Abstract
The hycu-ep32 gene of Hyphantria cunea NPV can inhibit Bombyx mori nucleopolyhedrovirus (BmNPV) multiplication in co-infected cells, but it is not known whether the overexpression of the hycu-ep32 gene has an antiviral effect in the silkworm, Bombyx mori. Thus, we constructed four transgenic vectors, which were under the control of the 39 K promoter of BmNPV (39 KP), Bombyx mori A4 promoter (A4P), hr3 enhancer of BmNPV combined with 39 KP, and hr3 combined with A4P. Transgenic lines were created via embryo microinjection using practical diapause silkworm. qPCR revealed that the expression level of hycu-ep32 could be induced effectively after BmNPV infection in transgenic lines where hycu-ep32 was controlled by hr3 combined with 39 KP (i.e., HEKG). After oral inoculation of BmNPV with 3 × 105 occlusion bodies per third instar, the mortality with HEKG-B was approximately 30% lower compared with the non-transgenic line. The economic characteristics of the transgenic lines remained unchanged. These results suggest that overexpression of an exogenous antiviral gene controlled by an inducible promoter and enhancer is a feasible method for breeding silkworms with a high antiviral capacity.
Introduction
Bombyx mori nucleopolyhedrovirus (BmNPV) is a member of the Baculoviridae family. Two virion phenotypes occur during the NPV infection cycle, i.e., occlusion-derived virus (ODV) and budded virus (BV). ODV causes contagion among individual silkworms (Bombyx mori), whereas BV induces systemic infection throughout the host body [1], [2]. In natural conditions, NPV infects host larvae mainly via the oral route by ODV. After the ODV nucleocapsids have invaded the host cells, the viral genes begin transcription in a temporal manner to initiate a primary infection [3]. The expression of the immediate early genes of Autographa californica multiplenucleopolyhedrovirus (AcMNPV), such as ie-1 and ie-2, begins within the first 6 hours post-infection (hpi) [4]. DNA replication can be detected initially at 6–8 hpi, before peaking at approximately 18 hpi [5]. The BV virions are produced next, which cause secondary infection at approximately 20 hpi [4]. The BmNPV life cycle is a slightly slower than that of AcMNPV. BmNPV is a primary pathogen of silkworms, which causes severe economic losses to sericulture each year. However, no effective strategy exists for controlling the virus.
Disease-resistant species can be constructed using transgenic technology. Various methods have been used to create disease-resistant species, such as knocking down the pathogenic genes via transgenic RNAi [6], [7], and overexpressing the endogenous anti-pathogen genes [8], [9], [10] or exogenous resistant genes [11], [12]. Several methods have been proposed to ensure the effectiveness of anti-disease factors and their high level expression in transgenic hosts, including the use of a constitutive promoter, an inducible promoter, a constitutive promoter combined with an enhancer, or an inducible promoter combined with an enhancer. Previous studies have reported the control of anti-disease factors by constitutive promoters [8], [9], [11], [12], which suggests that anti-disease factors could be expressed in a sustained manner, regardless of whether an infection is caused by a pathogen. However, a better method might be the increased expression of an anti-pathogen factor with an increase in the pathogen content after infection in the transgenic host, although there are no reports on this subject.
An ideal method of breeding antiviral silkworm lines would be the overexpression of an exogenous antiviral gene under the control of an inducible promoter and enhancer in the presence of the virus in transgenic silkworm. Baculovirus genomes contain homologous regions (hrs) formed of repeated sequences [13], [14]. The hrs site is the origin of viral DNA replication [15], [16] and certain hrs sits can enhance the activity of NPV early promoters, such as 39 K [13], [14], [17], p35 [18], [19], and ie-N [20]. Some hrs can also act as enhancers of non-viral promoters, such as the B. mori cytoplasmic actin3 gene (A3) promoter [21] and Drosophila hsp70 promoter [22]. The 39 K gene of AcMNPV is a delayed early gene with no transcriptional activity in uninfected cells, but it can be activated during co-transfection with a virus [17]. In the presence of an hrs upstream of the 39 K promoter, the activity of 39 K is enhanced significantly during transactivation with IE1 protein [13], [23]. The IE1 protein is the primary transcriptional regulator of NPV [24], [25], which increases the promoter activity significantly by binding to the palindromic 28 bp repeats of hrs [25], [26], [27], [28], [29].
If BmNPV and Hyphantria cunea NPV (HycuNPV) are co-infected in BmN-4 cells, the hycu-ep32 gene of HycuNPV induces a global protein synthesis shutdown that inhibits BmNPV proliferation [30]. Shirata et al. [30] found that hycu-ep32 is an early and nonessential gene that encodes a polypeptide containing 312 amino acids, with no characteristic motifs or domains. A homologue of hycu-ep32 gene is present in the Orgyia pseudotsugata multicapsid NPV (OpMNPV), but it is not known in any other organism [30]. To the best of our knowledge, this is the only report of an exogenous gene that can inhibit BmNPV proliferation.
In the current study, we selected the exogenous antiviral gene hycu-ep32 and we used the inducible 39 K promoter of BmNPV (39 KP) and the hr3 enhancer of BmNPV to construct a transgenic overexpression vector where hycu-ep32 was controlled by hr3 combined with 39 KP. The embryos of practical diapause silkworm strain “932” were used for transgenic microinjection. Exogenous hycu-ep32 was expressed successfully in the transgenic lines. The induced mRNA expression level of hycu-ep32 was significantly increased in transgenic lines after infection with BmNPV. Compared with the non-transgenic line, the transgenic lines had significantly enhanced resistance and unaffected economic characteristics. This is the first report of the overexpression of an exogenous gene to enhance the antiviral ability of a practical silkworm strain using an inducible promoter and enhancer.
Results
Overexpression Vector Construction and Screening of Transgenic Silkworm Lines
The nucleotide sequences of hycu-ep32 and 39 KP were cloned from the genomic DNA of HycuNPV and BmNPV, respectively. The overexpression vectors pb-EKG, pb-HEKG, pb-EAG, and pb-HEAG (Fig. 1) were constructed using the transgenic plasmid piggyBac [3×p3 EGFP afm], where hycu-ep32 was driven by 39 KP, 39 KP combined with hr3, A4 promoter (A4P), and A4P combined with hr3, respectively. Microinjection was performed using mixtures of the overexpression plasmid and helper vector pHA3PIG [10], [31], [32] in 932 nondiapause embryos. Table 1 shows the transformation results of the embryo microinjection. G0 moths were mated with each other or backcrossed to produce G1 offspring. The G1 broods were screened for EGFP-positive expression and transgenic G1 moths in the same broods were sibling-mated to generate offspring [10]. Finally, we obtained a transgenic line from each of the vectors pb-EKG, pb-EAG, and pb-HEAG, and we named the corresponding transgenic lines as EKG, EAG, and HEAG, respectively. Eight transgenic lines of pb-HEKG were obtained and two lines (HEKG-A and HEKG-B) were selected randomly for subsequent detection experiments. Each transgenic line was sibling-mated to generate offspring for each generation.
Figure 1. Schematic diagram of the transgenic overexpression vectors.
PiggyBac [3×p3 EGFP afm] was a basic transgenic vector with a report marker 3×P3-EGFP-SV40. PL and PR indicate the left and right terminal inverted repeats, respectively. 39 KP and A4P represent the 39 K promoter of BmNPV and A4 promoter of B. mori, respectively. Hr3 is an enhancer from BmNPV. Hycu-ep32 typifies the coding sequences of the hycu-ep32 gene, while SV40 indicates the polyadenylation signal.
Table 1. The results of embryo microinjection.
| Injectedvector | Injectedstrain | Injectedembryos | Hatchedlarvae | Efficiencyof hatching | G1 broods | Broods with EGFPpositive | Efficiency ofpositive |
| pb-EKG | 932 | 91 | 25 | 27.47% | 6 | 1 | 16.67% |
| pb-EAG | 932 | 67 | 18 | 26.87% | 5 | 1 | 20.00% |
| pb-HEKG | 932 | 183 | 47 | 25.68% | 13 | 8 | 61.54% |
| pb-HEAG | 932 | 51 | 11 | 21.57% | 2 | 1 | 50.00% |
Analysis of Insertion Sites in Transgenic Lines
The genomic DNA in each transgenic line was extracted from G1 male moths. Inverse PCR was performed to detect the insertion sites in these transgenic lines. The left part of the insert and the next left part of the genome sequence of the insertion site were amplified using the transposon-specific primer pBacL, while the right part of the insert and the next right part of the genome sequence of the insertion site were amplified using the transposon-specific primer pBacR. PCR amplification using pBacL and pBacR showed that each transgenic line contained only one band (data not shown). This suggested that only a single copy was inserted into each transgenic line. These results were also confirmed by sequencing of the PCR-amplified products (Fig. 2). Bioinformatics analysis demonstrated that the insertion site in EKG was located in an intergenic region, while the nearest genes to the left and right of the insertion site were located within 11 kb and 50 kb. The insertion site of HEKG-A was also in an intergenic region, while the nearest genes to the left and right of the insertion site were within 96 kb and 248 kb. The insertion site of HEKG-B was in the BGIBMGA004763 gene intron, which was a predicted gene that with no Expressed Sequence Tags (ESTs) [33], [34], [35] and no similar gene in NCBI. Thus, the predicted BGIBMGA004763 gene was probably not a legitimate B. mori gene and the insert might not have affected the expression of normal genes. The insertion site of EAG was in an intergenic region, while the nearest genes to the left and right of the insertion site were within 3 kb and 39 kb. The insertion site of HEAG was located an intergenic region while the nearest genes to the left and right of the insertion site were within 10 kb and 13 kb.
Figure 2. Analysis of the insertion sites of transgenic lines.
The insertion sites were detected using inverse PCR. The genomic DNA from each line was digested fully using Hae III, and then self-ligated. Two pairs of transposon-specific primers pBacL and pBacR were used for the PCR amplification reaction. The PCR products were cloned and sequenced.
Expression Profile of Hycu-ep32 in Transgenic Lines
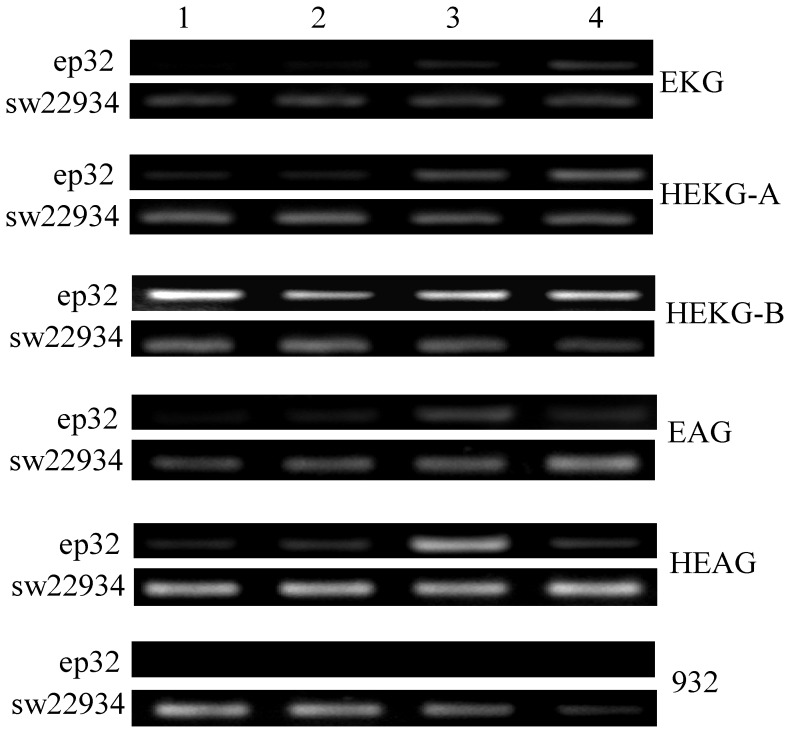
hycu-ep32 has no homologous gene in the silkworm genome [33], [35]. To test whether hycu-ep32 was expressed in each transgenic line, the cDNA templates of the third instar, third instar molt, fourth instar, and fourth instar molt larvae of EKG, HEKG-A, HEKG-B, EAG, HEAG, and 932 were used to detect hycu-ep32 transcription. The PCR-amplified cycles of primers ep32QRT and sw22934 were 28 and 25, respectively. The RT-PCR results showed that hycu-ep32 was expressed in most transgenic lines, but not in non-transgenic silkworm 932. The expression levels of hycu-ep32 in the fourth instar were higher than those of the third instar in the transgenic lines (Fig. 3). There was almost no expression of hycu-ep32 in the third instar and very low expression in the fourth instar in EKG. These results showed that 39 KP had little or no activity in silkworm larvae. The hycu-ep32 expression level in HEKG was higher than that in EKG, which suggested that hr3 could enhance the activity of 39 KP in silkworm larvae. There were no differences in the hycu-ep32 expression levels of EAG and HEAG. The hycu-ep32 expression level in HEKG-B was the highest among all the lines tested. (Fig. 3). These results indicated that the exogenous hycu-ep32 was successfully expressed in the transgenic lines.
Figure 3. Analysis of hycu-ep32 using RT-PCR in transgenic and non-transgenic silkworms.
RNA was extracted from different developmental stages of EKG, HEKG-A, HEKG-B, EAG, HEAG, and 932, and then reverse-transcribed to cDNA. cDNA templates were used for RT-PCR. The PCR-amplified cycles of ep32 and sw22934 were 28 and 25, which detected the hycu-ep32 gene and the silkworm house-keeping gene BGIBMGA003186, respectively. Points 1 to 4 represent third instar, third instar molt, fourth instar, and fourth instar molt larvae, respectively.
Determination of the Anti-BmNPV Capacity of Transgenic Lines
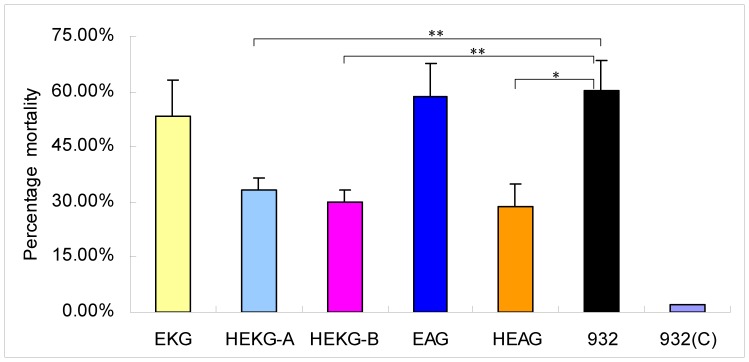
To investigate the capacity of these transgenic lines to protect against BmNPV infection, the newly exuviated third instar larvae of EKG, HEKG-A, HEKG-B, EAG, HEAG, and 932 were orally inoculated with BmNPV using 3 × 105 occlusion bodies (OB)/larva. The OBs were smeared on a piece of 1 cm diameter fresh mulberry leaf and any larvae that consumed an entire piece of OB-treated leaf were collected for continuous rearing in normal conditions. Each line was infected with BmNPV using three replicate groups, each of which contained 70 larvae. The mortality statistics were calculated each day until the 10th day postinfection (dpi). Compared with transgenic lines EKG and EAG, the transgenic lines HEKG-A, HEKG-B, and HEAG had a sustained and distinctive protective capacity (Fig. 4). The mortality of EKG, HEKG-A, HEKG-B, EAG, HEAG, and 932 were 53.08%, 33.11%, 30.00%, 58.42%, 28.64%, and 60.19%, respectively. There was little decrease in the mortality rate of EAG and EKG compared with 932. However, the mortality rates of HEKG-A, HEKG-B, and HEAG were decreased by 27.08%, 30.19%, and 31.55% compared with 932, respectively. Most untreated silkworms 932(C) survived (Fig. 4).
Figure 4. Mortality statistics after infection with BmNPV per os using third instar larvae.
Six silkworm lines, i.e., EKG, HEKG-A, HEKG-B, EAG, HEAG, and 932, were infected orally with 3 × 105 OB/larva using newly exuviated third instar larvae. The same quantity of OBs was applied to pieces of fresh mulberry leaves with a 1 cm diameter. Larvae that consumed an entire piece of leaf treated with OBs were selected for the subsequent rearing. The mortality of each line was the average of triplicate infection replicates. Each replicate consisted of 70 larvae. Mortality statistics were analyzed daily from the time of infection until 10 dpi. The accumulative mortality up to 10 dpi is shown for each line. 932(C) is the non-infected control. Bars represent the standard deviations. Statistically significant differences: * P<0.05, ** P<0.01.
The second determination also used third instar larvae (Fig. S1). After BmNPV oral infection using 3 × 105 OB/larva, the mortality of HEKG-A, HEKG-B, and HEAG was decreased by 37.53%, 37.31%, and 24.35% compared with 932, respectively. However, when the BmNPV infection dose was decreased to 2.5 × 105 OB/larva, the mortality rate of EAG and EKG was not decreased compared with 932 (Fig. S1).
The third determination used fourth instar larvae (Fig. S2). The mortality rates of HEKG-A, HEKG-B, and HEAG were decreased by 12.80%, 21.66%, and 18.38% compared with 932, respectively, after infection with BmNPV per os using 106 OB/larva. However, there were virtually no differences in the mortality rates of EKG, EAG, and 932 after infection using 8.3 × 105 OB/larva (Fig. S2).
The mortality statistics indicated that the HEKG-A, HEKG-B, and HEAG lines had significantly higher resistance to BmNPV compared with EKG, EAG, and the non-transgenic silkworms. According to the three comprehensive tests of resistance analysis, the antivirus capacity of HEKG-B was the highest among all of the lines tested.
Hycu-ep32 Expression after BmNPV Infection
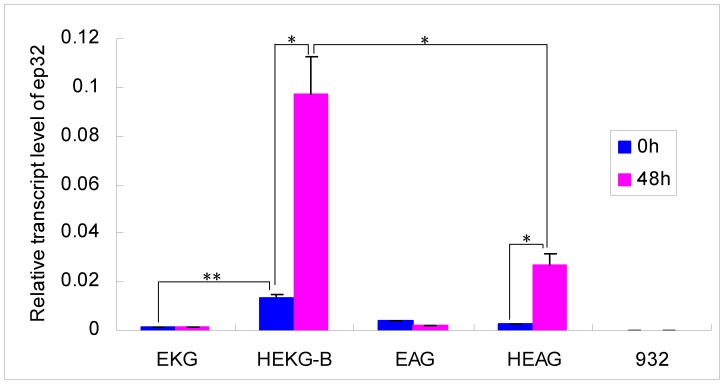
To confirm whether the expression of hycu-ep32 was changed after BmNPV infection in the transgenic lines, the cDNA templates of EKG, HEKG-B, EAG, HEAG, and 932 at 0 and 48 h postinfection were used for qPCR analysis. qPCR showed that the expression levels of hycu-ep32 in HEKG-B were the highest among all the lines, regardless of whether they were infected with BmNPV, whereas the expression levels of hycu-ep32 in EKG were very low (Fig. 5). With a normal physical status, the expression level of hycu-ep32 in HEKG-B was 11.92-fold that of EKG, while the expression level of hycu-ep32 in HEAG was no higher than that of EAG. These results indicated that hr3 could significantly enhance the transcription capacity of 39 KP in normal silkworm larvae. At 48 hpi, the expression levels of hycu-ep32 in EKG, HEKG-B, EAG, and HEAG were 1.32-fold, 7.41-fold, 0.52-fold, and 10.10-fold, respectively, compared with the insects before infection (Fig. 5). Thus, hr3 could significantly increase the transcription activity of 39 KP and A4P after BmNPV infection. After infection with BmNPV, the expression levels of antivirus factor hycu-ep32 were induced effectively in HEAG, particularly in HEKG-B (Fig. 5), which suggested that hr3 combined with 39 KP was the best of all the promoters tested. These results also showed that the higher transcription of hycu-ep32 mRNAs led to a higher anti-BmNPV capacity in the transgenic lines (Figs 4, S1, S2, 5).
Figure 5. qPCR analysis of hycu-ep32 expression after BmNPV infection of transgenic lines.
The same viral quantity was inoculated into newly exuviated third instar larvae. The cDNA was extracted from EKG, HEKG-B, EAG, HEAG, and 932 using 10 treated larvae at 0 and 48 hpi, respectively. Primers ep32QRT were used for the qPCR analysis reactions. The time that the infection process ended was set as time point 0 h. Bars represent the standard deviations. Statistically significant differences: * P<0.05, ** P<0.01.
qPCR Analysis of Virus Proliferation after Inoculation
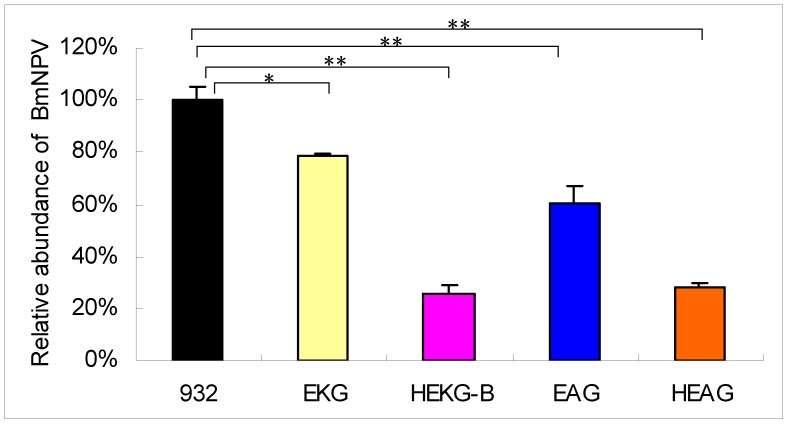
To further detect the antiviral capacity of each transgenic line against BmNPV, the number of BmNPV in the bodies of transgenic and non-transgenic silkworm lines were analyzed after infection. The viral dose ingested by each individual was the same as that used at the stage of newly exuviated third instar larvae. Total DNA was extracted from 10 treated larvae in each line at 48 hpi. The DNA-accumulated virus number was determined via qPCR using GP41 primers [10]. The virus content of the non-transgenic 932 was set to 100% and the values of the transgenic lines were normalized against this value. Thus, the levels of BmNPV DNA accumulated in EKG, HEKG-B, EAG, and HEAG were 78.43%, 25.48%, 60.72%, and 28.39%, respectively (Fig. 6). The virus content of all the transgenic lines was lower than that of the control at 2 dpi, particularly in the transgenic lines HEKG-B and HEAG. These results suggested that transgenic lines could inhibit BmNPV proliferation.
Figure 6. Analysis of BmNPV by qPCR in transgenic and non-transgenic lines after viral inoculation.
The ingested viral dose of each individual was the same as that used for the newly exuviated third instar larvae. Total DNA was obtained from 10 treated larvae in each line at 48 hpi. The DNA-accumulated numbers of BmNPV were detected using qPCR with GP41 primers. The average of non-transgenic 932 was set at 100% and the values for transgenic lines were normalized against that of 932. Bars represent the standard deviations. Statistically significant differences: * P<0.05, ** P<0.01.
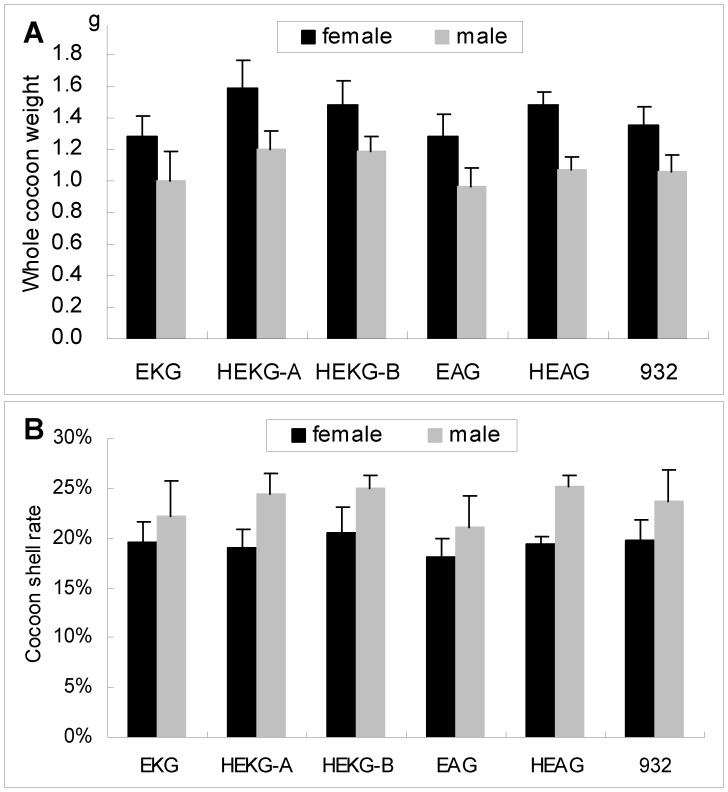
Economic Characteristic of the Transgenic Lines
The sericulture industry is very important for silk production, but it faces great challenges because of severe diseases. Traditional methods of breeding resistant silkworm varieties might enhance their antivirus capacity but reduce their economic characteristics. To investigate the effects of hycu-ep32 overexpression in transgenic lines on their economic characteristics, we investigated the overall cocoon weights and cocoon shell rates of the transgenic lines. The cocoon shell rate is the ratio of the cocoon shell weight to the overall whole cocoon weight. There were no obvious differences in the economic characteristics of the transgenic and non-transgenic lines (Fig. 7). This showed that hycu-ep32 overexpression did not affect the economic characteristics of transgenic silkworms.
Figure 7. Economic characteristics analysis.
The economic characteristics of EKG, HEKG-A, HEKG-B, EAG, HEAG, and 932 were investigated. Fifteen cocoons from male individuals and 15 cocoons from female individuals were selected randomly from each line. The weights of the entire cocoon and the cocoon shell were determined for each individual. The cocoon shell rate was the ratio of the cocoon shell weight to the overall cocoon weight. Each value was the average of 15 replicate tests. Bars represent the standard deviations.
Discussion
B. mori is a typical lepidopteran model and it is economically important in the silk industry [33], [34], [35]. However, BmNPV causes severe annual economic losses in the sericulture industry. In our previous study, we reported that transgenic silkworms had significantly improved anti-BmNPV capacity due to overexpression of the endogenous antiviral gene Bmlipase-1 [10]. In the current study, we further optimized the method of vector construction and induced the overexpression of an exogenous antiviral gene, before analyzing its anti-BmNPV capacity.
The hycu-ep32 gene of HycuNPV can restrict BmNPV proliferation in co-infected cells [30]. In this study, we constructed four overexpression vectors where hycu-ep32 was driven by 39 KP, hr3+39 KP, A4P, or hr3+A4P. The corresponding transgenic lines EKG, HEKG-A and HEKG-B, EAG, HEAG were generated to test their resistance. As expected, the expression level of exogenous hycu-ep32 was increased significantly after infection with BmNPV in HEKG-B, which had a significantly enhanced anti-BmNPV capacity. Importantly, the economic characteristics of the transgenic strains remained the same.
Hycu-ep32 was induced abundantly after BmNPV infection of HEKG-B and HEAG (Fig. 5) where hr3 was upstream of 39KP and A4P, which controlled the expression of hycu-ep32 (Fig. 1). It was assumed that hr3 was transactivated by the IE1 protein that was produced by the virus after BmNPV infection. The expression level of hycu-ep32 in HEKG-B (Fig. 5) and the resistance of HEKG-B (Figs 4, S1, S2) were the highest among all the detected transgenic lines, suggesting that hr3 combined with 39KP gave the best performance of all the promoters tested. Compared with target genes expressed stably by using constitutive promoters [8], [9], [11], [12], the expression of hycu-ep32 was appropriate with normal physical status, while it increased significantly as the virus content of HEKG-B increased.
The 39KP activity was very low in EKG (Figs 3, 5), which was only slightly enhanced after BmNPV infection (Fig. 5). However, the 39 K promoter of AcMNPV had no activity, although it could be activated by the virus in cells [17]. This difference may be attributable to differences in BmNPV and AcMNPV, or different individual larvae and cell lines. The transgenic lines EKG and EAG inhibited the multiplication of BmNPV to some extent (Fig. 6) via the expression of hycu-ep32 (Figs 3, 5). However, the expression of hycu-ep32 was not sufficiently high to decrease the mortality rate (Figs 4, 5, S1, S2). The anti-BmNPV capacity was enhanced significantly due to the high level expression of hycu-ep32 after infection in HEKG-B. Hycu-ep32 can induce a severe global protein synthesis shutdown, thereby suppressing BmNPV multiplication in co-infected cells [30]. We assumed that hycu-ep32 could suppress BmNPV DNA replication or protein synthesis in transgenic larvae.
The overexpression of anti-pathogen genes or downregulation of pathogen genes by transgenic RNAi in hosts are proven and effective disease-control strategies. However, there have been no previous reports of enhanced disease resistance in animals due to the overexpression of exogenous anti-pathogen genes. Resistance to BmNPV can be boosted by overexpressing endogenous Bmlipase-1 in transgenic silkworm [10] or knocking down the ie-1 gene using transgenic RNAi [7]. A combination of these methods would be more effective. In previous reports, transgenic silkworms were created using nondiapause or non-practical silkworm [6], [7], [10], [31], [32]. In the current study, we generated the first transgenic silkworm using a practical diapause strain.
In conclusion, we successfully generated transgenic practical silkworms with an exogenous antiviral gene via embryo microinjection. The expression level of the antiviral factor was significantly upregulated with an increase in the viral content after infection. The transgenic silkworms had an enhanced anti-BmNPV capacity with no negative effects on their economic characteristics, which means they could be applied in sericulture to enhance the resistance of silkworms. This is the first report of disease resistance via overexpression of an anti-pathogen factor using an inducible promoter and enhancer in animals, and it may pave the way for disease control studies in other organisms in the future.
Materials and Methods
Silkworm Strain and Virus
The practical diapause silkworm strain “932” was maintained at the Gene Resource Library of Domesticated Silkworm (Southwest University, Chongqing, China). The silkworm was reared on fresh mulberry leaves under standard conditions. The larvae underwent oral inoculation with wild BmNPV (Guangdong strain, China) and the OBs were harvested from larvae hemolymph before the larvae died.
Transgenic Vectors Construction
DNA was extracted from HycuNPV OBs and BmNPV OBs using a MiniBEST viral RNA/DNA Extraction Kit Ver. 3.0 (TaKaRa). The hycu-ep32 gene was amplified from the HycuNPV genomic DNA using the primers ep32F (5′-CGGGATCCATGAAGAACCAACAACAG-3′) and ep32R (5′-ATAGTTTAGCGGCCGCTTAATTTATTAACATATCAAAG-3′) [30]. The promoter sequence of the 39K gene (39KP) was amplified from the BmNPV genomic DNA using the primers 39KPF (5′-ACGCGTCGACCTTGACCCGAAGCGAAAT-3′) and 39KPR (5′-CGCGGATCCTGTTGCTCCGGCATGTTT-3′) (provisional Chinese Patent No. 201010231957.9, Pan et al.). The BmNPV hr3 enhancer (provisional Chinese Patent No. 201110423280.3, Xia et al.), B. mori A4 promoter (A4P), and termination signal SV40 were preserved in the laboratory. The 39KP, hycu-ep32, and SV40 were added to piggyBac [3×p3 EGFP afm] [31], [32] to generate transgenic vector piggyBac [39KP-ep32-SV40-3×p3 EGFP afm] (abbreviated as pb-EKG). hr3 was added to pb-EKG to generate the transgenic vector pb-HEKG. A4P, hycu-ep32, and SV40 were added to piggyBac [3×p3 EGFP afm] to generate the transgenic vector piggyBac [A4P-ep32-SV40-3×p3 EGFP afm] (abbreviated as pb-EAG). hr3 was added to pb-EAG to generate the transgenic vector pb-HEAG (Fig. 1).
Microinjection and Screening
The practical silkworm strain 932 was used for the transgenic embryo microinjection, which is a diapause strain. The 932 embryos were incubated at 15°C after acidic treatment and the larvae were fed with mulberry leaves under standard conditions [10]. After this processes, most of the embryos were nondiapaused in the next generation. Mixtures of transgenic vector (400 ng/µL) and helper plasmid pHA3PIG (400 ng/µL) were injected into the nondiapause 932 embryos within 2 h of oviposition [10], [31], [32]. The G1 embryos were screened for EGFP protein in the ocelli using a fluorescent microscope (Olympus) [10], [32]. EGFP-positive moths were then sibling-mated to generate offspring in each G1 brood for each transgenic vector. One transgenic line of pb-EKG (known as EKG), two transgenic lines of pb-HEKG (known as named HEKG-A and HEKG-B), one transgenic line of pb-EAG (known as EAG), and one transgenic line of pb-HEAG (known as HEAG) were screened.
Identifying the Insertion Site
Genomic DNA was extracted from EKG, HEKG-A, HEKG-B, EAG and HEAG in G1 male moths. There was a Hae III site in the left terminal inverted repeats (PL) and right terminal inverted repeats (PR) of piggyBac [3×p3 EGFP afm] vector, respectively. About 20 µg of the genomic DNA from each sample was fully digested using Hae III for 10 h at 37°C, then purified and self-ligated with Solution I (NEB) [10]. Each ligated product was PCR-amplified using the transposon-specific primers pBacL and pBacR (pBacL F: 5′-ATCAGTGACACTTACCGCATTGACA-3′, pBacL R: 5′-TGACGAGCTTGTTGGTGAGGATTCT-3′; pBacR F: 5′-TACGCATGATTATCTTTAACGTA-3′, pBacR R: 5′- GTACTGTCATCTGATGTACCAGG-3′), respectively. The PCR products were cloned and sequenced.
Analysis of the Expression Level of Hycu-ep32 by RT-PCR
Different developmental stages of EKG, HEKG-A, HEKG-B, EAG, HEAG, and non-transgenic 932 were used for RNA extraction. The total RNA was extracted using a total RNA (mini) kit (Watson) and digested with 20 U Rnase-free Dnase I (Promega). Approximately 4 µg of treated RNA was reverse-transcribed in a 25 µL reaction system using M-MLV reverse transcriptase (Promega). Each template was diluted with 100 µL, before 1 µL of cDNA was used for RT-PCR reactions using the primers ep32QRT (F: 5′-ACATCAGAATACCCATCACG-3′, R: 5′-ATTGTTCAATGGTAACTCCC-3′). The primers sw22934 for the housekeeping gene BGIBMGA003186 (F: 5′-TTCGTACTGGCTCTTCTCGT-3′, R: 5′-CAAAGTTGATAGCAATTCCCT-3′) were used as the controls.
Mortality Analysis
The mortality of different transgenic strains and non-transgenic strain were investigated after being infected with wild BmNPV per os. The infection testing of each line used three replicates and each replicate was performed with 70 larvae. Fresh mulberry leaves were cut into 1 cm or 1.5 cm diameter round pieces and then treated with solution containing OBs. Newly exuviated 3 third instar or fourth instar larvae were used in the test. Individuals in the one-repeat experiment were confirmed as consuming equal quantities of OBs by feeding them individually [10]. There were three replicates of non-infection and every replicate consisted of 70 larvae. The mortality rate was analyzed from the time of infection until 10 dpi.
Confirmation of the Change in Hycu-ep32 after Virus Induction by qPCR
Total RNA was extracted from EKG, HEKG-B, EAG, HEAG, and 932 using 10 treated larvae at 0 and 48 hpi. The time that the infection process ended was set as time point 0 h. cDNA templates from 10 biological materials were used for qPCR reactions with ep32QRT primers on the ABI StepOnePlus™ Real-Time PCR System (Applied Biosystems). BGIBMGA003186 was used as a reference gene to standardize the variance of the different templates. Each assay was performed three times.
qPCR Analysis of Virus after BmNPV Infection
Total DNA was obtained from the larvae of EKG, HEKG-B, EAG, HEAG, and 932 at 48 hpi. Each sample that was extracted from 10 treated larvae was used as templates for qPCR. The DNA templates (20 ng) were PCR-amplified using primers for the BmNPV GP41 gene (F: 5′-CGTAGTAGTAGTAATCGCCGC-3′, R: 5′-AGTCGAGTCGCGTCGCTTT-3′) [10], [36] using an ABI StepOnePlus™ Real-Time PCR System. BmGAPDH (F: 5′-CATTCCGCGTCCCTGTTGCTAAT-3′, R: 5′-GCTGCCTCCTTGACCTTTTGC-3′) was used as an internal control to standardize the variance of the different templates [10], [36]. The test was performed three times.
Comparison of the Economic Characteristics
B. mori is an important insect for the commercial production of silk. The economic characteristics of EKG, HEKG-A, HEKG-B, EAG, HEAG, and 932 were investigated to determine whether hycu-ep32 overexpression might affect the silk production of transgenic lines. Fifteen cocoons from male individuals and 15 cocoons from female individuals were selected randomly from each line for analysis. The weights of the entire cocoon and the cocoon shell were determined for each individual.
Supporting Information
The second time of resistance detection using third instar larvae. EKG, EAG, and 932 (white) were infected orally with 2.5 × 105 OB/larva using newly exuviated third instar larvae. HEKG-A, HEKG-B, HEAG, and 932 (black) were infected with 3 × 105 OB/larva per os at third instar larvae. The ingested viral dose of each individual in each replicate was the same. The mortality of each line was the average of triplicate infection replicates. Each replicate consisted of 70 larvae. Mortality statistics were analyzed daily from the time of infection until 10 dpi. The accumulative mortality up to 10 dpi is shown for each line. 932(C) is the non-infected control. Bars represent the standard deviations. Statistically significant differences: * P<0.05, ** P<0.01.
(TIF)
Mortality statistics after infection with BmNPV per os using fourth instar larvae. The solution of OBs was smeared on pieces of fresh mulberry leaf with 1.5 cm diameter. EKG, EAG, and 932 (white) were infected orally with 8.3 × 105 OB/larva using newly exuviated fourth instar larvae. HEKG-A, HEKG-B, HEAG, and 932 (black) were infected with 106 OB/larva per os at fourth instar larvae. The ingested viral dose of each individual in each replicate was the same. The mortality of each line was the average of triplicate infection replicates. Each replicate consisted of 70 larvae. Mortality statistics were analyzed daily from the time of infection until 10 dpi. The accumulative mortality up to 10 dpi is shown for each line. 932(C) is the non-infected control. Bars represent the standard deviations. Statistically significant differences: * P<0.05, ** P<0.01.
(TIF)
Acknowledgments
Thank Dr. Yong’an Zhang (Chinese Academy of Forestry) for gifting the HycuNPV virus.
Funding Statement
This work was funded by the National Basic Research Program of China (No. 2012CB114600), http://www.973.gov.cn/Default_3.aspx; the National Hi-Tech Research and Development Program of China (No. 2011AA100306), http://www.863.gov.cn/; the Foundation for Sci & Tech Research Project of Chongqing (CSTC2009AA1024), http://www.cstc.gov.cn/; the National Natural Science Foundation of China (No. 30901054), http://www.nsfc.gov.cn/Portal0/default152.htm; and the Production, Education & Research of Guangdong and Ministry of Education (No. 2010A090200079), http://cxy.gdstc.gov.cn/. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Keddie BA, Aponte GW, Volkman LE (1989) The pathway of infection of Autographa californica nuclear polyhedrosis virus in an insect host. Science 243: 1728–1730. [DOI] [PubMed] [Google Scholar]
- 2. Rahman MM, Gopinathan KP (2004) Systemic and in vitro infection process of Bombyx mori nucleopolyhedrovirus. Virus Res 101: 109–118. [DOI] [PubMed] [Google Scholar]
- 3. Huh NE, Weaver RF (1990) Categorizing some early and late transcripts directed by the Autographa californica nuclear polyhedrosis virus. J Gen Virol 71 (Pt 9): 2195–2200. [DOI] [PubMed] [Google Scholar]
- 4. Slack J, Arif BM (2007) The baculoviruses occlusion-derived virus: virion structure and function. Advances in Virus Research, Vol 69 69: 99–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tjia ST, Carstens EB, Doerfler W (1979) Infection of Spodoptera frugiperda cells with Autographa californica nuclear polyhedrosis virus II. The viral DNA and the kinetics of its replication. Virology 99: 399–409. [DOI] [PubMed] [Google Scholar]
- 6. Isobe R, Kojima K, Matsuyama T, Quan GX, Kanda T, et al. (2004) Use of RNAi technology to confer enhanced resistance to BmNPV on transgenic silkworms. Archives of Virology 149: 1931–1940. [DOI] [PubMed] [Google Scholar]
- 7. Kanginakudru S, Royer C, Edupalli SV, Jalabert A, Mauchamp B, et al. (2007) Targeting ie-1 gene by RNAi induces baculoviral resistance in lepidopteran cell lines and in transgenic silkworms. Insect Molecular Biology 16: 635–644. [DOI] [PubMed] [Google Scholar]
- 8. Nishiyama S, Shitara H, Nakada K, Ono T, Sato A, et al. (2010) Over-expression of Tfam improves the mitochondrial disease phenotypes in a mouse model system. Biochem Biophys Res Commun 401: 26–31. [DOI] [PubMed] [Google Scholar]
- 9. de Bilbao F, Arsenijevic D, Moll T, Garcia-Gabay I, Vallet P, et al. (2009) In vivo over-expression of interleukin-10 increases resistance to focal brain ischemia in mice. J Neurochem 110: 12–22. [DOI] [PubMed] [Google Scholar]
- 10. Jiang L, Wang G, Cheng T, Yang Q, Jin S, et al. (2012) Resistance to Bombyx mori nucleopolyhedrovirus via overexpression of an endogenous antiviral gene in transgenic silkworms. Arch Virol 157: 1323–1328. [DOI] [PubMed] [Google Scholar]
- 11. Ohshima M, Mitsuhara I, Okamoto M, Sawano S, Nishiyama K, et al. (1999) Enhanced resistance to bacterial diseases of transgenic tobacco plants overexpressing sarcotoxin IA, a bactericidal peptide of insect. J Biochem 125: 431–435. [DOI] [PubMed] [Google Scholar]
- 12. Wally O, Punja ZK (2010) Enhanced disease resistance in transgenic carrot (Daucus carota L.) plants over-expressing a rice cationic peroxidase. Planta 232: 1229–1239. [DOI] [PubMed] [Google Scholar]
- 13. Guarino LA, Gonzalez MA, Summers MD (1986) Complete Sequence and Enhancer Function of the Homologous DNA Regions of Autographa californica Nuclear Polyhedrosis Virus. J Virol 60: 224–229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guarino LA, Summers MD (1986) Interspersed Homologous DNA of Autographa californica Nuclear Polyhedrosis Virus Enhances Delayed-Early Gene Expression. J Virol 60: 215–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Morris TD, Miller LK (1992) Promoter influence on baculovirus-mediated gene expression in permissive and nonpermissive insect cell lines. J Virol 66: 7397–7405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kool M, Voeten JT, Goldbach RW, Tramper J, Vlak JM (1993) Identification of seven putative origins of Autographa californica multiple nucleocapsid nuclear polyhedrosis virus DNA replication. J Gen Virol 74 (Pt 12): 2661–2668. [DOI] [PubMed] [Google Scholar]
- 17. Guarino LA, Summers MD (1986) Functional mapping of a trans-activating gene required for expression of a baculovirus delayed-early gene. J Virol 57: 563–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Nissen MS, Friesen PD (1989) Molecular analysis of the transcriptional regulatory region of an early baculovirus gene. J Virol 63: 493–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Rodems SM, Friesen PD (1993) The hr5 transcriptional enhancer stimulates early expression from the Autographa californica nuclear polyhedrosis virus genome but is not required for virus replication. J Virol 67: 5776–5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carson DD, Summers MD, Guarino LA (1991) Transient expression of the Autographa californica nuclear polyhedrosis virus immediate-early gene, IE-N, is regulated by three viral elements. J Virol 65: 945–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lu M, Farrell PJ, Johnson R, Iatrou K (1997) A baculovirus (Bombyx mori nuclear polyhedrosis virus) repeat element functions as a powerful constitutive enhancer in transfected insect cells. J Biol Chem 272: 30724–30728. [DOI] [PubMed] [Google Scholar]
- 22. Viswanathan P, Venkaiah B, Kumar MS, Rasheedi S, Vrati S, et al. (2003) The homologous region sequence (hr1) of Autographa californica multinucleocapsid polyhedrosis virus can enhance transcription from non-baculoviral promoters in mammalian cells. J Biol Chem 278: 52564–52571. [DOI] [PubMed] [Google Scholar]
- 23. Gong M, Guarino LA (1994) Expression of the 39k promoter of Autographa californica nuclear polyhedrosis virus is increased by the apoptotic suppressor P35. Virology 204: 38–44. [DOI] [PubMed] [Google Scholar]
- 24. Kool M, Ahrens CH, Goldbach RW, Rohrmann GF, Vlak JM (1994) Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci U S A 91: 11212–11216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lin X, Chen Y, Yi Y, Zhang Z (2010) Baculovirus immediately early 1, a mediator for homologous regions enhancer function in trans. Virol J 7: 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu A, Carstens EB (1993) Immediate-early baculovirus genes transactivate the p143 gene promoter of Autographa californica nuclear polyhedrosis virus. Virology 195: 710–718. [DOI] [PubMed] [Google Scholar]
- 27. Pullen SS, Friesen PD (1995) Early transcription of the ie-1 transregulator gene of Autographa californica nuclear polyhedrosis virus is regulated by DNA sequences within its 5′ noncoding leader region. J Virol 69: 156–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Rodems SM, Pullen SS, Friesen PD (1997) DNA-dependent transregulation by IE1 of Autographa californica nuclear polyhedrosis virus: IE1 domains required for transactivation and DNA binding. J Virol 71: 9270–9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Guarino LA, Dong W (1994) Functional dissection of the Autographa california nuclear polyhedrosis virus enhancer element hr5. Virology 200: 328–335. [DOI] [PubMed] [Google Scholar]
- 30. Shirata N, Ikeda M, Kobayashi M (2010) Identification of a Hyphantria cunea nucleopolyhedrovirus (NPV) gene that is involved in global protein synthesis shutdown and restricted Bombyx mori NPV multiplication in a B. mori cell line. Virology 398: 149–157. [DOI] [PubMed] [Google Scholar]
- 31. Tamura T, Thibert C, Royer C, Kanda T, Abraham E, et al. (2000) Germline transformation of the silkworm Bombyx mori L. using a piggyBac transposon-derived vector. Nat Biotechnol 18: 81–84. [DOI] [PubMed] [Google Scholar]
- 32. Thomas JL, Da Rocha M, Besse A, Mauchamp B, Chavancy G (2002) 3xP3-EGFP marker facilitates screening for transgenic silkworm Bombyx mori L. from the embryonic stage onwards. Insect Biochem Mol Biol 32: 247–253. [DOI] [PubMed] [Google Scholar]
- 33. Xia Q, Zhou Z, Lu C, Cheng D, Dai F, et al. (2004) A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306: 1937–1940. [DOI] [PubMed] [Google Scholar]
- 34. Xia Q, Guo Y, Zhang Z, Li D, Xuan Z, et al. (2009) Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 326: 433–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Duan J, Li R, Cheng D, Fan W, Zha X, et al. (2010) SilkDB v2.0: a platform for silkworm (Bombyx mori ) genome biology. Nucleic Acids Res 38: D453–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bao YY, Tang XD, Lv ZY, Wang XY, Tian CH, et al. (2009) Gene expression profiling of resistant and susceptible Bombyx mori strains reveals nucleopolyhedrovirus-associated variations in host gene transcript levels. Genomics 94: 138–145. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The second time of resistance detection using third instar larvae. EKG, EAG, and 932 (white) were infected orally with 2.5 × 105 OB/larva using newly exuviated third instar larvae. HEKG-A, HEKG-B, HEAG, and 932 (black) were infected with 3 × 105 OB/larva per os at third instar larvae. The ingested viral dose of each individual in each replicate was the same. The mortality of each line was the average of triplicate infection replicates. Each replicate consisted of 70 larvae. Mortality statistics were analyzed daily from the time of infection until 10 dpi. The accumulative mortality up to 10 dpi is shown for each line. 932(C) is the non-infected control. Bars represent the standard deviations. Statistically significant differences: * P<0.05, ** P<0.01.
(TIF)
Mortality statistics after infection with BmNPV per os using fourth instar larvae. The solution of OBs was smeared on pieces of fresh mulberry leaf with 1.5 cm diameter. EKG, EAG, and 932 (white) were infected orally with 8.3 × 105 OB/larva using newly exuviated fourth instar larvae. HEKG-A, HEKG-B, HEAG, and 932 (black) were infected with 106 OB/larva per os at fourth instar larvae. The ingested viral dose of each individual in each replicate was the same. The mortality of each line was the average of triplicate infection replicates. Each replicate consisted of 70 larvae. Mortality statistics were analyzed daily from the time of infection until 10 dpi. The accumulative mortality up to 10 dpi is shown for each line. 932(C) is the non-infected control. Bars represent the standard deviations. Statistically significant differences: * P<0.05, ** P<0.01.
(TIF)