Abstract
Background
Cotton (Gossypium spp.) is a model system for the analysis of polyploidization. Although ascertaining the donor species of allotetraploid cotton has been intensively studied, sequence comparison of Gossypium chloroplast genomes is still of interest to understand the mechanisms underlining the evolution of Gossypium allotetraploids, while it is generally accepted that the parents were A- and D-genome containing species. Here we performed a comparative analysis of 13 Gossypium chloroplast genomes, twelve of which are presented here for the first time.
Methodology/Principal Findings
The size of 12 chloroplast genomes under study varied from 159,959 bp to 160,433 bp. The chromosomes were highly similar having >98% sequence identity. They encoded the same set of 112 unique genes which occurred in a uniform order with only slightly different boundary junctions. Divergence due to indels as well as substitutions was examined separately for genome, coding and noncoding sequences. The genome divergence was estimated as 0.374% to 0.583% between allotetraploid species and A-genome, and 0.159% to 0.454% within allotetraploids. Forty protein-coding genes were completely identical at the protein level, and 20 intergenic sequences were completely conserved. The 9 allotetraploids shared 5 insertions and 9 deletions in whole genome, and 7-bp substitutions in protein-coding genes. The phylogenetic tree confirmed a close relationship between allotetraploids and the ancestor of A-genome, and the allotetraploids were divided into four separate groups. Progenitor allotetraploid cotton originated 0.43–0.68 million years ago (MYA).
Conclusion
Despite high degree of conservation between the Gossypium chloroplast genomes, sequence variations among species could still be detected. Gossypium chloroplast genomes preferred for 5-bp indels and 1–3-bp indels are mainly attributed to the SSR polymorphisms. This study supports that the common ancestor of diploid A-genome species in Gossypium is the maternal source of extant allotetraploid species and allotetraploids have a monophyletic origin. G. hirsutum AD1 lineages have experienced more sequence variations than other allotetraploids in intergenic regions. The available complete nucleotide sequences of 12 Gossypium chloroplast genomes should facilitate studies to uncover the molecular mechanisms of compartmental co-evolution and speciation of Gossypium allotetraploids.
Introduction
Cotton is one of the world's most important crops for producing natural textile fiber. Upland cotton (Gossypium hirsutum L.) accounts for more than 90% of production. Three other cultivated species, namely, Sea Island cotton (G. barbadense L.), Asian cotton (G. arboreum L.) and African cotton (G. herbaceum L.), are used in commercial production as well. All the cotton plants and species are classified in taxonomy into the genus Gossypium (Malvaceae), a model system for the study of polyploidization in plants [1]. Gossypium is comprised of 51 species, including 5 allotetraploid species and 46 diploids [2], [3]. The 5 allotetraploid species are G. hirsutum L. AD1, G. barbadense L. AD2, G. tomentosum Nuttal ex Seemann AD3, G. mustelinum Miers ex Watt AD4, and G. darwinii Watt AD5, which are further classified into 3 lineages [4], [5]: the G. mustelinum AD4 lineage, G. barbadense AD2-G. darwinii AD5 lineage and G. hirsutum AD1-G. tomentosum AD3 lineage. The diploid Gossypium species are comprised of 8 genome groups, namely, A group through G group and K group. They could cluster into 3 major lineages, corresponding to 3 continental regions: 13 D-genome species from the American continent, 15 species in the African-Asian group (A-, B-, E-, and F group genomes) from African-Asian continent, and another clade of 18 species (C-, G- and K- group genomes) from Australia [4].
The donor species of allotetraploid cottons had been intensively studied [4]–[7], but the origin and evolution of Gossypium allotetraploids has remained controversial, with debate over the identity of the progenitor diploid species, the time and mechanism of allopolyploidization. Restriction enzyme digestion of chloroplast DNA (cpDNA) derived from the species of A-, D-, and AD group genomes revealed that chloroplast genomes of 5 tetraploid species were similar to those of G. arboreum A2 and G. herbaceum A1, leading to speculation that the maternal donor of the allotetraploid originated from A group genome [6]. Because nuclear genomes of G. herbaceum A1 and G. arboreum A2 differed from A subgenome of allotetraploid cotton through two and three reciprocal translocation, respectively, most researchers considered G. herbaceum A1 to be genetically closer than G. arboreum A2 to the allotetraploid [3], [7]. Comparative genetic mapping [8], [9] and analysis of DNA sequences [3], [10], [11] indicated that paternal donor of the allotetraploid was similar to extant D group genome in the Americas and the closest living species was G. raimondii D5. However, the evolutionary history of Gossypium suggested that G. gossypioides D6, rather than G. raimondii D5, was involved in the origin of allotetraploid cotton [4], [7], [12].
In the absence of a clear fossil record, the divergence of Gossypium was always estimated under a molecular clock model [3]. Based on the analysis of ndhF sequence and average variation rate in eukaryotes, divergence time between Gossypium and its sister-group Kokia-Gossypioides was calculated to be approximately 12.5 million years ago (MYA) [13]. The divergence of the diploid progenitors was estimated to have occurred around 6.8 MYA using data of some nuclear genes [14]–[16]. Molecular evolutionary data indicated that the Gossypium allotetraploid originated during the Pleistocene (0.3 to 2 MYA) [13], [14], [16], predating the evolution of New World humans.
The plastid chromosomes, which have relatively low numbers of genes and high levels of conservation, have provided valuable information to estimate phylogenetic relationship among plants and green algae [17]–[20]. The chloroplast genome sequences of G. hirsutum AD1 and G. barbadense AD2 published, providing strong reliable evidence for the detailed position of cotton in the Malvales [21], [22]. Though many taxonomic studies have advanced our understanding of Gossypium, few data from chloroplast genomes can be used to explore inter- and intra-specific differentiation of Gossypium. It is necessary to expand taxon sampling at a high taxonomic level.
To further understand the origin and evolution of Gossypium allotetraploids, we sequenced the chloroplast genomes of the following taxa: G. herbaceum var. africanum A1 and G. arboreum A2 from the A-genome lineage; G. raimondii D5 and G. gossypioides D6 from the D-genome lineage; 2 races of G. hirsutum AD1, G. hirsutum race hainansijimian AD1 and G. hirsutum race lanceolatum AD1, originating from Hainan Province of China and southern part of United States, respectively; G. barbadense AD2 and its 2 local races, G. barbadense race yuanmou AD2 and G. barbadense race kaiyuan AD2, from Yunnan Province of China; and 3 wild allotetraploids including G. tomentosum AD3, G. mustelinum AD4, and G. darwinii AD5. Our objectives were to assess dynamic process of the inter- and intra-specific sequence divergence of Gossypium chloroplast genomes, and to re-estimate the origin and evolution of Gossypium allotetraploids.
Results
Size, content and structure of Gossypium chloroplast genomes
Twelve Gossypium chloroplast genomes were sequenced in this study. Complete DNA sequences were deposited in GenBank under accession numbers HQ325740 through HQ325745 (G. arboreum A2, G. darwinii AD5, G. herbaceum var. africanum A1, G. mustelinum AD4, G. raimondii D5, and G. tomentosum AD3, respectively), and HQ901195 through HQ901200 (G. gossypioides D6, G. hirsutum race lanceolatum AD1, G. hirsutum race hainansijimian AD1, G. barbadense race yuanmou AD2, G. barbadense AD2, and G. barbadense race kaiyuan AD2, respectively). HQ901199 is a new version of the chloroplast genome sequence of G. barbadense AD2.
The 12 chloroplast genomes were typical circular chromosomes like those of most other higher plants, including the large single copy (LSC), the small single copy (SSC) and 2 IR regions (Figure 1). The whole genome size ranged between 159,959 bp (G. gossypioides D6) and 160,433 bp (G. tomentosum AD3). The length varied from 88,656 bp (G. raimondii D5) to 88,932 bp (G. tomentosum AD3) in the LSC region, from 20,004 bp (G. gossypioides D6) to 20,285 bp (G. herbaceum var. africanum A1) in the SSC region, and from 25,576 bp (G. gossypioides D6) to 25,650 bp (G. raimondii D5) in the IR region (Table 1).
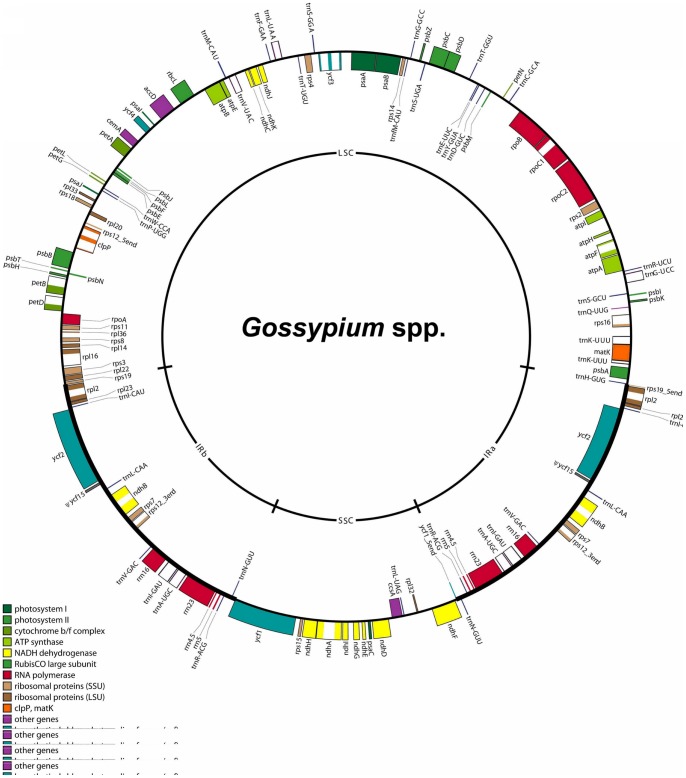
Figure 1. Gene map of Gossypium chloroplast genome.
The map includes the inverted repeats, IRa and IRb, small (SSC) and large (LSC) single copy regions. Genes exhibited on the inside of the map are transcribed in a clockwise direction, while genes on the outside of the map are transcribed in a reverse order.
Table 1. Size of 13 Gossypium chloroplast genomes with the LSC, SSC and IR regions.
| Species | Genome (bp) | LSC (bp) | SSC (bp) | IR (bp) |
| G. herbaceum var. africanum A1 | 160,315 | 88,790 | 20,285 | 25,620 |
| G. arboreum A2 | 160,230 | 88,722 | 20,274 | 25,617 |
| G. hirsutum AD1 | 160,301 | 88,816 | 20,269 | 25,608 |
| G. hirsutum race lanceolatum AD1 | 160,256 | 88,799 | 20,273 | 25,592 |
| G. hirsutum race hainansijimian AD1 | 160,265 | 88,782 | 20,279 | 25,602 |
| G. barbadense AD2 | 160302 | 88,849 | 20,267 | 25,593 |
| G. barbadense race kaiyuan AD2 | 160,289 | 88,836 | 20,267 | 25,593 |
| G. barbadense race yuanmou AD2 | 160,291 | 88,838 | 20,267 | 25,593 |
| G. tomentosum AD3 | 160,433 | 88,932 | 20,271 | 25,615 |
| G. mustelinum AD4 | 160,313 | 88,826 | 20,269 | 25,609 |
| G. darwinii AD5 | 160,378 | 88,906 | 20,266 | 25,603 |
| G. raimondii D5 | 160,161 | 88,656 | 20,205 | 25,650 |
| G. gossypioides D6 | 159,959 | 88,803 | 20,004 | 25,576 |
Note: G. hirsutum AD1[21].
The corresponding coding sequences accounted for 49.4% of the genome, 48.4%, 71.8%, and 40.8%of LSC, SSC and IR region, respectively. Each genome contained 112 unique functional genes, including 78 protein-coding and 34 RNA-coding genes (Table S1). Eighteen genes were located in IR regions, thus each genome harbored a total of 130 genes. And 34 RNA-coding genes consisted of 30 tRNA genes and 4 rRNA genes, in which 7 tRNA (trnAUGC, trnICAU, trnIGAU, trnLUAA, trnNGUU, trnRACG, and trnVGAC) and 4 rRNA genes (rrn16, rrn23, rrn5, and rrn4.5) were located in IR regions. And the other seven genes in IR regions were ndhB, rpl2, rps7, rps12, rpl23, ycf2, ans ycf15. It was found that accD, rpl23, ycf1, and ycf2 are often absent from plant species [23], but these four genes could be detected among 13 Gossypium chloroplast genomes. Similar to other higher plants, 2 pairs of genes had overlapping regions, namely, atpB-atpE (3-bp overlap) and psbC-psbD (53-bp overlap). All protein-coding genes used a standard plastid/bacterial genetic codon model which started with ATG, except ndhD with ACG. To transfer 20 kinds of amino acids, including 61 codons, because 30 tRNA genes were encoded by the chloroplast genome, the remaining 31 tRNAs had to be coded by nuclear genome. Among 112 unique genes, atpF, ndhA, ndhB, petB, petD, rpoC1, rpl2, rpl16, rps16, trnA-UGC, trnG-UCC, trnI-GAU, trnK-UUU, trnL-UAA, and trnV-UAC each had one intron, whereas 2 introns presented in clpP, rps12, and ycf3. Furthermore, matK was located within the intron of trnK-UUU.
The overall AT content was approximately 62.8%, which was very similar among the 13 Gossypium chloroplast genomes. However, different levels of AT content were found across regions, namely, 64.8%, 68.3%, and 57.0% AT content in the LSC, SSC, and IR regions, respectively. In addition protein-coding regions were found to have an AT content of 61.8% whereas non protein-coding regions had an AT content of 68.4%. tRNA and rRNA genes had a slightly lower AT content with 46.9% and 44.5%, respectively. Triple codon analysis demonstrated that the AT content also varied widely with 53.8%, 61.8%, and 69.7% at the first, second, and third positions, respectively (Table S2).
Although the overall structure, genome size, gene number, and gene order are well conserved, the junctions between IR and SSC regions are usually different in higher plant chloroplast genomes. Using cacao (Theobroma cacao L., Malvaceae), a close relative of Gossypium, as outgroup, a slight difference in junction positions was observed among 13 Gossypium chloroplast genomes (Figure 2). For example, rps19 extended into IRb with 55 bp in G. raimondii D5, while the whole rps19 was located within the LSC region in the other 12 Gossypium genomes. The distance of rps19 from the junction of LSC/IRb varied from 0 bp (G. mustelinum AD4) to 11 bp (G. hirsutum race lanceolatum AD1), similar to Nuphar, Ranunculus, and Nicotiana, where distances of 89 bp, 36 bp and 9 bp respectively have been reported. Gene ycf1 extended slightly into the IR region in Gossypium which partly explain why the SSC region in Gossypium is larger than that in other plants.
Figure 2. Chloroplast genome IR junctions of 13 Gossypium species with Theobroma cacao as an outgroup.
Genome substitutions and indels between different genomes
Global alignment of the 13 Gossypium chloroplast genomes was conducted. The produced alignment spanned over 161.6 kb. In the alignment, the total number of substitutions varied from 6 (AD2-AD2) to 1,000 bp (D6-AD1) (Table 2), and the nucleotide divergence between them was estimated in the range of 0.004%–0.62%. The ratio of transition to transversion (Si/Sv) ranged between 0.2 (AD2–AD2) and 1.2 (A1–A2), with an average value of 0.45 (Table S3). The base substitution type between C and G was significantly less than other types. The total length of substitutions within G. barbadense AD2 lineages varied from 6 to13 bp, while that withiniG. hirsutum AD1 lineages ranged from 25 to 43 bp. Within allotetraploid species, the total substitutions varied from 63 bp (AD2-AD5, with 0.039% divergence) to 175 bp (AD1–AD3, with 0.108% divergence), and from 147 (AD5–A2, with 0.091% divergence) to 231 bp (AD1–A1, with 0.143% divergence) between allotetraploid species and diploid A-genome specie. There were only 11-bp substitutions between A1 and A2, but 180-bp substitutions between D5 and D6. The total length of substitutions between A-genome species and D-genome species varied from 852 (A2–D5) to 916 bp (A1–D6), and the nucleotide divergence between them ranged from 0.527% to 0.567%.
Table 2. The substitutions and indel number between any two of 13 Gossypium chloroplast genomes.
| Gaf A1 | Ga A2 | Gh AD1 | Ghh AD1 | Ghl AD1 | Gb AD2 | Gbk AD2 | Gby AD2 | Gt AD3 | Gm AD4 | Gd AD5 | Gr D5 | Gg D6 | |
| Gaf A1 | 13 | 122 | 134 | 118 | 121 | 121 | 121 | 117 | 110 | 113 | 261 | 265 | |
| Ga A2 | 11 | 118 | 129 | 115 | 113 | 113 | 113 | 109 | 107 | 104 | 255 | 259 | |
| Gh AD1 | 224 | 219 | 18 | 22 | 84 | 83 | 82 | 90 | 90 | 82 | 260 | 266 | |
| Ghh AD1 | 231 | 226 | 25 | 32 | 93 | 91 | 92 | 100 | 94 | 92 | 264 | 269 | |
| Ghl AD1 | 221 | 216 | 35 | 43 | 81 | 80 | 80 | 85 | 83 | 80 | 255 | 257 | |
| Gb AD2 | 166 | 164 | 152 | 160 | 146 | 5 | 3 | 63 | 85 | 35 | 271 | 278 | |
| Gbk AD2 | 165 | 164 | 152 | 161 | 146 | 13 | 5 | 62 | 83 | 34 | 270 | 277 | |
| Gby AD2 | 162 | 161 | 149 | 157 | 143 | 6 | 9 | 63 | 84 | 33 | 270 | 277 | |
| Gt AD3 | 162 | 160 | 167 | 175 | 164 | 86 | 85 | 82 | 79 | 48 | 267 | 272 | |
| Gm AD4 | 152 | 147 | 164 | 174 | 168 | 89 | 89 | 86 | 84 | 75 | 260 | 264 | |
| Gd AD5 | 177 | 171 | 151 | 169 | 161 | 66 | 66 | 63 | 104 | 110 | 266 | 274 | |
| Gr D5 | 856 | 852 | 940 | 926 | 938 | 873 | 875 | 871 | 862 | 854 | 873 | 70 | |
| Gg D6 | 916 | 911 | 985 | 983 | 1,000 | 924 | 928 | 923 | 927 | 916 | 906 | 180 |
Note: Gaf, Ga, Gr, Gg, Gh, Ghh, Ghl, Gb, Gby, Gbk, Gt, Gm and Gd indicates G. herbaceum var. africanum A1, G. arboretum A2, G. raimondii D5, G. gossypioides D6, G. hirsutum AD1, G. hirsutum race hainansijimian AD1, G. hirsutum race lanceolatum AD1, G. barbadense AD2, G. barbadense race yuanmou AD2, G. barbadense race kaiyuan AD2, G. tomentosum AD3, G. mustelinum AD4 and G. darwinii AD5, respectively. They are the same hereinafter. The upper triangle shows the number of genome indels and the lower triangle shows the total substitutions of genome.
A comparison of indels between the chloroplast genomes was also carried out (Table 2). The number of indels ranged from 3 to 278, and thelengthier sizes varied from 5 to 1,863 bp (Table S3). The divergence due to indels between them was estimated up to 1.153% (the total length of indels divided by the total length of aligned). The number of indels varied from 33 (AD2-AD5) to 100 (AD1-AD3) and the size varying from 194 (AD2-AD5, with 0.120% divergence) to 559 bp (AD1-AD3, with 0.346% divergence) within allotetraploids. Additionally, the number of indels between allotetraploid species and diploid A-genome species varied from 104 (AD5-A2) to 134 (AD1-A1) with size from 457 (AD4-A2, with 0.283% divergence) to 711 bp (AD1-A1, with 0.440% divergence). Only 13 indels totaling 91 bp were identified between G. arboreum A2 and G. herbaceum var. africanum A1, much less than those within allotetraploid species. Seventy indels totaling 486 bp were identified between G. gossypioides D6 and G. raimondii D5. The number of indels was more than 255 between diploid D-genome species and other genome species. Taking nucleotide substitutions and indel divergence together, genomic divergence attributed to indels was estimated as 0.091% to 0.143% between allotetraploid species and diploid A-genome, while the nucleotide divergence was estimated as 0.283% to 0.440%. The total sequence divergence was estimated as 0.374% to 0.583%. And total sequence divergence within allotetraploid species was between 0.159% (AD2–AD5) and 0.454% (AD1–AD3). The indels displayed a higher degree of sequence divergence, about 3-fold larger than substitutions. In addition, we observed the correlation between the number of substitutions and the number of indel events (Table 2), and the ratio of nucleotide substitutions events to indels events (S/I) in different pairwise comparison was investigated. Between allotetraploid species and D-genome species, the S/I ratio varied from 3.2 (AD2–D5) to 3.9 (AD1–D5), but 1.3 (AD2–A1) to 1.9 (AD1–A2) between allotetraploid species and A-genome species. And the S/I ratio decreased to about 1.0 within allotetraploid species (AD2–AD4/AD3–AD4). These suggested that the S/I ratio increased when divergence time increased between genomes [24].
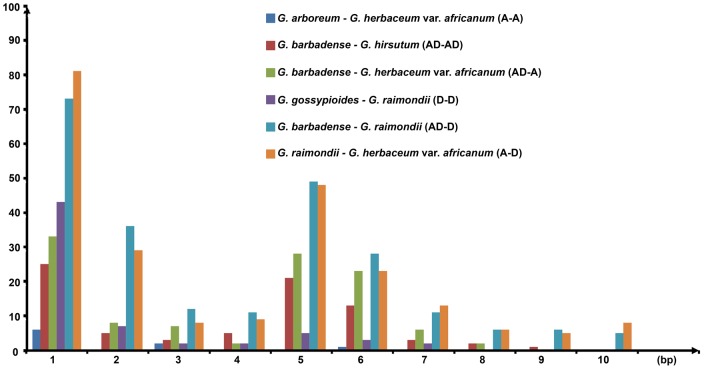
The size distribution of indels in our datasets was calculated (Figure 3). The number of short indels (1–10 bp) accounted for more than 90% of the total with more closely related species tending to have a higher proportion of short indels (Table S8A). As expected, single-nucleotide (1-bp) indels were the most common, accounting for approximately 32% of all indels. However, the number of short indels did not decrease with increasing length. The 5-bp indels, accounting for 16% to 24% of all indels, rather than 2-bp indels, were the second most abundant. The number of 5–6-bp indels was significantly more than the number of 3–4-bp indels (Table S8A). The results were consistent across almost all Gossypium species by pairwise comparison except for between the 2 A-genomes due to their close relationships.
Figure 3. The distribution of indel types in Gossypium chloroplast genomes.
We pairwise compared the indels of 13 chloroplast genomes. There was a marked tendency for all Gossypium species, except for between the 2 A genomes, to fluctuate wherein a point of inflection at the position of 5 bp. In this figure, A, D, and AD genome were represented by G. herbaceum var. africanum A1, G. raimondii D5, and G. barbadense AD2, respectively. The x-axis indicates the length of indel and y-axis represents the number of indels.
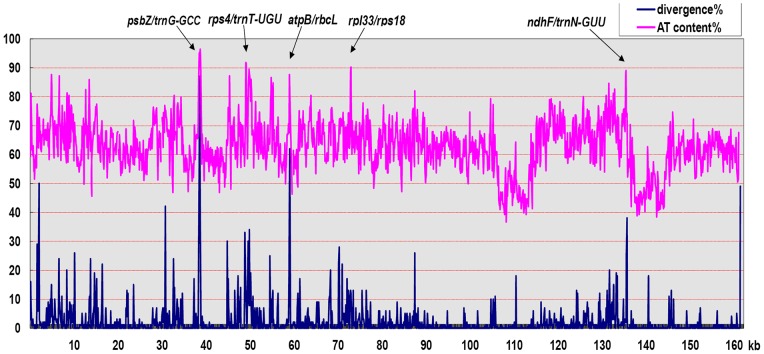
The substitutions and indels were analyzed separately for LSC, SSC, and IR regions (Table S4). The substitution rate (the number of substitutions per kb) was the highest in the SSC region and the lowest in the IR region. For example, 605, 201 and 24 substitutions were detected in the LSC (88.8 kb), SSC (20.3 kb), and IR (25.6 kb) regions between G. arboreum A2 and G. raimondii D5, with substitution rates of 6.8/kb, 9.9/kb and 0.9/kb, respectively. The substitution rate in SSC was about 11-fold larger than that in IR region. While the highest indel rate was in the LSC region and the lowest was in the IR region. For example, 212, 29 and 9 indels were identified in the LSC, SSC, and IR regions between G. arboreum A2 and G. raimondii D5, with indel rate (the number of indels per kb) of 2.3/kb, 1.4/kb and 0.4/kb, respectively. Specifically, the indel rate in the LSC region was about 6-fold larger than that in the IR region. Because high sequence divergence was usually accompanied by a high AT content as shown in Figure 4, the higher AT content in SSC region has contributed to a higher substitution rate (Table S2), but the fact that this region has a larger proportion of coding sequences appears to have constrained the indel rate for functional variation. Both the substitution and indel rates confirmed that the IR region was more conserved than single copy regions. And the conserved regions harbored relative lower AT content (Table S2, Figure 4).
Figure 4. The relationships between sequence divergence and the AT content in Gossypium chloroplast genome.
AT content was calculated using sliding sequence intervals of 100 bp, while divergence was measured by multiple genome alignment with a step size of 10 bp.
SSR variation in Gossypium chloroplast genomes
Simple Sequence Repeats (SSRs) are often used as molecular markers in genetics. SSRs usually have a higher rate of mutation compared to other neutral regions of DNA due to slipped strand mispairing (slippage) during DNA replication on a single DNA strand. A large number of SSR loci were found in Gossypium chloroplast genomes. The number of mononucleotide ≥8 bp, dinucleotide ≥8 bp, trinucleotide ≥9 bp, tetranucleotide ≥12 bp, and pentanucleotide ≥ 15 bp loci were counted. The number of SSRs ranged from 315 to 327, and the average rate was 2 SSRs/kb among the 13 genomes. Similarly, the number of mononucleotide ≥10 bp, dinucleotide ≥10 bp, trinucleotide ≥ 12 bp, tetranucleotide ≥16 bp, and pentanucleotide ≥20 bp loci were detected as well. The number of all SSRs varied from 58 to 70, and the average rate was 0.4 SSRs/kb between the 13 genomes (Table 3). Among the different SSRs motifs, they appeared at different frequencies, mononucleotide repeats were most abundant (50.97%–58.09%). The predominant mononucleotide repeats were A or T, which accounted for 95.14% to 97.21% of the mononucleotide repeats among the 13 genomes with a definition mononucleotide ≥8 bp. Most SSRs were located in intergenic spacers, and at least 68 intergenic spacers contained SSR loci among 13 Gossypium chloroplast genomes. More than 4 SSR loci were found in trnK-UUU/rps16, atpF/atpH, psbZ/trnG-GCC, atpH/atpI, rpoB/trnC-GCA, petN/psbM, psaA/ycf3, atpB/rbcL, psbE/petL, ycf4/cemA, 3′-rps12/trnV-GAC, petA/psbJ, psaJ/rpl33, and rpl32/ndhF. These regions could be utilized as targets for DNA polymorphism analysis.
Table 3. Number of SSRs identified in 13 chloroplast genomes.
| Species | Mononucleotide | Dinucleotide | Trinucleotide | Tetranucleotide | Pentanucleotide | Total | ||||||
| ≥8 | ≥10 | ≥8 | ≥10 | ≥9 | ≥12 | ≥12 | ≥16 | ≥15 | ≥20 | A | B | |
| G. herbaceum var. africanum A1 | 179 | 51 | 58 | 16 | 72 | 3 | 6 | 0 | 4 | 0 | 319 | 70 |
| G. arboreum A2 | 181 | 48 | 57 | 16 | 72 | 3 | 6 | 0 | 4 | 0 | 320 | 67 |
| G. hirsutum AD1 | 182 | 44 | 54 | 15 | 71 | 2 | 6 | 0 | 3 | 0 | 316 | 61 |
| G. hirsutum race hainansijimian AD1 | 183 | 41 | 54 | 15 | 70 | 2 | 6 | 0 | 2 | 0 | 315 | 58 |
| G. hirsutum race lanceolatum AD1 | 182 | 44 | 54 | 16 | 71 | 3 | 6 | 0 | 2 | 0 | 315 | 63 |
| G. barbadense AD2 | 184 | 44 | 55 | 15 | 72 | 3 | 5 | 0 | 2 | 0 | 318 | 62 |
| G. barbadense race yuanmou AD2 | 185 | 44 | 55 | 15 | 72 | 3 | 5 | 0 | 2 | 0 | 319 | 62 |
| G. barbadense race kaiyuan AD2 | 185 | 46 | 55 | 15 | 72 | 3 | 5 | 0 | 2 | 0 | 319 | 64 |
| G. tomentosum AD3 | 182 | 46 | 57 | 16 | 74 | 6 | 5 | 0 | 2 | 0 | 320 | 68 |
| G. mustelinum AD4 | 184 | 43 | 56 | 16 | 72 | 4 | 6 | 0 | 4 | 0 | 322 | 63 |
| G. darwinii AD5 | 183 | 44 | 56 | 16 | 72 | 4 | 6 | 0 | 2 | 0 | 319 | 64 |
| G. raimondii D5 | 186 | 48 | 58 | 12 | 73 | 2 | 7 | 0 | 3 | 0 | 327 | 62 |
| G. gossypioides D6 | 190 | 50 | 58 | 12 | 73 | 2 | 6 | 0 | 2 | 0 | 329 | 64 |
| Average | 184 | 46 | 56 | 15 | 72 | 3 | 6 | 0 | 3 | 0 | 320 | 64 |
| Conserved | 79 | 8 | 36 | 3 | 36 | 0 | 4 | 0 | 0 | 0 | 155 | 11 |
| Conserved rate | 0.43 | 0.18 | 0.64 | 0.20 | 0.50 | 0 | 0.69 | - | 0 | - | 0.48 | 0.17 |
Note: A: The total number of mononucleotide ≥8 bp, dinucleotide ≥8 bp, trinucleotide ≥9 bp, tetranucleotide ≥12 bp, and pentanucleotide ≥15 bp. B: The total number of mononucleotide ≥10 bp, dinucleotide ≥10 bp, trinucleotide ≥12 bp, tetranucleotide ≥16 bp, and pentanucleotide ≥20 bp.
The SSR polymorphism was estimated (Table 3). SSR variation came mainly from the mononucleotide SSRs. Among the 13 chloroplast genomes, 8 out of 46 (17.4%) mononucleotide SSR loci were completely conserved when the mononucleotide repeat unit was defined as ≥10 bp, 71 out of 138 mononucleotide SSR loci (51.4%) with lengths defined as 8 and 9 bp were found to be completely identical. Additionally, the polymorphism of dinucleotide repeats was about 36% in the size group ≥8 bp, which was much lower than that of mononucleotide repeats (57%). Of all analyzed SSR loci, 52% or 83% loci were polymorphic in Gossypium chloroplast genomes in the length groups ≥8 bp or ≥10 bp, respectively. These findings indicated that SSR polymorphism is very rich in the cpDNA of Gossypium and that the number of polymorphic SSRs would decrease along with SSR length decreasing or SSR motif elongation.
The numbers of polymorphic SSRs were quantified by pairwise comparison among the 13 chloroplast genomes (Table S5). There was wide variation in the number of polymorphic SSRs, ranging from a minimum of 2 loci (between G. barbadense race yuanmou AD2 and G. barbadense race kaiyuan AD2) to more than 115 loci (between allotetraploid species and D-genome species). Most of polymorphic SSRs generated 1–3-bp indels between genomes. We observed the relationships between the number of polymorphism SSR loci (Table S5) and the number of 1–3-bp indels (Table S8A), and the total number of 1–3-bp indels was very close to the number of polymorphism SSR loci. We concluded that 1–3-bp indels were mainly attributed to the SSRs polymorphism variation. Using the polymorphic SSR loci we performed the phylogenetic reconstruction. The tree clearly shows that the genomes of A, D, and AD were in different clades and roughly drew an acceptable topology of Gossypium (Figure S1).
Comparison of coding region among sequenced Gossypium species
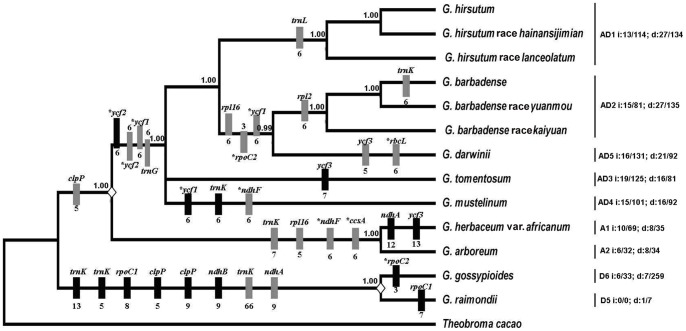
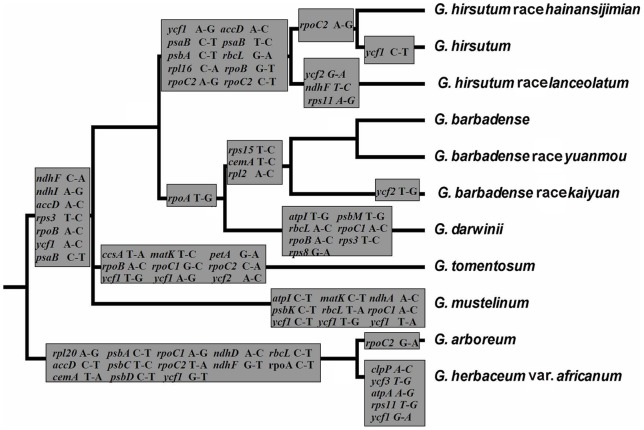
Although chloroplast DNA is, in general, highly conserved in coding region, there are still species differences. Each of the 13 Gossypium chloroplast genomes contained 78 unique protein-coding genes, of which 21 genes were completely identical at the DNA level and 19 genes carried only synonymous substitutions (Table S1). Most of these conserved sequences encoded components of the photosystems. The remaining 38 genes harbored amino acid changes between at least one homologous gene pair out of the compared. Seven genes (atpB, atpE, psbK, rpl2, psbM, rpl36 and rps2) contained only nonsynonymous substitution, and 6 genes had more than 12 mismatches (rpoB, matK, rbcL, rpoC2, ndhF and ycf1 with a maximum mismatch of 12, 13, 16, 21, 28 and 61, respectively). The comparative analysis demonstrated that ycf1 was the most variable coding region in the chloroplast genome. The nonsynonymous/synonymous substitution rate ratio (dN/dS) of ycf1 between the A genome and the D genome was significantly greater than 1.0, which could be considered as strong evidence for positive selection. Using Theobroma as an outgroup (See Method), 6 genes harbored indels, namely, rpoC2 (a 6-bp deletion in AD2 and AD5, and a 3-bp insertion in D6), rbcL (a 6-bp deletion in AD2), ycf2 (a 6-bp deletion and a 6-bp insertion in allotetraploids), ycf1 (a 6-bp deletion in all allotetraploids, a 6-bp deletion in AD2 and AD5, a 6-bp insertion in AD4), ccsA (a 6-bp deletion in A1 and A2) and ndhF (a 6-bp deletion in AD4, A1 and A2), but no frameshift mutation was detected in these genes. All the insertions and deletions found in the genes were mapped onto the phylogenetic tree (Figure 5). Sixteen genes (atpI, ccsA, cemA, matK, ndhF, psbK, psbM, rpl16, rpl2, rpoA, rpoB, rpoC1, rpoC2, rps8, ycf1, and ycf2) were diverged among 9 allotetraploid Gossypium at the protein level. These genes, having a faster rate of evolution could act be utilized to resolve phylogenetic issues ranging from lower taxonomic ranks to the relationships among genera and species, while those highly conserved genes could be used to help resolve issues at a high taxonomic level.
Figure 5. Phylogeny of Gossypium species or races.
The variable sequences were used to construct a phylogeny of Gossypium with Theobroma cacao as an outgroup. Bayesian phylogenetic analysis was performed using MrBayes program with GTR + inv + gamma model. The values at nodes represent Bayesian inference posterior probability. On the branch, the insertions (i) and deletions (d) of introns and genes (marked as *) are indicated by black and gray bars. The right tree shows the insertions (i) and deletions (d) of intergenic spacers happened on the branch from the node “<$>\raster="rg1"<$>” to common branches of corresponding genome. Before and after “/” represents the number and length of indels, respectively.
Using Theobroma as an outgroup, the substitutions of genes were mapped onto the phylogenetic tree as well (Figure 6). There were 13 genes with a total of 13-bp nucleotide substitutions which occurred in the A genome after allotetraploid formation. Seven genes with a total of 7-bp substitutions had occurred before the split of allotetraploid, while 10 genes, 4 genes, 8 genes, 7 genes, and 8 genes happened after the divergence of allotetraploid in AD1, AD2, AD3, AD4, and AD5, respectively. These suggested that the substitution of G. hirsutum AD1 was more than that of its sisters after the divergence of allotetraploid. When AD1 diverged, 1-bp, 2-bp and 3-bp nucleotide substitution occurred in G. hirsutum race hainansijimian, G. hirsutum and G. hirsutum race lanceolatum. Following the split of A genome, 6-bp and 1-bp substitution occurred in A1 and A2, respectively.
Figure 6. Distributions of the allotetraploid gene substitution.
The phylogenetic tree was the subtree of that in Figure 5.
To estimate the functional constraint of each gene group, the nonsynonymous substitution rate (dN), synonymous substitution rate (dS) and dN/dS ratio between Gossypium and Arabidopsis were calculated (Table 4). The mean dS value of the 13 Gossypium genomes varied among gene groups with the highest c-type cytochrome (ccsA gene) (0.463±0.0057) and the lowest conserved reading frames (ycf1 and ycf2) (0.0752±0.0004). Mean dN varied more widely than mean dS among the 16 gene groups, such as maturase (0.1911±0.0004) being about 21-fold larger than that of photosystem I (0.0089±0). The dN/dS ratio ranged from a minimum of 0.032±0.0002 for the 5 photosystem I genes, which suggested that these genes were under purifying selection [25], [26], to a maximum of 0.6355±0.0041 for conserved reading frames, which suggested that ycf1 and ycf2 experienced rate acceleration [27]. In addition, the dN/dS ratio of gene groups was always positively correlated with AT content. However, a strong positive correlation had not been detected among dN, dS, and dN/dS, which perhaps was due to a statistical bias from similar DNA sequences [28], [29].
Table 4. The dS, dN and dN/dS of chloroplast protein-coding gene groups between Gossypium and Arabidopsis.
| Gene group | No. of genes | Length (bp) | AT% | dS | dN | dN/dS |
| Photosystem I | 5 | 5,979 | 57.63 | 0.2794±0.0017 | 0.0089±0 | 0.0320±0.0002 |
| Photosystem II | 15 | 5,598 | 57.92 | 0.2675±0.0011 | 0.0113±0.0002 | 0.0422±0.0007 |
| Cytochrome b/f complex | 6 | 2,364 | 60.29 | 0.3797±0.0013 | 0.0217±0.0002 | 0.0572±0.0005 |
| ATP synthase | 6 | 4,929 | 58.35 | 0.3874±0.0001 | 0.0314±0.0001 | 0.0811±0.0004 |
| Assembly/stability of photosystem I | 2 | 1,056 | 61.11 | 0.3755±0.0038 | 0.0342±0 | 0.0910±0.0010 |
| RuBisCO large subunit | 1 | 1,440 | 56.03 | 0.2782±0.0023 | 0.0371±0.0003 | 0.1332±0.0016 |
| Ribosomal large subunit protein | 8 | 2,676 | 59.35 | 0.2689±0.0031 | 0.0374±0.0007 | 0.1392±0.0039 |
| Ribosomal small subunit protein | 12 | 4,998 | 61.35 | 0.3142±0.0011 | 0.0452±0.0002 | 0.1437±0.0007 |
| NADH genes | 11 | 10,316 | 64.08 | 0.3901±0.0007 | 0.0571±0.0004 | 0.1464±0.0011 |
| RNA polymerase genes | 4 | 10,423 | 61.42 | 0.3630±0.0043 | 0.0816±0.0007 | 0.2246±0.0009 |
| Carbon metabolism | 1 | 690 | 67.20 | 0.2963±0.0045 | 0.0827±0.0012 | 0.2792±0.0064 |
| Proteolysis subunit | 1 | 591 | 58.19 | 0.2173±0.0025 | 0.0613±0.0009 | 0.2820±0.0050 |
| c-type cytochrome | 1 | 962 | 66.98 | 0.4630±0.0057 | 0.1403±0.0004 | 0.3029±0.0043 |
| Acetyl-CoA carboxylase subunit | 1 | 1,494 | 65.38 | 0.4247±0.0071 | 0.1801±0.0006 | 0.4243±0.0073 |
| Maturase | 1 | 1,515 | 66.27 | 0.3734±0.0084 | 0.1911±0.0004 | 0.5119±0.0105 |
| Conserved reading frames | 2 | 12,570 | 65.15 | 0.0752±0.0004 | 0.0478±0.0002 | 0.6355±0.0041 |
Among all the rRNA genes, only rrn23 in G. gossypioides D6 contained a 1-bp substitution. Among all tRNA genes, trnM-CAU and trnP-UGG in G. raimondii D5 and G. gossypioides D6, and trnW-CCA in G. herbaceum var. africanum A1 showed a 1-bp substitution. These mutation sites could not be located currently in anti-codon region.
The protein-coding genes were identical at the protein level among the G. barbadense lineages as well as between G. hirsutum AD1 and G. hirsutum race hainansijimian AD1 (Table S6). The total length of substitutions ranged from 10 (AD2–AD5) to 22 bp (AD1–AD3/AD1–AD4) within allotetraploid species, accounting for 9.4% (AD1–AD2) to 21.4% (AD3–AD4) of whole genome substitutions divergence (Table S6 and Table 2). Between allotetraploid species and A-genome Gossypium, the total length of substitutions varied from 25 (AD2–A2) to 38 bp (AD1–A1), accounting for 14.2% (AD1–A2) to 22.4% (AD4–A1) of whole genome substitution divergence. Between D-genome species and other species the total length of substitutions varied from 241 (D6–AD2) to 259 bp (D5–AD1), accounting for 25.7% (D6–AD1) to 29.8% (D5–A1) of whole genome substitutions (Table S6 and Table 2). And between 2 D-genome species, the total length of substitutions was 60 bp, accounting for 33.3% of whole genome substitution divergence. It suggested a higher proportion of variation rate of the protein-coding sequences after divergence of G. raimondii D5 and G. gossypioides D6. In addition, the lower proportion of divergence due to protein-coding sequences indicated that the major part of the genomic divergence should be attributed to the non-coding sequences.
Comparison of non-coding sequences among 13 Gossypium chloroplast genomes
Although various functional elements were identified in intergenic spacer regions, e.g., promoter sequences and ribosome-binding sites [30], intergenic spacers were found to be more susceptible to variation than gene sequences. Only 21 out of 128 (16.4%) intergenic spacers were completely identical, however, none of them exceeded 326 bp in length. It suggests that the observed conservation of these intergenic regions can perhaps be attributed to their small size.
The total number of nucleotide substitutions of each pair varied from 5 bp (AD2–AD2) to 673 bp (D6–AD1). And Si/Sv values ranged from 0.20 (AD2–AD3) to 2.00 (AD2–AD2) with an average value of 0.39 (Table S7A), which were less than those of genes. The base substitution between G and T was the most frequent, whereas substition between G and C was the least frequent. The number of indels in each pair ranged from 3 (AD2–AD2) to 229 (D6–AD3/D6–AD2), and the length of a single indel ranged from 1 to 234 bp. The total length of indels varied from 4 (AD2–AD2) to 1642 bp (D6–AD5) (Table S7B), and the average length of an indel was from 1.0 to 7.4 bp. Using Theobroma as an outgroup, most indels in intergenic spacers of allotetraploids could be classified into insertions or deletions, and the indels that could not be polarized were mostly in the SSR loci. The number of insertions and deletions were plotted onto the phylogenetic tree (Figure 5). There were 4 insertions and 6 deletions which happened before allotetraploids divergence. After the formation of allotetraploid, 27, 27, 16, 16, and 21 deletions and 13, 15, 19, 15, and 16 insertions happened in AD1, AD2, AD3, AD4 and AD5, respectively, These suggested that AD1 and AD2 had more deletions than other allotetraploids, and most allotetraploids, except for G. tomentosum AD3, had the biases for deletions since their formation. Additionally, only 8 deletions and 6 insertions occurred on G. arboreum A2 after allotetraploid formation. These indicated that allotetraploid had more deletions and insertions than diploid A-genomes.
Seven intergenic spacers, trnT-UGU/trnL-UAA, psbZ/trnG-GCC, atpB/rbcL, trnK-UUU/matK, atpH/atpI, rpl32/ndhF, and trnL-UAG/rpl32, were found to be highly variable. The intergenic spacer psbZ/trnG-GCC was the most divergent region, containing SSRs, substitutions, and indels, one of which reached 74 bp in length. AT content was typically more than 70% in these intergenic spacers regions, more than the average value of all intergenic spacers, attaining 83% in the intergenic spacer psbZ/trnG-GCC. These findings confirmed that the variation, to a large extent, was associated with AT content. A 51-bp insertion was detected in petN/psbM of G. hirsutum lineages and G. darwinii AD5. It indicated that the 51-bp insertion may have occurred independently after the differentiation of AD1 and AD5. In addition, a 234-bp deletion was found in intergenic spacer trnL-UAG/rpl32 of G. gossypioides D6. The deletion was located approximately 820 bp upstream of the rpl32 start codon. An EST (ES814751) containing trnL-UAG/rpl32 and rpl32 was identified from NCBI database, suggesting that the intergenic spacer trnL-UAG/rpl32 was co-transcribed with rpl32. Consequently, the large indel in trnL-UAG/rpl32 was speculated to regulate rpl32.
Indels found in introns were partitioned into insertions and deletions using Theobroma as an outgroup (See Method). Ten of the 17 intron sequences harbored indels, including 11 deletions and 11 insertions, and they were: ndhA (a 9-bp deletion in D5 and D6, and a 12-bp insertion in A1), ndhB (a 6-bp insertion in D5 and D6), rpl2 (a 5-bp deletion in AD2), rpl16 (a 5-bp deletion in A1 and A2, and a 5-bp deletion in AD2 and AD5), rpoC1 (an 8-bp insertion in D5 and D6, and a 7-bp insertion in D5), trnK-UUU (a 13-bp and a 5-bp insertion in D5 and D6,a 6-bp deletion in AD2, a 66-bp deletion in D5 and D6, a 6-bp insertion in AD4, and a 7-bp deletion in A1 and A2), ycf3 (a 5-bp deletion in AD2, a 13-bp insertion in A1, and a 7-bp insertion in AD3), clpP (a 5-bp and a 9-bp insertion in D5 and D6, and a 5-bp deletion in A1 and A2), trnL-UAA (a 6-bp deletion in AD1), trnG-GCC (a 5-bp deletion in allotetraploids). Distributions of the intron insertions and deletions were mapped onto the phylogenetic tree (Figure 5). These data indicated that the trnK-UUU intron had more indels than other introns.
Relationship and origin time of allotetraploid cotton
For evaluation of the phylogenetic relationships of Gossypium allotetraploids, 3 groups of sequences, variable protein-coding genes sequences, intron sequences, and variable intergenic sequences, were used to reconstruct the topologies by two independent methods (Bayesian method or maximum-likelihood method), respectively (Figure S2). The 2 topologies from variable protein-coding genes sequences using both maximum-likelihood and Bayesian method showed that allotetraploid divided into four separate branches rapidly after the differentiate. The phylogenetic trees from intron sequences using both maximum-likelihood and Bayesian method, and from variable intergenic spacer sequences using maximum-likelihood, showed that G. mustelinum AD4 was the sister to G. tomentosum AD3. And the phylogenetic tree from variable intergenic sequences using Bayesian method showed that G. tomentosum AD3 was closer than G. mustelinum AD4 to G. hirsutum lineages. Although the 6 topologies showed slight differences, as a whole, they exhibited great similarity with strong bootstrap or posterior probabilities support, indicating that the constructed phylogenies were highly reliable. In addition, the phylogeny of cotton species or races was reconstructed by using variable sequences with Theobroma cacao as an outgroup (Figure 5). The tree suggested a closer relationship of allotetraploid cottons to the 2 A-genome species than to 2 D-genome species. And G. darwinii AD5 was the sister to G. barbadense lineages, which showed a similar topology as many results [3], [5], [10]. However, in contradiction, the tree showed that the G. mustelinum AD4 rather than G. hirsutum AD1 was the sister to G. tomentosum AD3, which was also described by Wendel et al. [31]. Totally, the phylogenies of Gossypium confirmed a close relationship between allotetraploid species and the ancestor of A genome. Also the tree revealed that the split of the common chloroplast genomes of allotetraploid was earlier than that of G. raimondii D5 and G. gossypioides D6.
Sixty-one common chloroplast protein-coding genes from 2 diploid cotton species, G. herbaceum var. africanum A1 and G. raimondii D5, and 11 other plant species were used to reconstruct the phylogeny (Figure S3). The average length of the 61 concatenated sequences was 42,746 bp. Monocots and eudicots were classified into 2 distinct groups, and the Malvales and Brassicales had a close relationship. Each clade was supported with high bootstrap values. Divergence time between monocots and eudicots was evaluated as 145 MYA in previous studies [32]–[34]. It was defined as the start point to estimate the divergence time within the cotton genus. The divergence time of nodes B, C, and D were estimated to be 127.9, 94.6, and 3.4 MYA, respectively. This suggested that G. herbaceum var. africanum A1 and G. raimondii D5 diverged about 3.4 MYA, which could be used as a reference point.
A-genome and D-genome type cytoplasms represent the 2 large clades of Gossypium, which in turn represent the diversification of Gossypium. Nucleotide substitutions of 61 protein-coding genes were used to estimate the synonymous rate (rs) and nonsynonymous substitution rate (ra) between G. herbaceum var. africanum A1 and G. raimondii D5. The results showed that rs and ra were 1.162×10−9 and 0.206×10−9, respectively. The divergence time between G. herbaceum var. africanum A1 and G. raimondii D5 was estimated based on Ka and Ks values of 78 protein-coding genes. Because the calculation using the Ks value was more similar to the branch length method (Table 5), the Ks value was adopted to calculate the divergence time of cotton species. Differentiation between A-genome and D-genome cytoplasms was estimated to have occurred around 3.89 MYA. Allotetraploid species originated 0.43 (AD2–A2) to 0.68 (AD1–A1) MYA and initially diverged 0.38 (AD1–AD3) MYA, G. darwinii AD5 diverged from G. barbadense lineages about 0.09 MYA. The split of the 2 A-genome species occurred approximately 0.13 MYA, following the formation of allotetraploid cottons. Consequently, it was the ancestor of A-genome Gossypium species that was the maternal source of allotetraploid species. The differentiation of chloroplast genomes between G. raimondii D5 and G. gossypioides D6 was estimated to have occurred around 0.77 MYA, earlier than the advent of allotetraploid cottons.
Table 5. Evolutionary distance (MY) between any two of 13 Gossypium calculated by chloroplast protein-coding gene.
| Gaf A1 | Ga A2 | Gh AD1 | Ghh AD1 | Ghl AD1 | Gb AD2 | Gbk AD2 | Gby AD2 | Gt AD3 | Gm AD4 | Gd AD5 | Gr D5 | Gg D6 | |
| Gaf A1 | 0.04 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.34 | 0.34 | |
| Ga A2 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.13 | 0.34 | 0.34 | |
| Gh AD1 | 0.68 | 0.60 | 0.04 | 0.04 | 0.09 | 0.09 | 0.09 | 0.13 | 0.09 | 0.09 | 0.34 | 0.34 | |
| Ghh AD1 | 0.64 | 0.56 | 0.04 | 0.04 | 0.09 | 0.09 | 0.09 | 0.09 | 0.09 | 0.09 | 0.34 | 0.34 | |
| Ghl AD1 | 0.68 | 0.60 | 0.04 | 0.04 | 0.09 | 0.09 | 0.09 | 0.13 | 0.09 | 0.09 | 0.34 | 0.34 | |
| Gb AD2 | 0.47 | 0.43 | 0.30 | 0.26 | 0.30 | 0.04 | 0 | 0.09 | 0.09 | 0.04 | 0.34 | 0.34 | |
| Gbk AD2 | 0.51 | 0.47 | 0.30 | 0.30 | 0.30 | 0.04 | 0.04 | 0.09 | 0.09 | 0.04 | 0.34 | 0.34 | |
| Gby AD2 | 0.47 | 0.43 | 0.30 | 0.26 | 0.30 | 0 | 0.04 | 0.09 | 0.09 | 0.04 | 0.34 | 0.34 | |
| Gt AD3 | 0.60 | 0.56 | 0.38 | 0.38 | 0.38 | 0.21 | 0.26 | 0.21 | 0.09 | 0.09 | 0.34 | 0.34 | |
| Gm AD4 | 0.51 | 0.47 | 0.30 | 0.30 | 0.30 | 0.13 | 0.17 | 0.13 | 0.26 | 0.09 | 0.34 | 0.34 | |
| Gd AD5 | 0.51 | 0.47 | 0.30 | 0.30 | 0.30 | 0.09 | 0.13 | 0.09 | 0.26 | 0.21 | 0.34 | 0.34 | |
| Gr D5 | 3.89 | 3.89 | 3.89 | 3.85 | 3.89 | 3.68 | 3.72 | 3.68 | 3.80 | 3.72 | 3.72 | 0.17 | |
| Gg D6 | 3.89 | 3.89 | 3.89 | 3.85 | 3.89 | 3.68 | 3.72 | 3.68 | 3.80 | 3.72 | 3.72 | 0.77 |
Note: The lower triangle shows the divergence time (million years) estimated by 78 chloroplast protein-coding genes, and the upper triangle shows the ± standard error.
Discussion
In this study we present the complete sequences of 12 Gossypium chloroplast genomes, including 2 A-genome lineages (G. herbaceum var. africanum A1 and G. arboreum A2), 2 D-genome lineages (G. raimondii D5 and G. gossypioides D6), and 8 allotetraploid cottons (G. hirsutum race hainansijimian AD1, G. hirsutum race lanceolatum AD1, G. barbadense AD2, G. barbadense race yuanmou AD2, G. barbadense race kaiyuan AD2, G. tomentosum AD3, G. mustelinum AD4 and G. darwinii AD5). The data obtained has permitted an evaluation of the relationship and evolution of Gossypium allotetraploids. The results indicate that although the Gossypium chloroplast genome is highly conserved, sequence diversification is evident. Comparison of 13 closely related chloroplast genomes including previously published and newly sequenced genomes provided a potentially powerful means to evaluate the relationship and evolution of Gossypium allotetraploids. In this study, most indels in both genes and intergenic spacers could be polarized into insertions or deletions, and evolutionary point at which they arose could be estimated using Theobroma cacao as an outgroup. It is interesting to find that 5-bp indels appear to be preferred in the Gossypium chloroplast genomes. It supported that the ancestor of A-genome Gossypium species was the maternal source of allotetraploid species and allotetraploids evolved from a common ancestor. The progenitor allotetraploid Gossypium originated approximately 0.43 to 0.68 MYA and initially diverged about 0.38 MYA.
Conservation of Gossypium chloroplast genome
Each of the 13 genomes were found to encode the same set of 112 unique genes in a uniform order with a sequence identity >98%. Thus the data demonstrates that the Gossypium chloroplast genome is highly conserved. Most genes did not show any variation among the 13 Gossypium chloroplast genomes. There was little sequence variation between the genomes of G. hirsutum AD1 and its 2 races as well as between G. barbadense AD2 and its 2 races (Table 2). And there were only 13 indels and 11 substitutions between G. herbaceum var. africanum A1 and G. arboreum A2. Although great phenotypic variation are observed among them, it is difficult to separate them using chloroplast markers. There were 70 indel events and 180 substitution events between diploid D5 and D6 genomes, while only 13 indels events and 11 substitution events occurred between A1 and A2 genomes. In our study, we seldom used the data of previously sequenced chloroplast genome of G. barbadense AD2 [22] (marked as G. barbadense 2006, hereinafter), and the chloroplast genomes of G. barbadense AD2 lineages were re-sequenced. By comparing the whole genome sequences of G. barbadense cv. Zhonghai 7 and G. barbadense 2006, we detected 284-bp substitutions and 103-bp indels, larger than that between allotetraploid and A-genome species.
Indels of Gossypium chloroplast genome
Many recent evolutionary variations, notably indels, were found by comparing 13 chloroplast genomes. These indel events were mainly attributed to the perfect or near-perfect repetition of an adjacent sequence, probably caused by slipped-strand mis-pairing in DNA replication [35]. Indels are thought to be a major driving force in sequence evolution [16], [36]. The number of indels decrease rapidly with increasing indel length [37]–[39]. However, the 5–6-bp indels, which was caused by adjacent 5–6-bp motif duplication or loss, are the second most common type in our study (Figure 3 and Table S8A). We also compare intraspecies or inter-subspecies of Acorus, Aethionema, Nicotiana, Oryza, Oenothera, Populas, and Solanaceae (Table S8B). Only Oenothera has similar, but much weaker results as Gossypium. The common ground between Gossypium and other species is the relatively few indels of 4-bp in length. It is necessary to further investigate whether a preference for 5-bp indels has happened in the nuclear genome of Gossypium.
The S/I ratio increased when divergence time increased between genomes. When we explained the increasing of genome S/I ratio as divergence time increased, one aspect may attribute to systematic underestimation of indels in more distantly related species [24]. The other reason may be that the indels scarcely appeared in chloroplast coding regions where substitutions tend to accumulate as divergence time increased. Nevertheless, we also observed that the S/I ratio increased as divergence time increased in just noncoding region (Table S7B).
Multiple closely related genomes were always required to distinguish insertions from deletions [40]. In our study, the number of all indels was counted by pairwise comparison, and most indels could be polarized into insertions and deletions using Theobroma cacao as outgroup (See Method). Insertions and deletions were mapped onto the phylogenetic tree (Figure 5). As shown in Figure 5, after the divergence of allotetraploids, the number of deletions was approximately 2-fold larger than that of insertions in the intergenic spacers of AD1 and AD2 genomes, and only deletions were detected in introns and coding sequences of AD1 and AD2 genomes. Although each genome experienced more deletions than insertions to various extents, the average length of single insertion was longer than that of deletion. During the period before the formation of allotetraploid and after the divergence of diploid A genome and D genome (data not shown in the Figure 5), 27 deletions (108 bp) and 19 insertions (179 bp) happened during the stage of 3.2 million years, while 18 deletions (132 bp) and 32 insertions (326 bp) happened before the split of diploid D5 and D6 genomes during the stage of 3.12 million years in whole genomes. Based on our evaluated divergence time and the number of indels, the indel ratio was standardized to the number of indels per genome per million years. These results showed that insertion ratio as well as deletion ratio was incongruent in the 2 branches after the split of A genome and D genomes, but the indel ratio converged to a fixed level for a substantial period of time.
Origin and evolution of Gossypium
A global molecular clock among species typically does not exist, and the rate of genome variation is known to change among inter-species specifically over time [45]. Because branch length method does not assume a strict clock, it is a more precise tool for estimating divergence time [32], [46]. Thus the branch length method was used to estimate the divergence time between Gossypium and other plant species. However, a likelihood ratio test of substitution rate constancy across species indicated that the protein-coding sequences of Gossypium chloroplast genomes had an equal evolutionary rate (P = 0.947). This suggested that a constant molecular clock method could be appropriate to date the divergence time of Gossypium. The divergence time between Gossypium and other dicotyledons based on our datasets was consistent with previous reports [33], [47]. Little incongruence on substitution rate among the 13 chloroplast genomes was observed in this experiment with the molecular clock method, and the estimated synonymous substitution rate (r = 1.162×10−9) was very close to previous estimates (r = 1.242×10−9) [48].
The synonymous substitution rate of the chloroplast genome was considerably different from that of the nuclear genome. The synonymous substitution rate for nuclear genes in plants was from 2.6×10−9 to 1.5×10−8 [49], which was 2 to 13 times higher than that in the chloroplast genome. Based on this synonymous substitution rate and using various nuclear gene sequence data, the formation of allotetraploids was estimated to have occurred approximately 1-2 MYA. In this study, the divergence time between A-genome species and allotetraploid cotton was estimated to be 0.43 to 0.68 MYA, which is very similar to previous reports [6], [14], [16]. Analysis of the ndhF sequence by Seelanan et al. [13] (assuming an average mutation rate of cpDNA genes r = 5×10−10), indicated that the divergence time between G. herbaceum A1 and G. hirsutum AD1 was arround 2 MYA much earlier than our estimation. These incongruent findings could be explained by that the substitution rate of ndhF sequence was the most rapid, which would not reflect the average substitution rate (r = 1.162×10−9).
Previous little was known regarding the divergence time of G. arboreum A2 and G. herbaceum A1 [3]. In this study, the data has shown that there is little variation between G. herbaceum var. africanum A1 and G. arboreum A2, only 13 indels events and 11 substitution events, suggesting that their divergence occurred at a similar time and thus inclusion of G. herbaceum A1 was probably merited. The lower level of sequence variation between G. herbaceum var. africanum A1 and G. arboreum A2 could possibly result in inaccurate estimations of the divergence time. However it is quite plausible that G. herbaceum var. africanum A1 and G. arboreum A2 diverged after the formation of allotetraploid taxa. In view of the highly conserved chloroplast genome sequences, it also seems likely that G. herbaceum A1 and G. arboreum A2 diverged after the formation of allotetraploids.
Although phylogenic analysis of Gossypium suggested that the split of the common chloroplast genomes of allotetraploid occurred slightly earlier than that of G. raimondii D5 and G. gossypioides D6, it could not suggest that the ancestor of the 2 D-genome species, rather than its offspring, were the close relatives of allotetraploids because of the intergenomic introgression between G. raimondii D5 and G. gossypioides D6 [3]. However, the time of divergence between G. gossypioides D6 and G. raimondii D5 was estimated to be slightly earlier than the formation of allotetraploids based on protein coding gene sequences. Altogether, it suggested that divergence of the chloroplast genomes between G. gossypioides D6 and G. raimondii D5 occurred at a time close to that of allotetraploid formation.
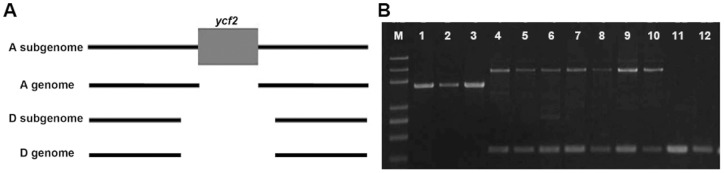
The allotetraploids shared 14 indels and 7-bp nucleotide substitutions in protein-coding genes (Figure 5 and Figure 6). The presence of unique indels and substitutions among all allotetraploids indicated that they possibly evolved from a common ancestor. Many studies had confirmed that cpDNA could frequently transfer to nuclear and mitochondrial genomes [41]–[43]. Although the information from chloroplast genomes could provide little evidence about the paternal donor of allotetraploids, the transfer of cpDNA to the nucleus could be helpful to recognize it. Grover et al. [44] found that a ycf2 fragment was inserted into the AT genome (A genome in tetraploids) but not into DT genome (D genome in tetraploids) of G. hirsutum AD1. Here additional Gossypium species were investigated for insertion of the ycf2 fragment which was found to be present in the AT genome of G. hirsutum AD1, G. hirsutum race hainansijimian AD1, G. barbadense AD2, G. barbadense race yuanmou AD2, G. barbadense race kaiyuan AD2, G. mustelinum AD4 and G. darwinii AD5, but was not detected in G. herbaceum A1, G. herbaceum var. africanum A1 and G. arboreum A2, G. raimondii D5 and G. gossypioides D6, or the DT genome. Furthermore, it was verified in G. robinsonii from C genome, G. somalense from E genome, and G. longicalyx from F genome as well, but none of them had this ycf2 fragment in nuclear genomes (Figure 7). The cpDNA ycf2 fragment was not found in nuclear genome of verified diploid cottons, including A, C, D, E, and F genomes, which suggested that the ycf2 fragment probably did not exist in the nucleus of the ancestor of A-genome species. So the ancestral allotetraploid lineage from inter-genomic hybridization initially lacked the ycf2 gene fragment in its nuclear genome. We could speculate that transfer of the ycf2 fragment occurred earlier than the divergence of allotetraploids, but later than tetraploid formation. Altogether, extant allotetraploid cottons probably share the same ancestor containing the cpDNA-derived ycf2 fragment from in its nuclear genome. It appears reasonable to infer that the probable paternal donor like the maternal donor, was monophyletic, though details regarding this are still vague.
Figure 7. The transfer of the ycf2 fragment to the nucleus.
A: Schematic view of the transfer of cpDNA ycf2 fragment; B: PCR amplification of the nuclear genome flanking the ycf2 insertion. M: D2000 plus; 1–12: G. herbaceum A1, G. herbaceum var. africanum A1, G. arboreum A2, G. hirsutum AD1, G. hirsutum race hainansijimian AD1, G. barbadense AD2, G. barbadense race yuanmou AD2, G. barbadense race kaiyuan AD2, G. mustelinum AD4, G. darwinii AD5, G. raimondii D5, and G. gossypioides D6, respectively.
Different divergence rate of allotetraploids
Indels play an important role in estimating genome evolution rate [16]. Indel ratio was standardized to the number of indels between genomes per million years (Table S9). Because indels scarcely appeared in chloroplast coding regions, it actually revealed the noncoding indel rates. Indel ratio between any two of allotetraploids varied from 92 to 323, while that was 33 to 34 between diploid A-genome species and diploid D-genome species and even much lower within A genome or D genome. This suggested that indel ratio was inconsistent within Gossypium species, and the chloroplast genomes of allotetraploids experienced higher indel ratio than those of diploids. In comparison with a relatively constant indel ratio of Gossypium species before the formation of allotetraploids (See above), the higher indel ratio of allotetraploids might be a result of a higher degree of evolutionary variability following the polyploidization [15], [16].
Whole genomes pairwise comparison among allotetraploids showed sequence variation, notably for G. hirsutum lineages being more variable (Table 2). So we tested the relative divergence rate between allotetraploid species and A-genome species with D-genome species as outgroup by Tajima's relative rate test [50]. According to the monophyletic theory, all allotetraploids should share a similar genetic distance to the A-genome group. Expectedly, the relative rates of 9 allotetraploid genomes showed no significant difference at the concatenated protein-coding gene level. However, a faster rate of variation was detected in G. hirsutum AD1 and its 2 races relative to other allotetraploids at the whole genome level (Table 6). It is probable that the incongruency of the rate was mainly determined by the presence of intergenic spacers.
Table 6. Tajima's relative rate test of chloroplast protein-coding genes and whole genome between Gossypium allotetraploid and A-genome diploid with G. raimondii D5 as an outgroup.
| Protein-coding genes | Whole genome | |||
| Species | G. herbaceum var. africanum A1 | G. arboreum A2 | G. herbaceum var. africanum A1 | G. arboreum A2 |
| G. hirsutum AD1 | 0.6219 | 0.3841 | 0.0052** | 0.0024** |
| G. hirsutum race hainansijimian AD1 | 0.7389 | 0.4795 | 0.0032** | 0.0014** |
| G. hirsutum race lanceolatum AD1 | 0.5164 | 0.3035 | 0.0184* | 0.0094** |
| G. barbadense AD2 | 0.3532 | 0.5485 | 0.6310 | 0.4640 |
| G. barbadense race yuanmou AD2 | 0.3532 | 0.5485 | 0.8711 | 0.6805 |
| G. barbadense race kaiyuan AD2 | 0.4652 | 0.6949 | 0.6310 | 0.4639 |
| G. tomentosum AD3 | 1 | 0.7150 | 0.5164 | 0.6801 |
| G. mustelinum AD4 | 1 | 0.7150 | 0.0707 | 0.1144 |
| G. darwinii AD5 | 0.8618 | 0.8527 | 0.2143 | 0.1343 |
Note: *p<0.05, **p<0.01.
The indel rate of allotetraploid species was also estimated. All allotetraploids had very similar indel rates at the protein-coding gene level compared to A-genome group, but G. hirsutum AD1 lineages experienced more indel events than other allotetraploids in intergenic spacers. Altogether, the intergenic spacers of G. hirsutum AD1 and its 2 races, G. hirsutum race hainansijimian AD1 and G. hirsutum race lanceolatum AD1, experienced faster divergence rates than other allotetraploids.
Conclusions
The data presented here represents a thorough analysis of the Gossypium chloroplast genomes utilizing both comparative genomics and phylogenetic analyse. The availability of complete nucleotide sequences of 13 Gossypium chloroplast genomes provides an opportunity to study the mechanisms under the origin and evolution of Gossypium allotetraploids. Although many valuable taxonomic studies have advanced our understanding of Gossypium, this is the first effort using such an extensive sample of chloroplast genomes to further understanding inter- and intra-specific differentiation of Gossypium. By comparing 13 Gossypium chloroplast genomes, species sequence variations have been detected, including substitutions and indels. Gossypium chloroplast genomes have a preference for 5–6-bp indels caused by adjacent 5–6-bp motif duplication or loss, whereas 1–3-bp indels are mainly attributed to the SSR polymorphisms. The genome S/I ratio increased as divergence time increased because the indels scarcely appeared in chloroplast coding regions where substitutions tended to accumulate as divergence time increased. This study offers support to the theory that the ancestor of A-genome Gossypium species is the maternal source of allotetraploid species. In addition, the transfer of cpDNA ycf2 to the nuclear genomes shows that the paternal donor is probably monophyletic. The differences of indel ratio among genomes reveal that allotetraploids experienced faster sequence gain or loss than diploids. In addition, our data show that the intergenic spacers of G. hirsutum lineages have experienced faster divergence rates than other allotetraploids. The availability of complete nucleotide sequences of 12 Gossypium chloroplast genomes should facilitate research to uncover the molecular mechanism of evolution of Gossypium allotetraploids.
All data obtained in this study but not shown in the manuscript are available upon request.
Materials and Methods
Plant taxa
Species or races chosen for this study were based on closely related lineages for Gossypium allotetraploid. Three wild allotetraploid species, G. tomentosum AD3, G. mustelinum AD4 and G. darwinii Watt AD5, as well as each 2 races of G. hirsutum (G. hirsutum race hainansijimian AD1 and G. hirsutum race lanceolatum AD1) and G. barbadense (G. barbadense race yuanmou AD2 and G. barbadense race kaiyuan AD2), were adopted for more datasets representing allopolyploid cottons. G. hirsutum AD1, G. hirsutum race hainansijimian AD1 and G. hirsutum race lanceolatum AD1 were considered as G. hirsutum lineages, while G. barbadense AD2, G. barbadense race yuanmou AD2 and G. barbadense race kaiyuan AD2 were treated as G. barbadense lineages. G. raimondii D5 and G. gossypioides D6 were chosen since they are the best candidates as living models of the D-genome donor. The 2 A-genome lineages (G. arboreum A2 and G. herbaceum var. africanum A1) were indispensable for the present purpose. In addition, unreliable phylogeny of G. barbadense AD2 was found based on published datasets, so G. barbadense AD2 cultivar ‘Zhonghai 7’ (The inbred line originated from G. barbadense AD2 cultivar ‘Xinhai 7’ bred in Xinjiang, China) was adopted to represent the important clade of allopolyploid cottons.
All the materials were collected from Cotton Research Institute, Chinese Academy of Agricultural Sciences, Anyang, Henan, China.
Chloroplast genome sequencing
CpDNA was prepared from fresh leaves following a previous published protocol [51]. CpDNA of G. raimondii D5 was sheared into random fragments of 35 kb to 50 kb, which were subsequently cloned into a pCC1FOS vector to generate a Fosmid library. Using 6 cpDNA markers (Table S10), 6 clones were screened accounting for the whole genome; 4 of 6 clones were then shotgun sequenced using an ABI 3730 sequencer (Applied Biosystems). Sequence coverage of each clone was more than 6-fold. The other 2 clones were sequenced using a general PCR method. The primers were designed according to chloroplast sequences of G. hirsutum AD1 [21].
The cpDNA of another 11 cotton species or races were sequenced with Solexa sequencing technology [52]. Each inverted repeat (IR) region was identified by 2 long PCR products (Table S10), each approximately 13 kb in length, and was purified for sequencing separately with Solexa.
Sequences were assembled with programs Phrap (http://www.phrap.com) and Velvet (hash length = 21, cov_cutoff = 30) [53]. Chloroplast genes were annotated using an online DOGMA tool (http://dogma.ccbb.utexas.edu/) [54] with G. hirsutum AD1 as a guide. Genome maps were drawn with OGDRAW [55].
Sequence divergence analyses
The chloroplast genome sequences of 12 Gossypium species or races were assembled by the authors, and those of G. hirsutum AD1 and G. barbadense 2006 AD2 were downloaded from GenBank, however the G. barbadense AD2 chloroplast genome was analyzed mainly based on our re-sequencing data. Sequence alignment including coding sequences, introns, and intergenic spacers were carried out by CLUSTALW. The chloroplast simple sequence repeats (SSRs) were searched by SSR Extractor under the repeat length definition ≥8 bp (http://www.aridolan.com/ssr/ssr.aspx).
The transfer of ycf2 to nuclear genome
For detecting the transfer of cpDNA to nuclear genome, we aligned the sequence of cpDNA across the genus using sliding sequence with a step size of 1000 bp. Finally, a ycf2 fragment was found in a bacterial artificial chromosome (BAC) of G. hirsutum AD1 [44]. We further investigated the transfer of ycf2 fragment by PCR with the following primer pairs: L2-sense (ACCAAAGATGAGCATAGCG); L2-antisense (GAATGGGACGAGAATACCGAA); ycf2-sense (CGTTTGTCTTTACCGCATTC); ycf2-antisense (CCCTACTGAGAGGTCCACTA). The primer L2 was designed to target the nuclear genome flanking the ycf2 insertion while primer ycf2 were used to verify the ycf2 gene.
Phylogenetic analyses and dating of Gossypium
The polymorphic SSRs were concatenated to construct a phylogenetic tree with the maximum-likelihood (ML) method. Likelihood analysis was performed in PhyML 3.0 [56] using the HKY model. Support values for nodes on the ML tree were estimated with 100 bootstrap replicates.
For validation of the phylogenetic relationships of Gossypium allotetraploids, phylogenetic analyses were performed under maximum-likelihood and Bayesian method using the following data respectively: variable protein-coding genes sequences, intron sequences and variable intergenic sequences. We also constructed a phylogeny of Gossypium using the variable sequences. To choose an appropriate outgroup for the phylogenetic analysis, the complete chloroplast genome sequence of Gossypium hirsutum was queried using BLAST against the publicly available databases. It showed the highest similarity to the whole-genome sequence of the chloroplast Theobroma cacao, another member of Malvaceae [57]. Thus Theobroma cacao was chosen as the outgroup for our analysis. Bayesian phylogenetic analyses were performed using MrBayes program [58]. The best-fitting model of nucleotide substitution was determined using both the Akaike information criterion (AIC) and the hierarchical likelihood ratio tests (hLRT) implemented in the program Modeltest 3.7 [59]. Both the AIC and hLRT suggested that the GTR + inv + gamma model was the most appropriate for our data. The Markov chain Monte Carlo (MCMC) algorithm ran for 200,000 generations and the trees sampled each 10 generations for data partition. The first 25% of trees were discarded and the remaining trees were used to construct majority-rule consensus tree for inferring Bayesian posterior probabilities of nodal supports. In all analyses, Theobroma cacao was set as outgroup. The Bayesian method was performed with the above-mentioned model. And maximum-likelihood (ML) trees were constructed in MEGA5 program [60] using a General Time Reversible (GTR) model and a rate of Gamma distributed with invariant site (G+I). Sixty-one concatenated protein-coding genes from the 13 taxa (Table S11), including 1 Ranunculaceae species [47], 3 Brassicaceae species [61], and 2 diploid Gossypium species representing dicotyledons; 1 Acoraceae species [62], 1 Typhaceae species [63], and 2 Poaceae species [64], [65], representing monocotyledons; and 2 species of basal angiosperms [47], [66] and 1 gymnosperm [67] forming outgroup, were aligned one by one. The 61 common genes and the analyses of multiple sequences followed previous methods [27], [32]. The divergence time of Gossypium, represented by G. herbaceum var. africanum A1 and G. raimondii D5 due to their relative divergent relationships, was calculated. Based on the protein-coding sequences, the values of Ks and Ka were calculated with PAML 4 [68].
To evaluate the constant molecular clock model, 78 concatenated protein-coding sequences were tested for the maximum likelihood-based phylogeny under the General Time Reversible model using likelihood ratio test method implemented in the MEGA5 package [60].
The polarization of indels
Indels were polarized into insertions and deletions using Theobroma cacao as an outgroup. The indels that occurred on the D genome after the split of A and D could be divided into two stages. If 2 A genomes and 9 allotetraploids shared sequence (gap) with the outgroup, then the gap (sequence) the 2 D genomes shared was considered as a deletion (insertion) within D genomes after the split of A genome and D genome and before the split of 2 D genomes, which was defined as the first stage of D-genome divergence. If only one of D genomes had the gap (sequence), then the deletion (insertion) happened after the split of two D genomes, which was defined as the second stage of D-genome divergence.
The indels that arose on the A genome after the divergence of A and D could be divided into three stages. If 2 D genomes had the same sequence (gap) as the outgroup, and the 2 A genomes and 9 allotetraploid genomes shared gap (sequence), then the shared gap (sequence) was considered as a deletion (insertion) in A genome after the divergence of A and D and before the formation of allotetraploids, which was defined as the first stage of A-genome divergence. Conversely, If 2 D genomes and 9 allotetraploid genomes shared sequence with the outgroup, then the shared gap (sequence) by 2 A genomes was considered as a deletion (insertion) in A genomes after the formation of allotetraploid and before the split of 2 A genomes, which was defined as the second stage of A-genome divergence. If only one of A genome existed the gap (sequence), then the deletion (insertion) happened after the split of two A genomes, which was defined as the third stage of A-genome divergence.
The indels in allotetraploids could be divided into two stages. If D genomes and A genomes shared sequence (gap) with the outgroup, then the shared gap (sequence) by allotetraploid genomes was considered as a deletion (insertion) at the first stage. If A genomes, D genomes and partial allotetraploids shared sequence (gap) with outgroup, then the gap (sequence) in other allotetraploids was considered as a deletion (insertion) at the second stage.
Supporting Information
Phylogenetic tree of 13 Gossypium species based on polymorphic SSRs. The polymorphic SSR loci were used to construct phylogenetic trees using maximum-likelihood method.
(TIFF)
The topologies of Gossypium allotetraploids. The 3 groups of sequences were used under 2 independent models (Maximum-likelihood model and Bayesian model), respectively. A: 39 variable protein-coding genes using maximum-likelihood method; B: 16 intron sequences using maximum-likelihood method; C: 92 variable intergenic sequences using maximum-likelihood method; D: 39 variable Protein coding genes using Bayesian method; E: 16 intron sequences using Bayesian method; F: 92 variable intergenic sequences using Bayesian method. Numbers above nodes were maximum likelihood bootstrap under Maximum-likelihood model or Bayesian inference posterior probability under Bayesian model.
(TIFF)
Phylogeny of 13 taxa. The common genes of 13 taxa were used to reconstruct the phylogeny. The number on each branch was the nucleotide substitutions per 100 sites.
(TIFF)
List of conserved and variable genes in Gossypium chloroplast genomes.
(DOC)
The AT content (%) of different partitions in 13 chloroplast genomes.
(DOC)
The Si/Sv values and total length of indels between genomes.
(DOC)
The substitutions and indel number in LSC, SSC and IR regions among 13 Gossypium chloroplast genomes.
(DOC)
The number of polymorphic SSRs between any two of 13 Gossypium chloroplast genomes.
(DOC)
The substitution number of protein-coding genes between any two of 13 Gossypium chloroplast genomes.
(DOC)
Substitutions and indels of intergenic spacers between any two of 13 Gossypium chloroplast genomes.
(DOC)
The number of indels in different length of chloroplast genomes.
(DOC)
The estimated indel ratio between Gossypium chloroplast genomes.
(DOC)
Primers employed for sequencing.
(DOC)
Chloroplast genomes of species adopted in current study.
(DOC)
Acknowledgments
We thank Dr James Doughty and Dr. Baoxiu Qi at University of Bath, UK for helpful critical reading of the manuscript and revising work, Dr. Ziding Zhang at China Agricultural University for providing computing platform, and Dr. Dingming Kang at China Agricultural University for helpful suggestions. We are also grateful to two anonymous reviewers for their helpful suggestions and comments.
Funding Statement
This work was supported by Training Program Foundation for the New Century Excellent Talents in University (NCET-06-0106) and National Natural Science Foundation of China (Grant No. 31171591) to J. Hua. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Soltis D, Soltis P, Tate J (2004) Advances in the study of polyploidy since plant speciation. New Phytol 161: 173–191. [Google Scholar]
- 2.Fryxell P (1979) The natural history of the cotton tribe: Texas A and M University Press. [Google Scholar]
- 3.Wendel JF, Brubaker CL, Seelanan T (2010) The origin and evolution of Gossypium. In: Stewat J, Oosterhuis D, Heitholt JJ, Mauney J, editors. Physiology of cotton. Springer. 1–18. [Google Scholar]
- 4. Wendel JF, Cronn RC (2003) Polyploidy and the evolutionary history of cotton. Adv Agron 78: 139–186. [Google Scholar]
- 5. Flagel LE, Wendel JF (2010) Evolutionary rate variation, genomic dominance and duplicate gene expression evolution during allotetraploid cotton speciation. New Phytol 186: 184–193. [DOI] [PubMed] [Google Scholar]
- 6. Wendel JF (1989) New World tetraploid cottons contain Old World cytoplasm. Proc Natl Acad Sci U S A 86: 4132–4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Brubaker CL, Paterson AH, Wendel JF (1999) Comparative genetic mapping of allotetraploid cotton and its diploid progenitors. Genome 42: 184–203. [Google Scholar]
- 8. Rong J, Abbey C, Bowers JE, Brubaker CL, Chang C, et al. (2004) A 3347-locus genetic recombination map of sequence-tagged sites reveals features of genome organization, transmission and evolution of cotton (Gossypium). Genetics 166: 389–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Grover CE, Kim HR, Wing RA, Paterson AH, Wendel JF (2007) Microcolinearity and genome evolution in the AdhA region of diploid and polyploid cotton (Gossypium). Plant J 50: 995–1006. [DOI] [PubMed] [Google Scholar]
- 10. Small RL, Ryburn JA, Cronn RC, Seelanan T, Wendel JF (1998) The tortoise and the hare: choosing between noncoding plastome and nuclear Adh sequences for phylogeny reconstruction in a recently diverged plant group. Am J Bot 85: 1301–1315. [PubMed] [Google Scholar]
- 11. Cronn RC, Small RL, Wendel JF (1999) Duplicated genes evolve independently after polyploid formation in cotton. Proc Natl Acad Sci U S A 96: 14406–14411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Endrizzi JE, Turcotte EL, Kohel RJ (1985) Genetics, cytology, and evolution of Gossypium . Adv Genet 23: 271–375. [Google Scholar]
- 13. Seelanan T, Schnabel A, Wendel JF (1997) Congruence and consensus in the cotton tribe (Malvaceae). Syst Bot 22: 259–290. [Google Scholar]
- 14. Cronn RC, Small RL, Haselkorn T, Wendel JF (2002) Rapid diversification of the cotton genus (Gossypium: Malvaceae) revealed by analysis of sixteen nuclear and chloroplast genes. Am J Bot 89: 707–725. [DOI] [PubMed] [Google Scholar]
- 15. Senchina DS, Alvarez I, Cronn RC, Liu B, Rong J, et al. (2003) Rate variation among nuclear genes and the age of polyploidy in Gossypium . Mol Biol Evol 20: 633–643. [DOI] [PubMed] [Google Scholar]
- 16. Grover CE, Yu Y, Wing RA, Paterson AH, Wendel JF (2008) A phylogenetic analysis of indel dynamics in the cotton genus. Mol Biol Evol 25: 1415–1428. [DOI] [PubMed] [Google Scholar]
- 17. Turmel M, Gagnon MC, O'Kelly CJ, Otis C, Lemieux C (2009) The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol Biol Evol 26: 631–648. [DOI] [PubMed] [Google Scholar]
- 18. Turmel M, Otis C, Lemieux C (2009) The chloroplast genomes of the green algae Pedinomonas minor, Parachlorella kessleri, and Oocystis solitaria reveal a shared ancestry between the Pedinomonadales and Chlorellales. Mol Biol Evol 26: 2317–2331. [DOI] [PubMed] [Google Scholar]
- 19. Chan CX, Gross J, Yoon HS, Bhattacharya D (2011) Plastid origin and evolution: new models provide insights into old problems. Plant Physiol 155: 1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Greiner S, Rauwolf U, Meurer J, Herrmann RG (2011) The role of plastids in plant speciation. Mol Ecol 20: 671–691. [DOI] [PubMed] [Google Scholar]
- 21. Lee SB, Kaittanis C, Jansen RK, Hostetler JB, Tallon LJ, et al. (2006) The complete chloroplast genome sequence of Gossypium hirsutum: organization and phylogenetic relationships to other angiosperms. BMC Genomics 7: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ibrahim RIH, Azuma JI, Sakamoto M (2006) Complete nucleotide sequence of the cotton (Gossypium barbadense L.) chloroplast genome with a comparative analysis of sequences among 9 dicot plants. Genes Genet Syst 81: 311–321. [DOI] [PubMed] [Google Scholar]
- 23. Wu FH, Kan DP, Lee SB, Daniell H, Lee YW, et al. (2009) Complete nucleotide sequence of Dendrocalamus latiflorus and Bambusa oldhamii chloroplast genomes. Tree Physiol 29: 847–856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chen JQ, Wu Y, Yang H, Bergelson J, Kreitman M, et al. (2009) Variation in the ratio of nucleotide substitution and indel rates across genomes in mammals and bacteria. Mol Biol Evol 26: 1523–1531. [DOI] [PubMed] [Google Scholar]
- 25. Matsuoka Y, Yamazaki Y, Ogihara Y, Tsunewaki K (2002) Whole chloroplast genome comparison of rice, maize, and wheat: implications for chloroplast gene diversification and phylogeny of cereals. Mol Biol Evol 19: 2084–2091. [DOI] [PubMed] [Google Scholar]
- 26. Guisinger MM, Kuehl JV, Boore JL, Jansen RK (2008) Genome-wide analyses of Geraniaceae plastid DNA reveal unprecedented patterns of increased nucleotide substitutions. Proc Natl Acad Sci U S A 105: 18424–18429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greiner S, Wang X, Rauwolf U, Silber MV, Mayer K, et al. (2008) The complete nucleotide sequences of the five genetically distinct plastid genomes of Oenothera, subsection Oenothera: I. Sequence evaluation and plastome evolution. Nucleic Acids Res 36: 2366–2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eric V, Bruce L (2007) Uncovering the mutation-fixation correlation in short lineages. BMC Evol Biol 7: 168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Stoletzki N, Eyre-Walker A (2011) The positive correlation between dN/dS and dS in mammals is due to runs of adjacent substitutions. Mol Biol Evol 28: 1371–1380. [DOI] [PubMed] [Google Scholar]
- 30. Greiner S, Wang X, Herrmann RG, Rauwolf U, Mayer K, et al. (2008) The complete nucleotide sequences of the 5 genetically distinct plastid genomes of Oenothera, subsection Oenothera: II. A microevolutionary view using bioinformatics and formal genetic data. Mol Biol Evol 25: 2019–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wendel JF, Rowley R, Stewart J (1994) Genetic diversity in and phylogenetic relationships of the Brazilian endemic cotton, Gossypium mustelinum (Malvaceae). Plant Syst Evol 192: 49–59. [Google Scholar]
- 32. Chaw SM, Chang CC, Chen HL, Li WH (2004) Dating the monocot-dicot divergence and the origin of core eudicots using whole chloroplast genomes. J Mol Evol 58: 424–441. [DOI] [PubMed] [Google Scholar]
- 33. Leebens-Mack J, Raubeson L, Cui L, Kuehl J, Fourcade M, et al. (2005) Identifying the basal angiosperm node in chloroplast genome phylogenies: sampling one's way out of the Felsenstein zone. Mol Biol Evol 22: 1948–1963. [DOI] [PubMed] [Google Scholar]
- 34. Bell CD, Soltis DE, Soltis PS (2010) The age and diversification of the angiosperms re-revisited. Am J Bot 97: 1296–1303. [DOI] [PubMed] [Google Scholar]
- 35. Leseberg CH, Duvall MR (2009) The complete chloroplast genome of Coix lacryma-jobi and a comparative molecular evolutionary analysis of plastomes in cereals. J Mol Evol 69: 311–318. [DOI] [PubMed] [Google Scholar]
- 36. Britten RJ, Rowen L, Williams J, Cameron RA (2003) Majority of divergence between closely related DNA samples is due to indels. Proc Natl Acad Sci U S A 100: 4661–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gu X, Li WH (1995) The size distribution of insertions and deletions in human and rodent pseudogenes suggests the logarithmic gap penalty for sequence alignment. J Mol Evol 40: 464–473. [DOI] [PubMed] [Google Scholar]
- 38. Yamane K, Yano K, Kawahara T (2006) Pattern and rate of indel evolution inferred from whole chloroplast intergenic regions in sugarcane, maize and rice. DNA Res 13: 197–204. [DOI] [PubMed] [Google Scholar]
- 39. McCluskey K, Wiest AE, Grigoriev IV, Lipzen A, Martin J, et al. (2011) Rediscovery by whole genome sequencing: classical mutations and genome polymorphisms in Neurospora crassa . G3: Genes, Genomes, Genetics 1: 303–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ma J, Bennetzen JL (2004) Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A 101: 12404–12410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baldauf SL, Palmer JD (1990) Evolutionary transfer of the chloroplast tufA gene to the nucleus. Nature 344: 262–265. [DOI] [PubMed] [Google Scholar]
- 42. Sugiura C, Kobayashi Y, Aoki S, Sugita C, Sugita M (2003) Complete chloroplast DNA sequence of the moss Physcomitrella patens: evidence for the loss and relocation of rpoA from the chloroplast to the nucleus. Nucleic Acids Res 31: 5324–5331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Jansen RK, Saski C, Lee SB, Hansen AK, Daniell H (2011) Complete plastid genome sequences of three rosids (Castanea, Prunus, Theobroma): evidence for at least two independent transfers of rpl22 to the nucleus. Mol Biol Evol 28: 835–847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Grover CE, Kim HR, Wing RA, Paterson AH, Wendel JF (2004) Incongruent patterns of local and global genome size evolution in cotton. Genome Res 14: 1474–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Heads M (2005) Dating nodes on molecular phylogenies: a critique of molecular biogeography. Cladistics 21: 62–78. [DOI] [PubMed] [Google Scholar]
- 46. Li WH, Tanimura M (1987) The molecular clock runs more slowly in man than in apes and monkeys. Nature 326: 93–96. [DOI] [PubMed] [Google Scholar]
- 47. Raubeson LA, Peery R, Chumley TW, Dziubek C, Fourcade HM, et al. (2007) Comparative chloroplast genomics: analyses including new sequences from the angiosperms Nuphar advena and Ranunculus macranthus . BMC Genomics 8: 174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Muse SV (2000) Examining rates and patterns of nucleotide substitution in plants. Plant Mol Biol 42: 25–43. [PubMed] [Google Scholar]
- 49.Gaut BS (1998) Molecular clocks and nucleotide substitution rates in higher plants. In: al HMKe, editor. Evolutionary biology. New York: Plenum Press. 93–120. [Google Scholar]
- 50. Tajima F (1993) Simple methods for testing the molecular evolutionary clock hypothesis. Genetics 135: 599–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Gong X, Yan L (1991) Improvement of the purification of chloroplast DNA from higher plants. Chinese Sci Bull 36: 467–469. [Google Scholar]
- 52. Cronn R, Liston A, Parks M, Gernandt DS, Shen R, et al. (2008) Multiplex sequencing of plant chloroplast genomes using Solexa sequencing-by-synthesis technology. Nucleic Acids Res 36: e122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wyman SK, Jansen RK, Boore JL (2004) Automatic annotation of organellar genomes with DOGMA. Bioinformatics 20: 3252–3255. [DOI] [PubMed] [Google Scholar]
- 55. Lohse M, Drechsel O, Bock R (2007) OrganellarGenomeDRAW (OGDRAW): a tool for the easy generation of high-quality custom graphical maps of plastid and mitochondrial genomes. Curr Genet 52: 267–274. [DOI] [PubMed] [Google Scholar]
- 56. Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59: 307–321. [DOI] [PubMed] [Google Scholar]
- 57. Argout X, Salse J, Aury JM, Guiltinan MJ, Droc G, et al. (2010) The genome of Theobroma cacao . Nature genetics 43: 101–108. [DOI] [PubMed] [Google Scholar]
- 58. Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574. [DOI] [PubMed] [Google Scholar]
- 59. Posada D, Buckley TR (2004) Model selection and model averaging in phylogenetics: advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol 53: 793–808. [DOI] [PubMed] [Google Scholar]
- 60. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sato S, Nakamura Y, Kaneko T, Asamizu E, Tabata S (1999) Complete structure of the chloroplast genome of Arabidopsis thaliana . DNA Res 6: 283–290. [DOI] [PubMed] [Google Scholar]
- 62. Goremykin VV, Holland B, Hirsch-Ernst KI, Hellwig FH (2005) Analysis of Acorus calamus chloroplast genome and its phylogenetic implications. Mol Biol Evol 22: 1813–1822. [DOI] [PubMed] [Google Scholar]
- 63. Guisinger MM, Chumley TW, Kuehl JV, Boore JL, Jansen RK (2010) Implications of the plastid genome sequence of Typha (Typhaceae, Poales) for understanding genome evolution in Poaceae. J Mol Evol 70: 149–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Maier RM, Neckermann K, Igloi GL, Kossel H (1995) Complete sequence of the maize chloroplast genome: gene content, hotspots of divergence and fine tuning of genetic information by transcript editing. J Mol Evol 251: 614–628. [DOI] [PubMed] [Google Scholar]
- 65. Ogihara Y, Isono K, Kojima T, Endo A, Hanaoka M, et al. (2002) Structural features of a wheat plastome as revealed by complete sequencing of chloroplast DNA. Mol Genet Genomics 266: 740–746. [DOI] [PubMed] [Google Scholar]
- 66. Goremykin VV, Hirsch-Ernst KI, W lfl S, Hellwig FH (2003) Analysis of the Amborella trichopoda chloroplast genome sequence suggests that Amborella is not a basal angiosperm. Mol Biol Evol 20: 1499–1505. [DOI] [PubMed] [Google Scholar]
- 67. Wakasugi T, Tsudzuki J, Ito S, Nakashima K, Tsudzuki T, et al. (1994) Loss of all ndh genes as determined by sequencing the entire chloroplast genome of the black pine Pinus thunbergii . Proc Natl Acad Sci U S A 91: 9794–9798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 69. Yang Z, Nielsen R (2000) Estimating synonymous and nonsynonymous substitution rates under realistic evolutionary models. Mol Biol Evol 17: 32–43. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic tree of 13 Gossypium species based on polymorphic SSRs. The polymorphic SSR loci were used to construct phylogenetic trees using maximum-likelihood method.
(TIFF)
The topologies of Gossypium allotetraploids. The 3 groups of sequences were used under 2 independent models (Maximum-likelihood model and Bayesian model), respectively. A: 39 variable protein-coding genes using maximum-likelihood method; B: 16 intron sequences using maximum-likelihood method; C: 92 variable intergenic sequences using maximum-likelihood method; D: 39 variable Protein coding genes using Bayesian method; E: 16 intron sequences using Bayesian method; F: 92 variable intergenic sequences using Bayesian method. Numbers above nodes were maximum likelihood bootstrap under Maximum-likelihood model or Bayesian inference posterior probability under Bayesian model.
(TIFF)
Phylogeny of 13 taxa. The common genes of 13 taxa were used to reconstruct the phylogeny. The number on each branch was the nucleotide substitutions per 100 sites.
(TIFF)
List of conserved and variable genes in Gossypium chloroplast genomes.
(DOC)
The AT content (%) of different partitions in 13 chloroplast genomes.
(DOC)
The Si/Sv values and total length of indels between genomes.
(DOC)
The substitutions and indel number in LSC, SSC and IR regions among 13 Gossypium chloroplast genomes.
(DOC)
The number of polymorphic SSRs between any two of 13 Gossypium chloroplast genomes.
(DOC)
The substitution number of protein-coding genes between any two of 13 Gossypium chloroplast genomes.
(DOC)
Substitutions and indels of intergenic spacers between any two of 13 Gossypium chloroplast genomes.
(DOC)
The number of indels in different length of chloroplast genomes.
(DOC)
The estimated indel ratio between Gossypium chloroplast genomes.
(DOC)
Primers employed for sequencing.
(DOC)
Chloroplast genomes of species adopted in current study.
(DOC)