Abstract
The synthesis of virulence factors by pathogenic bacteria is highly regulated and occurs in response to diverse environmental cues. An array of two component systems (TCSs) serves to link perception of different cues to specific changes in gene expression and/or bacterial behaviour. Those TCSs that regulate functions associated with virulence represent attractive targets for interference in anti-infective strategies for disease control. We have previously identified PA2572 as a putative response regulator required for full virulence of Pseudomonas aeruginosa, the opportunistic human pathogen, to Galleria mellonella (Wax moth) larvae. Here we have investigated the involvement of candidate sensors for signal transduction involving PA2572. Mutation of PA2573, encoding a probable methyl-accepting chemotaxis protein, gave rise to alterations in motility, virulence, and antibiotic resistance, functions which are also controlled by PA2572. Comparative transcriptome profiling of mutants revealed that PA2572 and PA2573 regulate expression of a common set of 49 genes that are involved in a range of biological functions including virulence and antibiotic resistance. Bacterial two-hybrid analysis indicated a REC-dependent interaction between PA2572 and PA2573 proteins. Finally expression of PA2572 in the PA2573 mutant background restored virulence to G. mellonella towards wild-type levels. The findings indicate a role for the orphan chemotaxis sensor PA2573 in the regulation of virulence and antibiotic tolerance in P. aeruginosa and indicate that these effects are exerted in part through signal transduction involving PA2572.
Introduction
Bacterial genomes encode a multitude of sensory systems that allow them to detect alterations in their extracellular and intracellular environments and to respond appropriately. Two-component signal transduction systems (TCS) comprise a sensor kinase, which in many cases is directly responsible for signal perception, and a response regulator that has a receiver domain and an output domain that exerts a particular regulatory action. Chemosensory two-component systems contain additional machinery for signal transduction such as methyl-accepting chemotaxis proteins (MCPs). Importantly, some TCSs have been shown to play a role in the virulence of diverse pathogenic bacteria including the human pathogen Pseudomonas aeruginosa [1]–[3]. As no such systems have been found in mammals, TCSs could provide attractive targets for anti-infective therapies [4]. Such a strategy for P. aeruginosa requires identification of those signalling systems that are key for virulence from the inventory of 26 MCPs and 47 other TCSs encoded by the genome [5].
Although most response regulators contain DNA-binding output domains and serve as transcriptional regulators, other classes of output domain are seen [6]. These classes include enzymes involved in the turnover of the nucleotide second messenger cyclic di-GMP [7], [8]; GGDEF domain proteins are diguanylate cyclases (DGCs) catalyzing cyclic di-GMP synthesis whereas EAL and HD-GYP domain proteins are phosphodiesterases (PDEs) catalyzing degradation of cyclic di-GMP. Such regulators can exert their effects via an influence on cyclic di-GMP levels but also through protein-protein interactions [9]–[11]. Of particular interest here are the proteins of P. aeruginosa with an HD-GYP domain. The P. aeruginosa PAO1 proteome contains three HD-GYP domain proteins. PA4108 and PA4781 are enzymatically active phosphodiesterases, whereas PA2572 encodes a non-canonical variant YN-GYP and does not have detectable activity in cyclic di-GMP degradation. Despite these differences, all three HD-GYP domain proteins play a role in the regulation of biofilm formation, production of virulence factors and virulence in the Galleria mellonella model of infection [12]. Both PA4781 and PA2572 have an N-terminal Receiver (REC) domain, which suggests that they are regulatory components of signal transduction pathways associated with virulence. In this study we examined the involvement of candidate sensory proteins in signal transduction involving PA2572.
Genes flanking PA2572 on the PAO1 chromosome encode candidate sensors for signal transduction involving PA2572; PA2571 is a probable two-component sensor kinase, and PA2573 is a probable methyl-accepting chemotaxis protein (Fig. S1). Although the three genes are predicted to be in separate transcriptional units, PA2573 is co-expressed with PA2572 under several conditions including aerobic growth in the presence of nitrate and in bacteria co-cultured with respiratory epithelia cells [13]–[15]. We investigated the potential interplay between these sensors and the regulator PA2572 utilizing bacterial two-hybrid assay to assess protein-protein interactions together with phenotypic and comparative transcriptome profiling analysis of mutants. The findings indicate a role for the orphan chemotaxis sensor PA2573 in the regulation of virulence and antibiotic tolerance in P. aeruginosa and suggest that these effects are exerted in part through signal transduction involving PA2572.
Materials and Methods
Bacterial Strains, Plasmids, Primers and Culture Conditions
Pseudomonas aeruginosa and Escherichia coli strains were cultured in Luria-Bertani (LB) broth at 37°C. For solid media, 1.5% agar was added. Bacterial strains used in this study are listed in Table S1. Antibiotics, obtained from Sigma Chemical Company, gentamycin (Gm), chloramphenicol (Cm) and tetracycline (Tc) were added where appropriate. Oligonucleotide primers were synthesized by MWG (Germany) and are listed in Table S2.
Construction and Complementation of PA2571, PA2572, and PA2573 Mutants
Gene disruption mutants were created using the suicide plasmid pEX18Gm (Table S1). In short, central fragments of each gene were amplified using primers (Table S2) and cloned into pEX18Gm. Restriction enzymes and T4 ligase used for cloning purposes were obtained from Roche Diagnostics and used according to the manufacturer’s instructions. This construct was isolated using the Qiagen QIAprep Spin Miniprep Kit and subsequently introduced into the wild-type P. aeruginosa PAO1 strain by electroporation. Integration results in the formation of a stable GmR mutant and was confirmed by colony PCR using primers detailed in Table S2. Complementation clones were assembled in pBBR1MCS using wild-type gene amplifications from genomic DNA with primers listed in Table S2. The resulting constructs were introduced into the appropriate disruption mutants by electroporation. These strains were selected on LB agar medium containing Gm (20 µg/ml) and Cm (50 µg/ml) and confirmed by colony PCR.
Motility
Swimming and swarming phenotypes of P. aeruginosa strains were assessed on LB media with 0.3% agar and Eiken media with 0.5% agar, respectively. Motility plates were inoculated by sterile tips using freshly streaked cultures grown on LB media. Plates were incubated at 37°C for 24 to 48 hours.
Pyoverdine and Pyocyanin Production
For pyoverdine production, a conical flask containing 30 ml of F broth, comprised of 20 g/l bacteriological peptone (Oxoid), 1.5 g/l magnesium sulphate hydrated (Fluka), 1.5 g/l dipotassium phosphate (Sigma), and 10 ml/l glycerol, was inoculated with P. aeruginosa strains (1∶1000). OD420 measurements were taken from supernatants removed from cultures grown to an OD600 of 2.0. Pyoverdine production was calculated as OD420/OD600. For production of pyocyanin, 30 ml of P broth, comprised of 20 g/l bacteriological proteose peptone no. 3 (Oxoid), 1.4 g/l magnesium chloride anhydrous (Sigma), 10 g/l potassium sulphate (Sigma) and 10 ml/l glycerol, was inoculated with P. aeruginosa strains (1∶1000). Cultures were grown at 37°C for 24 to 48 hours and 3 ml chloroform was added to supernatant removed from 5 ml of culture. This suspension was vortexed for 10 sec and centrifuged for 5 min at 5,000 rpm. 1.5 ml of chloroform was removed and samples were re-extracted with 2 ml of 0.2N HCl, vortexed for 3 sec and left still to separate. OD520 measurements were taken from the top phase and the concentration of pyocyanin determined as described previously [16].
Antibiotic Susceptibility
Strains were subcultured and grown to logarithmic phase (0.6−0.7 OD600) at 37°C, washed twice and resuspended in phosphate buffered saline (PBS) (Sigma). 300 µl of cell suspensions were spread onto 150×20 mm Petri dishes (Sarstedt) containing 60 ml of LB agar and left to dry for 10 min. Antibiotic E-test strips (Biomerieux) were overlaid and plates were incubated at 37°C for 24 hours. Minimum inhibitory concentrations (MIC) (µg/ml) for each antibiotic were recorded in triplicate in two separate experiments for each strain.
Galleria mellonella Virulence Assay
Virulence of P. aeruginosa strains was assessed as previously described by Ryan et al. [12]. Briefly, bacteria from overnight cultures were washed twice and re-suspended in PBS. Serial 10-fold dilutions were made in PBS and 10 µL aliquots were injected into each larva, with 10 larvae inoculated for each dilution. A minimum of three different dilutions were inoculated per strain in three independent experiments. Uninoculated larvae and larvae injected with PBS alone served as control groups for each experiment. Larvae were incubated in the dark at 37°C in Petri dishes lined with Whatman paper. Minimum inoculum required to kill all 10 larvae was determined for each strain.
RNA Extraction
Three independent cultures of each P. aeruginosa strain was sub-cultured and grown to logarithmic phase (0.6−0.7 OD600) at 37°C in LB broth without selection. 800 µl of RNA protect (Qiagen) was added to 400 µl culture and incubated at room temperature for 5 min. Cell suspensions were centrifuged, the supernatant was disregarded, and pellets were stored at −80°C. After thawing, 100 µl TE-lysozyme (400 µg/ml) was added and samples were incubated at room temperature. Total RNA was isolated using the RNeasy Mini Kit (Qiagen) whereby cells were homogenized utilizing a 20-gauge needle and syringe. Samples were treated with DNase (Ambion) according to manufacturer’s instructions and the removal of DNA contamination was confirmed by PCR.
Transcriptome Profiling and Analysis
RNA samples were analysed using the Affymetrix GeneChip technology platform at University College Dublin Conway Institute of Biomolecular and Biomedical Research. Analysis of microarray data was achieved using GeneSpring Multi-Omic Analysis Version 11.5 (Agilent Technologies). Two-fold changes in gene expression between mutant and the wild-type parental P. aeruginosa PAO1 strain were considered to be significant (p≤0.05). The microarray data set has been deposited into ArrayExpress Database under the accession number [pending].
Semi-quantitative and Quantitative Real-time PCR
Semi-quantitative and quantitative RT-PCRs were used to validate microarray data. Reverse transcription PCR was achieved using a cDNA synthesis kit (Promega) according to the manufacturer’s instructions. Specific RT-PCR primers (Table S2) were used to amplify central fragments of approximately 200 bp in length from different genes. Semi-quantitative RT-PCRs were completed using 250 ng/µl cDNA template and PCR Mastermix (Promega) for 24–36 cycles. For qRT-PCRs, quantification of gene expression and melting curve analysis were completed using a LightCycler (Roche) and Platinum SYBR Green qPCR Supermix-UGD (Invitrogen) was used according to manufacturer’s instructions. The constitutively expressed housing keeping gene, 16S rRNA was used as a reference to standardize all samples and replicates.
Bacterial Two-hybrid Analysis
BacterioMatch II Two-Hybrid System Vector Kit (Agilent Technologies) was used to evaluate protein-protein interactions of P. aeruginosa PAO1 proteins in E. coli. This analysis was carried out according to manufacturer’s instructions. The pBT bait plasmid and the pTRG target plasmid containing partial and full-length gene fragments were constructed using primers listed in Table S2. A co-transformant containing empty pBT and pTRG vectors served as a negative control and a co-transformant containing pBT-LGF2 and pTRG-GAL11P served as a positive control, validated by growth of colonies on M9+ His-dropout media containing 5 mM 3-amino-1,2,4, triazole (3-AT). Co-transformations for detecting interactions between bait and target proteins were plated on selective screening medium (SSM) (5 mM 3-AT) and on non-selective screening medium (NSSM) (without 3-AT) to serve as a control.
Expression of Individual Domains and Full Length PA2572 in PA2572 and PA2573 Mutant Backgrounds
PA2572 complementation clones were created using pME6032 plasmid (Table S1). Briefly, primers (Table S2) were used to amplify DNA encoding the REC domain, the YN-GYP domain and the full length PA2572 gene. The amplicons were sequenced and were cloned into pME6032. These three constructs were transformed into Top 10 E. coli cells (Invitrogen) and isolated using the Qiagen QIAprep Spin Miniprep Kit. Constructs were subsequently introduced into the PA2572 and PA2573 mutant strains and into the parental PAO1 strain by electroporation. As an additional control, the empty pME6032 vector was introduced into all strains. The presence of pME6032 containing the relevant DNA fragment was confirmed using primers (Table S2) corresponding to the multiple cloning site of the plasmid.
Results
Mutation of PA2572 and PA2573 but not PA2571 Influences Virulence of P. aeruginosa in the Greater Wax Moth Larvae Model
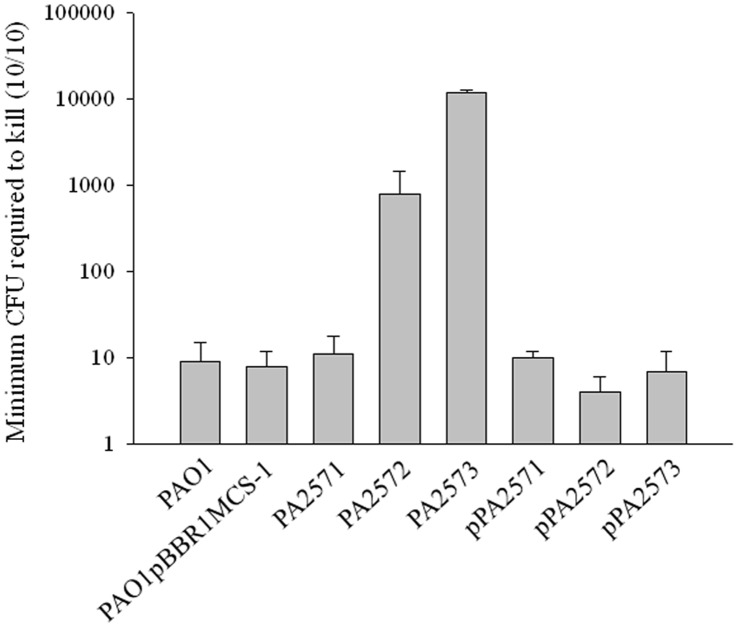
Virulence of P. aeruginosa strains in the G. mellonella larvae model was assessed by determination of the minimum CFU required to kill all of a group of 10 larvae in which each member received an inoculum from a 10-fold serial dilution series (see Materials and Methods). This minimum inoculum for the parental PAO1 strain was 9±6 CFU (n = 8) (Fig. 1). The minimum inoculum for the PA2571 mutant was similar to that of the wild-type, suggesting no effect on virulence (Fig. 1). In contrast, considerably higher numbers of the PA2572 and PA2573 mutant strains were required, indicating that both PA2572 and PA2573 contribute to the full virulence of P. aeruginosa in the G. mellonella model of infection. These findings for PA2572 confirm our earlier published work [12]. Importantly, complementation of the PA2572 and PA2573 mutants with wild-type copies of the appropriate genes restored virulence to wild-type levels (Fig. 1).
Figure 1. Pathogenesis of strains in the Wax Moth model of infection.
Effects of mutation inactivating genes encoding PA2571, PA2572 and PA2573 on virulence in the Galleria mellonella model 24 hours post-infection. Virulence potential of P. aeruginosa strains was assessed based on the number of CFUs required to kill a group of ten larvae. Groups injected with PBS and groups without any inoculation served as negative controls. Mutation of PA2572 and PA2573 led to reduced virulence compared to wild-type PAO1 or wild-type with the cloning vector pBBR1MCS-1, which were equivalent. Complemented strains (indicated by pPA2572 and pPA2573) had wild-type virulence. Mutation of PA2571 had no effect. Error bars represent mean ± standard deviation of at least three experiments.
Bacterial Two-hybrid Analysis Reveals Direct Interaction between PA2572 and PA2573
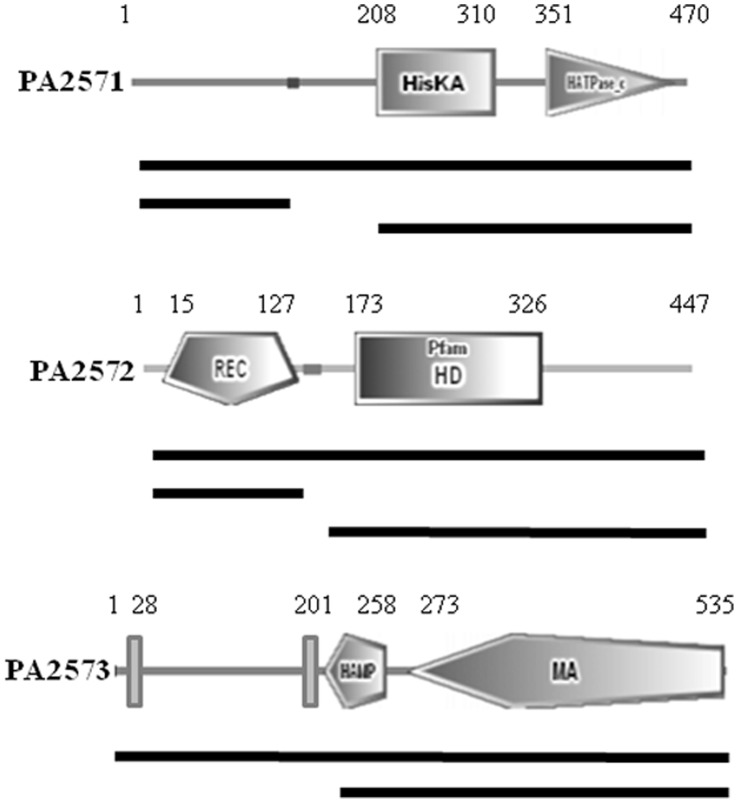
The BacterioMatch II Two-Hybrid System was used to evaluate potential protein-protein interactions between the putative regulator PA2572 and the predicted sensor proteins PA2571 and PA2573. For these experiments, the pBT bait plasmid and the pTRG target plasmid expressing full-length PA2571, PA2572, and PA2573 or different domains of the proteins were constructed following PCR amplification of the appropriate DNA fragments using primers listed in Table S2. The domain structures of the full-length proteins and protein fragments used in this analysis are illustrated in Fig. 2. Detection of interactions between bait and target proteins was based on growth of co-transformant colonies on SSM media containing 5 mM 3-AT (Table 1).
Figure 2. Domain organizations of PA2571, PA2572 and PA2573.
Domains were predicted by SMART with amino acid positions indicated. Domain abbreviations are as follows; HisKA (Histidine Kinase A phosphoacceptor domain), HATPase_c (Histidine kinase-like ATPase), REC (Receiver domain), HD (superfamily with predicted or known phosphohydrolase activity), HAMP (Histidine kinase, Adenylyl cyclase, Methyl binding protein, Phosphatase domain), and MA (Methyl-accepting chemotaxis-like domain). Vertical bars represent predicted transmembrane domains (SOUSI). Black lines below figures represent constructs cloned into either pBT or pTRG vectors to assess potential protein-protein interactions using the bacterial two-hybrid assay.
Table 1. Bacterial two-hybrid analysis investigating potential protein-protein interactions.
| Target Protein (pTRG vector) | ||||||
| Bait Protein (pBT vector) | PA2571 RECdomain | PA2571 HKdomain | PA2571 Full length | PA2573 - TMdomain | PA2573 Fulllength | PA4781 REC domain |
| PA2572 REC domain | − | − | − | + | + | ND |
| PA2572 YN-GYP domain | − | − | − | − | − | ND |
| PA2572 Full length | − | − | − | + | + | ND |
| PA2573 - TM domain | ND | ND | ND | ND | ND | − |
(+) indicates a positive protein-protein interaction based on growth of colonies on Selective Screening Media (SSM) (M9+ His-dropout media containing 5 mM 3-AT). (−) indicates a negative interaction based on lack of growth on SSM. (-TM) represents a PA2573 construct without the N-terminal transmembrane domains. Results were observations from three independent experiments.
No interaction was detected between PA2572 and PA2571 in experiments using either the full-length proteins or any of the domains of these proteins. In contrast, protein-protein interactions between PA2572 and the predicted methyl-accepting chemotaxis protein PA2573 were detected in three independent experiments. Furthermore, interactions between PA2572 and the predicted cytoplasmic portion of PA2573 (comprising the HAMP and methyl-accepting domains) and between the REC domain of PA2572 and full-length PA2573 were detected (Fig. 2). No protein-protein interaction was detected between PA2573 and the REC domain of PA4781, an enzymatically-active HD-GYP domain protein, suggesting a specificity of interaction with PA2572. Taken together, these results suggest an interaction between PA2572 and PA2573 mediated by the REC domain of PA2572 and the cytoplasmic portion of PA2573, but no interaction between PA2572 and PA2571. These findings and the virulence phenotypes of different mutants outlined above suggest no role for PA2571 in virulence-related signal transduction involving PA2572. As a consequence, subsequent experiments focussed on the role of PA2573.
Comparison of Phenotypic Effects of Mutation of PA2572 and PA2573
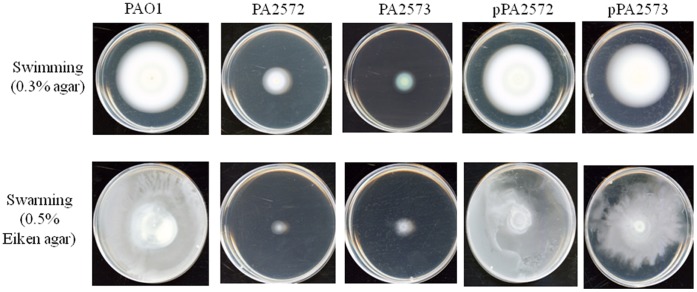
The PA2572 and PA2573 mutant strains were assessed for phenotypes associated with P. aeruginosa virulence including production of pyocyanin and the siderophore pyoverdine as well as motility. Mutation of PA2572 and PA2573 resulted in strains with a reduced ability to produce pyoverdine (Fig. 3A). Complementation of the mutations with wild-type copies of the genes cloned into the pBBR1MCS vector restored pyoverdine levels to near wild-type. Mutation of PA2573 also led to a reduction of pyocyanin production, although this effect was not seen in the PA2572 mutant (Fig. 3B). Complementation of the PA2573 mutations restored pyocyanin levels to wild-type (Fig. 3B). Disruption of PA2572 and PA2573 had a notable influence on both swimming and swarming motility; mutant strains displayed a significant reduction in motility compared to PAO1 (Fig. 4). Complementation of mutant strains restored both swarming and swimming motility to wild-type levels.
Figure 3. Production of virulence factors.
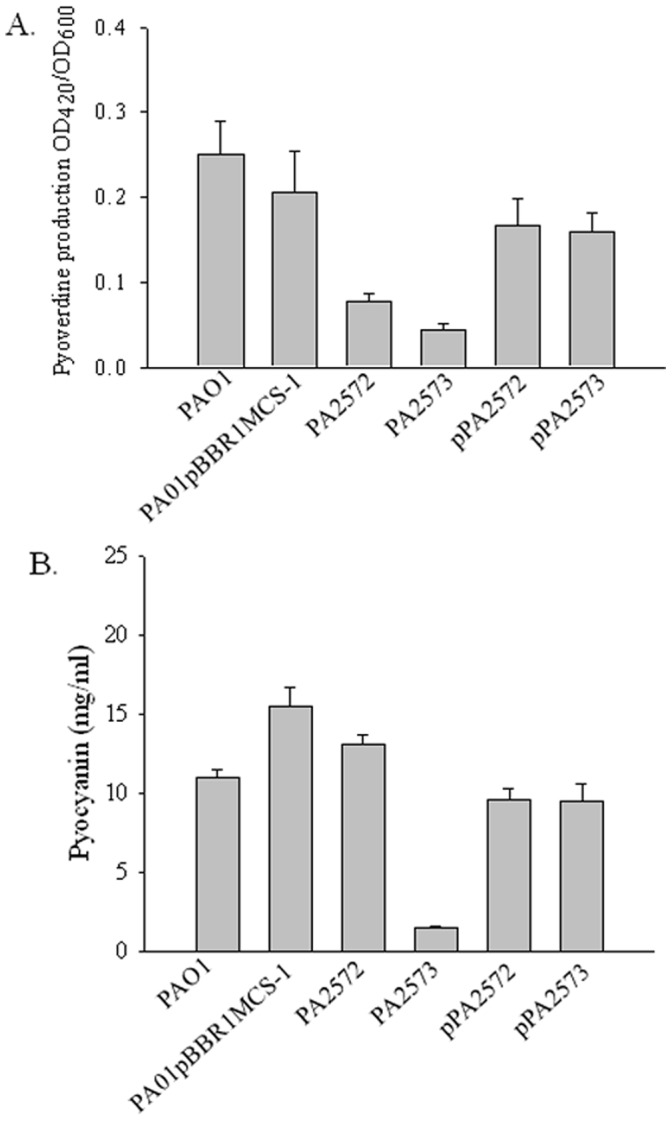
Effects of mutations of PA2572 and PA2573 on production of pyoverdine (A) and pyocyanin (B) in P. aeruginosa. Complemented strains (indicated by pPA2572 and pPA2573) had wild-type levels of the factors. Phenotypic effects of mutations were complemented with reintroduction of genes into mutant strains. Error bars represent the mean ± standard deviation of three independent experiments in triplicate.
Figure 4. Motility of strains.
Motility phenotypes of P. aeruginosa for swimming (0.3% agar) and swarming (0.5% Eiken agar) after 24 hours at 37°C. Mutant strains (indicated by PA2572 and PA2573) have reduced swimming and swarming, whereas complemented strains (indicated by pPA2572 and pPA2573) had wild-type levels of motility. Each plate is representative of the motility phenotype observed for strains in triplicate in three independent experiments.
Comparative Global Gene Expression Profiles in PA2572 and PA2573 Mutant Strains
To compare the influence of PA2572 and PA2573 on gene expression, the transcriptomes of PA2572 and PA2573 mutant strains and wild-type were compared through microarray analysis using the P. aeruginosa Affymetrix GeneChip. For these experiments, RNA was isolated from bacterial strains growing in exponential phase in LB medium. Transcriptome profile analysis revealed that expression of 152 genes was significantly altered (≥2.0-fold) by the disruption of PA2572 (Fig. 5). Disruption of PA2573 appeared to have a broader influence on global gene expression as 450 genes were significantly differentially expressed in the mutant compared to PAO1. PA2572 and PA2573 influenced expression of a common set of 49 genes: 24 genes with an increase in expression and 25 genes with a decrease in expression (Fig. 5). These genes were involved in a range of biological functions including virulence, membrane transport, multidrug resistance, amino acid biosynthesis and signal transduction. Additionally, microarray analysis revealed 12 genes whose expression levels were altered in opposite directions following mutation of PA2572 and PA2573. These genes included pqsA involved in the biosynthesis of the Pseudomonas quinolone signal (PQS) [17] and several genes of the lipopolysaccharide modification operon arnBCADTEF [18], [19]. Differentially expressed genes in mutant strains compared to PAO1 with a fold change ≥3.0 are listed in Tables S3 and Table S4 along with their assigned annotations.
Figure 5. Venn diagrams representing unique and overlapping gene regulation in PA2572 and PA2573 mutants.
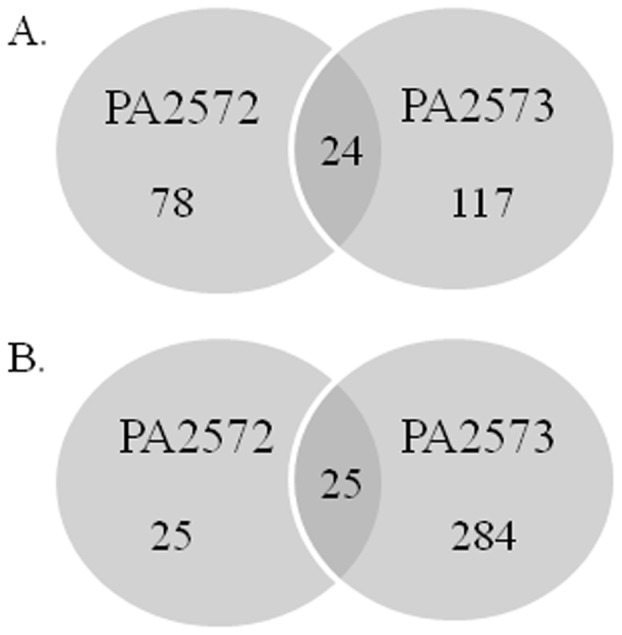
Mutant strains were compared to the parental PAO1 strain during exponential growth phase (0.6–0.7 OD600) in LB media. Genes were considered to have significant alteration in expression based on two-fold changes compared with wild-type. The numbers of genes regulated in an upward direction (A) and a downward direction (B) are shown.
Notably, expression of several genes encoding components of the P. aeruginosa type III secretion system were down-regulated in both PA2572 and PA2573 mutants compared to the wild-type. These virulence genes included those encoding PopB and PopD, which are essential for the translocation process, the chaperone PcrH, and the secreted toxin ExoS [20], [21]. Disruption of PA2572 and PA2573 also led to common expression of a number of genes involved in signal transduction including PA4781 encoding an HD-GYP domain phosphodiesterase [12], PA3271 encoding a probable two-component sensor, and pctC encoding a chemotactic transducer.
Both semi-quantitative and qRT-PCR methods were used to confirm alterations in gene expression revealed by microarray analysis (Table 2). The genes selected for these analyses represented those with a range of fold change of expression and of diverse functional classes. The relative expression levels of seven genes measured using qRT-PCR and ten genes using semi-quantitative RT-PCR reflected in each case the differences in gene expression observed by transcriptome analysis (Table 2).
Table 2. Validation of transcriptome data using quantitative and semi-quantitative real-time PCR.
| Transcriptome | qRT-PCR | Transcriptome | qRT-PCR | ||
| Gene | Description | Fold changePA2572/PAO1 (>2.0) | Fold changePA2572/PAO1 | Fold changePA2573/PAO1(>2.0) | Fold changePA2573/PAO1 |
| PA1425 | probable ATP-binding component of ABCtransporter | 4.51 | 2.89 | NS | −0.49 |
| PA2019 | mexX, RND multidrug efflux membranefusion protein precursor | 16.92 | 36.5 | 12.66 | 12.8 |
| PA3530 | conserved hypothetical protein | 5.09 | 9.03 | 4.66 | 2.31 |
| PA4290 | probable chemotaxis transducer | 2.86 | 5.72 | NS | 1.65 |
| PA4307 | pctC, chemotactic transducer PctC | −2.20 | 0.95 | −3.73 | −9.06 |
| PA4527 | pilC, still frameshift type 4 fimbrialbiogenesis protein PilC | NS | 0.62 | −4.83 | −8.12 |
| PA4825 | mgtA, Mg(2+) transport ATPase, P-type 2 | 10.89 | 14.1 | NS | −0.23 |
| Transcriptome | Semi-q RT-PCR | Transcriptome | Semi-q RT-PCR | ||
| Gene | Description | Fold change PA2572/PAO1 (>2.0) | Fold change PA2572/PAO1 | Fold change PA2573/PAO1(>2.0) | Fold change PA2573/PAO1 |
| PA0517 | nirC, probable c-type cytochrome precursor | NS | NC | −14.71 | − |
| PA0830 | hypothetical protein | −4.12 | – | −3.77 | − |
| PA1326 | ilvA2, threonine dehydratase | 11.80 | + | 5.57 | + |
| PA1710 | exsC, ExsC, exoenzyme S synthesis proteinC precursor | NS | NC | −4.35 | − |
| PA1718 | pscE, type III export protein PscE | NS | NC | −30.95 | − |
| PA1797 | hypothetical protein | 56.20 | + | NS | NC |
| PA2513 | antB, anthranilate dioxygenase small subunit | NS | NC | 29.35 | + |
| PA3190 | probable binding protein component(ABC sugar transporter) | −6.65 | − | NS | NC |
| PA4825 | mgtA, Mg(2+) transport ATPase | 10.89 | + | NS | NC |
| PA5471 | hypothetical protein | 7.01 | + | 3.14 | + |
Abbreviations and symbols: NS (Not significant, fold change was less than 2.0), RND (Resistance-Nodulation-Cell Division), + (represents increased gene expression in mutant strain based on increased abundance of PCR amplicon compared to PAO1), − (represents decreased gene expression in mutant strain based on decreased abundance of PCR amplicon compared to PAO1), NC (no apparent change in abundance was observed).
PA2572 and PA2573 Regulate Expression of Genes Involved in the Antibiotic Tolerance of P. aeruginosa
Transcriptome profiling revealed that both PA2572 and PA2573 regulate a number of genes associated with antibiotic tolerance (Table 3). Significant increases in expression (∼9- to 17-fold) were observed for mexX and mexY that encode Resistance-Nodulation-Cell Division (RND) multidrug efflux proteins involved in resistance to aminoglycoside antibiotics in P. aeruginosa [22], [23]. The oprD gene, that encodes an outer membrane porin that facilitates the uptake of carbapenem antibiotics [24], was also seen to be downregulated in both mutant strains compared to the wild-type. Conversely, disruption of PA2572 and PA2573 resulted in the differential regulation of pmrA, a gene encoding a two-component regulator associated with resistance to cationic antimicrobial peptides [18]; expression of pmrA increased in the PA2572 mutant but decreased in the PA2573 mutant when compared to wild-type. Additionally, mutation of PA2572 also led to the up-regulation of pmrB encoding the sensor kinase component of this two-component system.
Table 3. Transcriptome profiles highlight regulation of genes related to antibiotic resistance in PA2572 and PA2573 mutants.
| Transcriptome | Transcriptome | ||||
| Gene | Description | Fold change PA2572/PAO1 (>2.0) | Fold change PA2573/PAO1 (>2.0) | Antibiotic* | Ref. |
| PA0958 | oprD, Basic amino acid, basic peptide andimipenem outer membrane porin OprD precursor | −3.24 | −2.01 | Meropenem | [33], [34] |
| PA1797 | hypothetical protein harboring beta-lactamaseconserved domain (Pfam) | 56.20 | NS | β-lactam (Ceftazidime)/Polymyxin B | [19] |
| PA2018 | mexY, Resistance-Nodulation-Cell Division (RND)multidrug efflux transporter | 11.14 | 9.04 | Amikacin/Tobramycin | [22], [23], [35] |
| PA2019 | mexX, Resistance-Nodulation-Cell Division (RND) multidrugefflux membrane fusion protein precursor | 16.92 | 12.66 | Amikacin/Tobramycin | [22], [23], [35] |
| PA4200 | hypothetical protein harboring metallo Beta-lactamase_Band B_2 domains (Pfam) | −3.82 | NS | β-lactam (Ceftazidime) | |
| PA4599 | mexC, Resistance-Nodulation-Cell Division (RND) multidrug efflux membrane fusion protein MexC precursor | 4.45 | NS | Ciprofloxacin | [35], [36] |
| PA4776 | pmrA, two-component regulator system responseregulator PmrA | 3.60 | −2.01 | Polymyxin B/Colistin | [18], [37] |
| PA4777 | pmrB, two-component regulator system signalsensor kinase PmrB | 4.36 | NS | Polymyxin B/Colistin | [18], [37] |
Antibiotics in which the expression of listed genes has been implemented in resistance or susceptibility. References are indicated.
These effects on gene expression prompted us to examine the antibiotic tolerance of the different P. aeruginosa strains. Minimum inhibitory concentrations (MIC) of antibiotics towards P. aeruginosa strains grown to exponential phase were assessed using E-test strips (Biomerieux) after 24 hours (Table 4). Both PA2572 and PA2573 mutants demonstrated substantially increased tolerance to the aminoglycosides amikacin and tobramycin compared to the parental PAO1 strain (Table 4). Complementation of the mutations restored susceptibility to both aminoglycosides towards wild-type levels. Mutation of PA2572 led to increased tolerance to the cationic antimicrobial peptides, polymyxin B and colistin, although mutation of PA2573 had the opposite effect. Intriguingly, genes involved in resistance appeared to be divergently transcribed. Tolerance to ceftazidime, meropenem and ciprofloxacin was minimally affected by disruption of PA2572 and PA2573.
Table 4. Minimum inhibitory concentrations of antibiotics toward P. aeruginosa strains grown on LB agar.
| MIC (µg/ml) | |||||||
| Strain | Amikacin | Tobramycin | Polymixin B | Colistin | Ceftazidime | Ciprofloxacin | Meropenem |
| PAO1 | 6.67 | 1.41 | 2.83 | 4.17 | 4.33 | 0.21 | 0.25 |
| PA2572 | >254 | 32 | 4.67 | 10 | 2.33 | 0.21 | 0.29 |
| PA2573 | 122.67 | 14 | 1.83 | 1.67 | 3.83 | 0.25 | 0.38 |
| pPA2572 | 11.33 | 1.83 | 4 | 7.33 | 4.33 | 0.21 | 0.46 |
| pPA2573 | 10 | 2 | 2.67 | 4 | 4 | 0.29 | 0.38 |
MICs were measured using Antibiotic E-test strips (Biomerieux) and results shown are the average of 6 independent experiments.
Expression of the PA2572 YN-GYP Domain Partially Restores Virulence of the PA2573 Mutant
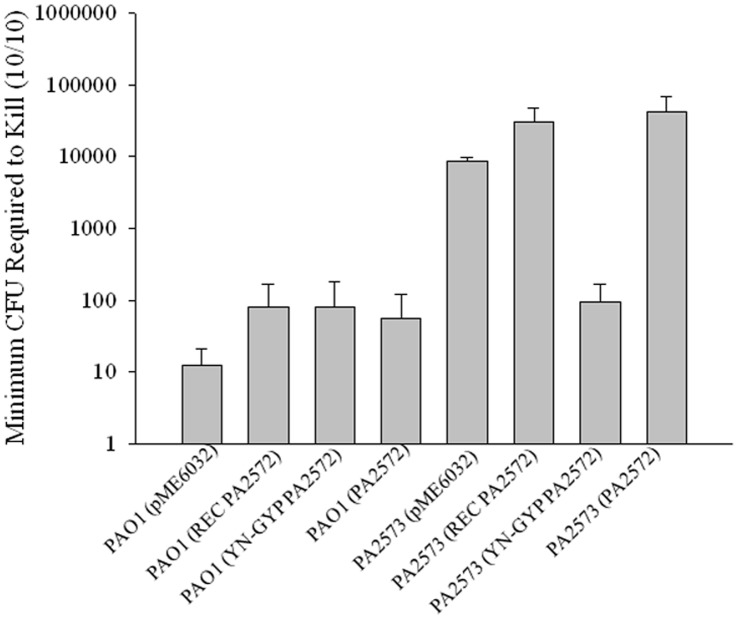
The findings described above indicate a role for the PA2573 sensor in the regulation of virulence and antibiotic tolerance in P. aeruginosa. The overlap in regulatory influence of PA2572 and PA2573 at the level of gene transcription and on various phenotypes including virulence suggests that these effects may be exerted in part through signal transduction involving PA2572. For a number of two-component systems, it has been shown that over-expression of the response regulator can restore the phenotypes of a mutant lacking the related sensor to wild-type phenotypes [25], [26]. Accordingly, we assessed the effects of expression of the full length PA2572, and the component REC or YN-GYP domains on the virulence of the PA2573 mutant. For this experiment the DNA fragments were cloned into pME6032 and introduced into PA2573 mutant and wild-type, as a control. Subsequently, all strains were assessed for their virulence potential in the Galleria mellonella model of infection (Fig. 6, S2). The empty pME6032 vector had no effect on the virulence of PA2573 mutant or wild-type strain (Fig. 6). While expression of the REC domain and the full length PA2572 in the PA2573 mutant had a small negative influence on virulence, introduction of the YN-GYP domain resulted in a significant increase in virulence (Fig. 6).
Figure 6. Pathogenesis of strains expressing PA2572.
Effects of expressing partial and full-length fragments of PA2572 in the PA2573 mutant and in the wild-type on virulence in the Galleria mellonella model. Expression of the empty pME6032 vector in strains served as a control. Virulence potential of P. aeruginosa strains was assessed based on the number of CFU required to kill a group of ten larvae 24 hours post-infection. Error bars represent mean ± standard deviation of at least three experiments.
Discussion
The work in this paper had the aim of identifying signalling partners for the putative response regulator PA2572, which we have previously shown to contribute to the virulence of P. aeruginosa to Galleria mellonella larvae. Two candidates were examined: the probable two-component sensor kinase PA2571 and probable methyl-accepting chemotaxis protein PA2573. It has been shown previously that PA2573 is co-expressed with PA2572 under several conditions including aerobic growth in the presence of nitrate and in bacteria co-cultured with respiratory epithelia cells [13]–[15]. Here we present a number of lines of evidence that suggest regulatory interplay between PA2573 and PA2572. We have shown that PA2572 and PA2573 are both involved in the regulation of motility, virulence and antibiotic tolerance and influence the expression of a common set of 49 genes. Furthermore, we have demonstrated using bacterial two-hybrid analysis that protein-protein interactions can occur between PA2573 and PA2572 and that these involve the REC domain of PA2572. In contrast, PA2571 does not influence virulence and no interaction with PA2572 was evident in bacterial two-hybrid analysis. The findings indicate for the first time a role for the orphan chemotaxis sensor PA2573 in the regulation of virulence and antibiotic tolerance in P. aeruginosa and suggest that the action of PA2573 is exerted in part through signal transduction involving PA2572.
For a number of two-component systems it has been shown that over-expression of the response regulator can restore to wild-type the phenotypes of a mutant lacking the related sensor. Overproduction of response regulators is often thought to mimic the physiological phosphorylation response. For many two-component regulators, interactions between the unphosphorylated receiver domain and the effector domain prevent effector domain activity by restricting it in an unfavorable conformation; phosphorylation of the REC domain relieves this inhibition [27]. However over-expression of PA2572 had a small but negative effect on the virulence of the PA2573 mutant. Intriguingly, over-expression of the YN-GYP domain of PA2572 gave a substantial increase in virulence, which was restored towards wild-type levels. We speculate that this effect is due to release of the YN-GYP effector domain from a negative influence of the REC domain.
While genes encoding cognate two-component signal transduction proteins are often found flanking one another on the genome, there is evidence that a number of signalling systems employ orphan histidine kinases (HK) and response regulators not encoded in the genomic vicinity of their partners [28]–[32]. Under the conditions tested, PA2571 does not appear to have a role in virulence of P. aeruginosa and there was no evidence for an interaction between PA2571 and PA2572. While it is possible that an orphan HK is part of the machinery for this proposed signalling pathway involving PA27573 and PA2572, we cannot exclude that that regulatory interplay between PA2571 and PA2572 occurs under different environmental conditions.
Mutation of PA2573 influenced the expression of a broader range of genes than mutation of PA2572, suggesting that PA2573 may interact with additional response regulators in other signaling pathways. However mutation of either PA2573 or PA2572 leads to enhanced expression of genes associated with antibiotic tolerance, which is reflected in increased MICs for the aminoglycoside antibiotics amikacin and tobramycin in these mutant strains. Mutation of PA2573 or PA2572 both lead to the down-regulation of several genes encoding virulence factors including those involved in type III secretion (popB, popD, pcrH, as well as the secreted effector exoS (Tables S3 and S4). This is reflected in a decreased virulence of both mutants.
Several laboratories have described modified chemotaxis systems that are linked to alterations in the levels of nucleotide second messengers in P. aeruginosa. The Wsp system functions to regulate cellular processes such as EPS production and biofilm formation [7], [8]. Transduction of environmental signals through the MCP WspA and a soluble sensor kinase WspE leads to phosphorylation of the response regulator WspR, activating this GGDEF domain protein for the synthesis of cyclic di-GMP. The Chp chemosensory system is linked to cAMP-dependent virulence response in P. aeruginosa through modulation of adenylate kinase activity [2]. In a similar fashion, we propose that a novel signalling pathway that includes an orphan chemosensory protein component (PA2573) and a response regulator (PA2572) with a variant HD-GYP domain acts in regulation of factors promoting the virulence and antibiotic tolerance of P. aeruginosa. Although the mechanisms by which PA2572 exerts a regulatory action remain obscure, the discovery of a role of PA2573 in virulence in model organisms suggests that further studies of this system are warranted.
Supporting Information
Organization of PA2572 and flanking genes in the P. aeruginosa PAO1 chromosome. Region is drawn approximately to scale and arrows represent gene orientation.
(TIF)
Pathogenesis of the PA2572 mutant expressing PA2572. Effects of expressing partial and full-length fragments of PA2572 in the PA2572 mutant on virulence in the Galleria mellonella model. Expression of the empty pME6032 vector served as a control. Virulence potential of P. aeruginosa strains was assessed based on the number of CFU required to kill a group of ten larvae 24 hours post-infection. Error bars represent mean ± standard deviation of at least three experiments.
(TIF)
Bacterial strains and plasmids used in this study.
(DOCX)
Primers used in this study.
(DOCX)
Genes regulated in the PA2572 mutant compared to the wild-type PAO1strain during exponential growth in LB media (>3.0-fold).
(DOCX)
Genes regulated in the PA2573 mutant compared to the wild-type PAO1strain during exponential growth in LB media (>3.0-fold).
(DOCX)
Funding Statement
This work has been supported by grants from Science Foundation of Ireland (SFI07/NI.1/B955, SFI09/SSIRG/B1654) through an investigator award (to J.M.D) and by the Wellcome Trust (WT093314MA) through a project grant (to J.M.D and R.P.R), which are gratefully acknowledged. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Gooderham WJ, Hancock RE (2009) Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol Rev 33: 279–294. [DOI] [PubMed] [Google Scholar]
- 2. Fulcher NB, Holliday PM, Klem E, Cann MJ, Wolfgang MC (2010) The Pseudomonas aeruginosa Chp chemosensory system regulates intracellular cAMP levels by modulating adenylate cyclase activity. Mol Microbiol 76: 889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Jimenez PN, Koch G, Thompson JA, Xavier KB, Cool RH, et al. (2012) The multiple signaling systems regulating virulence in Pseudomonas aeruginosa. Microbiol Mol Biol Rev 76: 46–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barrett JF, Hoch JA (1998) Two-component signal transduction as a target for microbial anti-infective therapy. Antimicrob Agents Chemother 42: 1529–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stover CK, Pham XQ, Erwin AL, Mizoguchi SD, Warrener P, et al. (2000) Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406: 959–964. [DOI] [PubMed] [Google Scholar]
- 6. Galperin MY (2006) Structural classification of bacterial response regulators: diversity of output domains and domain combinations. J Bacteriol 188: 4169–4182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Guvener ZT, Harwood CS (2007) Subcellular location characteristics of the Pseudomonas aeruginosa GGDEF protein, WspR, indicate that it produces cyclic-di-GMP in response to growth on surfaces. Mol Microbiol 66: 1459–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hickman JW, Tifrea DF, Harwood CS (2005) A chemosensory system that regulates biofilm formation through modulation of cyclic diguanylate levels. Proc Natl Acad Sci U S A 102: 14422–14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hengge R (2009) Principles of c-di-GMP signalling in bacteria. Nat Rev Microbiol 7: 263–273. [DOI] [PubMed] [Google Scholar]
- 10. Romling U (2011) Cyclic di-GMP, an established secondary messenger still speeding up. Environ Microbiol. [DOI] [PubMed]
- 11. Ryan RP, Tolker-Nielsen T, Dow JM (2012) When the PilZ don’t work: effectors for cyclic di-GMP action in bacteria. Trends in Microbiology. 1–8. [DOI] [PubMed]
- 12. Ryan RP, Lucey J, O’Donovan K, McCarthy Y, Yang L, et al. (2009) HD-GYP domain proteins regulate biofilm formation and virulence in Pseudomonas aeruginosa. Environ Microbiol 11: 1126–1136. [DOI] [PubMed] [Google Scholar]
- 13. Filiatrault MJ, Wagner VE, Bushnell D, Haidaris CG, Iglewski BH, et al. (2005) Effect of anaerobiosis and nitrate on gene expression in Pseudomonas aeruginosa. Infect Immun 73: 3764–3772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schuster M, Lostroh CP, Ogi T, Greenberg E (2003) Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J Bacteriol 185: 2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chugani S, Greenberg E (2007) The influence of human respiratory epithelia on Pseudomonas aeruginosa gene expression. Microbial pathogenesis 42: 29–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Drenkard E (2003) Antimicrobial resistance of Pseudomonas aeruginosa biofilms. Microbes Infect 5: 1213–1219. [DOI] [PubMed] [Google Scholar]
- 17. McGrath S, Wade DS, Pesci EC (2004) Dueling quorum sensing systems in Pseudomonas aeruginosa control the production of the Pseudomonas quinolone signal (PQS). FEMS microbiology letters 230: 27–34. [DOI] [PubMed] [Google Scholar]
- 18. McPhee JB, Lewenza S, Hancock REW (2003) Cationic antimicrobial peptides activate a two-component regulatory system, PmrA-PmrB, that regulates resistance to polymyxin B and cationic antimicrobial peptides in Pseudomonas aeruginosa. Mol Microbiol 50: 205–217. [DOI] [PubMed] [Google Scholar]
- 19. Fernandez L, Gooderham WJ, Bains M, McPhee JB, Wiegand I, et al. (2010) Adaptive resistance to the” last hope” antibiotics polymyxin B and colistin in Pseudomonas aeruginosa is mediated by the novel two-component regulatory system ParR-ParS. Antimicrob Agents Chemother 54: 3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schoehn G, Di Guilmi AM, Lemaire D, Attree I, Weissenhorn W, et al. (2003) Oligomerization of type III secretion proteins PopB and PopD precedes pore formation in Pseudomonas. The EMBO journal 22: 4957–4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nicas TI, Bradley J, Lochner JE, Iglewski BH (1985) The role of exoenzyme S in infections with Pseudomonas aeruginosa. J Infect Dis 152: 716–721. [DOI] [PubMed] [Google Scholar]
- 22. Aires JR, Kohler T, Nikaido H, Plesiat P (1999) Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob Agents Chemother 43: 2624–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Westbrock-Wadman S, Sherman DR, Hickey MJ, Coulter SN, Zhu YQ, et al. (1999) Characterization of a Pseudomonas aeruginosa efflux pump contributing to aminoglycoside impermeability. Antimicrob Agents Chemother 43: 2975–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Trias J, Nikaido H (1990) Outer membrane protein D2 catalyzes facilitated diffusion of carbapenems and penems through the outer membrane of Pseudomonas aeruginosa. Antimicrob Agents Chemother 34: 52–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dong YH, Zhang XF, An SW, Xu JL, Zhang LH (2008) A novel two-component system BqsS-BqsR modulates quorum sensing-dependent biofilm decay in Pseudomonas aeruginosa. Commun Integr Biol 1: 88–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D (2001) Molecular basis of thermosensing: a two-component signal transduction thermometer in Bacillus subtilis. EMBO J 20: 1681–1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gao R, Stock AM (2010) Molecular strategies for phosphorylation-mediated regulation of response regulator activity. Curr Opin Microbiol 13: 160–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ulrich LE, Zhulin IB (2010) The MiST2 database: a comprehensive genomics resource on microbial signal transduction. Nucleic acids research 38: D401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petters T, Zhang X, Nesper J, Treuner-Lange A, Gomez-Santos N, et al. (2012) The orphan histidine protein kinase SgmT is a c-di-GMP receptor and regulates composition of the extracellular matrix together with the orphan DNA binding response regulator DigR in Myxococcus xanthus. Mol Microbiol 84: 147–165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Wang W, Shu D, Chen L, Jiang W, Lu Y (2009) Cross-talk between an orphan response regulator and a noncognate histidine kinase in Streptomyces coelicolor. FEMS Microbiol Lett 294: 150–156. [DOI] [PubMed] [Google Scholar]
- 31. Pan X, Ge J, Li M, Wu B, Wang C, et al. (2009) The orphan response regulator CovR: a globally negative modulator of virulence in Streptococcus suis serotype 2. J Bacteriol 191: 2601–2612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wuichet K, Cantwell BJ, Zhulin IB (2010) Evolution and phyletic distribution of two-component signal transduction systems. Curr Opin Microbiol 13: 219–225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Köhler T, Michea-Hamzehpour M, Epp SF, Pechere JC (1999) Carbapenem activities against Pseudomonas aeruginosa: respective contributions of OprD and efflux systems. Antimicrob Agents Chemother 43: 424–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Quale J, Bratu S, Gupta J, Landman D (2006) Interplay of efflux system, ampC, and oprD expression in carbapenem resistance of Pseudomonas aeruginosa clinical isolates. Antimicrob Agents Chemother 50: 1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aeschlimann JR (2003) The role of multidrug efflux pumps in the antibiotic resistance of Pseudomonas aeruginosa and other gram-negative bacteria. Insights from the Society of Infectious Diseases Pharmacists. Pharmacotherapy 23: 916–924. [DOI] [PubMed] [Google Scholar]
- 36. Poole K, Gotoh N, Tsujimoto H, Zhao Q, Wada A, et al. (1996) Overexpression of the mexC-mexD-oprJ efflux operon in nfxB-type multidrug-resistant strains of Pseudomonas aeruginosa. Mol Microbiol 21: 713–724. [DOI] [PubMed] [Google Scholar]
- 37. Moskowitz SM, Ernst RK, Miller SI (2004) PmrAB, a two-component regulatory system of Pseudomonas aeruginosa that modulates resistance to cationic antimicrobial peptides and addition of aminoarabinose to lipid A. J Bacteriol. 186: 575. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Organization of PA2572 and flanking genes in the P. aeruginosa PAO1 chromosome. Region is drawn approximately to scale and arrows represent gene orientation.
(TIF)
Pathogenesis of the PA2572 mutant expressing PA2572. Effects of expressing partial and full-length fragments of PA2572 in the PA2572 mutant on virulence in the Galleria mellonella model. Expression of the empty pME6032 vector served as a control. Virulence potential of P. aeruginosa strains was assessed based on the number of CFU required to kill a group of ten larvae 24 hours post-infection. Error bars represent mean ± standard deviation of at least three experiments.
(TIF)
Bacterial strains and plasmids used in this study.
(DOCX)
Primers used in this study.
(DOCX)
Genes regulated in the PA2572 mutant compared to the wild-type PAO1strain during exponential growth in LB media (>3.0-fold).
(DOCX)
Genes regulated in the PA2573 mutant compared to the wild-type PAO1strain during exponential growth in LB media (>3.0-fold).
(DOCX)