Abstract
The pond snail Lymnaea stagnalis is among several mollusc species that have been well investigated due to the simplicity of their nervous systems and large identifiable neurons. Nonetheless, despite the continued attention given to the physiological characteristics of its nervous system, the genetic information of the Lymnaea central nervous system (CNS) has not yet been fully explored. The absence of genetic information is a large disadvantage for transcriptome sequencing because it makes transcriptome assembly difficult. We here performed transcriptome sequencing for Lymnaea CNS using an Illumina Genome Analyzer IIx platform and obtained 81.9 M of 100 base pair (bp) single end reads. For de novo assembly, five programs were used: ABySS, Velvet, OASES, Trinity and Rnnotator. Based on a comparison of the assemblies, we chose the Rnnotator dataset for the following blast searches and gene ontology analyses. The present dataset, 116,355 contigs of Lymnaea transcriptome shotgun assembly (TSA), contained longer sequences and was much larger compared to the previously reported Lymnaea expression sequence tag (EST) established by classical Sanger sequencing. The TSA sequences were subjected to blast analyses against several protein databases and Aplysia EST data. The results demonstrated that about 20,000 sequences had significant similarity to the reported sequences using a cutoff value of 1e-6, and showed the lack of molluscan sequences in the public databases. The richness of the present TSA data allowed us to identify a large number of new transcripts in Lymnaea and molluscan species.
Introduction
The pond snail Lymnaea stagnalis has large identifiable neurons and a simple central nervous system (CNS). Many researchers have therefore used this animal model to investigate the cellular and molecular mechanisms related to various behaviors, such as respiration, feeding, learning and memory [1]–[3]. Despite the continued attention given to the physiological characteristics of the identified neurons, the genetic information of Lymnaea has not yet been fully explored. Thus, for molecular biological investigations, researchers have made numerous efforts to identify new genes before studying their function [4]–[6]. Previously, two Lymnaea expression sequence tag (EST) databases were established by classical Sanger sequencing [7], [8]. Nevertheless, they are still insufficient to perform transcriptome analysis and improved transcriptome data is continuously needed.
A recently developed technology, deep RNA sequencing (RNA-seq), produces at least 100 to 1,000 times higher throughput than classical Sanger sequencing [9]. Commercially available RNA-seq technologies, such as the Illumina Genome Analyzer and Roche Genome Sequencer FLX system (GS FLX), are now widely applied for RNA and genome sequencing, and the features of several RNA-seq platforms have been well defined [10], [11]. Simply put, GS FLX provides longer reads and has been reported to enable more efficient assembly compared to the Illumina sequencer. By contrast, the Illumina sequencer was reported to produce large amounts of data with short reads at a lower cost. More recent studies have reported that longer read data (>75 bp) can be obtained using an Illumina sequencer and the developed assembly programs enable researchers to perform de novo transcriptome sequencing at a low cost [12], [13]. In this study, we examined the RNA-seq method and several de novo assembly programs for their ability to provide transcriptome data of Lymnaea CNS using an Illumina sequencer.
Materials and Methods
Snails
Specimens of Lymnaea stagnalis with a 25 mm shell were maintained in tap water and fed on Komatsuna, Japanese mustard spinach, on a 12∶12 light-dark cycle at 20°C. Snails were anesthetized by 25% Listerine® before dissection. For RNA extraction, the isolated CNSs were frozen in liquid nitrogen.
RNA Extraction and Transcriptome Sequencing
The Lymnaea CNSs were dissected and immediately frozen in liquid nitrogen. RNA was extracted from 80 dissected CNSs using a FastPure RNA kit (Takara Bio, Shiga, Japan) and treated with DNase I (Takara). The quality and quantity were assessed on a 2100 Bioanalyzer using an RNA 6000 Nano kit (Agilent Technologies, Palo Alto, CA). After the measurement of RNA concentration, equal volumes of total RNA from each group were pooled and used for library preparation. Libraries were prepared using a TruSeq™ RNA sample preparation kit with a Low-Throughput protocol (Illumina Inc., San Diego, CA) according to the manufacturer’s instructions. The DNA concentration of the cDNA library was measured using a 2100 Bioanalyzer using a DNA 1000 kit (Agilent Technologies) and diluted to 4 nM, and a 120 µl aliquot was used to generate clusters on a Single-Read flow cell using the cBOT (Illumina) and sequenced on the GAIIx using the SBS 36-cycle Sequencing Kit v5. One lane for each group was sequenced as 100-bp reads, and image analysis and base calling were performed with SCS2.8/RTA1.8 (Illumina). FASTQ file generation and the removal of failed reads were performed by CASAVA ver.1.8.2 (Illumina).
Application of Published de novo Methods
Reads obtained using an Illumina sequencer were de novo assembled using ABySS ver. 1.3.2 [14], Velvet ver. 1.1.07 [15], OASES ver. 0.2.01 [16], Trinity 2011-08-20 [17], [18] or Rnnotator ver. 2.4.12 [19]. ABySS and Velvet were run using different k-mer lengths of 31 to 95 along with other default parameters. OASES was run using k-mers of 31 to 95 followed by merging the results by running the first stage of Velvet analysis. Trinity and Rnnotator were run with the default parameters.
BLAST Annotation
With the dataset of 82,127 contigs (>200 bp) assembled by Rnnotator, the BLAST program [20] was performed for the blastx homology search among the databases: Swiss Protein (Swissprot Release_12_2011), the NCBI Protein Reference sequences (Refseq, Release_14_01_2012) and the Invertebrate Protein Reference sequences (Invertebrate Refseq, Relase50_ 08_11_2011), respectively. For similarity searches of the molluscan ESTs, the available Lymnaea stagnalis and Aplysia californica EST sequences were downloaded from GenBank [8], [21]. The present contigs identified by Rnnotator (116,355 contigs, >100 bp) and the previous Lymnaea EST dataset (11,697 sequences) were used for blastn search bidirectionally with an e-value cutoff of 1e-100. For comparison with Aplysia ESTs, the contigs identified by Rnnotator (116,355 contigs, >100 bp) were used to perform blastn and tblastx homology searches in the Aplysia EST database (255,605 sequences) with an e-value cutoff of 1e-6.
Gene Ontology Annotation
Using the 82,127 sequences of Rnnotator assembled contigs (>200 bp), assignment of gene ontology (GO) terms was performed by importing the GO-numbers obtained by a blastx search against the Swissprot database into BLAST2GO (version 2.5.0; http://www.blast2go.org/) [22]. Categorization of the BLAST matches and construction of column bar graphs was conducted using the standard graph configurations including level two GO-terms only.
The Phylogenetic Analyses of Newly Identified Proteins
The computer program MEGA4 [23] was used to construct a neighbor-joining phylogeny from Poisson amino acid distances. Bootstrap values are given for each branch. The accession numbers for the sequences of tyrosine hydroxylase included in the tree are as follows: Mus musculus: NP_033403; Homo sapiens: AAI04968; Bos taurus: NP_776309; Gallus gallus: NP_990136; Xenopus laevis: NP_001091392; Danio rerio: NP_571224; Branchiostoma floridae: AAZ32939; Strongylocentrotus purpuratus: XP_786206; Pediculus humanus corporis: XP_002430839; Apis mellifera: NP_001011633; Bombus impatiens: XP_003486645; Tribolium castaneum: NP_001092299; Bombyx mori: ADV56714; Manduca sexta: ABQ95973; Caenorhabditis elegans: P90986. The accession numbers for the sequences of dopa decarboxylase included in the tree are as follows: Homo sapiens: NP_000781; Bos taurus: NP_776332; Mus musculus: NP_057881; Monodelphis dome: XP_001379620; Anolis carolinensis: XP_003222345; Meleagris gallopavo: XP_003204897; Gallus gallus: XP_419032; Xenopus laevis: NP_001104211; Danio rerio: NP_998507; Pediculus humanus corporis: XP_002426339; Bombus terrestrius: XP_003399661; Bombyx mori: NP_001037174; Aedes aegypti: XP_001648263; Apis mellifera: XP_394115; Drosophila melanogaster: P05031; Caenorhabditis elegans tyrosine decarboxylase: NP_495744. The accession numbers for the sequences of dopamine beta-hydroxylase included in the tree are as follows: Bos taurus: NP_851338; Homo sapiens: NP_000778; Mus musculus: NP_620392; Ornithorhynchus anatinus: XP_001505587; Gallus gallus: XP_415429; Xenopus tropicalis: XP_002942324; Danio rerio: NP_001103164; Dugesia japonica: BAG86630; Aedes aegypti: XP_001661114; Drosophila melanogaster: NP_788884; Bombyx mori: NP_001243923; Acyrthosiphon pisum: XP_001951145; Pediculus humanus corporis: XP_002425605; Tribolium castaneum: XP_974169; Nasonia vitripennis: XP_001602880; Apis mellifera: NP_001071292; Caenorhabditis elegans: Q9XTQ6.
Results
Lymnaea CNS Transcriptome Assembly
The messenger RNA samples were prepared from pooled Lymnaea CNSs of 80 animals, and sequencing was performed in two lanes of a flow cell using an Illumina Genome Analyzer IIx. To obtain better assembly for Lymnaea CNS transcriptome, we here examined five de novo assembly tools, ABySS [14], Velvet [15], OASES [16], Trinity [17], [18] and Rnnotator [19].
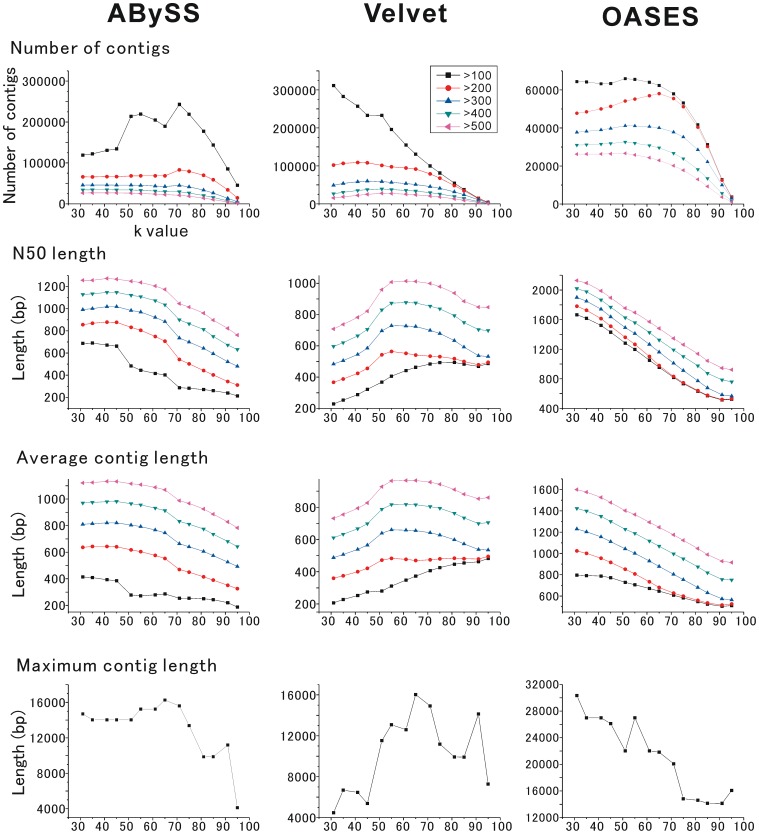
First, to choose the best k-mer length for de novo assembly, we applied different k-mer lengths for ABySS, Velvet and OASES. We first used a dataset from one lane with 41 M reads to save the memory usage. The results with distinct k-mer values were compared using the performance criteria, contig number, N50 length, average contig length and maximum contig length. The N50 length was calculated by adding the lengths of contigs from long to short until the summed length exceeded 50% of the total length of all contigs. It is generally accepted that larger values of these criteria indicate better assembly performance. The result showed that the values were extensively varied according to the length of k-mer in each assembly (Fig. 1), and the data of contigs longer than 200 bp were chosen to assess the assembly quality. This was because the results of the three programs consistently revealed that the data including short contigs (>100 bp) yielded less accurate results than the data with longer contigs. For example, the trial using the program ABySS (Fig. 1, left column) revealed that the criteria values obtained using short contigs (>100 bp, black lines) largely differed from those obtained using long contigs (>200 bp, colored lines).
Figure 1. Comparison of de novo assembly with a distinct k-value.
Three assembly programs, ABySS, Velvet and OASES, were tested with a distinct k-mer from 31 to 95. In each assembly program, the number of contigs, the N50 length, and the average and maximum contig length were calculated using the assembled contigs longer than 100 bp (black), 200 bp (orange), 300 bp (blue), 400 bp (green) and 500 bp (pink).
In the result of ABySS assembly, with a k-mer length of 65, the criteria values of the number of contigs, N50 length and average contig length seemed to plateau, and the maximum contig length was longest. Velvet assembly produced the best values of N50 length, average contig length and maximum contig length, with a k-mer length of 55. In the case of OASES, the data included a large number of variants, and we used the dataset with the longest variants. The results clearly demonstrated that the criteria values were better with a shorter length of k-mer for OASES, and seemed to plateau with a k-mer length of 31. According to the obtained results, the best k-mer lengths were determined as 65 for ABySS, 55 for Velvet and 31 for OASES, respectively.
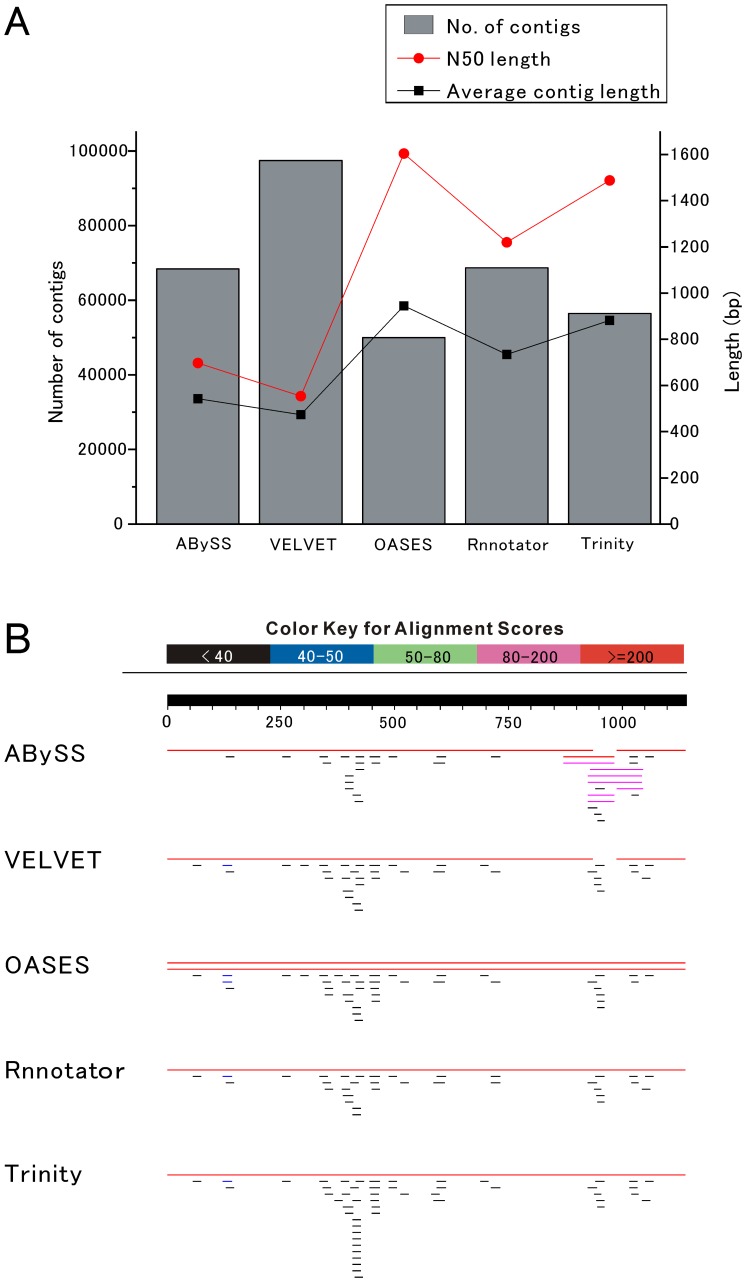
We then performed assemblies using each of the five programs and compared the results using the contigs longer than 200 bp (Fig. 2A). Because splicing variants were included in the results of the OASES, Trinity and Rnnotator assemblies, the longest variant for a singular transcript was extracted and used as the data for the comparison. As a result, the assemblies of the OASES, Trinity and Rnnotator programs appeared to be better than those of ABySS and Velvet. The N50 lengths in the ABySS and Velvet data were obviously shorter than those yielded by the programs OASES, Trinity and Rnnotator (707 for ABySS; 564 for Velvet; 1,614 for OASES; 1,230 for Rnnotator; 1,497 for Trinity) and the values of average contig length showed a similar tendency (553 for ABySS; 483 for Velvet; 955 for OASES; 744 for Rnnotator; 892 for Trinity). In contrast, Velvet produced the largest number of contigs among the programs, and ABySS also produced a larger contig number than OASES and Trinity (68,378 for ABySS; 97,492 for Velvet; 50,021 for OASES; 68,734 for Rnnotator; 56,446 for Trinity). So far, these results indicated that ABySS and Velvet produced much greater numbers of shorter contigs than OASES, Trinity and Rnnotator.
Figure 2. Comparison of de novo assembly quality among the different programs.
(A) Overall comparison of the results from five assembly programs. The bars indicate the number of contigs longer than 200 bp (left axis). The red lines indicate the N50 length and the black lines indicate average contig length in bp (right axis). (B) Blast requests of LymCREB2 CDS (1,140 bp) for differently assembled contigs. The black line above represents the query sequence and the colored lines below represent the results of the similarity of hit contigs in the databases. The alignment scores are indicated by five colors in the label at the top.
On the other hand, a previous report showed that the values of the contig number, N50 length and average contig length are not in themselves sufficient to assess the accuracy of assembly [24]. Therefore, BLAST methodology was additionally examined to check each contig length and the coverage of known sequences (Fig. 2B). We here applied a single open reading frame (ORF) of LymCREB2 [2], and observed that it was covered with a single contig in the datasets of OASES, Trinity and Rnnotator. By contrast, it was covered with 3 and 2 shorter contigs in the datasets of ABySS and Velvet, respectively. The sequence coverage with a known sequence also indicated that the ABySS and Velvet programs were inferior to OASES, Trinity and Rnnotator in assembly performance.
Among the programs OASES, Trinity and Rnnotator, the results so far do not show a consistent advantage or any significant superiority for any particular program. The contig number was the largest in the dataset of Rnnotator and the N50 lengths and average lengths of contigs were longer in the datasets of OASES and Trinity. However, it should be noted that the datasets of OASES, Trinity and Rnnotator include plural variants for a singular transcript and the numbers of variants in OASES and Trinity datasets were much larger than that of Rnnotator dataset (OASES: 54,601; Trinity: 44,062; Rnnotator: 2,675). For the following gene ontology (GO) analysis, a dataset including redundant sequences is not suitable. Therefore, the Rnnotator program was selected for assembly of the full dataset.
The dataset from two lanes of a flow cell (81.9 M reads) was then assembled with Rnnotator and the results are summarized in Table 1. The redundant read and low quality sequences were removed at the preprocessing step, and 20 M reads were used for assembly. As a result, we obtained 116,355 non-redundant sequences as a Lymnaea transcriptome shotgun assembly (TSA) and deposited the TSA data in DDBJ (the DNA Data Bank of Japan, accession number FX180119 to FX296473). The data had a total size of about 76.4 Mbp, N50 length of 1,438 bp, and average contig length of 656 bp. We further confirmed that the longest contig (26,147 bp, accession number: FX180119), coding a homologue of titin protein, had a large open reading frame of 25,191 bp without any frame shift or stop codon (data not shown), indicating that Rnnotator successfully assembled the present sequence data. Rnnotator assembly also produced the value of reads per kilo per million (rpkm) as an index of the expression level [19]. The present data showed a maximum rpkm value of 64707.57 and an average rpkm value of 8.59.
Table 1. The Lymnaea TSA obtained from Rnnotator assembly.
| Total number of reads | 81,851,004 |
| Number of used reads | 20,020,897 |
| Number of unused reads | 61,830,107 |
| Number of non-redundant transcripts | 116,355 |
| Number of transcript isoforms | 4,222 |
| Total size of transcriptome (bp) | 76,435,832 |
| N50 length (bp) | 1,438 |
| Average contig length (bp) | 656 |
| Largest contig length (bp) | 26,147 |
| Average coverage (rpkm) | 8.594 |
Gene Ontology Mapping
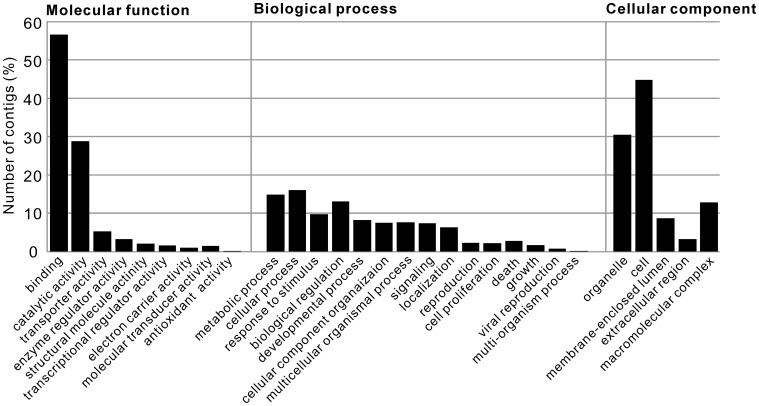
We then classified the Lymnaea TSA sequences associated with GO terms at the protein level annotation using BLAST2GO [25]. The 82,127 contigs longer than 200 bp were subjected to blastx analysis against the Swiss Protein (Swissprot) database using a cutoff e-value of 1e-6. As a result, 16,534 sequences (20.1%) had significant matches in the Swissprot database, and these were functionally annotated with GO terms by BLAST2GO. A domain analysis was also performed with InterProScan, and 1,454 sequences without balstx hits were further combined. Then, 16,002 sequences with annotated GO terms were remapped on a reduced set of GO categories (GO-slim) and classified into three categories: molecular function, biological process and cellular component. Figure 3 is the summary of the classification of annotated Lymnaea TSA sequences at level 2. In the molecular function classification, the clusters relating to “binding” and “catalytic activity” were enriched (56.7% and 28.8%, respectively). In the biological process classification, no cluster was specifically enriched. For the cellular component classification, the cluster sizes of “cell” (30.5%) and “organelle” (44.8%) were relatively large compared to those of “membrane-enclosed lumen,” “extracellular region” and “macromolecular complex.”
Figure 3. Gene ontology distribution for the Lymnaea TSA.
Gene ontology distribution of the Lymnaea TSA derived from BLAST2GO. The results are summarized as molecular functions, biological processes and cellular components. The x-axis represents the percentage of contigs divided by the total number of cells counted with the given level 2 GO terms.
Blast Search in Public Protein Databases
Next, the Lymnaea TSA (>200 bp) was subjected to blastx analysis against three different databases, Swissprot, NCBI Protein Reference sequences (Refseq) and Invertebrate Protein Reference sequences (Invertebrate Refseq). The results with several cutoff values are summarized in Table 2. Interestingly, the results were not largely varied between blast analyses against invertebrate protein and universal protein databases. For instance, using cutoff values of 1e-6, 16,534 contigs (20.1%) of Lymnaea TSA sequences showed significant hits with an existing entry in Swissprot, and 20,642 and 20,578 contigs had significant hits to the Refseq and Invertebrate Refseq databases, respectively. With a strict cutoff value of 0, the numbers of contigs having blast hits was not largely varied (1,261 for the Swissprot; 1,510 for the Refseq; 1,430 for the Invertebrate Refseq database).
Table 2. Blast hits of the Lymnaea TSA to different protein databases.
| e-value | Number of contigs havinga blast hit in Swissprot | (%) | Number of contigs having ablast hit in Refseq | (%) | Number of contigs having a blasthit in Invertebrate Refseq | (%) |
| 0 | 1,261 | 1.5 | 1,510 | 1.8 | 1,430 | 1.7 |
| 1.00E-150 | 1,799 | 2.2 | 2,120 | 2.6 | 2,018 | 2.5 |
| 1.00E-100 | 3,344 | 4.1 | 3,925 | 4.8 | 3,767 | 4.6 |
| 1.00E-50 | 6,670 | 8.1 | 7,685 | 9.4 | 7,510 | 9.1 |
| 1.00E-20 | 11,314 | 13.8 | 13,202 | 16.1 | 12,957 | 15.8 |
| 1.00E-10 | 14,474 | 17.6 | 17,523 | 21.3 | 17,318 | 21.1 |
| 1.00E-06 | 16,534 | 20.1 | 20,642 | 25.1 | 20,578 | 25.1 |
| total | 82,127 | 82,127 | 82,127 |
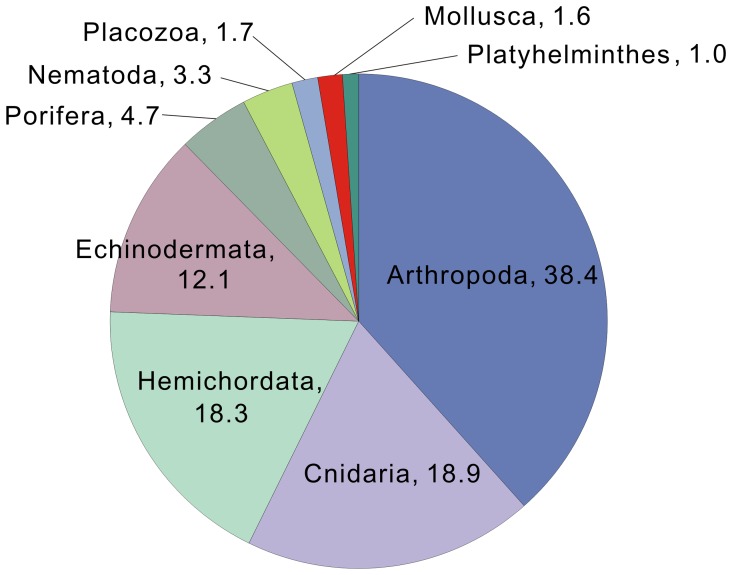
We further compared the phylum distribution of top hit species in a blastx search and observed that the percentages of mollusca phylum were clearly lower than those of the other phyla. Using a cutoff value of 1e-10 for the blastx analysis against Invertebrate Refseq, 38.4% of total hit sequences had similarity to arthropoda sequences (Fig. 4). On the other hand, only 1.6% had blast hits to mollusca sequences. Moreover, the phylum distributions did not vary among the results when more stringent cutoff values of 1e-50 and 1e-150 were used (43.6% and 40.9% for arthropoda; 2.8% and 5.5% for mollusca, respectively). These results demonstrated that the number of molluscan sequences in the public databases is not sufficient to analyze the transcriptome data.
Figure 4. Phylum distributions of top blast hit species in the invertebrate protein database.
The percentages of phylum for top blast hit species are shown for the results with a cutoff e-value of 1e-10.
Comparison with the Previously Reported Lymnaea EST Sequences
To compare the present TSA data with previously reported Lymnaea EST sequences, we next performed a bidirectional blastn analysis (Table 3). In GenBank, 11,697 EST from Lymnaea CNS [8] are registered. They include sequences shorter than 100 bp, and thus we can use the whole dataset including short contigs (116,355 contigs, >100 bp) in this analysis. As a result, over 80% of the previous EST data had blast hits to the present TSA data using a stringent cutoff value of 1e-100. By contrast, only 5% of the present TSA sequences had hits to the previous EST data with the same cutoff value. The results clearly demonstrated that about 95% of the present TSA, 110,000 sequences, were the newly identified transcripts.
Table 3. Comparison of the present Lymnaea TSA and the previously reported Lymnaea EST.
| e-value | Number of the previous Lymnaea ESThaving a blast hit in the present TSA | (%) | Number of the present Lymnaea TSA having ablast hit in the previous EST | (%) |
| 0 | 8,398 | 71.8 | 4,520 | 3.9 |
| 1.00E-150 | 8,783 | 75.1 | 4,936 | 4.2 |
| 1.00E-100 | 9,388 | 80.3 | 6,049 | 5.2 |
| total | 11,697 | 116,355 |
We further noted that the numbers of hit sequences differed according to the direction of the blast search. Using a strict cutoff value of 0, 8,398 sequences of the previous EST and 4,520 sequences of the present TSA showed blast hits against the data of the other. Closer inspection revealed that multiple sequences in the previous EST showed blast hits to a single contig in the present TSA. In addition, the average lengths of these 8,398 EST sequences were approximately three times shorter than those of the 4,520 TSA sequences (the EST, 823 bp; the TSA, 2,487 bp). The results indicated that multiple short sequences of the previous EST had similarity to different parts of a single long contig in the present TSA. On the other hand, 4,520 contigs of the present TSA had blast hits to independent sequences of the previous EST. These results clearly indicated that the present RNA-seq analysis largely improved the coverage and the contig length of transcriptome data from Lymnaea.
Comparison with Aplysia EST Sequences
To assess transcriptome coverage of the present data, we further performed a blastn analysis against Aplysia californica EST (255,605 sequences), which is the largest molluscan EST available in the public databases [21]. As a result, with a cutoff value of 1e-6, only 4,806 sequences (4.1%) of Lymnaea TSA showed blast hits (Table 4). A tblastx analysis was also performed to compare the translated sequences of TSA. Using a lenient cutoff value of 1e-6, 18,275 sequences (15.7%) of Lymnaea TSA data showed tblastx hits to the Aplysia EST data.
Table 4. Comparison of the Lymnaea TSAs and Aplysia EST.
| e-value | Number of contigs having a blastn hitin Aplysia EST | (%) | Number of contigs having a tblastx hitin Aplysia EST | (%) |
| 0 | 11 | 0.0 | 13 | 0.0 |
| 1.00E-150 | 24 | 0.0 | 245 | 0.2 |
| 1.00E-100 | 124 | 0.1 | 2,050 | 1.8 |
| 1.00E-50 | 768 | 0.7 | 5,669 | 4.9 |
| 1.00E-20 | 2,262 | 1.9 | 10,314 | 8.9 |
| 1.00E-10 | 3,733 | 3.2 | 14,437 | 12.4 |
| 1.00E-06 | 4,806 | 4.1 | 18,275 | 15.7 |
| total | 116,355 | 116,355 |
The tblastx result was further compared with the result of blast analysis against Invertebrate Refseq (Table 2). Using the same dataset (>200 bp) and a stringent cutoff value of 0, 1,430 contigs showed blast hits to Invertebrate Refseq, while only 13 contigs did so to Aplysia EST. The number of contigs showing similarity to Aplysia EST was clearly less than that showing similarity to sequences of other phylum species. The results revealed that the present Lymnaea TSA data were not comparable to the present Aplysia EST data.
Newly Identified Transcripts in Molluscan Species
Finally, to verify the accuracy of assembly, we observed sequences of particular contigs with a single complete open reading frame (ORF) as representative examples. According to the annotated result, we here isolated the sequences of monoamine synthesis enzymes that have not yet been identified in molluscan species. Because monoamines (e.g. dopamine, octopamine and serotonin) play critical functions in neuroregulatory mechanisms in animal species, the genes of monoamine synthesis enzymes should be expressed in molluscan nervous system.
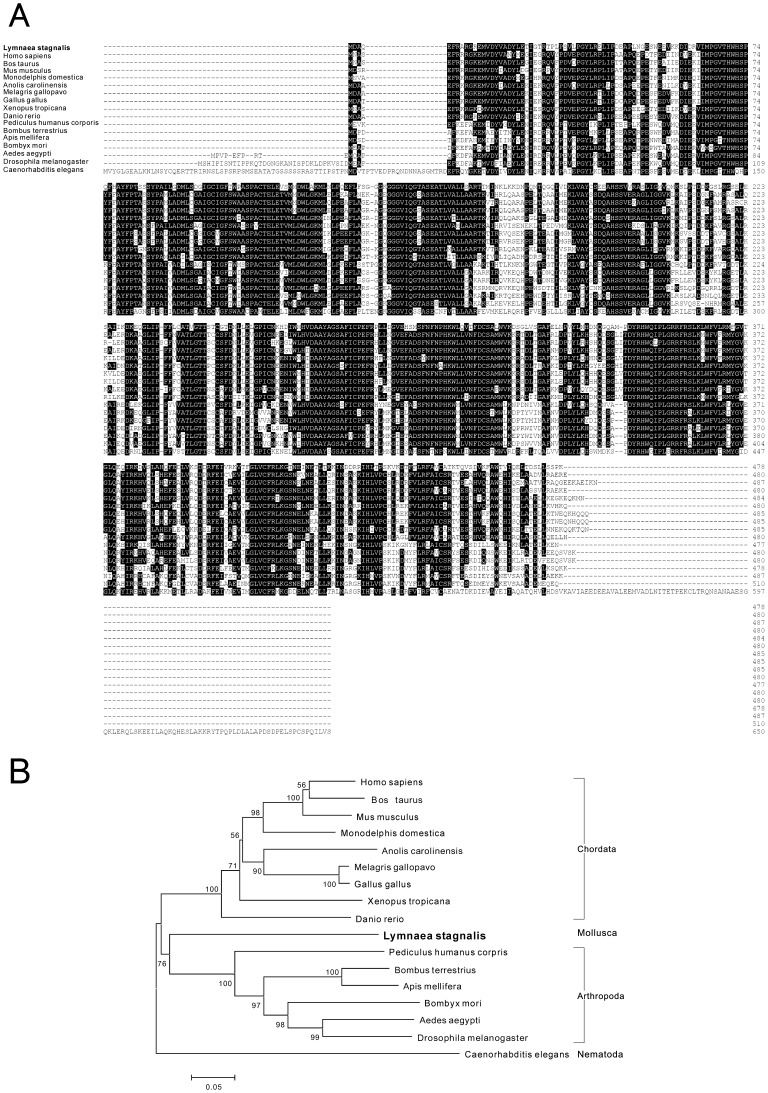
First, a homolog sequence of Lymnaea tyrosine hydroxylase (TH) was isolated according to the blast search result. The enzyme TH catalyzes the conversion of L-tyrosine to L-dihydroxyphenylalanine (L-DOPA), the rate-limiting step in the dopamine synthesis pathway. The Lymnaea TH sequence (DDBJ accession number: FX186872) had a length of 2,331 bp and encoded 502 amino acids. Amino acid alignment was performed with hit sequences identified by blastx searches in public databases, including the Swissprot, GenBank and Refseq protein databases. Using the ClustalW program in the MEGA4 software package [23], multiple alignments were performed and the result revealed amino acid sequences of TH were highly conserved among animal species (Fig. 5A). In addition, a phylogenetic tree, constructed by the neighbor-joining method, showed that Lymnaea TH was distant from the other phylum species THs, and forms a deep phylogenetic branch (Fig. 5B). The relatively high bootstrap value for each node supported the result.
Figure 5. Protein sequence alignment and phylogenetic tree of tyrosine hydroxylase.
Protein sequence alignment of tyrosine hydroxylase (A) and phylogenetic analyses (B) were conducted by the neighbor-joining method using the MEGA4 program with the C. elegans tyrosine hydroxylase as outgroup. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches. The scale bars indicate the estimated evolutionary distance in the units of the number of amino acid substitutions per site.
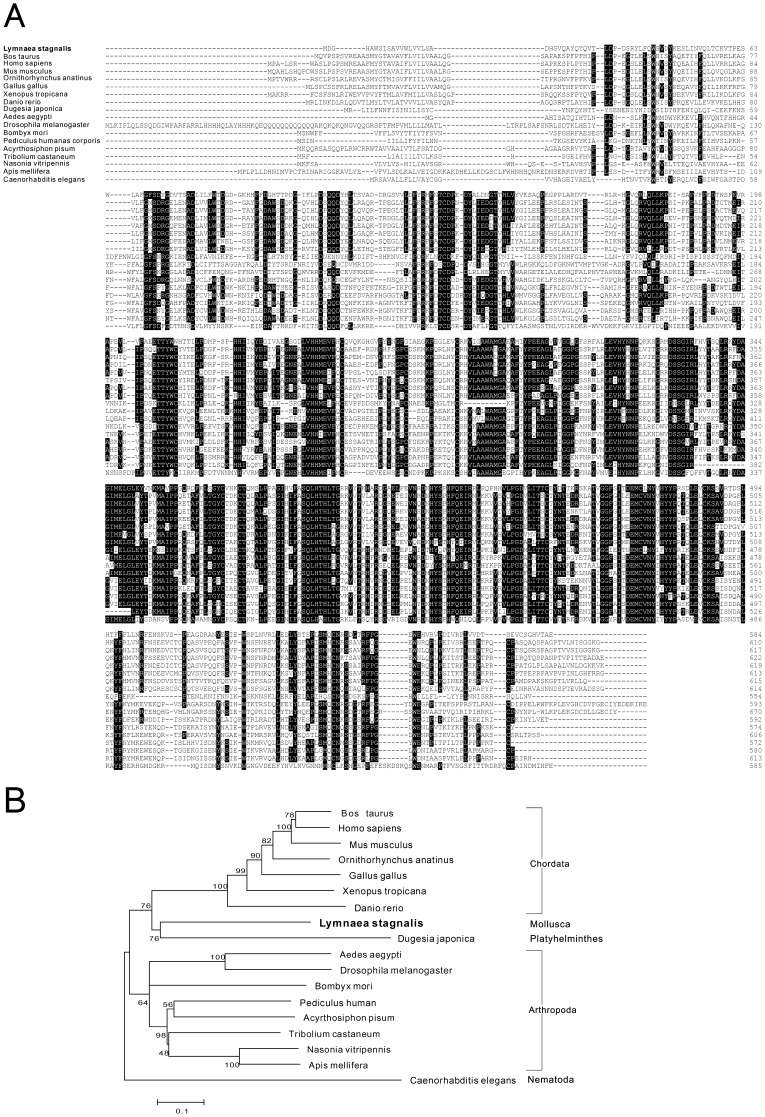
Next, we isolated a contig encoding dopa decarboxylase (DDC), which catalyzes the final step in the synthesis of dopamine and serotonin. The Lymnaea DDC sequence (DDBJ accession number: FX190337) had a length of 1,763 bp and encoded 478 amino acids. Multiple alignments were performed and the result revealed highly conserved sequences of DDC among animal species (Fig. 6A). The phylogenetic tree with the neighbor-joining method showed that Lymnaea DDC, an only molluscan sequence in the tree, was distant from the DDCs of arthropoda and chordata species (Fig. 6B). The relatively high bootstrap value for each node further supported the notion that these were apparent taxa in this tree.
Figure 6. Protein sequence alignment and phylogenetic tree of dopa decarboxylase.
Protein sequence alignment of dopa decarboxylase (A) and phylogenetic analyses (B) were conducted by the neighbor-joining method using the MEGA4 program with the C. elegans tyrosine decarboxylase as outgroup. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches. The scale bars indicate the estimated evolutionary distance in the units of the number of amino acid substitutions per site.
We further identified a sequence of Lymnaea tyramine beta hydroxylase (TBH), a homolog of dopamine beta hydroxylase (DBH), which catalyzes the last step in the synthesis of octopamine. The Lymnaea TBH sequence (DDBJ accession number: FX185559) had a length of 2,644 bp and encoded 584 amino acids. Protein sequences of other species TBH/DBH were identified by a blast search and the sequence alignment showed conserved positions among species (Fig. 7A). The amino acid sequence conservation of TBH was relatively low compared to the alignment results of TH and DDC (Figs. 5A and 6A), which was consistent with the previous report of DBH structural analysis [26]. A phylogenetic analysis clearly demonstrated that Lymnaea TBH of mollusca, as well as the freshwater planarian Dugesia japonica TBH of platyhelminthes, was distant from arthropoda and chordate TBH/DBH (Fig. 7B) and the relatively high bootstrap values also supported these results.
Figure 7. Protein sequence alignment and phylogenetic tree of tyramine beta hydroxylase.
Protein sequence alignment of tyramine beta hydroxylase (A) and phylogenetic analyses (B) were conducted by the neighbor-joining method using the MEGA4 program with the C. elegans of tyramine beta hydroxylase as outgroup. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown next to the branches. The scale bars indicate the estimated evolutionary distance in the units of the number of amino acid substitutions per site.
For these three sequences, we performed tblastx searches against GenBank, and confirmed that any nucleotide sequence in molluscan species has not been reported yet. Blastn searches against the public EST database were further performed and several molluscan sequences were found (7 sequences for DDC and 1 for TBH), but they did not code the whole ORFs. Therefore, the sequences were the first reported homologues of TH, DDC and TBH in molluscs. The cDNA sequences for Lymnaea TH, DDC and TBH were identified by RT-PCR experiments (Fig. S1 and Protocol S1) and the cloned sequences were compared to the TSA sequences. As a result, we found no, or very few, nucleotide differences between the cDNA and TSA sequences (Figs. S2 to S4). We further examined the read mapping with Bowtie [27] and confirmed that the positions of different nucleotide showed some mismatches between the contig and reads (data not shown). Thus, the nucleotide differences were possibly caused by single nucleotide polymorphisms (SNPs), or sequence specific errors (SSEs) as previously reported [28]. The obtained cDNA sequences have been deposited in DDBJ (accession numbers AB733115 to AB733117). These results further supported the accuracy of assembly in the present RNA-seq analysis.
Discussion
In this study, we performed de novo transcriptome sequencing of the central nervous system (CNS) in Lymnaea stagnalis using an Illumina sequencer. First, we compared the five assembly programs, ABySS, Velvet, OASES, Trinity and Rnnotator, with respect to several well-known criteria, i.e., N50 length, maximum contig length and contig number (Figs. 1 and 2A). In addition, the coverage of known sequences was used as another criterion (Fig. 2B). The results showed that ABySS and Velvet were clearly inferior to OASES, Trinity and Rnnotator in the assembly of long transcriptome sequences. The large difference seems to have been caused by the different assembly strategies for transcriptome or genome analysis because transcriptome data includes a large amount of redundant sequence information, unlike genomic sequence data. ABySS and Velvet are the programs developed for de novo genome assembly with the de Bruijn graph method [14], [15]. In contrast, OASES, Trinity and Rnnotator were developed as de novo transcriptome assemblers with a strategy that merges the contigs obtained by the first assembly. It is also consistent with the previous report that Velvet itself is not sufficient for assembly of long contigs in de novo transcriptome sequencing with an Illumina Genome Analyzer [29].
Among OASES, Trinity and Rnnotator, we did not see a consistent difference in the criteria values (Fig. 2), and the only striking difference was the number of variants (OASES: 54,601; Trinity: 44,062; Rnnotator: 2,675). The datasets of OASES and Trinity included a large number of variants because they were designed to finely detect the variants as splicing products [16], [18]. Longer transcripts seemed to have a larger number of variants and longer variants as possible assemblies. In the comparison using the longest variants shown in Figure 2A, the reason of longer N50 and average contig lengths in OASES and Trinity can be considered to be the result of possible assembly. On the other hand, Rnnotator produced a small number of variants because of the error correction program at the post-processing step [19]. The effects of splicing variants or duplicate genes on Rnnotator data have not been studied yet. Even if the genetic variants are compressed into a small number of contigs, the redundant reads of variants are counted as the expression level with reads per kilo base per million (rpkm) values. Thus, we here selected the data of Rnnotator, including the smallest number of variants, for GO analysis. However, OASES and Trinity were also useful to obtain long transcripts. According to the purpose of research, these programs can be used singly or jointly as in the previous reports [24], [30].
The present results demonstrated that RNA-seq analysis using an Illumina sequencer was more efficient than the classical Sanger sequencing, in terms of both sequence quantity and quality, for de novo transcriptome sequencing. Rnnotator assembly produced 116,355 sequences as Lymnaea transcriptome shotgun assembly (TSA) and the data was compared to the previous Lymnaea expression sequence tag (EST) data (Table 3). The blast comparison demonstrated that about 5% of the present TSA data covered 80% of the previous EST, and about 110,000 sequences were proved to be newly obtained Lymnaea transcripts. The good assembly of the present RNA-seq analysis was also confirmed because a single contig of the present TSA was equivalent to several shorter sequences of the previous EST [8]. In GO analysis, the distributions of GO terms showed low similarity to the results of the previous study [8], and the large difference was caused by the different size of the two datasets.
The present study also demonstrated that the reported molluscan data are insufficient to cover the whole transcriptome data. The small number of molluscan sequences in the blast analyses (Table 2) and the phylum distribution of top hit species (Fig. 4) strongly supported this idea. These results did not indicate that Lymnaea sequences are more similar to the sequences of other phylum species, but rather that there is a lack of molluscan sequences in the public database. In addition, the present Lymnaea TSA showed low similarity to Aplysia EST (Table 4). A similar observation was made in a previous study, in which the EST obtained from juvenile Aplysia did not show similarity to the currently available Aplysia EST database [31]. Our findings, combined with the previous observations, suggest that the present data obtained by RNA-seq enlarge the molluscan transcriptome database substantially.
As representative examples of contig sequences, Lymnaea homologues of tyrosine hydroxylase (TH), dopamine decarboxylase (DDC) and tyramine beta hydroxylase (TBH) were selected (Figs. 5 to 7). We also confirmed the accuracy of their sequences by DNA sequencing of the cloned cDNA sequences (Figs. S1, S2, S3, S4). Those enzymes catalyze the synthesis of monoamines that play critical roles in various behavioural regulations [32]–[35]. The amino acid alignments showed that the predicted sequences of Lymnaea TH, DDC and TBH were reliable for conserved sequences among species. Phylogenetic analyses also revealed that the evolutional distances were reasonable because of the high boot strap values. The identification of these sequences further supported the accuracy of de novo transcriptome assembly in this study.
The development of techniques for RNA-seq has now enabled researchers to perform de novo sequencing of mollusc species. Recently, transcriptome sequencing in the pond snail Radix balthica [29] and genomic sequencing of the pearl oyster Pinctada fucata [36] were reported. The Illumina short read sequencer, when used together with a good assembly program, is highly useful, and its low cost-performance will speed up de novo sequencing. Further, the available genetic information in public databases will be valuable for future sequencing. Several studies have demonstrated that the genome of a closely related species can be used as a reference for de novo transcriptome assembly of non-model organisms [13], [37], [38]. In addition to raw sequencing data, the assembled sequences can now be updated in public databases, such as TSA in DDBJ and GenBank. Reliable data of good assembly sequences is continuously needed to enlarge the genetic database, not only for mollusc species but for all non-model species, as we demonstrated in this study.
Conclusions
Using an Illumina short read sequencer and well developed assembly programs, we successfully performed de novo transcriptome sequencing. The assembly program Rnnotator produced 116,355 transcript sequences as a transcriptome shotgun assembly (TSA) in Lymnaea CNS. The present data improved the previous Lymnaea transcriptome data and further helped to enlarge the public database of mollusc species. Moreover, the present study using the RNA-seq method will facilitate the future genetic analyses of non-model animals.
Supporting Information
Molecular cloning of Lymnaea TH, DDC and TBH cDNAs. A. RT-PCR was performed using primers that were designed according to the TSA sequences for Lymnaea TH, DDC and TBH. The ORFs are depicted as the colored boxes, and locations and directions of primers are shown by arrowheads. B. Electrophoresis gel showing RT-PCR products generated from cDNA sample of Lymnaea CNS. Used primer set with expected product length is shown for each lane. DNA ladder markers (Fermentas, Hanover, Germany) in the left and right lanes are labeled.
(PDF)
Sequence alignments of coding regions of the isolated cDNA and TSA for LymTH. The cDNA sequence is determined based on analysis of RT-PCR using a cDNA library sample derived from a single Lymnaea CNS. Nucleotide differences are shaded in black, and positions with codon differences are indicated by underlines when causing amino acid changes.
(PDF)
Sequence alignments of coding regions of the isolated cDNA and TSA for LymDDC. The cDNA sequence is determined based on analysis of RT-PCR using a cDNA library sample derived from a single Lymnaea CNS. One nucleotide difference without amino acid sequence changing was found.
(PDF)
Sequence alignments of coding regions of the isolated cDNA and TSA for LymTBH. The cDNA sequence is determined based on analysis of RT-PCR using a cDNA library sample derived from a single Lymnaea CNS. No nucleotide difference was found between LymTBH cDNA and TSA sequences.
(PDF)
Method for molecular cloning of Lymnaea TH, DDC and TBH cDNAs.
(PDF)
Acknowledgments
We wish to thank Professor Etsuro Ito for his kind support during our work on this paper.
Funding Statement
This work was supported by grants from the Japan Society for the Promotion of Science (21770081 and 24570091 to HS) and Senryaku Research from the Ministry of Education, Culture, Sports, Science and Technology. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kojima S, Nanakamura H, Nagayama S, Fujito Y, Ito E (1997) Enhancement of an inhibitory input to the feeding central pattern generator in Lymnaea stagnalis during conditioned taste-aversion learning. Neurosci Lett 230: 179–182. [DOI] [PubMed] [Google Scholar]
- 2. Sadamoto H, Azami S, Ito E (2004) The expression pattern of CREB genes in the central nervous system of the pond snail Lymnaea stagnalis . Acta Biol Hung 55: 163–166. [DOI] [PubMed] [Google Scholar]
- 3. Kemenes G, Benjamin PR (2009) Lymnaea . Curr Biol 19: R9–11. [DOI] [PubMed] [Google Scholar]
- 4. van Nierop P, Bertrand S, Munno DW, Gouwenberg Y, van Minnen J, et al. (2006) Identification and functional expression of a family of nicotinic acetylcholine receptor subunits in the central nervous system of the mollusc Lymnaea stagnalis . J Biol Chem 281: 1680–1691. [DOI] [PubMed] [Google Scholar]
- 5. Sadamoto H, Kitahashi T, Fujito Y, Ito E (2010) Learning-Dependent Gene Expression of CREB1 Isoforms in the Molluscan Brain. Front Behav Neurosci 4: 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ribeiro M, Schofield M, Kemenes I, Benjamin PR, O’Shea M, et al. (2010) A typical guanylyl cyclase from the pond snail Lymnaea stagnalis: cloning, sequence analysis and characterization of expression. Neuroscience 165: 794–800. [DOI] [PubMed] [Google Scholar]
- 7. Davison A, Blaxter ML (2005) An expressed sequence tag survey of gene expression in the pond snail Lymnaea stagnalis, an intermediate vector of trematodes. Parasitology 130: 539–552. [DOI] [PubMed] [Google Scholar]
- 8. Feng ZP, Zhang Z, van Kesteren RE, Straub VA, van Nierop P, et al. (2009) Transcriptome analysis of the central nervous system of the mollusc Lymnaea stagnalis . BMC Genomics 10: 451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Metzker ML (2010) Sequencing technologies - the next generation. Nat Rev Genet 11: 31–46. [DOI] [PubMed] [Google Scholar]
- 10. Dames S, Durtschi J, Geiersbach K, Stephens J, Voelkerding KV (2010) Comparison of the Illumina Genome Analyzer and Roche 454 GS FLX for resequencing of hypertrophic cardiomyopathy-associated genes. J Biomol Tech 21: 73–80. [PMC free article] [PubMed] [Google Scholar]
- 11. Suzuki S, Ono N, Furusawa C, Ying BW, Yomo T (2011) Comparison of sequence reads obtained from three next-generation sequencing platforms. PLoS One 6: e19534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kawahara Miki R, Wada K, Azuma N, Chiba S (2011) Expression profiling without genome sequence information in a non-model species, Pandalid shrimp (Pandalus latirostris), by next-generation sequencing. PLoS One 6: e26043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ward JA, Ponnala L, Weber CA (2012) Strategies for transcriptome analysis in nonmodel plants. Am J Bot 99: 267–276. [DOI] [PubMed] [Google Scholar]
- 14. Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, et al. (2009) ABySS: a parallel assembler for short read sequence data. Genome Res 19: 1117–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zerbino DR, Birney E (2008) Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Schulz MH, Zerbino DR, Vingron M, Birney E (2012) Oases: robust de novo RNA-seq assembly across the dynamic range of expression levels. Bioinformatics 28: 1086–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Knowles DG, McLysaght A (2009) Recent de novo origin of human protein-coding genes. Genome Res 19: 1752–1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, et al. (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Martin J, Bruno VM, Fang Z, Meng X, Blow M, et al. (2010) Rnnotator: an automated de novo transcriptome assembly pipeline from stranded RNA-Seq reads. BMC Genomics 11: 663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. McGinnis S, Madden TL (2004) BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 32: W20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Moroz LL, Edwards JR, Puthanveettil SV, Kohn AB, Ha T, et al. (2006) Neuronal transcriptome of Aplysia: neuronal compartments and circuitry. Cell 127: 1453–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Conesa A, Gotz S, Garcia Gomez JM, Terol J, Talon M, et al. (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676. [DOI] [PubMed] [Google Scholar]
- 23. Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599. [DOI] [PubMed] [Google Scholar]
- 24. Garg R, Patel RK, Tyagi AK, Jain M (2011) De novo assembly of chickpea transcriptome using short reads for gene discovery and marker identification. DNA Res 18: 53–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gotz S, Garcia Gomez JM, Terol J, Williams TD, Nagaraj SH, et al. (2008) High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res 36: 3420–3435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kapoor A, Shandilya M, Kundu S (2011) Structural insight of dopamine beta-hydroxylase, a drug target for complex traits, and functional significance of exonic single nucleotide polymorphisms. PLoS One 6: e26509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakamura K, Oshima T, Morimoto T, Ikeda S, Yoshikawa H, et al. (2011) Sequence-specific error profile of Illumina sequencers. Nucleic Acids Res 39: e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Feldmeyer B, Wheat CW, Krezdorn N, Rotter B, Pfenninger M (2011) Short read Illumina data for the de novo assembly of a non-model snail species transcriptome (Radix balthica, Basommatophora, Pulmonata), and a comparison of assembler performance. BMC Genomics 12: 317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Iorizzo M, Senalik DA, Grzebelus D, Bowman M, Cavagnaro PF, et al. (2011) De novo assembly and characterization of the carrot transcriptome reveals novel genes, new markers, and genetic diversity. BMC Genomics 12: 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Fiedler TJ, Hudder A, McKay SJ, Shivkumar S, Capo TR, et al. (2010) The transcriptome of the early life history stages of the California Sea Hare Aplysia californica . Comp Biochem Physiol Part D Genomics Proteomics 5: 165–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vehovszky A, Hiripi L, Elliott CJ (2000) Octopamine is the synaptic transmitter between identified neurons in the buccal feeding network of the pond snail Lymnaea stagnalis . Brain Res 867: 188–199. [DOI] [PubMed] [Google Scholar]
- 33. Vehovszky A, Elliott CJ (2002) Heterosynaptic modulation by the octopaminergic OC interneurons increases the synaptic outputs of protraction phase interneurons (SO, N1L) in the feeding system of Lymnaea stagnalis . Neuroscience 115: 483–494. [DOI] [PubMed] [Google Scholar]
- 34. Kemenes I, O’Shea M, Benjamin PR (2011) Different circuit and monoamine mechanisms consolidate long-term memory in aversive and reward classical conditioning. Eur J Neurosci 33: 143–152. [DOI] [PubMed] [Google Scholar]
- 35. Filla A, Hiripi L, Elekes K (2004) Serotonergic and dopaminergic influence of the duration of embryogenesis and intracapsular locomotion of Lymnaea stagnalis L. Acta Biol Hung. 55: 315–321. [DOI] [PubMed] [Google Scholar]
- 36. Takeuchi T, Kawashima T, Koyanagi R, Gyoja F, Tanaka M, et al. (2012) Draft genome of the pearl oyster Pinctada fucata: a platform for understanding bivalve biology. DNA Res 19: 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Poelchau MF, Reynolds JA, Denlinger DL, Elsik CG, Armbruster PA (2011) A de novo transcriptome of the Asian tiger mosquito, Aedes albopictus, to identify candidate transcripts for diapause preparation. BMC Genomics 12: 619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Birzele F, Schaub J, Rust W, Clemens C, Baum P, et al. (2010) Into the unknown: expression profiling without genome sequence information in CHO by next generation sequencing. Nucleic Acids Res 38: 3999–4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Molecular cloning of Lymnaea TH, DDC and TBH cDNAs. A. RT-PCR was performed using primers that were designed according to the TSA sequences for Lymnaea TH, DDC and TBH. The ORFs are depicted as the colored boxes, and locations and directions of primers are shown by arrowheads. B. Electrophoresis gel showing RT-PCR products generated from cDNA sample of Lymnaea CNS. Used primer set with expected product length is shown for each lane. DNA ladder markers (Fermentas, Hanover, Germany) in the left and right lanes are labeled.
(PDF)
Sequence alignments of coding regions of the isolated cDNA and TSA for LymTH. The cDNA sequence is determined based on analysis of RT-PCR using a cDNA library sample derived from a single Lymnaea CNS. Nucleotide differences are shaded in black, and positions with codon differences are indicated by underlines when causing amino acid changes.
(PDF)
Sequence alignments of coding regions of the isolated cDNA and TSA for LymDDC. The cDNA sequence is determined based on analysis of RT-PCR using a cDNA library sample derived from a single Lymnaea CNS. One nucleotide difference without amino acid sequence changing was found.
(PDF)
Sequence alignments of coding regions of the isolated cDNA and TSA for LymTBH. The cDNA sequence is determined based on analysis of RT-PCR using a cDNA library sample derived from a single Lymnaea CNS. No nucleotide difference was found between LymTBH cDNA and TSA sequences.
(PDF)
Method for molecular cloning of Lymnaea TH, DDC and TBH cDNAs.
(PDF)