Abstract
The RAD3 gene of Saccharomyces cerevisiae is required for excision of pyrimidine dimers and is essential for viability. We present the nucleotide sequence of the RAD3 protein coding region and its flanking regions, and the deduced primary structure of the RAD3 protein. In addition, we have mapped the 5' end of RAD3 mRNA. The predicted RAD3 protein contains 778 amino acids with a calculated molecular weight of 89,779. A segment of the RAD3 protein shares homology with several adenine nucleotide binding proteins, suggesting that RAD3 protein may react with ATP. The twenty carboxyl terminal amino acids of RAD3 protein are predominantly acidic; however, deletion of this acidic region has no obvious effect on viability or DNA repair.
Full text
PDF


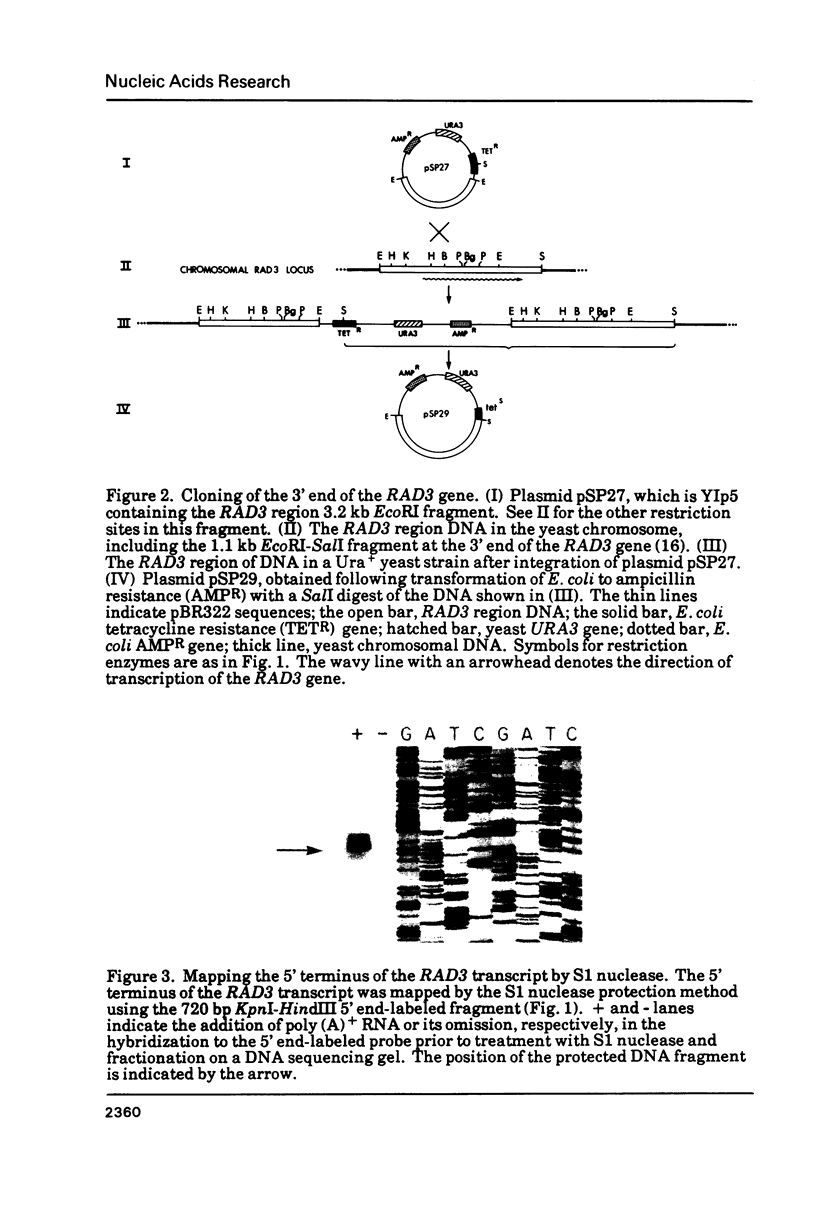
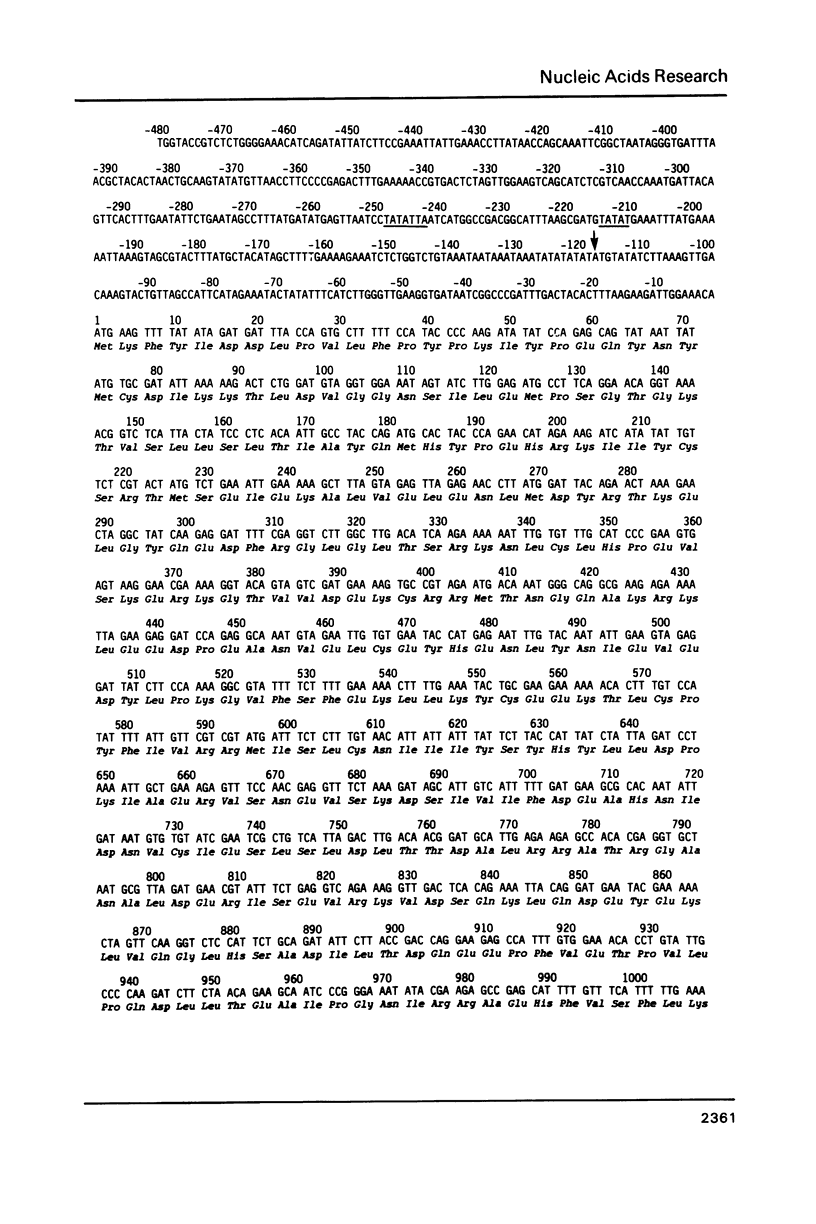


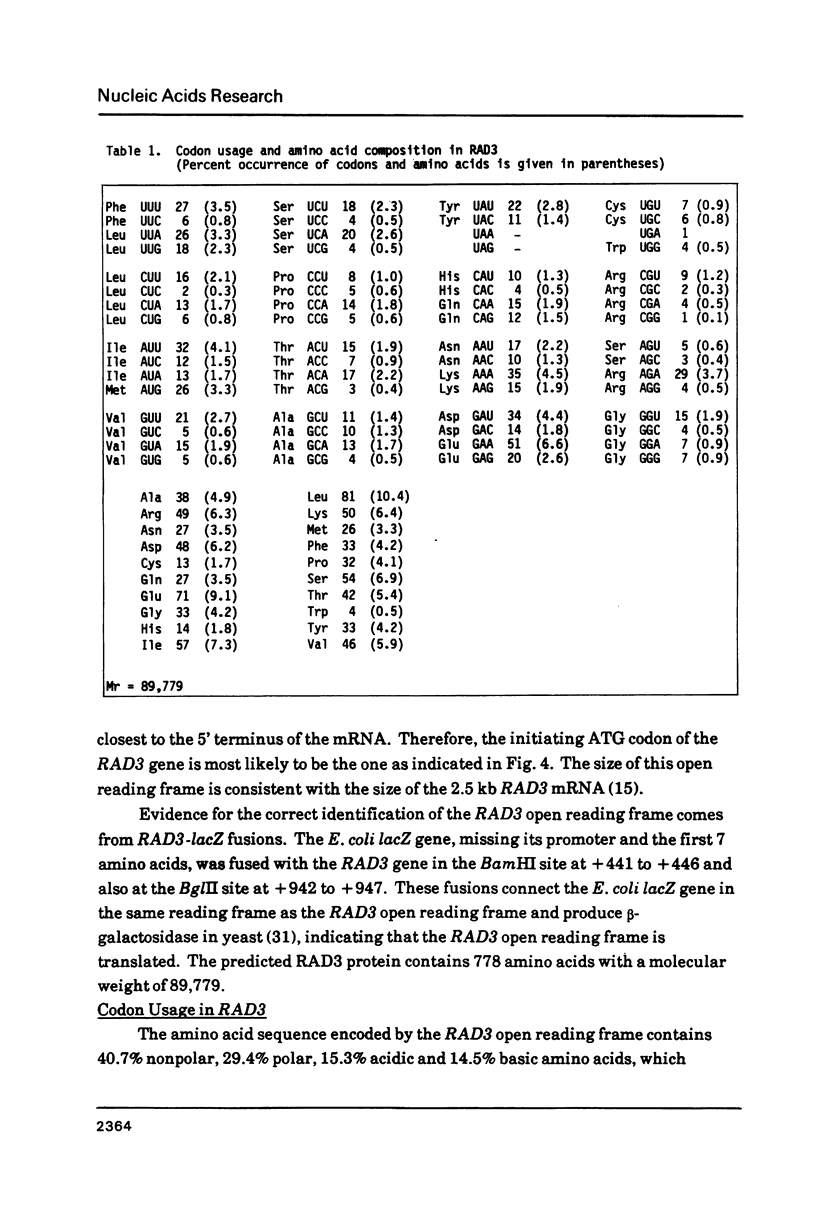






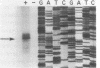
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bennetzen J. L., Hall B. D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase. J Biol Chem. 1982 Mar 25;257(6):3018–3025. [PubMed] [Google Scholar]
- Benoist C., O'Hare K., Breathnach R., Chambon P. The ovalbumin gene-sequence of putative control regions. Nucleic Acids Res. 1980 Jan 11;8(1):127–142. doi: 10.1093/nar/8.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Spliced early mRNAs of simian virus 40. Proc Natl Acad Sci U S A. 1978 Mar;75(3):1274–1278. doi: 10.1073/pnas.75.3.1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggin M. D., Gibson T. J., Hong G. F. Buffer gradient gels and 35S label as an aid to rapid DNA sequence determination. Proc Natl Acad Sci U S A. 1983 Jul;80(13):3963–3965. doi: 10.1073/pnas.80.13.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breathnach R., Chambon P. Organization and expression of eucaryotic split genes coding for proteins. Annu Rev Biochem. 1981;50:349–383. doi: 10.1146/annurev.bi.50.070181.002025. [DOI] [PubMed] [Google Scholar]
- Burke R. L., Tekamp-Olson P., Najarian R. The isolation, characterization, and sequence of the pyruvate kinase gene of Saccharomyces cerevisiae. J Biol Chem. 1983 Feb 25;258(4):2193–2201. [PubMed] [Google Scholar]
- Dobson M. J., Tuite M. F., Roberts N. A., Kingsman A. J., Kingsman S. M., Perkins R. E., Conroy S. C., Fothergill L. A. Conservation of high efficiency promoter sequences in Saccharomyces cerevisiae. Nucleic Acids Res. 1982 Apr 24;10(8):2625–2637. doi: 10.1093/nar/10.8.2625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donahue T. F., Farabaugh P. J., Fink G. R. The nucleotide sequence of the HIS4 region of yeast. Gene. 1982 Apr;18(1):47–59. doi: 10.1016/0378-1119(82)90055-5. [DOI] [PubMed] [Google Scholar]
- Faye G., Leung D. W., Tatchell K., Hall B. D., Smith M. Deletion mapping of sequences essential for in vivo transcription of the iso-1-cytochrome c gene. Proc Natl Acad Sci U S A. 1981 Apr;78(4):2258–2262. doi: 10.1073/pnas.78.4.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finch P. W., Emmerson P. T. The nucleotide sequence of the uvrD gene of E. coli. Nucleic Acids Res. 1984 Jul 25;12(14):5789–5799. doi: 10.1093/nar/12.14.5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzgerald M., Shenk T. The sequence 5'-AAUAAA-3'forms parts of the recognition site for polyadenylation of late SV40 mRNAs. Cell. 1981 Apr;24(1):251–260. doi: 10.1016/0092-8674(81)90521-3. [DOI] [PubMed] [Google Scholar]
- Gannon F., O'Hare K., Perrin F., LePennec J. P., Benoist C., Cochet M., Breathnach R., Royal A., Garapin A., Cami B. Organisation and sequences at the 5' end of a cloned complete ovalbumin gene. Nature. 1979 Mar 29;278(5703):428–434. doi: 10.1038/278428a0. [DOI] [PubMed] [Google Scholar]
- Grosschedl R., Birnstiel M. L. Identification of regulatory sequences in the prelude sequences of an H2A histone gene by the study of specific deletion mutants in vivo. Proc Natl Acad Sci U S A. 1980 Mar;77(3):1432–1436. doi: 10.1073/pnas.77.3.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S., Kelly J. D., Cohen E. H. Transcription terminates in yeast distal to a control sequence. Cell. 1983 Jun;33(2):607–614. doi: 10.1016/0092-8674(83)90441-5. [DOI] [PubMed] [Google Scholar]
- Higgins D. R., Prakash L., Reynolds P., Prakash S. Isolation and characterization of the RAD2 gene of Saccharomyces cerevisiae. Gene. 1984 Oct;30(1-3):121–128. doi: 10.1016/0378-1119(84)90112-4. [DOI] [PubMed] [Google Scholar]
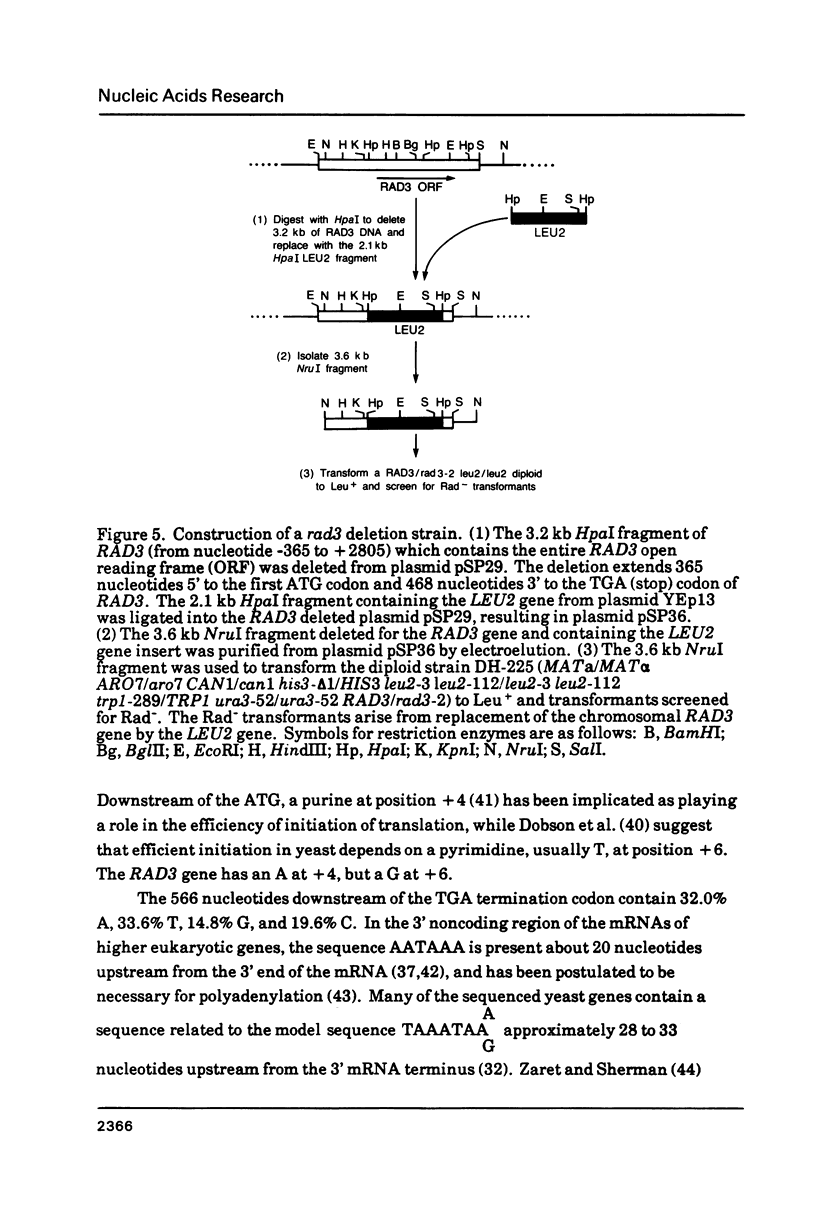
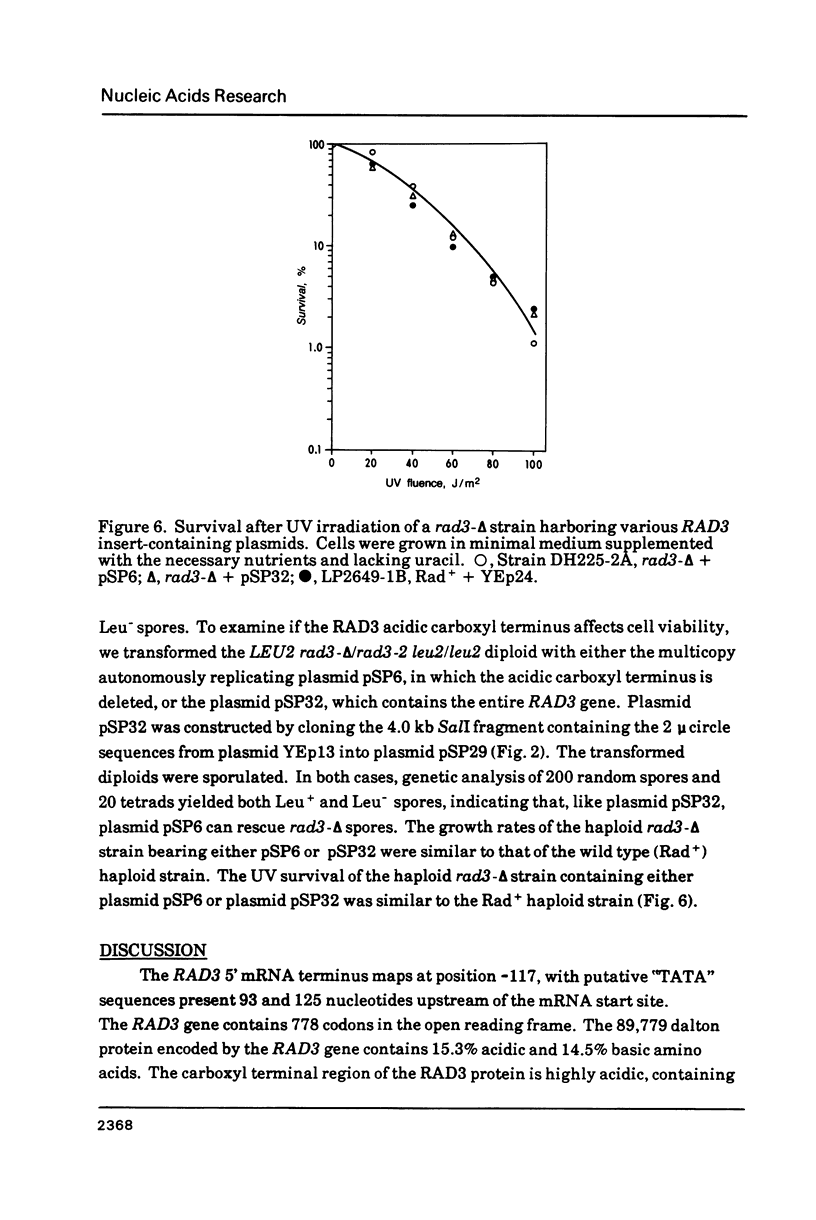
- Higgins D. R., Prakash S., Reynolds P., Polakowska R., Weber S., Prakash L. Isolation and characterization of the RAD3 gene of Saccharomyces cerevisiae and inviability of rad3 deletion mutants. Proc Natl Acad Sci U S A. 1983 Sep;80(18):5680–5684. doi: 10.1073/pnas.80.18.5680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins D. R., Prakash S., Reynolds P., Prakash L. Molecular cloning and characterization of the RAD1 gene of Saccharomyces cerevisiae. Gene. 1983 Dec;26(2-3):119–126. doi: 10.1016/0378-1119(83)90181-6. [DOI] [PubMed] [Google Scholar]
- Horii Z., Clark A. J. Genetic analysis of the recF pathway to genetic recombination in Escherichia coli K12: isolation and characterization of mutants. J Mol Biol. 1973 Oct 25;80(2):327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- Jachymczyk W. J., von Borstel R. C., Mowat M. R., Hastings P. J. Repair of interstrand cross-links in DNA of Saccharomyces cerevisiae requires two systems for DNA repair: the RAD3 system and the RAD51 system. Mol Gen Genet. 1981;182(2):196–205. doi: 10.1007/BF00269658. [DOI] [PubMed] [Google Scholar]
- Klinkert M. Q., Klein A., Abdel-Monem M. Studies on the functions of DNA helicase I and DNA helicase II of Escherichia coli. J Biol Chem. 1980 Oct 25;255(20):9746–9752. [PubMed] [Google Scholar]
- Kozak M. Possible role of flanking nucleotides in recognition of the AUG initiator codon by eukaryotic ribosomes. Nucleic Acids Res. 1981 Oct 24;9(20):5233–5252. doi: 10.1093/nar/9.20.5233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuemmerle N. B., Masker W. E. An in vitro complementation assay for the Escherichia coli uvrD gene product. Nucleic Acids Res. 1983 Apr 11;11(7):2193–2204. doi: 10.1093/nar/11.7.2193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuemmerle N. B., Masker W. E. Effect of the uvrD mutation on excision repair. J Bacteriol. 1980 May;142(2):535–546. doi: 10.1128/jb.142.2.535-546.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumura K., Sekiguchi M. Identification of the uvrD gene product of Escherichia coli as DNA helicase II and its induction by DNA-damaging agents. J Biol Chem. 1984 Feb 10;259(3):1560–1565. [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Clark S., Modrich P. Methyl-directed repair of DNA base-pair mismatches in vitro. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4639–4643. doi: 10.1073/pnas.80.15.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magaña-Schwencke N., Henriques J. A., Chanet R., Moustacchi E. The fate of 8-methoxypsoralen photoinduced crosslinks in nuclear and mitochondrial yeast DNA: comparison of wild-type and repair-deficient strains. Proc Natl Acad Sci U S A. 1982 Mar;79(6):1722–1726. doi: 10.1073/pnas.79.6.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mathis D. J., Chambon P. The SV40 early region TATA box is required for accurate in vitro initiation of transcription. Nature. 1981 Mar 26;290(5804):310–315. doi: 10.1038/290310a0. [DOI] [PubMed] [Google Scholar]
- Miller R. D., Prakash L., Prakash S. Defective excision of pyrimidine dimers and interstrand DNA crosslinks in rad7 and rad23 mutants of Saccharomyces cerevisiae. Mol Gen Genet. 1982;188(2):235–239. doi: 10.1007/BF00332681. [DOI] [PubMed] [Google Scholar]
- Miller R. D., Prakash L., Prakash S. Genetic control of excision of Saccharomyces cerevisiae interstrand DNA cross-links induced by psoralen plus near-UV light. Mol Cell Biol. 1982 Aug;2(8):939–948. doi: 10.1128/mcb.2.8.939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. A DNA repair gene required for the incision of damaged DNA is essential for viability in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1983 Aug;80(15):4818–4821. doi: 10.1073/pnas.80.15.4818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. Molecular cloning of eucaryotic genes required for excision repair of UV-irradiated DNA: isolation and partial characterization of the RAD3 gene of Saccharomyces cerevisiae. J Bacteriol. 1982 Oct;152(1):323–331. doi: 10.1128/jb.152.1.323-331.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naumovski L., Friedberg E. C. Saccharomyces cerevisiae RAD2 gene: isolation, subcloning, and partial characterization. Mol Cell Biol. 1984 Feb;4(2):290–295. doi: 10.1128/mcb.4.2.290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash L. Defective thymine dimer excision in radiation-sensitive mutants rad10 and rad16 of Saccharomyces cerevisiae. Mol Gen Genet. 1977 Apr 29;152(3):125–128. doi: 10.1007/BF00268808. [DOI] [PubMed] [Google Scholar]
- Prakash L., Dumais D., Polakowska R., Perozzi G., Prakash S. Molecular cloning of the RAD10 gene of Saccharomyces cerevisiae. Gene. 1985;34(1):55–61. doi: 10.1016/0378-1119(85)90294-x. [DOI] [PubMed] [Google Scholar]
- Prakash L., Prakash S. Three additional genes involved in pyrimidine dimer removal in Saccharomyces cerevisiae: RAD7, RAD14 and MMS19. Mol Gen Genet. 1979 Nov;176(3):351–359. doi: 10.1007/BF00333097. [DOI] [PubMed] [Google Scholar]
- Prakash L. Repair of pyrimidine dimers in nuclear and mitochondrial DNA of yeast irradiated with low doses of ultraviolet light. J Mol Biol. 1975 Nov 15;98(4):781–795. doi: 10.1016/s0022-2836(75)80010-6. [DOI] [PubMed] [Google Scholar]
- Prakash L. Repair of pyrimidine dimers in radiation-sensitive mutants rad3, rad4, rad6 and rad9 of Saccharomyces cerevisiae. Mutat Res. 1977 Oct;45(1):13–20. doi: 10.1016/0027-5107(77)90038-0. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Resnick M. A., Setlow J. K. Repair of pyrimidine dimer damage induced in yeast by ultraviolet light. J Bacteriol. 1972 Mar;109(3):979–986. doi: 10.1128/jb.109.3.979-986.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds P., Weber S., Prakash L. RAD6 gene of Saccharomyces cerevisiae encodes a protein containing a tract of 13 consecutive aspartates. Proc Natl Acad Sci U S A. 1985 Jan;82(1):168–172. doi: 10.1073/pnas.82.1.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R. J., Friedberg E. C. Molecular mechanisms of pyrimidine dimer excision in Saccharomyces cerevisiae: incision of ultraviolet-irradiated deoxyribonucleic acid in vivo. J Bacteriol. 1981 May;146(2):692–704. doi: 10.1128/jb.146.2.692-704.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds R. J. Removal of pyrimidine dimers from Saccharomyces cerevisiae nuclear DNA under nongrowth conditions as detected by a sensitive, enzymatic assay. Mutat Res. 1978 Apr;50(1):43–56. doi: 10.1016/0027-5107(78)90059-3. [DOI] [PubMed] [Google Scholar]
- Rothman R. H., Clark A. J. Defective excision and postreplication repair of UV-damaged DNA in a recL mutant strain of E. coli K-12. Mol Gen Genet. 1977 Oct 24;155(3):267–277. doi: 10.1007/BF00272805. [DOI] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sancar G. B., Sancar A., Rupp W. D. Sequences of the E. coli uvrC gene and protein. Nucleic Acids Res. 1984 Jun 11;12(11):4593–4608. doi: 10.1093/nar/12.11.4593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer S., Davis R. W. Replacement of chromosome segments with altered DNA sequences constructed in vitro. Proc Natl Acad Sci U S A. 1979 Oct;76(10):4951–4955. doi: 10.1073/pnas.76.10.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F., Stewart J. W., Schweingruber A. M. Mutants of yeast initiating translation of iso-1-cytochrome c within a region spanning 37 nucleotides. Cell. 1980 May;20(1):215–222. doi: 10.1016/0092-8674(80)90249-4. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Unrau P., Wheatcroft R., Cox B. S. The excision of pyrimidine dimers from DNA of ultraviolet irradiated yeast. Mol Gen Genet. 1971;113(4):359–362. doi: 10.1007/BF00272336. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Runswick M. J., Gay N. J. Distantly related sequences in the alpha- and beta-subunits of ATP synthase, myosin, kinases and other ATP-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1(8):945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters R., Moustacchi E. The disappearance of ultraviolet-induced pyrimidine dimers from the nuclear DNA of exponential and stationary phase cells of Saccharomyces cerevisiae following various post-irradiation treatments. Biochim Biophys Acta. 1974 Jul 24;353(4):407–419. doi: 10.1016/0005-2787(74)90048-3. [DOI] [PubMed] [Google Scholar]
- Weaver R. F., Weissmann C. Mapping of RNA by a modification of the Berk-Sharp procedure: the 5' termini of 15 S beta-globin mRNA precursor and mature 10 s beta-globin mRNA have identical map coordinates. Nucleic Acids Res. 1979 Nov 10;7(5):1175–1193. doi: 10.1093/nar/7.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcox D. R., Prakash L. Incision and postincision steps of pyrimidine dimer removal in excision-defective mutants of Saccharomyces cerevisiae. J Bacteriol. 1981 Nov;148(2):618–623. doi: 10.1128/jb.148.2.618-623.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]