Abstract
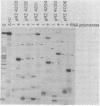
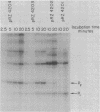
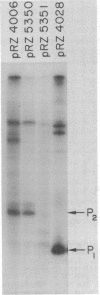
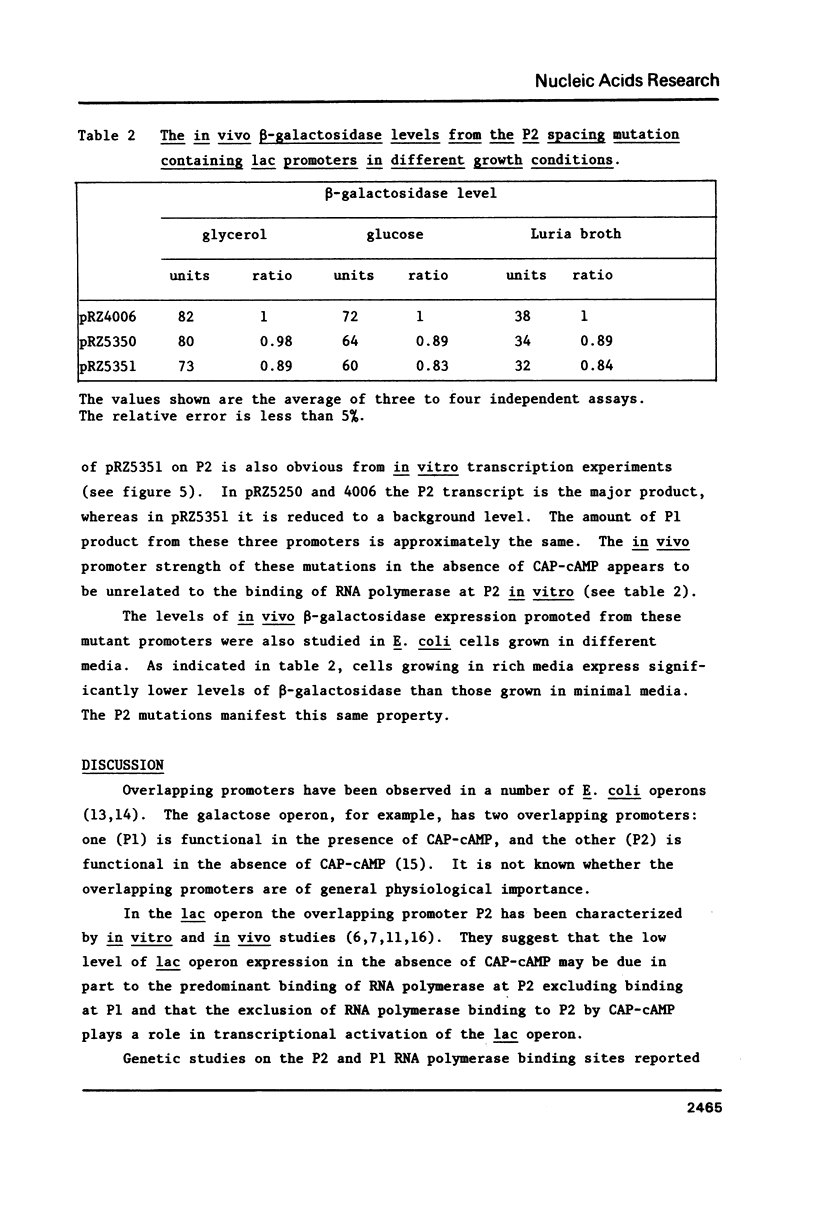
The Escherichia coli lactose (lac) operon transcription control region includes at least two sequences which are recognized by RNA polymerase holoenzyme in vitro, the normal lac promoter (termed P1) and an overlapping upstream promoter (termed P2). The structure of the P2 and the effect of RNA polymerase interaction at P2 on the association of RNA polymerase with P1 was analyzed by the isolation and characterization of various mutations at P2. A set of deletions with varying lengths of DNA between the lac P2 -10 region and a "-35 region" contributed by the vector DNA were constructed. In vitro studies indicate that as the spacing between the -10 region and "-35 region" is increased from 16 to 22 base pairs (bp), the steady state occupancy as measured by exonuclease III protection experiments and the ability to initiate transcripts from P2 decrease. Studies were also conducted using a single base pair insertion and a two base pair deletion between the natural -35 and -10 regions of P2. The mutation which decreases the in vitro occupancy and transcription initiation potential of P2 does not significantly affect the steady state in vitro occupancy of P1 nor the in vivo expression of the lac operon. These results are not consistent with the model that RNA polymerase occupancy at P2 competes with the P1 expression and therefore that this competition plays a role in cAMP bound catabolite gene activator protein (CAP-cAMP) control of the lac operon.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maizels N. M. The nucleotide sequence of the lactose messenger ribonucleic acid transcribed from the UV5 promoter mutant of Escherichia coli. Proc Natl Acad Sci U S A. 1973 Dec;70(12):3585–3589. doi: 10.1073/pnas.70.12.3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majors J. Initiation of in vitro mRNA synthesis from the wild-type lac promoter. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4394–4398. doi: 10.1073/pnas.72.11.4394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malan T. P., McClure W. R. Dual promoter control of the Escherichia coli lactose operon. Cell. 1984 Nov;39(1):173–180. doi: 10.1016/0092-8674(84)90203-4. [DOI] [PubMed] [Google Scholar]
- Maquat L. E., Reznikoff W. S. In vitro analysis of the Escherichia coli RNA polymerase interaction with wild-type and mutant lactose promoters. J Mol Biol. 1978 Nov 15;125(4):467–490. doi: 10.1016/0022-2836(78)90311-x. [DOI] [PubMed] [Google Scholar]
- Musso R. E., Di Lauro R., Adhya S., de Crombrugghe B. Dual control for transcription of the galactose operon by cyclic AMP and its receptor protein at two interspersed promoters. Cell. 1977 Nov;12(3):847–854. doi: 10.1016/0092-8674(77)90283-5. [DOI] [PubMed] [Google Scholar]
- Spassky A., Busby S., Buc H. On the action of the cyclic AMP-cyclic AMP receptor protein complex at the Escherichia coli lactose and galactose promoter regions. EMBO J. 1984 Jan;3(1):43–50. doi: 10.1002/j.1460-2075.1984.tb01759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. M., Reznikoff W. S. Deletion analysis of the CAP-cAMP binding site of the Escherichia coli lactose promoter. Nucleic Acids Res. 1984 Jul 11;12(13):5449–5464. doi: 10.1093/nar/12.13.5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Berg E., Zwetsloot J., Noordermeer I., Pannekoek H., Dekker B., Dijkema R., van Ormondt H. The structure and function of the regulatory elements of the Escherichia coli uvrB gene. Nucleic Acids Res. 1981 Nov 11;9(21):5623–5643. doi: 10.1093/nar/9.21.5623. [DOI] [PMC free article] [PubMed] [Google Scholar]