Abstract
CpG island promoters often lack canonical core promoter elements such as the TATA box, and have dispersed transcription initiation sites. Despite the prevalence of CpG islands associated with mammalian genes, the mechanism of transcription initiation from CpG island promoters remains to be clarified. Here we investigate the mechanism of transcription initiation of the CpG island-associated gene, insulin-degrading enzyme (IDE). IDE is ubiquitously expressed, and has dispersed transcription initiation sites. The IDE core promoter locates within a 32-bp region, which contains three CGGCG repeats and a nuclear respiratory factor 1 (NRF-1) binding motif. Sequential mutation analysis indicates that the NRF-1 binding motif is critical for IDE transcription initiation. The NRF-1 binding motif is functional, because NRF-1 binds to this motif in vivo and this motif is required for the regulation of IDE promoter activity by NRF-1. Furthermore, the NRF-1 binding site in the IDE promoter is conserved among different species, and dominant negative NRF-1 represses endogenous IDE expression. Finally, TATA-box binding protein (TBP) is not associated with the IDE promoter, and inactivation of TBP does not abolish IDE transcription, suggesting that TBP is not essential for IDE transcription initiation. Our studies indicate that NRF-1 mediates IDE transcription initiation in a TBP-independent manner, and provide insights into the potential mechanism of transcription initiation for other CpG island-associated genes.
Introduction
DNA methylation, an epigenetic modification that regulates chromatin structure and gene expression [1], [2], occurs predominantly at cytosines of CpG dinucleotides in vertebrates [3], [4]. There are three striking features of CpG sites in mammalian genomes. First, the majority (60%–90%) of CpG sites are methylated [5]. Second, mammalian genomes are depleted of CpG sites, a phenomenon called CG suppression. The observed rate of CpG sites is approximately one-fifth of the rate expected based on the GC content [6]. This rarity of CpG sites arises from the spontaneous mutation of methylated cytosines to thymidines by deamination, which converts CpG to TpG dinucleotides [7], [8]. Thirdly, unmethylated CpG sites cluster to form CpG islands [9]. CpG islands, which have a high GC content and a high observed-to-expected CpG ratio relative to the bulk genome, are often associated with mammalian gene promoters, including all housekeeping genes and some tissue-specific genes [10]. Actually, CpG islands serve as an important criterion for gene prediction [11], [12], [13]. In human, it was estimated that approximately 72% of genes are associated with CpG islands [14].
The core promoter is the minimal promoter region necessary to direct transcription initiation. Several core promoter elements, including the TATA box, the Inr (initiator) element, the downstream core promoter element (DPE), the motif ten element (MTE) and TFIIB recognition elements (BREs), have been identified for RNA polymerase II-transcribed genes, and they function individually or cooperatively in recruiting the transcription apparatus to the promoter region [15]. Promoters with these core promoter elements initiate transcription at a focused start site, which varies by only a few nucleotides. However, CpG island promoters often lack these canonical core promoter elements and have dispersed transcription initiation sites over 50–150 nucleotides [16]. Although genes with focused promoters are well studied, the mechanism for transcription apparatus recruitment to CpG island promoters remains to be clarified.
Insulin-degrading enzyme (IDE) is a ubiquitously expressed zinc metalloprotease that degrades several substrates [17], including insulin and β-amyloid (Aβ) [18], [19]. IDE is closely related to diabetes mellitus (DM) and Alzheimer’s disease (AD), because insulin and Aβ play critical roles in the pathogenesis of DM and AD, respectively. GK rats, which are a well-characterized model for type II DM, have IDE missense mutations, and exhibit impaired insulin and Aβ degradation [20]. IDE knockout mice also show accumulation of endogenous Aβ, hyperinsulinemia and glucose intolerance [21]. However, the exact role of IDE in DM pathogenesis seems a little elusive. A recent study indicates that the diabetic phenotype of IDE knockout mice is an emergent compensatory response to chronic hyperinsulinemia, but not a direct consequence of IDE deficiency [22]. In addition, human genetic studies suggest that IDE polymorphisms are associated with the pathogenesis of both type II DM [23] and AD [24]. These results imply that IDE dysfunction may cause DM or AD and underline the importance of characterizing the transcriptional regulation of IDE.
In this article, we focus on the mechanism of IDE transcription initiation. The mouse IDE promoter contains a CpG island and has dispersed transcription initiation sites. Promoter deletion analysis indicates that the IDE core promoter contains a nuclear respiratory factor 1 (NRF-1) binding motif which is essential for transcription initiation. We further demonstrated that NRF-1 binds to the IDE promoter and dominant negative NRF-1 represses IDE transcription. Finally, we showed that TBP is not essential for IDE transcription initiation.
Results
Mapping the Core Promoter Region of Mouse IDE
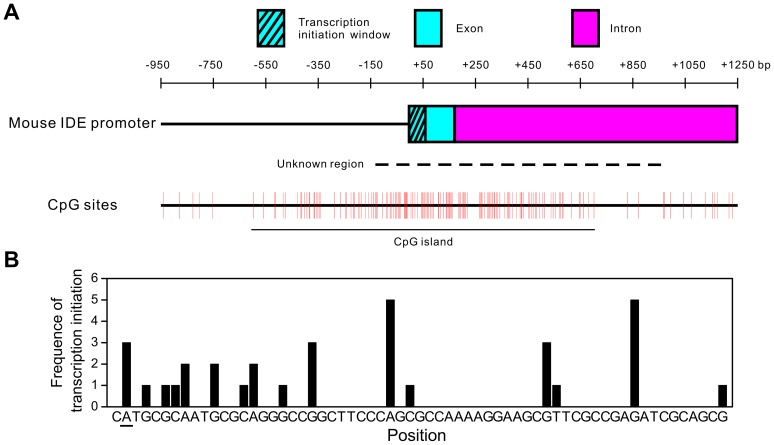
Because the sequence of the mouse IDE promoter was incomplete in the NCBI genomic database (Figure 1A), we cloned and sequenced the unknown region [Genbank: JN038396]. The mouse IDE promoter has a high CpG frequency and contains a CpG island approximately 1300 bp long (Figure 1A). The transcription initiation sites of mouse IDE were determined using the 5′-rapid amplification of cDNA ends (5′-RACE) method. The IDE promoter possesses dispersed transcription initiation sites located within a 62-bp region (Figure 1 A and B). IDE transcription starts with a purine at position +1 at a frequency of 91% and with a pyrimidine-purine dinucleotide at position −1,+1 at a frequency of 67%, which is consistent with previous studies showing that RNA polymerase II-mediated transcription is preferentially initiated at a pyrimidine-purine dinucleotide at position −1,+1 in mammals [16].
Figure 1. The mouse IDE promoter contains a CpG island and has dispersed transcription initiation sites.
(A) Representation of the mouse IDE promoter. The dashed line indicates the unknown region which was cloned and sequenced in this study. The mouse IDE promoter contains a CpG island with a length of approximately 1300 bp. (B) The transcription initiation sites of mouse IDE. The frequency of transcription initiation from different sites is shown. The mouse IDE promoter has dispersed transcription initiation sites located within a window of 62 bp. The first transcription initiation site is underlined.
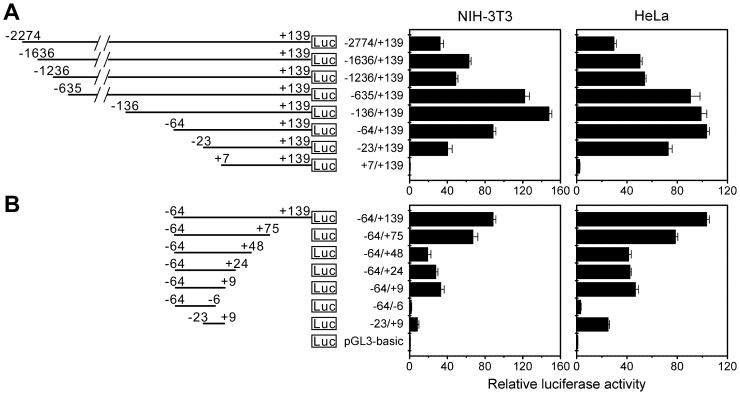
Searching the mouse IDE promoter revealed no TATA box, Inr or other canonical core promoter elements around the transcription initiation sites. To determine the region that is essential for IDE transcription initiation, we constructed luciferase reporter plasmids of the mouse IDE promoter with different truncations at the 5′ or 3′ terminus. For convenience, we set the first transcription initiation site that we identified (Figure 1B) as position +1. Deleting the 5′ terminus of the mouse IDE promoter from −2774 to −23 retained its transcriptional activity, while further deletion to +7 abolished its transcriptional activity (Figure 2A). On the other hand, deleting the 3′ terminus of the mouse IDE promoter from +139 to +9 retained its transcriptional activity, while further deletion to −6 abolished its transcriptional activity (Figure 2B). These results indicate that the region between −23 and +9 of the mouse IDE promoter is essential for transcription initiation and behaves as the core promoter. Indeed, this region was capable of driving transcription initiation in both NIH-3T3 and HeLa cells (Figure 2B).
Figure 2. Mapping the core promoter region of mouse IDE.
(A) and (B) The region between −23 and +9 of the mouse IDE promoter is essential for transcription initiation and behaves as the core promoter. Different truncations of the mouse IDE promoter were cloned into the pGL3-basic vector. NIH-3T3 or HeLa cells were transiently transfected with luciferase reporter plasmids of the mouse IDE promoter (0.8 µg) and Renilla luciferase reporter plasmid (pCMV-RL, 8 ng). Twenty-four hours after transfection, cells were lysed, and the luciferase activity was determined. Firefly luminescence signal was normalized based on the Renilla luminescence signal. Data are represented as fold of the firefly luciferase activity of the pGL3-basic vector.
The IDE Core Promoter Contains a Functional NRF-1 Binding Site that is Essential for Transcription Initiation
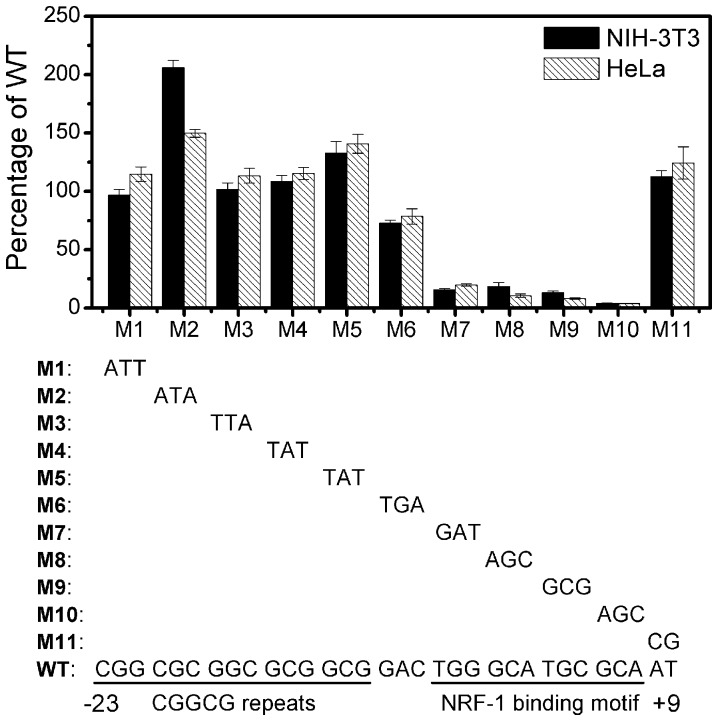
Analysis of the region between −23 and +9 of the mouse IDE promoter revealed three CGGCG repeats and a NRF-1 binding motif (Figure 3). To identify which element is essential for IDE transcription initiation, we sequentially mutated this region and detected changes in its transcriptional activity. Mutations within the NRF-1 binding motif abolished transcription initiation, while mutations in other regions resulted in little or no decrease in the transcriptional activity (Figure 3), suggesting that the NRF-1 binding motif is critical for IDE transcription initiation.
Figure 3. Sequential mutation of the mouse IDE core promoter.
The mouse IDE core promoter contains three CGGCG repeats and a NRF-1 binding motif. The core promoter region with sequential mutations (M1 to M11) was cloned into the pGL3-basic vector. Luciferase activity of the reporter plasmids in NIH-3T3 and HeLa cells is represented as a percentage of the wild-type (WT) reporter plasmid.
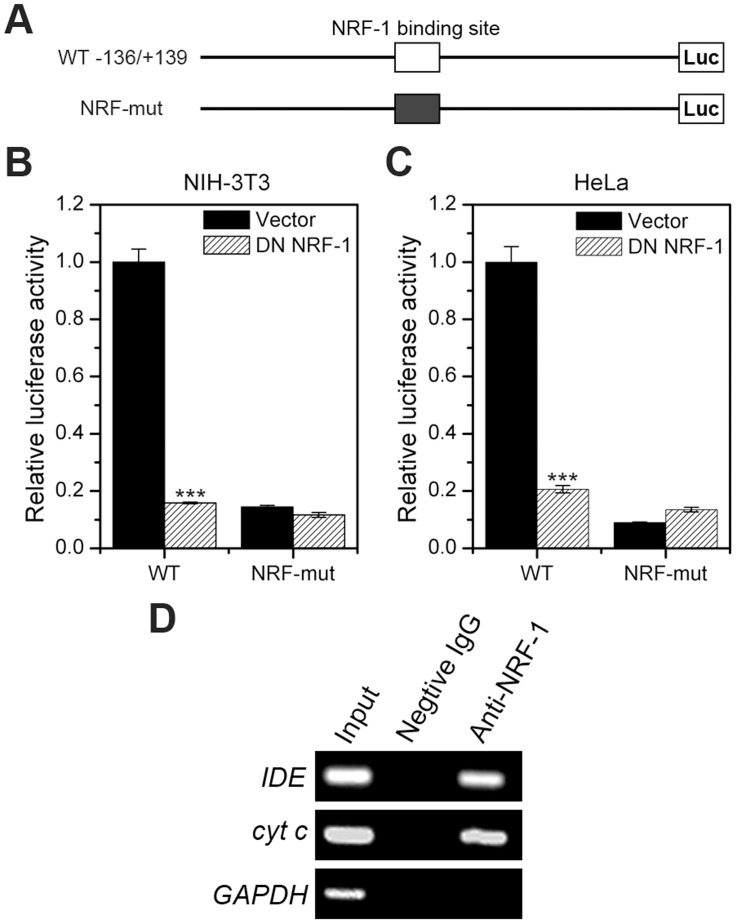
Next, we aimed to demonstrate whether this NRF-1 binding motif is functional. Mammalian cells are not sensitive to NRF-1 stimulation [25], [26]. To determine the effect of NRF-1 on IDE transcription, we employed a dominant negative form of NRF-1which lacks the trans-activating domain [27]. In both NIH-3T3 and HeLa cells, dominant negative NRF-1 repressed IDE promoter activity dramatically (Figure 4 A–C). When the NRF-1 binding motif was mutated, the effect of dominant negative NRF-1 on IDE transcription was abolished (Figure 4 A–C). We subsequently tested whether NRF-1 binds to the predicted NRF-1 binding motif in vivo by chromatin immunoprecipitation assays (ChIP). Indeed, NRF-1 binds to this site and the promoter of a positive control gene, cytochrome c, but not to the promoter of a negative control gene, GAPDH (Figure 4D). PCR products were cloned and sequenced to verify the specificity of the primers. Therefore, the NRF-1 binding motif in the mouse IDE core promoter is functional.
Figure 4. The NRF-1 binding motif in the mouse IDE promoter is functional.
(A) Representation of wild-type (−136/+139 WT) and the NRF-1 binding site-mutated (NRF-mut) luciferase reporter plasmids of the mouse IDE promoter. (B) and (C) Dominant negative NRF-1 represses IDE promoter activity. NIH-3T3 and HeLa cells were transiently co-transfected with wild-type (-136/+139 WT) or NRF-1 binding site-mutated (NRF-mut) IDE reporter plasmids (0.4 µg) and Renilla luciferase plasmid (4 ng) along with or without dominant negative (DN) NRF-1 expression plasmids (0.4 µg). Twenty-four hours after transfection, cells were lysed, and the luciferase activity was examined. Firefly luminescence signal was normalized based on the Renilla luminescence signal. (D) ChIP. NRF-1 binding to the IDE promoter in NIH-3T3 cells was determined by ChIP. The promoter of cytochrome c (cyt c) is used as a positive control for NRF-1 binding, while the promoter of GAPDH acts as a negative control.
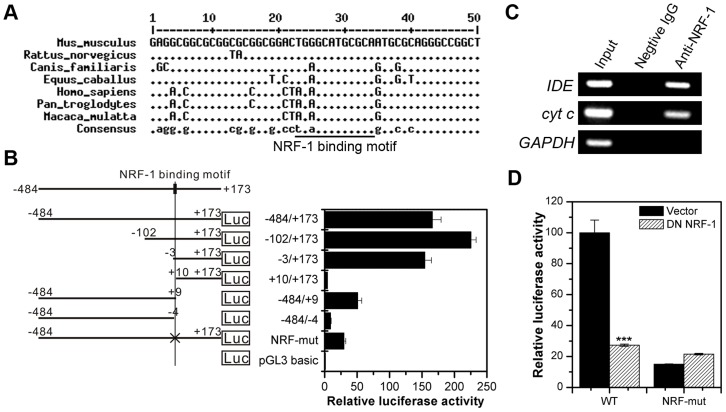
The NRF-1 Binding Site in the IDE Promoter is Conserved between Mouse and Human
The NRF-1 binding site in the IDE promoter is conserved among different species (Figure 5A). The human IDE promoter contains a NRF-1 binding motif, which locates between −3 and +9 relative to the first transcription initiation site (Figure 5B). Deleting the 5′ terminus of the human IDE promoter from −484 to −3 retained its transcriptional activity, while further deletion to +10 abolished its transcriptional activity (Figure 5B). On the other hand, deleting the 3′ terminus of the human IDE promoter from +173 to +9 retained its transcriptional activity, while further deletion to −4 abolished its transcriptional activity (Figure 5B). Furthermore, mutation of the NRF-1 binding motif reduced the human IDE promoter activity by approximately 70% (Figure 6B). These results indicate that the NRF-1 binding motif is critical for human IDE transcription initiation. ChIP experiments showed that NRF-1 binds to the human IDE promoter in HeLa cells (Figure 5C). Furthermore, dominant negative NRF-1 repressed the human IDE promoter activity dramatically, which was abolished by the mutation of the NRF-1 binding motif (Figure 5D). Therefore, the NRF-1 binding motif in the human IDE promoter is functional.
Figure 5. The NRF-1 binding site in the IDE promoter is conserved among different species.
(A) The NRF-1 binding site in the IDE promoter is conserved among different species. The underlined region indicates the conserved NRF-1 binding motif. (B) The NRF-1 binding motif is critical for human IDE transcription initiation. Different truncations of the human IDE promoter were cloned into the pGL3-basic vector. Luciferase activity of the reporter plasmids in HeLa cells is represented as fold of the pGL3-basic vector. (C) ChIP. NRF-1 binding to the IDE promoter in HeLa cells was determined by ChIP. The promoter of cytochrome c (cyt c) is used as a positive control for NRF-1 binding, while the promoter of GAPDH acts as a negative control. (D) The NRF-1 binding motif is essential for the effect of dominant negative NRF-1 on human IDE promoter activity. HeLa cells were transiently co-transfected with wild-type (−484/+173) or NRF-1 binding site-mutated (NRF-mut) human IDE reporter plasmids (0.4 µg) and Renilla luciferase plasmid (4 ng) along with or without dominant negative (DN) NRF-1 expression plasmids (0.4 µg). Twenty-four hours after transfection, cells were lysed, and the luciferase activity was examined. Firefly luminescence signal was normalized based on the Renilla luminescence signal.
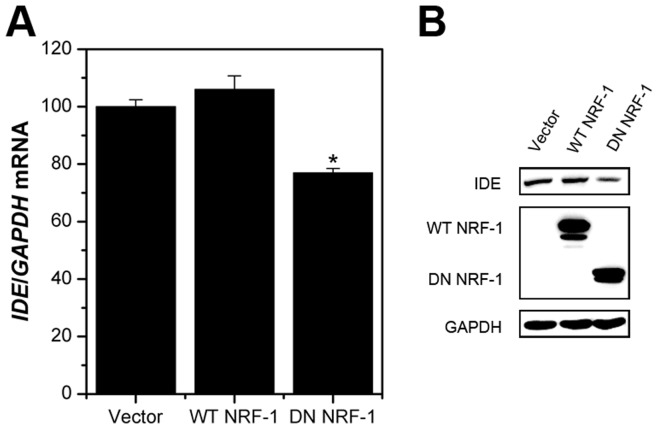
Figure 6. Dominant negative NRF-1 represses endogenouse IDE expression.
HeLa cells were transiently transfected with wild-type (WT) or dominant negative (DN) NRF-1 expression plasmids or the empty vector (4 µg). Twenty-four hours later, total RNA were prepared and analyzed by quantitative real-time RT-PCR (A). GAPDH was used as an internal control. Alternatively, the IDE protein levels were detected by Western immublotting (B). GAPDH was used as an internal control. Overexpression of NRF-1 was confirmed by His-tag antibodies. *p<0.05 as compared with the vector control.
To test whether NRF-1 regulates endogenous IDE expression, HeLa cells were transiently transfected with wild-type or dominant negative NRF-1 expression plasmids. Both the mRNA and protein levels of IDE were not sensitive to wild-type NRF-1 stimulation (Figure 6 A and B). However, enforced expression of dominant negative NRF-1 decreased IDE mRNA levels by 25%, and also reduced IDE protein levels, as compared with the vector control (Figure 6 A and B).
TBP is not Essential for IDE Transcription Initiation
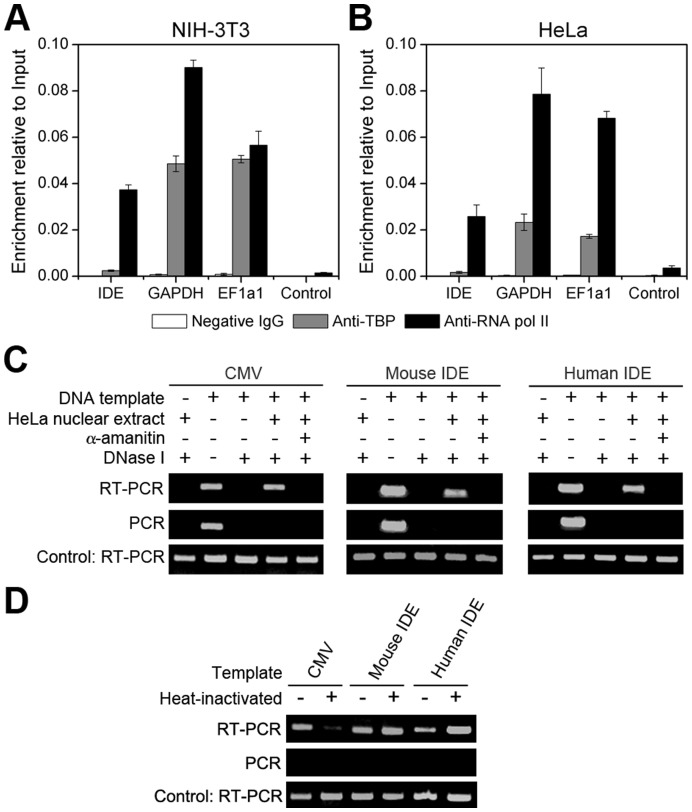
The IDE promoter is CpG-rich and contains no TATA box. Therefore, we wondered whether TBP is required for IDE transcription initiation. ChIP assays showed that TBP is not associated with the IDE promoter in both NIH-3T3 (Figure 7A) and HeLa (Figure 7B) cells. By contrast, TBP is recruited to the promoters of GAPDH and elongation factor 1 α 1 (EF1α1), which contain the TATA box. These results suggest that TBP is not essential for IDE transcription initiation.
Figure 7. TBP is not required for IDE transcription initiation.
(A) and (B) TBP is not associated with the IDE promoter. Binding of TBP and RNA polymerase II to the IDE promoter in NIH-3T3 (A) and HeLa (B) cells was tested by ChIP assays. Promoters of GAPDH and EF1α1 were used as positive controls for TBP and RNA polymerase II binding. Negative controls were also included. Data are represented as relative enrichment to the input. (C) In vitro transcription assays. DNA template for the CMV, mouse IDE or human IDE promoter was incubated with HeLa nuclear extracts and ribonucleotides. The resulting transcripts were purified, digested with DNase I to eliminate the DNA template and detected by RT-PCR. An internal control RNA was included to indicate the purification efficiency of different samples. Complete elimination of the DNA template was confirmed by PCR. Transcription from all the three DNA templates proceeded only when both the DNA template and HeLa nuclear extracts existed, and was inhibited by 1 µg/mL of α-amanitin. (D) Heat-inactivation of TBP does not block IDE transcription. HeLa nuclear extracts were heated at 47°C for 15 min before in vitro transcription assays.
To further address this question, we carried out in vitro transcription assays. The CMV promoter, which contains the TATA box, was used as a control. In vitro transcription occurred from all three DNA templates of the CMV, mouse IDE and human IDE promoters, which was abolished by α-amanitin (Figure 7C). When the DNA template or the nuclear extract was omitted, no transcripts were detected (Figure 7C). TBP was reported to be specifically inhibited by being heated at 47°C for 15 minutes, which was rescued by adding back of TBP [28]. In vitro transcription assays showed that transcription from the CMV promoter was inhibited by heat-inactivation of TBP, while transcription from the mouse or human IDE promoters was activated (Figure 7D), indicating that TBP is not required for IDE transcription initiation.
Discussion
Despite the prevalence of CpG islands associated with mammalian promoters, the mechanism of transcription initiation from this type of promoters is not well understood. CpG island promoters often lack the TATA box or other common core promoter elements. It has been proposed that Sp1 may participate in the transcription initiation of CpG island promoters, because Sp1 binds to a consensus sequence of GGGCGG which is present in many CpG islands due to their high GC content [29]. NRF-1 is a transcription factor that activates nuclear genes required for respiration as well as genes regulating heme biosynthesis and mitochondrial DNA transcription and replication. NRF-1 has been reported to play an important role in the transcriptional regulation of several genes [30], [31], [32], [33]. Interestingly, NRF-1 binding motifs are frequently found around the transcription initiation sites of genes [27], [34], typically those without the TATA box [35], suggesting that NRF-1 may function in regulating the transcription initiation of TATA box-less genes.
Here we studied the mechanism of transcription initiation of the ubiquitously expressed gene, IDE, which has a CpG island promoter and dispersed transcription initiation sites. We mapped the core promoter region of IDE, and showed that the IDE core promoter contains a NRF-1 binding motif that is essential for transcription initiation. We further demonstrated that NRF-1 binds to this motif and dominant negative NRF-1 represses IDE transcription. Our studies suggest that, besides Sp1, NRF-1 may be another important transcription factor that regulates the transcription initiation from CpG island promoters. These results are suggestive for the mechanism of transcription initiation from other CpG island promoters. Since CpG island promoters often lack canonical core promoter elements, some transcription factors, such as Sp1 and NRF-1, may function in recruiting the transcription apparatus to the promoter region without the assistance of other core promoter elements. In this hypothesis, the transcription apparatus is not “fixed” to the promoter through its own interactions to the promoter, which may explain the fact that CpG island promoters often have dispersed transcription initiation sites.
TBP was originally regarded as a universal factor that participated in the transcription initiation of genes transcribed by all three RNA polymerases in eukaryotes [36]. However, emerging studies have indicated the existence of TBP-independent transcription by RNA polymerase II [37], [38]. In addition, A TBP-free TAFII-containing complex was reported to initiate transcription from both TATA box-free and TATA box-containing promoters [39]. The IDE promoter contains no TATA box, and is free of TBP binding. Furthermore, inactivation of TBP does not block IDE transcription. These results suggest that TBP is not required for IDE transcription initiation. It is fascinating to demonstrate whether TBP is essential for the transcription initiation from other CpG island promoters which do not contain the TATA box.
IDE is a zinc metalloprotease that typically degrades insulin and Aβ, and is associated with both type II DM and AD. However, the mechanism of the transcriptional regulation of IDE has not been studied in detail. Here, we showed that NRF-1 acts as a central transcription factor that regulates IDE transcription. Thus, NRF-1 dysfunction may result in a down-regulation of IDE expression and DM or AD pathogenesis. Indeed, human genetic studies suggest that NRF-1 polymorphisms are associated with the pathogenesis of type II DM [40], [41]. The transcriptional regulation of IDE by NRF-1 reported here provides a mechanism that links NRF-1 to type II DM. Although IDE is predominantly distributed in the cytosol, an isoform of IDE generated by alternative translation initiation is targeted to mitochondria [42]. NRF-1 is an important transcription factor that mediates nuclear-mitochondrial interactions. The transcriptional regulation of IDE by NRF-1 may be of significance to the mitochondrial function.
In conclusion, our studies indicate that NRF-1 mediates IDE transcription initiation in a TBP-independent manner, and provide insights into the potential mechanism of the transcription initiation from other CpG island promoters. NRF-1 has been reported to regulate the transcription of a variety of genes. It is fascinating to investigate whether NRF-1 is essential for the transcription initiation of these genes. In addition, NRF-1 interacts with the peroxisome proliferator-activated receptor γ coactivator 1 (PGC-1) family of proteins, including PGC-1α [43], PGC-1β [44] and PGC-1-related coactivator (PRC) [45]. It remains to be clarified whether NRF-1 regulates IDE transcription initiation through these coactivators.
Materials and Methods
Plasmid Constructs
Genome DNA from C57/BL6 mice or HeLa cells was used as the template to construct the promoter reporter plasmids. Different truncations of the mouse and human IDE promoter were cloned into the Xho I and Hind III restriction sites of the pGL3-basic vector (Promega, Madison, WI). The primers used during the plasmid construction are shown in Table S1 and S2. The NRF-1 binding sites in the mouse and human IDE promoter were mutated from TGGGCATGCGCA and AGAGCATGCGCA to TGGGCAGCGACA and AGATAGGTAGCA, respectively. The expression plasmids for wild-type NRF-1and dominant negative NRF-1 which contains 1–304 amino acids of full-length NRF-1 [27] were constructed by cloning the corresponding sequences into the Bam HI and Xba I restriction sites of the pcDNA 3.1 myc-His A vector (Invitrogen, Carlsbad, CA).
Cell Culture and Cell Transfection
NIH-3T3 and HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (Thermo Scientific HyClone, Logan, UT) containing 10% fetal bovine serum (Thermo Scientific HyClone). HeLa cells were seeded into 6-well plates at a density of 4×105 cells per well in antibiotic-free medium the day before transfection. Each well of cells were transiently transfected with 4 µg of pcDNA 3.1, wild-type or dominant negative NRF-1 expression plasmids using Lipofectamine™ 2000 (Invitrogen). Twenty-four hours later, cells were collected, and analyzed by quantitative real-time RT-PCR or Western immunoblotting.
5'-RACE
5′-RACE was carried out using the SMART™ RACE cDNA Amplification Kit (Clontech, Shiga, Japan) according to the manufacturer’s instructions, to determine the transcription initiation sites of mouse IDE. 5′-RACE-ready cDNA was synthesized from total RNA from NIH-3T3 cells. The 5′-flanking region of IDE was amplified using the universal primer A mix (Clontech) and an IDE-specific primer: 5′-TCTTCAACGTGCAATAACCCT-3′. The resulting PCR products were used as a template for a nested PCR reaction using the nested universal primer A (Clontech) and a nested IDE-specific primer: 5′-TGTGGTGGGATCGCTGATGAGAAG-3′. The nested PCR products were then cloned into the pMD19-T vector (Takara Bio Inc., Shiga, Japan), and a total of 33 positive clones were sequenced.
Dual-luciferase Reporter Assays
NIH-3T3 and HeLa cells were seeded into 24-well plates at a density of 8×104 cells per well in antibiotic-free culture medium the day before transfection. Each well of cells were transiently co-transfected with 0.8 µg of IDE reporter plasmids and 8 ng of Renilla reporter plasmid (pCMV-RL, Promega) as an internal control using Lipofectamine™ 2000. Alternatively, cells were co-transfected with 0.4 µg of IDE reporter plasmids, 4 ng of pCMV-RL plasmid and 0.4 µg of pcDNA 3.1 or NRF-1 expressing plasmids. Twenty-four hours later, cells were lysed and the luciferase acitvity was detected using the Dual-Luciferase Reporter Assay System (Promega). Firefly luminescence signal was normalized based on the Renilla luminescence signal.
RNA Isolation and Quantitative Real-time RT-PCR
RNA was isolated using Trizol reagent (Invitrogen), and treated with DNase I (Promega) before cDNA synthesis. cDNA was synthesized with 2 µg of total RNA with anchored oligo (dT)20 primers using the Transcript First-Strand cDNA Synthesis Kit (Transgene Biotechnology Inc., Beijing, China). Quantitative real-time PCR was performed using the UltraSYBR mixture (Cwbiotech, Beijing, China) in the Stratagene Mx3000P™ Real-Time PCR System (Agilent Technologies, La Jolla, CA). PCR primers for human IDE were forward: 5′-AAAGACAAAGAGAGGCCACGGGG-3′ and reverse: 5′-TGGCAACCCGGACATTTTCTGGTC-3′. PCR primers for human GAPDH were forward: 5′-ACCGTCAAGGCTGAGAACGGGA-3′ and reverse: 5′-CCTGCAAATGAGCCCCAGCCTT-3′. Both the IDE and GAPDH primers are intron-spanning. Melting curves were performed on the products, which confirmed the specificity of the primers. The thermal cycling conditions were as follows: initial denaturation at 95°C for 10 min, followed by 40 cycles of denaturing at 95°C for 15 s, annealing at 60°C for 30 s and extension at 72°C for 30 s.
Western Immunoblotting
Cells were collected and lysed with the cell lysis buffer (Beyotime, Shanghai, China). Protein concentrations were determined using the BCA method. Whole cell extracts were boiled for 4 min, resolved by 10% SDS-PAGE and electrophoretically transferred to polyvinylidene difluoride membranes (Millipore, Schwalbach, Germany). The membranes were blocked with 5% non-fat dry milk in Tris-buffered saline containing 0.05% Tween 20 at room temperature for 1 h, and then incubated with mouse monoclonal anti-IDE (1∶1000, Abcam), mouse monoclonal anti-His tag (1∶2000, Cwbiotech) or rabbit polyclonal anti-GAPDH (1∶1000, Cwbiotech) antibodies at 4°C overnight, followed by an incubation with the appropriate horseradish peroxidase-conjugated secondary antibodies (1∶10000, Cwbiotech) at room temperature for 1 h. The chemiluminescence reaction was performed using ECL reagent (Thermo Scientific).
ChIP
ChIP was performed using the Magnetic Chromatin Immunoprecipitation Kit (Active Motif, Carlsbad, CA) as described in the manufacturer’s protocol. Briefly, NIH-3T3 or HeLa cells were washed once with phosphate buffered saline, and fixed in culture medium containing 1% formaldehyde at room temperature for 10 min. The cells were then collected and lysed to release the nucleus. The nucleus was digested with the enzyme mix at 37°C for 10 min to shear the chromatin into small segments. The sheared chromatin was then immunoprecipitated with 2 µg of NRF-1 antibodies (Abcam), 2 µg of TBP antibodies (Abcam), 2 µg of RNA polymerase II antibodies (Active Motif), or a negative control IgG at 4°C for 4 h. The pulled-down chromatin was washed, reverse-crosslinked and purified. Semi-quantitative or quantitative real-time PCR was performed using the primers shown in Table S3.
In vitro Transcription
In vitro transcription was performed using the HeLaScribe® Nuclear Extract in vitro Transcription System (Promega) with some modifications. DNA template of the mouse IDE promoter was amplified from mouse genomes using primers forward: 5′-TGCCGAGACGACGACCCACC-3′ and reverse: 5′-TGAGAAGCGGTTTCCCACGAC-3′. DNA template of the human IDE promoter was amplified from human genomes using primers forward: 5′- CTCACAGTCAGACACACGTCGCCACC -3′ and reverse: 5′-CCACACGGTCCTGGAAACTCAGTGCC-3′. DNA template of the CMV promoter was amplified from a mouse peroxisome proliferator-activated receptor α expression plasmid drived by the CMV promoter using primers forward: 5′-TGTTGGAGGTCGCTGAGTAGTGC-3′ and reverse: 5′-AGGCGGGTTGTTGCTGGTCT-3′. The DNA template (100 ng) was incubated with HeLa nuclear extracts and four types of ribonucleotides at 37°C for 30 minutes. The resulting transcripts were purified, digested with DNase I (Promega) at 37°C for 1 h to eliminate the DNA template, and then used as RT-PCR template. An internal control RNA was included to indicate the purification efficiency of different samples. RT-PCR was performed using the PrimeScript® One Step RT-PCR Kit Ver.2 (Takara), using the following primers–forward: 5′-GCACCTTGCGCTCCATCCTCG-3′ and reverse: 5′-CCGCTGCACTTGTGGCGTTCTC-3′ for mouse IDE, forward: 5′-CCAGCACCTTCCGCTCAGTCC-3′ and reverse: 5′-GGTGACTCTGTCCGCCCCTGC-3′ for human IDE, and forward: 5′-GACCTGGAAAGTCCCTTATCT-3′ and reverse: 5′-AGCCCTTACAGCCTTCACAT-3′ for CMV. Elimination of DNA template was confirmed by PCR. To confirm that IDE is transcribed by RNA polymerase II, 1 µg/mL of α-amanitin was included in the in vitro transcription system. To determine the role of TBP in transcription initiation, HeLa nuclear extracts were heated at 47°C for 15 min before the in vitro transcription assay.
Statistical Analysis
Results are represented as means ± S.E.M from three independent experiments. Statistical significance was determined by one-way ANOVA, followed by the post-hoc Tukey multiple comparison test.
Supporting Information
PCR primers used for constructing the reporter plasmids of the mouse IDE promoter.
(DOC)
PCR primers used for constructing the reporter plasmids of the human IDE promoter.
(DOC)
PCR primers used for ChIP assays.
(DOC)
Funding Statement
This work was financially supported by the National Basic Research Program (973 Project) of China (No. 2007CB507406), and the Tsinghua University Initiative Scientific Research Program (20111081143). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Jones PA, Takai D (2001) The role of DNA methylation in mammalian epigenetics. Science 293: 1068–1070. [DOI] [PubMed] [Google Scholar]
- 2. Geiman TM, Robertson KD (2002) Chromatin remodeling, histone modifications, and DNA methylation-how does it all fit together? J Cell Biochem 87: 117–125. [DOI] [PubMed] [Google Scholar]
- 3. Grippo P, Iaccarino M, Parisi E, Scarano E (1968) Methylation of DNA in developing sea urchin embryos. J Mol Biol 36: 195–208. [DOI] [PubMed] [Google Scholar]
- 4. Pech M, Streeck RE, Zachau HG (1979) Patchwork structure of a bovine satellite DNA. Cell 18: 883–893. [DOI] [PubMed] [Google Scholar]
- 5. Cooper DN (1983) Eukaryotic DNA methylation. Hum Genet 64: 315–333. [DOI] [PubMed] [Google Scholar]
- 6. Swartz MN, Trautner TA, Kornberg A (1962) Enzymatic synthesis of deoxyribonucleic acid. XI. Further studies on nearest neighbor base sequences in deoxyribonucleic acids. J Biol Chem 237: 1961–1967. [PubMed] [Google Scholar]
- 7. Coulondre C, Miller JH, Farabaugh PJ, Gilbert W (1978) Molecular basis of base substitution hotspots in Escherichia coli. Nature 274: 775–780. [DOI] [PubMed] [Google Scholar]
- 8. Bird AP (1980) DNA methylation and the frequency of CpG in animal DNA. Nucleic Acids Res 8: 1499–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cooper DN, Taggart MH, Bird AP (1983) Unmethylated domains in vertebrate DNA. Nucleic Acids Res 11: 647–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gardiner-Garden M, Frommer M (1987) CpG islands in vertebrate genomes. J Mol Biol 196: 261–282. [DOI] [PubMed] [Google Scholar]
- 11. Bird AP (1987) Cpg Islands as Gene Markers in the Vertebrate Nucleus. Trends in Genetics 3: 342–347. [Google Scholar]
- 12. Antequera F, Bird A (1993) Number of CpG islands and genes in human and mouse. Proc Natl Acad Sci U S A 90: 11995–11999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bajic VB, Tan SL, Suzuki Y, Sugano S (2004) Promoter prediction analysis on the whole human genome. Nature Biotechnology 22: 1467–1473. [DOI] [PubMed] [Google Scholar]
- 14. Saxonov S, Berg P, Brutlag DL (2006) A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc Natl Acad Sci U S A 103: 1412–1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Juven-Gershon T, Hsu JY, Theisen JW, Kadonaga JT (2008) The RNA polymerase II core promoter - the gateway to transcription. Curr Opin Cell Biol 20: 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, et al. (2006) Genome-wide analysis of mammalian promoter architecture and evolution. Nat Genet 38: 626–635. [DOI] [PubMed] [Google Scholar]
- 17. Authier F, Posner BI, Bergeron JJ (1996) Insulin-degrading enzyme. Clin Invest Med 19: 149–160. [PubMed] [Google Scholar]
- 18. Kurochkin IV, Goto S (1994) Alzheimers Beta-Amyloid Peptide Specifically Interacts with and Is Degraded by Insulin Degrading Enzyme. Febs Letters 345: 33–37. [DOI] [PubMed] [Google Scholar]
- 19. Qiu WQ, Walsh DM, Ye Z, Vekrellis K, Zhang JM, et al. (1998) Insulin-degrading enzyme regulates extracellular levels of amyloid beta-protein by degradation. J Biol Chem 273: 32730–32738. [DOI] [PubMed] [Google Scholar]
- 20. Farris W, Mansourian S, Leissring MA, Eckman EA, Bertram L, et al. (2004) Partial loss-of-function mutations in insulin-degrading enzyme that induce diabetes also impair degradation of amyloid beta-protein. Am J Pathol 164: 1425–1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Farris W, Mansourian S, Chang Y, Lindsley L, Eckman EA, et al. (2003) Insulin-degrading enzyme regulates the levels of insulin, amyloid beta-protein, and the beta-amyloid precursor protein intracellular domain in vivo. Proc Natl Acad Sci U S A 100: 4162–4167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Abdul-Hay SO, Kang D, McBride M, Li L, Zhao J, et al. (2011) Deletion of insulin-degrading enzyme elicits antipodal, age-dependent effects on glucose and insulin tolerance. PLoS One 6: e20818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. McCarthy MI, Zeggini E (2009) Genome-wide association studies in type 2 diabetes. Curr Diab Rep 9: 164–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Serretti A, Olgiati P, De Ronchi D (2007) Genetics of Alzheimer's disease. A rapidly evolving field. Journal of Alzheimers Disease 12: 73–92. [DOI] [PubMed] [Google Scholar]
- 25. Ramachandran B, Yu G, Gulick T (2008) Nuclear respiratory factor 1 controls myocyte enhancer factor 2A transcription to provide a mechanism for coordinate expression of respiratory chain subunits. J Biol Chem 283: 11935–11946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Herzig RP, Scacco S, Scarpulla RC (2000) Sequential serum-dependent activation of CREB and NRF-1 leads to enhanced mitochondrial respiration through the induction of cytochrome c. J Biol Chem 275: 13134–13141. [DOI] [PubMed] [Google Scholar]
- 27. Gugneja S, Virbasius CM, Scarpulla RC (1996) Nuclear respiratory factors 1 and 2 utilize similar glutamine-containing clusters of hydrophobic residues to activate transcription. Mol Cell Biol 16: 5708–5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakajima N, Horikoshi M, Roeder RG (1988) Factors involved in specific transcription by mammalian RNA polymerase II: purification, genetic specificity, and TATA box-promoter interactions of TFIID. Mol Cell Biol 8: 4028–4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Antequera F (2003) Structure, function and evolution of CpG island promoters. Cell Mol Life Sci 60: 1647–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Asangani IA, Rasheed SA, Leupold JH, Post S, Allgayer H (2008) NRF-1, and AP-1 regulate the promoter of the human calpain small subunit 1 (CAPNS1) gene. Gene 410: 197–206. [DOI] [PubMed] [Google Scholar]
- 31. Blesa JR, Prieto-Ruiz JA, Abraham BA, Harrison BL, Hegde AA, et al. (2008) NRF-1 is the major transcription factor regulating the expression of the human TOMM34 gene. Biochem Cell Biol 86: 46–56. [DOI] [PubMed] [Google Scholar]
- 32. Mahishi L, Usdin K (2006) NF-Y, AP2, Nrf1 and Sp1 regulate the fragile X-related gene 2 (FXR2). Biochem J 400: 327–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tokusumi Y, Zhou S, Takada S (2004) Nuclear respiratory factor 1 plays an essential role in transcriptional initiation from the hepatitis B virus x gene promoter. J Virol 78: 10856–10864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, et al. (2005) Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature 434: 338–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Gomez-Cuadrado A, Martin M, Noel M, Ruiz-Carrillo A (1995) Initiation binding repressor, a factor that binds to the transcription initiation site of the histone h5 gene, is a glycosylated member of a family of cell growth regulators [corrected]. Mol Cell Biol 15: 6670–6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hernandez N (1993) TBP, a universal eukaryotic transcription factor? Genes Dev 7: 1291–1308. [DOI] [PubMed] [Google Scholar]
- 37. Martianov I, Viville S, Davidson I (2002) RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science 298: 1036–1039. [DOI] [PubMed] [Google Scholar]
- 38. Muller F, Lakatos L, Dantonel J, Strahle U, Tora L (2001) TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr Biol 11: 282–287. [DOI] [PubMed] [Google Scholar]
- 39. Wieczorek E, Brand M, Jacq X, Tora L (1998) Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature 393: 187–191. [DOI] [PubMed] [Google Scholar]
- 40. Cho YM, Shin HD, Park BL, Kim JH, Park KS, et al. (2005) Association between polymorphisms in the nuclear respiratory factor 1 gene and type 2 diabetes mellitus in the Korean population. Diabetologia 48: 2033–2038. [DOI] [PubMed] [Google Scholar]
- 41. Liu Y, Niu N, Zhu X, Du T, Wang X, et al. (2008) Genetic variation and association analyses of the nuclear respiratory factor 1 (nRF1) gene in Chinese patients with type 2 diabetes. Diabetes 57: 777–782. [DOI] [PubMed] [Google Scholar]
- 42. Leissring MA, Farris W, Wu X, Christodoulou DC, Haigis MC, et al. (2004) Alternative translation initiation generates a novel isoform of insulin-degrading enzyme targeted to mitochondria. Biochem J 383: 439–446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lin JD, Handschin C, Spiegelman BM (2005) Metabolic control through the PGC-1 family of transcription coactivators. Cell Metabolism 1: 361–370. [DOI] [PubMed] [Google Scholar]
- 44. Lin J, Tarr PT, Yang R, Rhee J, Puigserver P, et al. (2003) PGC-1beta in the regulation of hepatic glucose and energy metabolism. J Biol Chem 278: 30843–30848. [DOI] [PubMed] [Google Scholar]
- 45. Andersson U, Scarpulla RC (2001) Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol Cell Biol 21: 3738–3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR primers used for constructing the reporter plasmids of the mouse IDE promoter.
(DOC)
PCR primers used for constructing the reporter plasmids of the human IDE promoter.
(DOC)
PCR primers used for ChIP assays.
(DOC)