Abstract
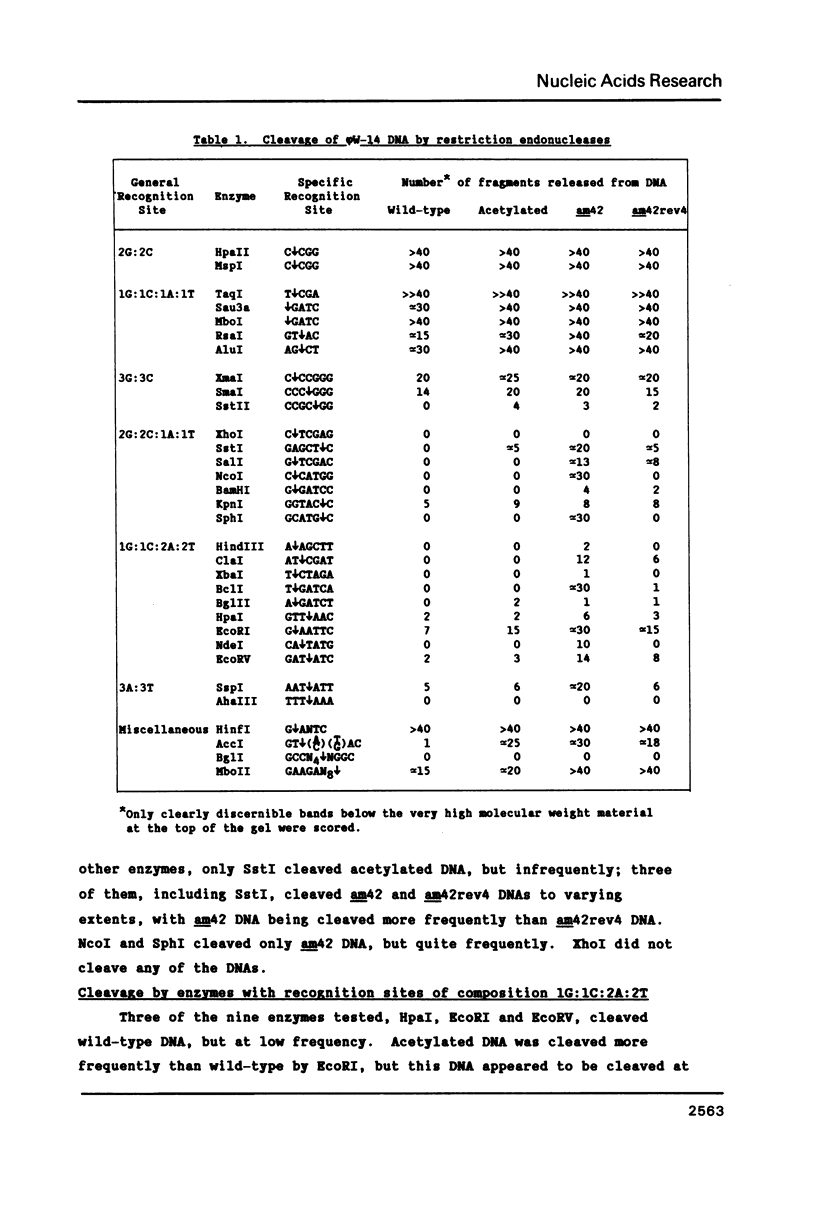
The modified base alpha-putrescinylthymine (putT) in phi W-14 DNA blocks cleavage of the DNA by 17 of 32 Type II restriction endonucleases. The enzymes cleaving the DNA do so to widely varying extents. The frequencies of cleavage of three altered forms of the DNA show that putT blocks recognition sites either when it occurs within the site or when it is in a sequence flanking the site. The blocking is dependent on both charge and steric factors. The charge effects can be greater than the steric effects for some of the enzymes tested. All the enzymes cleaving phi W-14 DNA release discrete fragments, showing that the distribution of putT is ordered. The cleavage frequencies for different enzymes suggest that the sequence CAputTG occurs frequently in the DNA. Only TaqI of the enzymes tested appeared not to be blocked by putT, but it was slowed down. TaqI generated fragments are joinable by T4 DNA ligase.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Armstrong K., Bauer W. R. Preferential site-dependent cleavage by restriction endonuclease PstI. Nucleic Acids Res. 1982 Feb 11;10(3):993–1007. doi: 10.1093/nar/10.3.993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azorin F., Hahn R., Rich A. Restriction endonucleases can be used to study B-Z junctions in supercoiled DNA. Proc Natl Acad Sci U S A. 1984 Sep;81(18):5714–5718. doi: 10.1073/pnas.81.18.5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. An assay for the rates of cleavage of specific sites in DNA by restriction endonucleases: its use to study the cleavage of phage lambda DNA by EcoRI and phage P22 DNA containing thymine or 5-bromouracil by HindIII. Anal Biochem. 1983 Mar;129(2):446–456. doi: 10.1016/0003-2697(83)90575-4. [DOI] [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. EcoRI cleavage and methylation of DNAs containing modified pyrimidines in the recogintion sequence. J Biol Chem. 1977 May 25;252(10):3185–3193. [PubMed] [Google Scholar]
- Berkner K. L., Folk W. R. The effects of substituted pyrimidines in DNAs on cleavage by sequence-specific endonucleases. J Biol Chem. 1979 Apr 10;254(7):2551–2560. [PubMed] [Google Scholar]
- Bodnar J. W., Zempsky W., Warder D., Bergson C., Ward D. C. Effect of nucleotide analogs on the cleavage of DNA by the restriction enzymes AluI, DdeI, HinfI, RsaI, and TaqI. J Biol Chem. 1983 Dec 25;258(24):15206–15213. [PubMed] [Google Scholar]
- Bron S., Luxen E., Venema G. Resistance of bacteriophage H1 to restriction and modification by Bacillus subtilis R. J Virol. 1983 Jun;46(3):703–708. doi: 10.1128/jvi.46.3.703-708.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao X. M., Huang L. H., Farnet C. M., Ehrlich M. Ligation of highly modified bacteriophage DNA. Biochim Biophys Acta. 1983 Nov 17;741(2):237–243. doi: 10.1016/0167-4781(83)90064-7. [DOI] [PubMed] [Google Scholar]
- Ehrlich M., Ehrlich K. C. A novel, highly modified, bacteriophage DNA in which thymine is partly replaced by a phosphoglucuronate moiety covalently bound to 5-(4',5'-dihydroxypentyl)uracil. J Biol Chem. 1981 Oct 10;256(19):9966–9972. [PubMed] [Google Scholar]
- Forsblom S., Rigler R., Ehrenberg M., Philipson L. Kinetic studies on the cleavage of adenovirus DNA by restriction endonuclease Eco RI. Nucleic Acids Res. 1976 Dec;3(12):3255–3269. doi: 10.1093/nar/3.12.3255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick C. A., Grable J., Melia M., Samudzi C., Jen-Jacobson L., Wang B. C., Greene P., Boyer H. W., Rosenberg J. M. Kinked DNA in crystalline complex with EcoRI endonuclease. Nature. 1984 May 24;309(5966):327–331. doi: 10.1038/309327a0. [DOI] [PubMed] [Google Scholar]
- Gerhard B., Warren R. A. Reactivity of the alpha-putrescinylthymine amino groups in phi W-14 deoxyribonucleic acid. Biochemistry. 1982 Oct 26;21(22):5458–5462. doi: 10.1021/bi00265a012. [DOI] [PubMed] [Google Scholar]
- Halford S. E., Johnson N. P., Grinsted J. The EcoRI restriction endonuclease with bacteriophage lambda DNA. Kinetic studies. Biochem J. 1980 Nov 1;191(2):581–592. doi: 10.1042/bj1910581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Nakanishi K., Brandon C., Marmur J. Structure and synthesis of dihydroxypentyluracil from bacteriophage SP-15 deoxyribonucleic acid. J Am Chem Soc. 1973 Dec 26;95(26):8749–8757. doi: 10.1021/ja00807a041. [DOI] [PubMed] [Google Scholar]
- Hofer B., Köster H. On the influence of thymidine analogues on the activity of phage fd promoters in vitro. Nucleic Acids Res. 1980 Dec 20;8(24):6143–6162. doi: 10.1093/nar/8.24.6143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang L. H., Farnet C. M., Ehrlich K. C., Ehrlich M. Digestion of highly modified bacteriophage DNA by restriction endonucleases. Nucleic Acids Res. 1982 Mar 11;10(5):1579–1591. doi: 10.1093/nar/10.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito J., Kawamura F., Duffy J. J. Susceptibility of non-thymine containing DNA to four bacterial restriction endonucleases. FEBS Lett. 1975 Jul 15;55(1):278–281. doi: 10.1016/0014-5793(75)81011-8. [DOI] [PubMed] [Google Scholar]
- Jack W. E., Terry B. J., Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc Natl Acad Sci U S A. 1982 Jul;79(13):4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan D. A., Nierlich D. P. Cleavage of Nonglucosylated Bacteriophage T4 deoxyribonucleic acid by Restriction Endonuclease Eco RI. J Biol Chem. 1975 Mar 25;250(6):2395–2397. [PubMed] [Google Scholar]
- Kim R., Modrich P., Kim S. H. 'Interactive' recognition in EcoRI restriction enzyme-DNA complex. Nucleic Acids Res. 1984 Oct 11;12(19):7285–7292. doi: 10.1093/nar/12.19.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kropinski A. M., Bose R. J., Warren R. A. 5-(4-Aminobutylaminomethyl)uracil, an unusual pyrimidine from the deoxyribonucleic acid of bacteriophage phiW-14. Biochemistry. 1973 Jan 2;12(1):151–157. doi: 10.1021/bi00725a025. [DOI] [PubMed] [Google Scholar]
- Lu A. L., Jack W. E., Modrich P. DNA determinants important in sequence recognition by Eco RI endonuclease. J Biol Chem. 1981 Dec 25;256(24):13200–13206. [PubMed] [Google Scholar]
- Maltman K. L., Neuhard J., Lewis H. A., Warren R. A. Synthesis of thymine and alpha-putrescinylthymine in bacteriophage phi W-14-infected Pseudomonas acidovorans. J Virol. 1980 May;34(2):354–359. doi: 10.1128/jvi.34.2.354-359.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchionni M. A., Roufa R. J. Digestion of 5-bromodeoxyuridine-substituted lambda-DNA by restriction endonucleases. J Biol Chem. 1978 Dec 25;253(24):9075–9081. [PubMed] [Google Scholar]
- Marusyk R., Sergeant A. A simple method for dialysis of small-volume samples. Anal Biochem. 1980 Jul 1;105(2):403–404. doi: 10.1016/0003-2697(80)90477-7. [DOI] [PubMed] [Google Scholar]
- Miller P. B., Maltman K. L., Warren R. A. Isolation and preliminary characterization of amber mutants of bacteriophage phi W-14 defective in DNA synthesis. J Virol. 1982 Jul;43(1):67–72. doi: 10.1128/jvi.43.1.67-72.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller P. B., Scraba D. G., Leyritz-Wills M., Maltman K. L., Warren R. A. Formation and possible functions of alpha-putrescinylthymine in bacteriophage phi W-14 DNA: analysis of bacteriophage mutants with decreased levels of alpha-putrescinylthymine in their DNAs. J Virol. 1983 Sep;47(3):399–405. doi: 10.1128/jvi.47.3.399-405.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petruska J., Horn D. Sequence-specific responses of restriction endonucleases to bromodeoxyuridine substitution in mammalian DNA. Nucleic Acids Res. 1983 Apr 25;11(8):2495–2510. doi: 10.1093/nar/11.8.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scraba D. G., Bradley R. D., Leyritz-Wills M., Warren R. A. Bacteriophage phi W-14: the contribution of covalently bound putrescine to DNA packing in the phage head. Virology. 1983 Jan 15;124(1):152–160. doi: 10.1016/0042-6822(83)90298-2. [DOI] [PubMed] [Google Scholar]
- Thomas M., Davis R. W. Studies on the cleavage of bacteriophage lambda DNA with EcoRI Restriction endonuclease. J Mol Biol. 1975 Jan 25;91(3):315–328. doi: 10.1016/0022-2836(75)90383-6. [DOI] [PubMed] [Google Scholar]
- Vardimon L., Rich A. In Z-DNA the sequence G-C-G-C is neither methylated by Hha I methyltransferase nor cleaved by Hha I restriction endonuclease. Proc Natl Acad Sci U S A. 1984 Jun;81(11):3268–3272. doi: 10.1073/pnas.81.11.3268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren R. A. Modified bases in bacteriophage DNAs. Annu Rev Microbiol. 1980;34:137–158. doi: 10.1146/annurev.mi.34.100180.001033. [DOI] [PubMed] [Google Scholar]
- Wiatr C. L., Witmer H. J. Selective protection of 5' ... GGCC ... 3' and 5' ... GCNGC ... 3' sequences by the hypermodified oxopyrimidine in Bacillus subtilis bacteriophage SP10 DNA. J Virol. 1984 Oct;52(1):47–54. doi: 10.1128/jvi.52.1.47-54.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodhead J. L., Malcolm A. D. Non-specific binding of restriction endonuclease EcoR1 to DNA. Nucleic Acids Res. 1980 Jan 25;8(2):389–402. doi: 10.1093/nar/8.2.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zacharias W., Larson J. E., Kilpatrick M. W., Wells R. D. HhaI methylase and restriction endonuclease as probes for B to Z DNA conformational changes in d(GCGC) sequences. Nucleic Acids Res. 1984 Oct 25;12(20):7677–7692. doi: 10.1093/nar/12.20.7677. [DOI] [PMC free article] [PubMed] [Google Scholar]


