, for a total of 140 features. Agreement between baseline and follow-up scan ROI feature values was assessed by Bland–Altman analysis for each feature; the range spanned by the 95% limits of agreement of feature value differences was calculated and normalized by the average feature value to obtain the normalized range of agreement (nRoA). Features with small nRoA were considered “registration-stable.” The normalized bias for each feature was calculated from the feature value differences between baseline and follow-up scans averaged across all ROIs in every patient. Because patients had “normal” chest CT scans, minimal change in texture feature values between scan pairs was anticipated, with the expectation of small bias and narrow limits of agreement.
Results: Registration with demons reduced the Euclidean distance between landmarks such that only 9% of landmarks were separated by ≥1 mm, compared with rigid (98%), affine (95%), and B-splines (90%). Ninety-nine of the 140 (71%) features analyzed yielded nRoA > 50% for all registration methods, indicating that the majority of feature values were perturbed following registration. Nineteen of the features (14%) had nRoA < 15% following demons registration, indicating relative feature value stability. Student's t-tests showed that the nRoA of these 19 features was significantly larger when rigid, affine, or B-splines registration methods were used compared with demons registration. Demons registration yielded greater normalized bias in feature value change than B-splines registration, though this difference was not significant (p = 0.15).
Conclusions: Demons registration provided higher spatial accuracy between matched anatomic landmarks in serial CT scans than rigid, affine, or B-splines algorithms. Texture feature changes calculated in healthy lung tissue from serial CT scans were smaller following demons registration compared with all other algorithms. Though registration altered the values of the majority of texture features, 19 features remained relatively stable after demons registration, indicating their potential for detecting pathologic change in serial CT scans. Combined use of accurate deformable registration using demons and texture analysis may allow for quantitative evaluation of local changes in lung tissue due to disease progression or treatment response.
Keywords: texture analysis; image registration, lung, CT
INTRODUCTION
For patients with lung disease, periodic computed tomography (CT) scans are often acquired to assess disease progression or treatment response. Detection of diffuse lung disease change, however, may be difficult for physicians due to unclear disease boundaries on CT. Furthermore, variability may exists among physicians evaluating diffuse lung disease.1 Quantitative measures of radiologic changes are being investigated to aid physicians with clinical decisions. A number of groups have developed quantitative image-based texture analysis techniques to identify and classify lung disease, improve consistency of measurements, and enhance accuracy in patient diagnosis. Chabat et al.2 developed a method to classify lung disease into three types (centrilobular emphysema, panlobular emphysema, and constrictive obliterative bronchiolitis) using a set of first-order, co-occurrence matrix, and gray-level run length features. Uchiyama et al.3 used morphological operations to highlight patterns characteristic of diffuse lung disease among six categories: ground-glass opacities, reticular and linear opacities, nodular opacities, honeycombing, emphysematous change, and consolidation. Uppaluri et al.4 developed the adaptive multiple feature method (AMFM), a tool that identifies specific lung disease patterns using a combination of first-order, co-occurrence matrix, gray-level run length, and fractal texture features.
While these methods can serve as a metric to assess overall changes in the extent of diffuse lung disease between serial CT scans, they cannot be used to track local changes in lung anatomy throughout treatment. Analysis of local differences in lung texture becomes important when disease severity is nonuniform due to localized treatment such as radiation therapy. In these cases, measuring changes in anatomically matched regions of the lungs between serial CT scans would allow for closer examination of differences in the appearance of disease or radiation-induced toxicities. To compare lung texture over time, it is necessary to perform registration of CT images prior to measuring differences in texture between the two scans. Rigid and affine registrations are the most simple and commonly applied registration algorithms. These algorithms, however, may not be adequate for registration of lung CT scans due to positioning and diaphragmatic differences between serial scans. Thus, a number of fully automated deformable registration algorithms applied to registration of lung CT images are based on optical flow (demons deformable registration)5, 6, 7 or control point matching (splines deformable registration).8, 9, 10 More recently, the B-splines deformable registration algorithm has been used to investigate CT pixel value changes following radiotherapy, thereby allowing for direct correlation between lung injury and dose to a local region of the lung.11
Although image registration has the potential to improve quantitative lung CT image analysis by allowing for direct comparison of differences between serially acquired scans, little has been done to investigate the effects of registration on image texture values. While some feature differences between registered temporally sequential CT scans result from an actual change in disease status, other differences may be falsely introduced by image matching and interpolation techniques used during the registration process or changes in imaging parameters, such as the concentration of intravenous contrast. Palma et al.12 demonstrated that CT scans acquired during the same imaging session and registered using B-splines showed variations in mean lung density due to differences in contrast level and the level of inspiration; no other CT features or registration methods were investigated. In the present study, we analyzed images from patients with two “normal” chest CT scans, as determined by an attending radiologist. Because these CT scans contained no lung pathology, we hypothesized that differences in the texture features between registered images of a scan pair were introduced by the registration algorithm or by variations in imaging parameters rather than by a change in the disease status of the patient. We examined four commonly used registration algorithms: rigid registration, affine registration, demons deformable registration, and B-splines deformable registration. Our goal was to identify a set of texture features that resulted in relatively stable feature values following registration with a particular algorithm, allowing for their combined use for future studies of patient response to treatment.
METHODS
Database selection
Two clinically indicated de-identified helical thoracic CT scans acquired from 1 week to 2 years apart at the University of Chicago Medical Center between November 2005 and January 2011 from each of 29 patients (Table 1) were retrospectively obtained through the Human Imaging Research Office (HIRO)13 under institutional review board (IRB) approval. All scans were determined to have no lung abnormalities, defined by the absence of acute disease or micronodules exceeding 4 mm, by an experienced radiologist. During rigid and affine registration of CT images, two patients were excluded from the study due to gross mis-registrations that may have been due to large differences in patient orientation between the two scans. Scans were acquired after patients were instructed to inspire and hold their breath using multislice Philips Brilliance CT scanners (Brilliance 16, Brilliance 16P, Brilliance 64, or Brilliance iCT256) and reconstructed at 1 mm slice thickness using identical high-resolution lung reconstruction and smoothing kernels. A gray-level thresholding technique combined with morphological operations and volume thresholding was used to extract lung masks from the CT images.
Table 1.
Summary of patient demographic information and CT scan acquisition parameters for the patient database.
| Patient data | Std. Dev. | |
|---|---|---|
| Number of patients (male:female) | 27 (21:6) | … |
| Median patient age (range) [yr] | 49 (18–68) | 14 |
| Contrast enhanced scan pairs | 24 | … |
| Number of scan pairs acquired with different scanners | ||
| 12 | … | |
| Slice thickness/spacing [mm] | 1 | 0 |
| kVp | 120 | 0 |
| Mean exposure (range) [mAs] | 235 (144–351) | 40 |
| Mean exposure difference between paired scans (range) [mAs] | ||
| 33 (0–156) | 45 | |
| Mean pixel spacing (range) [mm] | 0.67 (0.58–0.87) | 0.06 |
| Mean pixel spacing difference between paired scans (range) [mm] | ||
| 0.04 (0–0.18) | 0.04 | |
| Median time between paired scans (range) [days] | ||
| 126 (7–719) | 179 |
Image registration
Two open-source software packages were used to perform registration: Plastimatch for rigid, affine, and demons registrations and elastix for B-splines registration.6, 14 Computations were performed on a multicore computing cluster (Scientific Image Reconstruction and Analysis Facility, SIRAF) at the University of Chicago.
Rigid and affine registrations
Rigid and affine registrations are global registration methods whereby transformations are applied across the entire image. Rigid registration allows for six degrees of freedom (three translational directions and three rotational directions) in image motion, and affine registration allows for 12 degrees of freedom (translation, rotation, scaling, and shear in three directions). During this study, both rigid and affine registrations were performed in multiple resolution stages to allow for image matching at low resolution prior to matching at high resolution. Image similarity at each iteration was assessed using the mean-squared intensity difference between baseline and registered follow-up scans.
Demons deformable registration
Demons deformable registration was initially proposed by Thirion and is modeled after the thermodynamics paradox termed Maxwell's “demons.”15 In this technique, a moving image is allowed to diffuse through the fixed image until matching is achieved. The force and direction of motion at each iteration of the diffusion process are calculated using an optical flow equation. Demons registration as used in this study again proceeded as a multistage process. All scans were registered using identical parameters, and the number of iterations at each resolution stage was optimized by minimizing the global mean-squared intensity difference between the baseline and deformed follow-up scans.
B-splines deformable registration
During B-splines registration, a grid of control points identified within a moving image is shifted to match points in a fixed image. These control points determine the coefficients of cubic B-spline basis functions that in turn define a deformation field throughout the image.16 Registration using elastix was performed with the registration parameters optimized to deform lung CT images for the Evaluation of Methods for Pulmonary Image Registration 2010 (EMPIRE10) challenge.17 The registration was a multistage process and used the normalized correlation coefficient penalized for bending energy to assess image similarity at each iteration.
Landmark matching
Registration accuracy was evaluated using an open source semi-automated landmark selection and matching program (iX v. 1.2.0.0) to calculate the Euclidean distance between landmark points in the baseline scan and each of the four corresponding registered follow-up scans (i.e., the results of registering the follow-up scan using the four considered registration methods).18 One hundred fifty landmark points were chosen automatically in each baseline CT scan based on two factors: (1) the pixel-value gradient in a four-connected neighborhood exceeded 150 HU/pixel, and (2) the points were uniformly spaced throughout the three-dimensional scan. For at least 20 of the 150 landmark points automatically selected in the baseline scan, an observer [A.R.C.] manually identified the corresponding location in each of the four variants of the registered follow-up scan. A thin-plate splines interpolation technique was then used to estimate the location of the remaining landmark points in each of the registrations of the follow-up scan based on these points. When adequate matches could not be identified through the thin-plate splines technique due to registration inaccuracies, landmark matching was performed manually. All matched landmarks were visually reviewed by the observer and revised if the observer's subjective assessment indicated an improper match.
Region-of-interest (ROI) selection and texture feature analysis
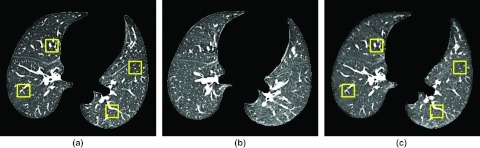
An automated method was used to randomly place approximately 2000 (range: 1515–2479) nonoverlapping ROIs that were fully contained within the lungs in each baseline scan [Fig. 1a]. Spatial image coordinates located within the lung boundaries in each CT scan section were randomly chosen as the center of a 32 × 32-pixel ROI. The ROI was accepted provided it (1) existed entirely within the lung boundaries and (2) did not intersect with a previously defined ROI. A maximum of 10 ROIs were selected in each axial CT section. Due to registration inaccuracies, some ROIs lying entirely within the lungs of the baseline scan contained background pixels when mapped to the registrations of the follow-up scan; these ROI pairs were not included for analysis, resulting in 1432 ROIs on average (range: 959–1936) per patient.
Figure 1.
(a) A slice taken from the baseline CT scan of a patient with 32 × 32-pixel ROIs superimposed. (b) A slice taken from the corresponding follow-up scan located in approximately the same anatomic location as (a). (c) A slice taken from a demons-registered follow-up scan with ROIs mapped from the baseline scan superimposed. Note the improved anatomic agreement with (a).
Using in-house texture analysis software,19 we calculated a series of first-order, fractal, Fourier, Laws’ filter, and gray-level co-occurrence matrix (GLCM) texture features in all baseline ROIs and in the corresponding ROIs mapped to the registered follow-up scans generated from each of the four registration methods (Fig. 1). The 140 calculated features are summarized below.
First-order histogram features
First-order histogram features20, 21, 22 describe characteristics of the gray-level histogram of an image region. Nineteen first-order features were calculated: mean, median, maximum, minimum, mean absolute deviation, range, interquartile range, standard deviation, skewness, kurtosis, energy, entropy, binned entropy (calculated after sorting data into 256 histogram bins), 5%, 30%, 60%, and 95% histogram quantiles, and balance of the inner 40% and inner 90% of the gray-level histogram.
Fractal features
The fractal dimension characterizes the self-similarity of a region at different scales and is an indicator of region detail. Three methods to compute the fractal dimension were used: the blanket method,23 the Brownian motion method,24 and the box-counting method.22, 25 In addition, the coarse and fine aspects of the box-counting dimension were calculated.26
Fourier features
Features extracted from the Fourier transform of an image region characterize the spatial frequency components of the region. The first moment of the power spectrum and root-mean-squared variation were measured.27 Using the rotationally invariant Fourier transform of a region, the energy of the transformed region and the energy of several subspaces representing specific frequency components were computed. These subspaces were the high- and low-frequency rings formed when the region was divided into two subsections; the low, moderately low, moderately high, and high frequency rings formed when the region was divided into four subsections; and the eight sectors formed when the region was divided into 45° slices.20
Laws’ filter features
Laws’ filters emphasize region microstructure, specifically spot, wave, ripple, edge, and level surfaces.28 Each filter is convolved with a region of interest, and features are calculated on the filtered regions. Six features were calculated on 14 rotationally invariant Laws’ filtered regions: mean, energy, entropy (after sorting into 256 histogram bins), maximum, minimum, and standard deviation.
Gray-level co-occurrence matrix features
Also referred to as Haralick features, GLCM features20, 21, 29 quantify the spatial relationship of gray-level values in a region. A GLCM is constructed to represent and count all gray-level pairs separated by a distance d and at angle θ in a selected region. Features are calculated from the GLCM to reveal underlying region structure. Fourteen features were calculated: correlation, inertia, absolute value, inverse difference, energy, entropy, contrast, sum of squares variance, sum average, sum variance, sum entropy, difference average, difference variance, and difference entropy. Four directions were examined (θ = {0°, 45°, 90°, and 135°}), and each feature was calculated by taking the average over all directions; a single pixel distance was considered: d = 1 pixel.
Statistical analysis
Bland–Altman 95% limits of agreement30 for the case of multiple measurements were used to assess agreement between feature values calculated in ROIs of the baseline scan and each of the four registrations of the follow-up scan. Biases in the values of features in registered ROIs were calculated, and upper and lower bounds of agreement between feature values were generated. For each of the 140 features, the bias and range of the 95% limits of agreement were compared to the feature value itself. The normalized bias and normalized range of agreement (nRoA) were calculated according to
where nROIs is the total number of ROIs across all patients. A nRoA close to zero indicated low variability in feature value change between baseline and registered follow-up scans, indicating that the feature was relatively “registration stable.” For registration methods that consistently achieved different nRoA values across features compared with the other algorithms, two-sided paired Student's t-tests were performed to test the significance of the observed differences. To maintain an overall model significance level of 0.05, significance levels for the individual t-tests were adjusted according to the Bonferroni method, resulting in significance at α = 0.017.
Due to the retrospective nature of data collection, parameters such as exposure, pixel size, or contrast level at the time of image acquisition varied between scans (see Table 1), resulting in inevitable variations in image texture between sequential CT scans. In addition to these image acquisition-related variations, the registration method itself could introduce global changes in feature values, causing consistent feature value changes between baseline and registered follow-up scans. To characterize the extent of feature value change introduced by the registration methods rather than image-acquisition differences, the registration direction was reversed so the baseline scan was registered to match the follow-up scan. ROI placement, texture analysis, and Bland–Altman analysis as detailed above were repeated. Feature value change was calculated by subtracting feature values in the fixed follow-up scan from feature values in matched ROIs from the registered baseline scan. For image acquisition-dependent feature value changes, reversing the registration direction was expected to reverse the sign of normalized bias while preserving the magnitude, whereas biases with similar magnitudes and signs were expected for feature changes introduced by registration irrespective of the registration direction. The normalized biases obtained using forward and reverse registrations were averaged to yield an overall bias introduced by registration for each feature. This reverse ordering analysis was limited to the deformable registration methods. Two-sided paired Student's t-tests were performed to test if the average bias in feature value change across all features was significantly different between the two deformable registration methods.
RESULTS
Landmark matching
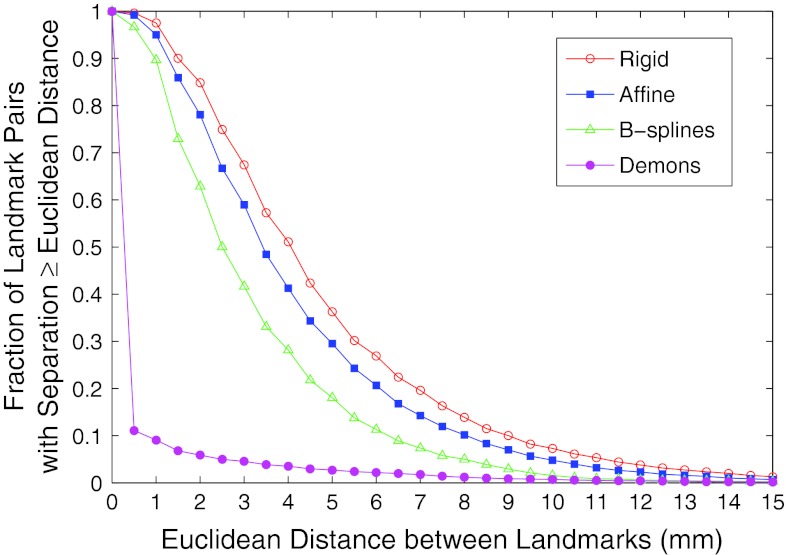
Landmark matches were automatically identified in the follow-up scans for 61% of the landmarks in the baseline scans when demons registration was used, as compared with 9%, 23%, and 24%, respectively, for B-splines, affine, and rigid registration. For individual patients, the mean Euclidean distance between corresponding landmarks ranged from 2.0 to 7.8 mm for rigid registration, from 1.6 to 9.9 mm for affine registration, from 1.5 to 5.6 mm for B-splines registration, and from 0.004 to 2.2 mm for demons registration. After combining all patient data, the mean Euclidean distance between landmarks and the 95% range of distances were calculated for the four registration methods (Table 2). The mean for demons surpassed the next best algorithm (B-splines) by 2.8 mm.
Table 2.
Mean Euclidean distances and 95% range of distances between matched landmarks for four registration methods.
| Registration method | Mean (mm) | 95% range of data (mm) |
|---|---|---|
| Rigid | 4.76 | (0.99, 13.30) |
| Affine | 4.16 | (0.65, 11.70) |
| B-splines | 3.16 | (0, 9.20) |
| Demons | 0.39 | (0, 5.27) |
The percentage of landmarks separated by a range of Euclidean distances was plotted for the four registration methods (Fig. 2). When all patient landmarks were considered, demons registration reduced the percentage of matched landmark points separated by at least 1 mm to 9% compared with rigid (98%), affine (95%), and B-splines registration (90%). Demons registration outperformed B-splines registration, B-splines outperformed affine registration, and affine outperformed rigid registration, as shown by the noncrossing curves in Fig. 2. This trend did not necessarily hold for individual patients. When comparing the percentage of landmarks matched to within 1 mm, rigid registration outperformed affine registration for five patients, and affine registration outperformed B-splines registration for three patients. In all patients, however, demons registration achieved better matching than any of the other registration methods.
Figure 2.
Comparison of the Euclidean distance between landmarks for four registration algorithms.
Texture feature analysis
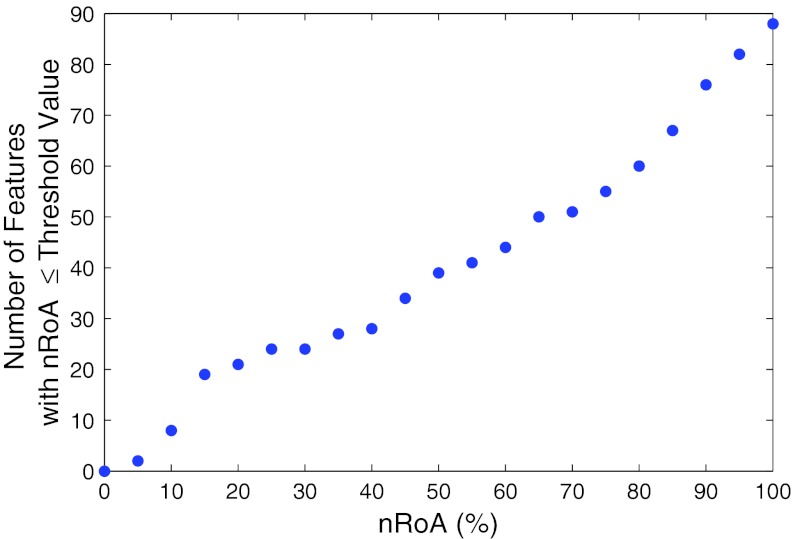
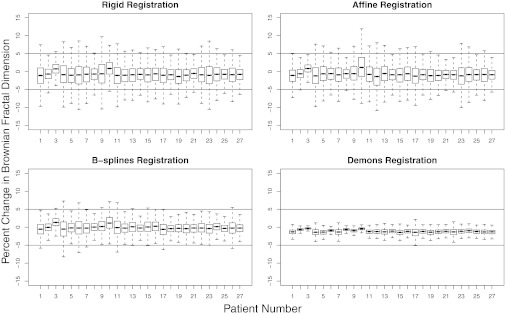
Fifty-two of the 140 features exhibited a high degree of variability between the registered scan pairs, with nRoA greater than 100% for all four registration methods. Figure 3 depicts a plot of the number of features with nRoA less than or equal to a given value ranging from 0% to 100% in 5% increments. This plot was generated with nRoA resulting from demons registration because of its superior landmark matching accuracy compared with the other registration algorithms. Visual inspection of this figure revealed inflection points located approximately at nRoA = 15% and nRoA = 75%. To identify registration-stable features, further analysis was focused on the 19 features with nRoA ≤ 15%. This cutoff was chosen both because it was low in value and because the number of features per nRoA increment declined once nRoA exceeded 15% (i.e., the slope in Fig. 3 declined). With the exception of three of the 19 features identified, nRoA was larger for rigid, affine, and B-splines registration than for demons registration due to the higher relative registration accuracy of demons registration. Furthermore, all features with nRoA ≤ 15% for rigid, affine, or B-splines registration also had nRoA ≤ 15% using demons registration. Two-sided paired Student's t-tests (Table 3) showed that for the 19 features identified, the mean nRoA for demons registration was significantly smaller than the mean nRoA calculated using each of the other registration methods (the maximum p-value among these three comparisons was p = 4 × 10−5). The bias was generally lowest when B-splines registration was used, though the variance in feature value change between baseline and registered scans was higher than for demons due to the inferior anatomic matching achieved (Fig. 4). Boxplots displaying the percent change in the values of the Brownian fractal dimension across all 27 patients for the four registration methods are displayed in Fig. 5, demonstrating that the lowest variance in feature value change was achieved using demons registration.
Figure 3.
The cumulative number of features with at most a given nRoA. For visualization purposes, only the features with nRoA < 100% are displayed.
Table 3.
Range of agreement and bias normalized to the mean feature value as a function of the registration algorithm used to register the follow-up to the baseline CT scan. All features with nRoA ≤ 15% for at least one registration method are listed.
| Demons |
B-splines |
Affine |
Rigid |
|||||
|---|---|---|---|---|---|---|---|---|
| nRoA (%) | Bias (%) | nRoA (%) | Bias (%) | nRoA (%) | Bias (%) | nRoA (%) | Bias (%) | |
| First-order features | ||||||||
| Minimum | 9.92 | 3.07 | 12.70 | −3.62 | 8.17 | 1.18 | 8.26 | 1.13 |
| Mean | 7.56 | 0.01 | 16.91 | −0.27 | 18.50 | −0.36 | 19.76 | −0.41 |
| Median | 7.28 | −0.11 | 11.40 | −0.44 | 11.83 | −0.32 | 11.57 | −0.37 |
| Entropy (binned) | 14.68 | −3.86 | 19.47 | −0.26 | 21.18 | −1.74 | 22.92 | −1.74 |
| Entropy (unbinned) | 10.33 | −4.30 | 12.54 | 0.95 | 13.26 | −1.85 | 14.34 | −1.84 |
| 5% quantile | 11.04 | 4.30 | 10.44 | −0.07 | 11.33 | 2.45 | 11.37 | 2.39 |
| 30% quantile | 7.49 | 1.37 | 9.73 | −0.36 | 9.86 | 0.59 | 9.59 | 0.54 |
| 70% quantile | 9.10 | −1.50 | 19.03 | −0.50 | 22.11 | −1.13 | 24.02 | −1.18 |
| Fractal features | ||||||||
| Box-counting dimension | 11.80 | −5.49 | 19.41 | −0.41 | 21.22 | −3.65 | 23.00 | −3.68 |
| Coarse dimension | 14.64 | −2.34 | 33.22 | −0.06 | 37.35 | −1.54 | 38.89 | −1.62 |
| Brownian dimension | 4.50 | −1.15 | 9.44 | −0.09 | 12.15 | −0.76 | 13.11 | −0.77 |
| Laws’ filter features | ||||||||
| E5L5 entropy | 10.79 | −2.16 | 24.09 | 0.25 | 26.42 | −1.44 | 30.02 | −1.46 |
| R5L5 entropy | 14.41 | −3.53 | 20.89 | −0.74 | 22.09 | −4.79 | 23.80 | −4.79 |
| S5L5 entropy | 11.70 | −3.26 | 24.83 | 0.22 | 27.34 | −2.57 | 30.66 | −2.58 |
| W5L5 entropy | 12.69 | −4.05 | 22.31 | −0.07 | 24.31 | −3.84 | 26.77 | −3.86 |
| GLCM features | ||||||||
| Difference average | 11.47 | 0.19 | 13.98 | −4.12 | 10.44 | −0.90 | 10.73 | −0.95 |
| Sum of squares variance | 1.47 | −0.29 | 3.75 | −0.01 | 4.43 | −0.25 | 5.06 | −0.24 |
| Sum average | 5.78 | −1.27 | 12.51 | 1.35 | 12.75 | −0.74 | 13.83 | −0.75 |
| Sum variance |
11.67 |
−2.52 |
25.78 |
2.73 |
26.54 |
−1.46 |
28.83 |
−1.49 |
| Mean nRoA | 9.91 | 16.97 | 17.96 | 19.29 | ||||
| p1 | … | 1 × 10−5 | 4 × 10−5 | 3 × 10−5 | ||||
Compared with mean nRoA for demons registration.
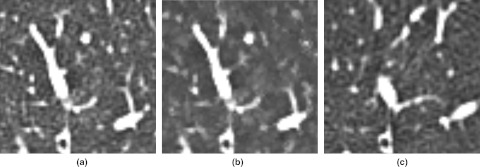
Figure 4.
Noise smoothing was observed when demons deformable registration was used. (a) A baseline CT image region. (b) A matched region in the demons deformed follow-up scan. Note the change in lung parenchymal texture. (c) A matched region in the B-splines deformed follow-up scan. Lung parenchymal texture is similar to the texture observed in (a), although anatomic matching is inferior to (b).
Figure 5.
For each registration method, boxplots display the percent change in the Brownian fractal dimension measured in baseline and registered follow-up CT scans for all 27 patients. Statistical outliers, comprising approximately 5% of the data, were excluded for visualization purposes. Horizontal lines are drawn at ±5% to also aid in visualization.
Of the 20 first-order features considered, eight had nRoA ≤ 15% for demons deformable registration (Table 3). Five features also had nRoA ≤ 15% for the other registration methods. For gray-level entropy, there was a clear negative bias in the data for demons registration that was not present for other registration methods, indicating that gray-level entropy tended to decrease due to demons deformable registration. Three of the five fractal features considered had nRoA ≤ 15%. Negative bias in fractal feature value change was observed due to all registration algorithms, though this bias was consistently larger when demons registration was used due to the noise smoothing introduced by demons (Fig. 4). The entropy of rotationally invariant Laws’ E5L5-, R5L5-, S5L5-, and W5L5-filtered microstructure regions yielded nRoA ≤ 15%. A negative bias resulted from demons, affine, and rigid registrations, indicating a decrease in microstructure entropy from baseline to follow-up scans due to these registration algorithms.
Four of the 14 average GLCM features were observed to have nRoA ≤ 15%. nRoA was comparatively small across all registration methods and generally smallest for demons registration and largest for rigid or affine registrations. One exception was the difference average of the GLCM, which yielded a slightly smaller nRoA with rigid and affine registrations than with B-splines or demons registration. Small nRoA did not, however, indicate that the scans were well registered, since landmark matching demonstrated the superior anatomic matching achieved by demons registration (Fig. 2). Rather, the difference average feature may not be appropriate for detecting texture change in this particular application, resulting in similar feature values regardless of changes between the two regions.
All of the Fourier features had nRoA > 15%, indicating a high degree of variability between feature values measured in the baseline and registered follow-up scans. nRoA was smallest for the first moment of the power spectrum and root-mean-squared variation (nRoA ≈ 40%). The relatively high degree of variability in texture value change indicated that these Fourier features were too sensitive to differences between serial scans in healthy patients and would therefore not be useful in the detection of actual pathologic changes in serial imaging studies.
Reverse image registration texture feature analysis
The extent of feature value change introduced specifically by deformable registration was measured for the 19 features with nRoA ≤ 15%. This evaluation was based on the average bias calculated using Bland–Altman analysis when forward and reverse registrations were used (Table 4). The average bias between forward and reverse registrations reflected the overall bias in feature value change introduced by deformable registration itself rather than by differences in image acquisition parameters, for which the sign of the bias depended on the registration direction.
Table 4.
Bias introduced by demons and B-splines deformable registration when forward and reverse image registrations were used.
| Demons |
B-splines |
|||||
|---|---|---|---|---|---|---|
| Forward bias (%) | Reverse bias (%) | Average bias (%) | Forward bias (%) | Reverse bias (%) | Average bias (%) | |
| First-order features | ||||||
| Median | −0.11 | 0.50 | 0.20 | −0.44 | 0.57 | 0.07 |
| Mean | 0.01 | 0.46 | 0.24 | −0.27 | 0.57 | 0.15 |
| 70% quantile | −1.50 | −1.38 | −1.44 | −0.50 | 0.18 | −0.16 |
| 30% quantile | 1.37 | 2.37 | 1.87 | −0.36 | 1.06 | 0.35 |
| Minimum | 3.07 | 3.84 | 3.46 | −3.62 | −2.38 | −3.00 |
| Entropy (binned) | −3.86 | −4.37 | −4.12 | −0.26 | −0.02 | −0.14 |
| Entropy (unbinned) | −4.30 | −5.24 | −4.77 | 0.95 | 0.27 | 0.61 |
| 5% quantile | 4.30 | 5.61 | 4.96 | −0.07 | 1.80 | 0.87 |
| Fractal features | ||||||
| Brownian dimension | −1.15 | −1.42 | −1.29 | −0.09 | −0.55 | −0.32 |
| Coarse dimension | −2.34 | −3.09 | −2.72 | −0.06 | −0.88 | −0.47 |
| Box-counting dimension | −5.49 | −6.69 | −6.09 | −0.41 | −2.24 | −1.33 |
| Laws’ filter features | ||||||
| E5L5 entropy | −2.16 | −2.99 | −2.58 | 0.25 | −0.37 | −0.06 |
| S5L5 entropy | −3.26 | −4.22 | −3.74 | 0.22 | −1.07 | −0.43 |
| R5L5 entropy | −3.53 | −4.65 | −4.09 | −0.74 | −3.47 | −2.11 |
| W5L5 entropy | −4.05 | −4.86 | −4.46 | −0.07 | −1.98 | −1.03 |
| GLCM features | ||||||
| Difference average | 0.19 | 0.25 | 0.22 | −4.12 | −3.92 | −4.02 |
| Sum of squares variance | −0.29 | −0.41 | −0.35 | −0.01 | −0.18 | −0.10 |
| Sum average | −1.27 | −1.28 | −1.28 | 1.35 | 1.40 | 1.38 |
| Sum variance |
−2.52 |
−2.54 |
−2.53 |
2.73 |
2.83 |
2.78 |
| Mean average bias | … | … | −1.50 | … | … | −0.37 |
| p1 | … | … | … | … | … | 0.15 |
Compared with average bias for demons registration.
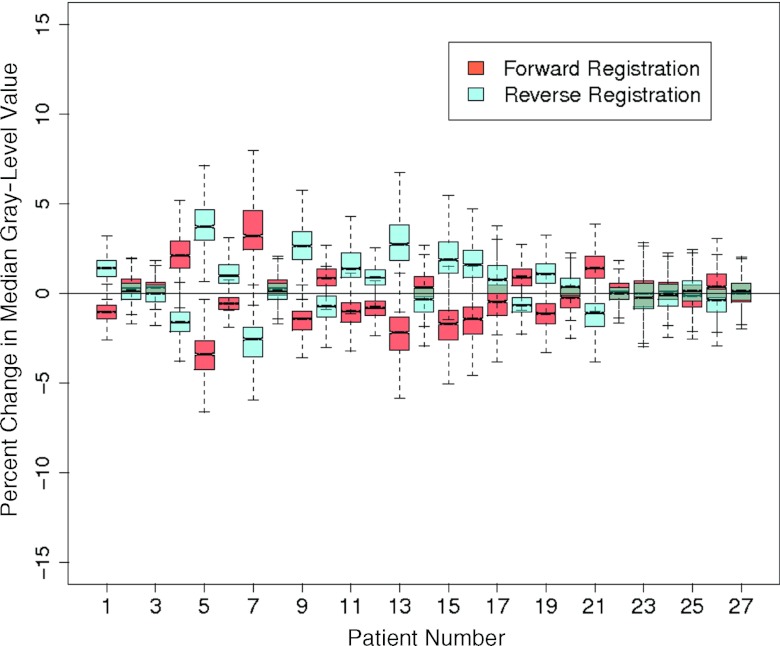
The extent of average bias in feature values introduced by deformation ranged from 0.20% (median gray-level value) to 6.09% (box-counting fractal dimension) using demons registration and from 0.06% (Law's filter E5L5 entropy) to 4.02% (difference average) using B-splines. Using a paired Student's t-test, the average bias for the 19 features was not significantly different between demons and B-splines registration methods (p = 0.15), though this value suggests the existence of a trend (Table 4). For both registration methods, mean, median, and sum of squares variance achieved average biases <1%, indicating that they were minimally affected by changes introduced by registration. Nine additional features achieved biases <1% using B-splines registration; using demons, however, only one other feature had bias <1% (difference average). As noted earlier (see Fig. 4), demons registration resulted in a decrease of high spatial frequencies in the registered images, yielding lower values for features that measure image roughness such as entropy and fractal dimension. This smoothing also resulted in narrowing of the image histogram, causing registration-dependent changes in the 30% and 70% histogram quantile features. Figure 6 displays boxplots representing percent feature value change between registered and fixed images across all 27 patients for median gray-level value. Although overall bias was low, feature values were affected by image acquisition parameters such as the exposure and pixel spacing and patient-dependent parameters such as the level of inspiration and contrast uptake at the time of image acquisition, resulting in feature value differences that were similar in magnitude but opposite in sign between forward and reverse registration directions.
Figure 6.
Boxplots display the percent change in median gray-level value between fixed and deformed scans for all patients using demons deformable registration.
DISCUSSION
Evaluation of registration algorithm accuracy
Demons deformable registration provided higher accuracy for landmark matching of masked lungs in serially acquired chest CT scans compared with B-splines, affine, or rigid registration. Analysis of landmark matching averaged over 27 patients with “normal” chest CT scans demonstrated that rigid, affine, and B-splines registrations failed to accurately match landmarks to within 1 mm for at least 90% of the landmarks, whereas demons registration resulted in matching to within 1 mm for all but 9% of the landmarks. The semi-automated landmark matching program successfully identified matched landmark locations for 61% of the points using demons, compared with 9% using B-splines. The low automatic matching percentage for B-splines may have occurred because the vector field interpolation technique introduced distortions in the appearance (e.g., size or shape) of landmark matches and/or their surrounding neighborhoods. Although manual matching may have affected the accuracy with which landmarks were placed, resulting in systematic over- or underestimates of the average Euclidean distances measured for B-splines compared with demons, the landmark matching data indicates that demons achieved better registration accuracy than B-splines.
Several studies have been performed to evaluate the registration accuracy achieved using B-splines and demons registration algorithms. Kashani et al.31 found that although three B-splines algorithms and one demons algorithm achieved similar mean landmark matching error when they were used to deform scans acquired with a deformable phantom, the maximal error was between 0.9 mm and 4.8 mm higher for B-splines than for demons. A multi-institutional study by Brock32 found demons registration algorithms achieved higher average landmark matching accuracy between phases of 4D CT scans than all but one of the B-splines algorithms, though the average landmark displacement difference between B-splines- and demons-registered scans remained small (<0.6 mm). For these studies, landmarks were identified based on 10–21 embedded markers or recognizable anatomic landmarks (i.e., bronchial bifurcations and aortic calcifications), whereas the present study selected 150 landmarks automatically based on pixel-value gradients. Additionally, these previous studies were performed using controlled deformation environments (i.e., phantom studies or clinical 4D scans performed on the same day and scanner) so that the extent of true landmark displacement due to deformation could be measured and compared with the displacement during registration. In the present study, clinical CT scans acquired during separate imaging sessions were deformed, thereby increasing the complexity of the registration task since several factors (e.g., patient position and scanner parameters) were not identical. When registering CT scans acquired three months apart, Palma et al.12 achieved results similar to those observed in this study, measuring mean B-splines inaccuracies ranging from 3.1 mm to 8.0 mm depending on the location of landmarks in the lungs.
Although demons registration has been evaluated using 4D CT datasets,5, 8 it has not been widely applied to serial CT scan registration. The present results demonstrate that demons registration has the potential to improve registration accuracy for studies of serially acquired clinical CT scans. The fact that demons deformable registration outperformed B-splines registration may be due to the larger number of degrees of freedom for demons registration, providing this method with a superior ability to achieve matching of lung volumes. While CT scans at our institution are routinely acquired using a breath-hold technique, no direct control on the level of inspiration is utilized, potentially complicating the registration problem.
Although demons deformable registration achieved superior anatomic matching relative to the other registration algorithms that were investigated, it introduced texture artifacts that were absent or less severe when B-splines registration was used. For example, demons registration caused noise smoothing (Fig. 4) that lowered gray-level entropy and fractal feature values. While this bias in feature value change existed to some extent when using B-splines, it was most extreme as a result of demons registration (Table 4). Although bias was consistently introduced for many of the features due to demons registration, the variance of texture feature value change between baseline and registered follow-up scans was lower for demons registration than for B-splines, affine, and rigid registrations. While this study used “normal” CT scans, the low variability in texture change between normal lung CT scans achieved using demons registration could facilitate its use to detect local changes in registered scans, thereby allowing for analysis of disease status change in serial scans. Further studies in patients with nonhealthy lungs are needed to determine the utility of combining demons registration with texture feature analysis.
Registration-stable texture features
Nineteen features were identified that remained relatively stable when deformable registration was used to register “normal” lung CT scans. Some of these features may have been ill-suited to measure the patterns in lung CT images, yielding consistent values regardless of actual changes present in the images. These features would appear to be minimally affected by registration when in fact they were insensitive to real change in CT images due to, for example, disease progression. Such features were identified by comparing texture feature value change across the four registration methods used. Due to the inferior anatomic matching achieved by rigid and affine registrations, a higher degree of variability between baseline and follow-up texture values introduced by registration inaccuracies would exist for these registration methods than for deformable registration. For three features (minimum gray-level value, 5% histogram quantile, and difference average of the GLCM), this trend was not observed, indicating that these particular features were ill-suited for detecting real changes in lung CT images as they remained invariant even to large registration inaccuracies. However, for 16 of these 19 features, lower variability existed between baseline and follow-up texture values when registration was performed with demons or B-splines than with rigid or affine registration. Moreover, the variability in feature value change for these relatively “registration-stable” features was lowest with demons registration, indicating that the combination of demons registration and texture analysis has the potential to detect true changes in lung parenchymal texture caused by disease progression. It should be noted that in future experiments, the cutoff of nRoA ≤ 15% may be modified based on the magnitude of change to be detected.
Future applications of texture analysis and deformable registration
We have identified 16 features that achieved low variability of texture feature value change between baseline and registered follow-up CT scans when demons deformable registration was used. For several features, demons registration introduced bias in feature values. Knowledge of such bias is vital for application of deformable image registration to texture feature analysis since in future work, this bias could be removed from texture changes to separate changes in disease status from artifacts introduced by the registration process. Texture change that is atypical of known registration effects may serve as an indicator of actual change in lung disease status.
For some features, changes may be introduced as a result of differences in the images themselves rather than by differences due to the registration process. Palma et al.12 demonstrated that differences in the concentration of intravenously injected contrast media or breathing phase at the time of image acquisition resulted in differences in the mean HU value of otherwise similar CT scans, though differences were accounted for on a per-scan basis by normalizing mean values by the average HU value in the contralateral lung. In addition to mean, other first-order features may be influenced by variations in image acquisition parameters or contrast uptake. Image acquisition-dependent changes may complicate the detection of texture changes due to a change in disease status. This complication may be remedied by normalizing to the average feature value in nondiseased lung regions. Rather than raw feature values, normalized feature values in baseline and deformed follow-up scans may then be compared to determine changes in lung disease status. Alternatively, it may be possible to normalize intravenous contrast-dependent feature values by the average gray-level value in the aorta to account for contrast injection differences (e.g., concentration and timing), though the utility of this approach remains to be investigated.
This study used only CT scans from subjects with no noted lung pathology. The registration task becomes more difficult when widespread pathologic differences are present between two scans, as is the case for changes due to diffuse lung disease or other pathologies. The application of the algorithms used in this study for registration of pathologically different lung CT images remains to be investigated. Lu et al.33 incorporated landmark-guided registration with demons deformable registration, and this registration method may be more appropriate in the presence of gross pathologic changes. Additionally, the utility of the identified features and deformation algorithms remains to be investigated when images are acquired using different CT scanners and/or technical parameters, as these factors have the potential to alter image texture.
CONCLUSIONS
This study investigated the accuracy of four image registration algorithms (rigid, affine, B-splines, and demons) and their effects on texture feature values using a set of 140 features distributed among first-order, fractal, Fourier, Laws’ filter, and GLCM classes. Of the four registration methods, the highest accuracy of spatially matched landmarks was achieved using demons. Nineteen features demonstrated values that exhibited low variation (nRoA ≤ 15%) between baseline and registered follow-up feature values as a result of demons deformable registration. nRoA was significantly larger when the other three registration methods were used.
The extent of feature value change introduced by deformable registration was characterized by measuring the bias between the values of the 19 “registration-stable” features in registered follow-up and baseline scans. Demons deformable registration introduced registration-dependent bias greater than 1% in the majority of the features, and knowledge of the magnitude of these changes would allow for better distinction between texture differences caused by disease progression and texture differences from registration-induced artifacts. Texture differences introduced by image-dependent factors were also observed.
This research creates an avenue for deformable image registration and texture feature analysis to be combined, and future work will target the utility of this combination to accurately detect and categorize local changes in lung disease status using serially acquired CT scans.
ACKNOWLEDGMENTS
This work was supported, in part, by The Coleman Endowment through The University of Chicago Comprehensive Cancer Center, National Science Foundation Research Experience for Undergraduates (NSF REU) Award No. 1062909, and National Institutes of Health (NIH) Grant Nos. S10 RR021039, P30 CA14599, and T32 EB002103-21. The authors would like to thank Gregory C. Sharp, Massachusetts General Hospital Department of Radiation Oncology, for his assistance with the Plastimatch demons registration algorithm, Neal Corson, The University of Chicago Department of Radiology, for his help in acquiring the patient database, Nick Gruszauskas, The University of Chicago Human Imaging Research Office, for providing CT scanner and imaging protocol information, and Kristen Wroblewski, and Chuanhong Liao, The University of Chicago Department of Health Studies, for their guidance in statistical analysis.
Presented in part at the 2011 Annual Meeting of the AAPM.
References
- Aziz Z. A., Wells A. U., Hansell D. M., Bain G. A., Copley S. J., Desai S. R., Ellis S. M., Gleeson F. V., Grubnic S., Nicholson A. G., Padley S. P. G., Pointon K. S., Reynolds J. H., Robertson R. J. H., and Rubens M. B., “HRCT diagnosis of diffuse parenchymal lung disease: Inter-observer variation,” Thorax 59, 506–511 (2004). 10.1136/thx.2003.020396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabat F., Yang G.-Z., and Hansell D. M., “Obstructive lung diseases: Texture classification for differentiation at CT,” Radiology 228, 871–877 (2003). 10.1148/radiol.2283020505 [DOI] [PubMed] [Google Scholar]
- Uchiyama Y., Katsuragawa S., Abe H., Shiraishi J., Li F., Li Q., Zhang C.-T., Suzuki K., and Doi D., “Quantitative computerized analysis of diffuse lung disease in high-resolution computed tomography,” Med. Phys. 30, 2440–2454 (2003). 10.1118/1.1597431 [DOI] [PubMed] [Google Scholar]
- Uppaluri R., Hoffman E. A., Sonka M., Hartley P. G., Hunninghake G. W., and McLennan G.,“Computer recognition of regional lung disease patterns,” Am. J. Respir. Crit. Care Med. 160, 648–654 (1999). [DOI] [PubMed] [Google Scholar]
- Wang H., Dong L., O’Daniel J., Mohan R., Garden A. S., Ang K. K., Kuban D. A., Bonnen M., Chang J. Y., and Cheung R., “Validation of an accelerated ‘demons’ algorithm for deformable image registration in radiation therapy,” Phys. Med. Biol. 50, 2887–2905 (2005). 10.1088/0031-9155/50/12/011 [DOI] [PubMed] [Google Scholar]
- Sharp G. C., Kandasamy N., Singh H., and Folkert M., “GPU-based streaming architectures for fast cone-beam CT image reconstruction and demons deformable registration,” Phys. Med. Biol. 52, 5771–5783 (2007). 10.1088/0031-9155/52/19/003 [DOI] [PubMed] [Google Scholar]
- Samant S. S., Xia J., Muyan-Ozcelik P., and Owens J. D., “High performance computing for deformable image registration: Towards a new paradigm in adaptive radiotherapy,” Med. Phys. 35, 3546–3553 (2008). 10.1118/1.2948318 [DOI] [PubMed] [Google Scholar]
- Wu Z., Rietzel E., Boldea V., Sarrut D., and Sharp G. C., “Evaluation of deformable registration of patient lung 4D CT with subanatomical region segmentations,” Med. Phys. 35, 775–781 (2008). 10.1118/1.2828378 [DOI] [PubMed] [Google Scholar]
- Stewart C. V., Lee Y.-L., and Tsai C.-L., “An uncertainty-driven hybrid of intensity-based and feature-based registration with application to retinal and lung CT images,” in Medical Image Computing and Computer-Assisted Intervention MICCAI 2004, Lecture Notes in Computer Science (Springer-Verlag, Berlin: 2004), Vol. 3217, pp. 870–877. [Google Scholar]
- Coselmon M. M., Balter J. M., McShan D. L., and Kessler M. L., “Mutual information based CT registration of the lung at exhale and inhale breathing states using thin-plate splines,” Med. Phys. 31, 2942–2948 (2004). 10.1118/1.1803671 [DOI] [PubMed] [Google Scholar]
- Palma D. A., Van Sornsen de Koste J., Verbakel W. F., Vincent A., and Senan S., “Lung density changes after stereotactic radiotherapy: A quantitative analysis in 50 patients,” Int. J. Radiat. Oncol. Biol. Phys. 81, 974–978 (2011). 10.1016/j.ijrobp.2010.07.025 [DOI] [PubMed] [Google Scholar]
- Palma D. A., Van Sornsen de Koste J. R., Verbakel W. F., and Senan S., “A new approach to quantifying lung damage after stereotactic body radiation therapy,” Acta Oncol. 50, 509–517 (2011). 10.3109/0284186X.2010.541934 [DOI] [PubMed] [Google Scholar]
- S. G.ArmatoIII, Gruszauskas N. P., MacMahon H., Torno M. D., Li F., Engelmann R. M., Starkey A., Pudela C. L., Marino J. S., Chang P. J., and Giger M. L., “Research imaging in an academic medical center,” Acad. Radiol. 19, 762–771 (2012). 10.1016/j.acra.2012.02.002 [DOI] [PubMed] [Google Scholar]
- Klein S., Staring M., Murphy K., Viergever M. A., and Pluim J. P. W., “Elastix: A toolbox for intensity-based medical image registration,” IEEE Trans. Med. Imaging 29, 196–205 (2010). 10.1109/TMI.2009.2035616 [DOI] [PubMed] [Google Scholar]
- Thirion J.-P., “Image matching as a diffusion process: an analogy with Maxwell's demons,” Med. Image Anal. 2, 243–260 (1998). 10.1016/S1361-8415(98)80022-4 [DOI] [PubMed] [Google Scholar]
- Rueckert D., Sonoda L. I., Hayes C., Hill D. L. G., Leach M. O., and Hawkes D. J., “Nonrigid registration using free-form deformations: Application to breast MR images,” IEEE Trans. Med. Imaging 18, 712–721 (1999). 10.1109/42.796284 [DOI] [PubMed] [Google Scholar]
- Staring M., Klein S., Reiber J. H. C., Niessen W. J., and Stoel B. C., “Pulmonary image registration with elastix using a standard intensity-based algorithm,” in Medical Image Analysis for the Clinic: A Grand Challenge, in Proceedings of the Workshop of MICCAI, Beijing, China, 2010.
- Murphy K., van Ginneken B., Klein S., Staring M., de Hoop B. J., Viergever M. A., and Pluim J. P. W., “Semi-automatic construction of reference standards for evaluation of image registration,” Med. Image Anal. 15, 71–84 (2011). 10.1016/j.media.2010.07.005 [DOI] [PubMed] [Google Scholar]
- Sensakovic W. F., Starkey A., and S. G.ArmatoIII, “Two-dimensional extrapolation methods for texture analysis on CT scans,” Med. Phys. 34, 3465–3472 (2007). 10.1118/1.2760307 [DOI] [PubMed] [Google Scholar]
- Pratt W. K., “Image feature extraction” in Digital Image Processing, 3rd ed. (Wiley, New York, 2001), pp. 509–550. [Google Scholar]
- Wagner T., “Texture analysis” in Handbook of Computer Vision and Applications, edited by Jahne B., Hanssecker H., and Geissler P. (Academic, San Diego, 1999), Vol. 2, pp. 275–308. [Google Scholar]
- Li H., Giger M. L., Huo Z., Olopade O. I., Lan L., Weber B. L., and Bonta I., “Computerized analysis of mammographic parenchymal patterns for assessing breast cancer risk: Effect of ROI size and location,” Med. Phys. 31, 549–555 (2004). 10.1118/1.1644514 [DOI] [PubMed] [Google Scholar]
- Peleg S., Naor J., Hartley R., and Avnir D., “Multiple resolution texture analysis and classification,” IEEE Trans. Pattern Anal. Mach. Intell. 6, 518–523 (1984). 10.1109/TPAMI.1984.4767557 [DOI] [PubMed] [Google Scholar]
- Chen C.-C., DaPonte J. S., and Fox M. D., “Fractal feature analysis and classification in medical imaging,” IEEE Trans. Med. Imaging 8, 133–142 (1989). 10.1109/42.24861 [DOI] [PubMed] [Google Scholar]
- Byng J. W., Boyd N. F., Fishell E., Jone R. A., and Yaffe M. J., “Automated analysis of mammographic densities,” Phys. Med. Biol. 41, 909–923 (1996). 10.1088/0031-9155/41/5/007 [DOI] [PubMed] [Google Scholar]
- Creutzburg R. and Ivanov E., “Fast algorithm for computing fractal dimensions of image segments” in Recent Issues in Pattern Analysis and Recognition, Lecture Notes in Computer Science, edited by Cantoni V., Creutzburg R., Levialdi S., and Wolf G. (Springer, Berlin, 1989), Vol. 399, pp. 42–51. [Google Scholar]
- Katsuragawa S., Doi K., and MacMahon H., “Image feature analysis and computer-aided diagnosis in digital radiography: Detection and characterization of interstitial lung disease in digital chest radiographs,” Med. Phys. 15(3), 311–319 (1988). 10.1118/1.596224 [DOI] [PubMed] [Google Scholar]
- Laws K. I., “Textured image segmentation,” USCIPI Technical Report No. 940 (University of Southern California, 1980).
- Haralick R. M., Shanmugam S., and Dinstein I., “Texture features for image classification,” IEEE Trans. Syst. Man Cybern. 3, 610–621 (1973). 10.1109/TSMC.1973.4309314 [DOI] [Google Scholar]
- Bland J. M. and Altman D. G., “Agreement between methods of measurement with multiple observations per individual,” J. Biopharm. Stat. 17, 571–582 (2007). 10.1080/10543400701329422 [DOI] [PubMed] [Google Scholar]
- Kashani R., Hub M., Balter J. M., Kessler M. L., Dong L., Zhang L., Xing L., Xie Y., Hawkes D., Schnabel J. A., McClelland J., Joshi S., Chen Q., and Lu W., “Objective assessment of deformable image registration in radiotherapy: A multi-institution study,” Med. Phys. 35(12), 5944–5953 (2008). 10.1118/1.3013563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brock K. K., “Results of a multi-institution deformable registration accuracy study (MIDRAS),” Int. J. Radiat. Oncol. Biol. Phys. 76, 583–596 (2010). 10.1016/j.ijrobp.2009.06.031 [DOI] [PubMed] [Google Scholar]
- Lu H., Cattin P. C., and Reyes M., “A hybrid multimodal non-rigid registration of MR images based on diffeomorphic demons,” in Proceedings of the International Conference of the IEEE Engineering in Medicine and Biology Society (IEEE, Buenos Aires, Argentina, 2010), pp. 5951–5954. [DOI] [PubMed]