Abstract
A disintegrin and metalloprotease 10 (ADAM10) is a key regulator of cellular processes by shedding extracellular domains of transmembrane proteins. We have previously demonstrated that deletion of B cell expressed ADAM10 results in changes in lymphoid tissue architecture and impaired germinal center (GC) formation. In this study, mice were generated in which ADAM10 is deleted in B cells following class switch recombination (ADAM10Δ/ΔIgG1-cre+/− mice). Despite normal GC formation, antibody responses were impaired in ADAM10Δ/ΔIgG1-cre+/− mice, implicating ADAM10 in post-GC and extrafollicular B cell terminal differentiation. Surprisingly, plasma cell (PC) numbers were normal in ADAM10Δ/ΔIgG1-cre+/− mice when compared to controls. However, PCs isolated from ADAM10Δ/ΔIgG1-cre+/− mice exhibited decreased expression of transcription factors important for PC function: Prdm1, Xbp1 and Irf4. Bcl6 is a GC transcriptional repressor that inhibits the PC transcriptional program and thus must be downregulated for PC differentiation to occur. Bcl6 expression was increased in PCs isolated from ADAM10Δ/ΔIgG1-cre+/− mice at both the mRNA and protein level. These results demonstrate that ADAM10 is required for proper transcription factor expression in PCs and thus, for normal PC function.
Introduction
Key features of antibody-mediated immune responses are the generation of antigen-specific plasma cells (PCs) and memory B cells. Plasma cells (PCs) are antibody factories and memory B cells can rapidly differentiate into PCs after reencountering antigen. Two general types of PCs are known. Short-lived PCs arise from extrafollicular responses while long-lived PCs are derived primarily from germinal center (GC) B cells [1], [2]. Within GCs, antigen-activated B cells undergo class-switch recombination (CSR), somatic hypermutation (SHM) and affinity maturation [3]. The transition from GC B cell to PC requires changes in the transcriptional program. The transcription factors that are generally required for PC differentiation are B lymphocyte-induced maturation protein 1 (Blimp1), interferon regulatory factor 4 (IRF4) and X-box binding protein 1 (Xbp1) [4]–[7]. GC B cells express Bcl6, a known suppressor of Prdm1, the gene encoding Blimp1. Moreover, Blimp1 is also able to repress Bcl6. Therefore, as long as Bcl6 is expressed in GC B cells, Blimp1 expression and plasmacytic differentiation are inhibited. If Blimp1 is expressed, however, Bcl6 will be repressed thus allowing for PC differentiation to occur [8]–[10]. Therefore, downregulation of Bcl6 and Blimp1 upregulation is essential for PC differentiation and optimal humoral responses [1], [2], [11]. Consistent with this idea, study of transgenic mice that constitutively express Bcl6 in B cells showed a decreased number of class-switched PCs [3], [12].
ADAMs (A disintegrin and metalloproteases) are membrane-bound proteins that mediate ectodomain shedding and regulated intramembrane proteolysis (RIP) of transmembrane proteins. Ectodomain shedding releases soluble fragments into the extracellular space, possibly downregulating events that depend on transmembrane receptor expression or activating paracrine signaling by soluble products derived from ADAMs' substrates. ADAMs carry out a wide range of functions, including but not limited to, paracrine signaling, cell adhesion, and intracellular signaling [4]–[7], [13]. ADAM10 is a proteolytically active ADAM family member that is critical for many important biological processes [8]–[10], [14]. Furthermore, as recently described, the intracellular domain of ADAM10 can itself be shed, allowing for the ADAM10 intracellular domain (ICD) to translocate to the nucleus and modulate gene expression [15].
ADAM10 is a key regulator of lymphocyte development [16]. We and others have demonstrated that ADAM10 is essential for T cell and marginal zone B cell development [17], [18]. We recently published that ADAM10 is highly expressed in GC B cells. Interestingly, mice that lack ADAM10 in all peripheral B cells (ADAM10B−/− mice) fail to generate GCs and have severely impaired humoral responses. Furthermore, the defects in antibody production are accompanied by changes in lymphoid architecture [19]. Whether the defects in GC formation and antibody production observed in ADAM10B−/− mice are secondary to the changes in lymphoid architecture or whether ADAM10 plays a role in GC formation and/or antibody production independently of these changes remains to be determined.
In order to investigate the involvement of ADAM10 in PC development and function, ADAM10 was deleted post-isotype switching by crossing ADAM10-floxxed (ADAM10Δ/Δ) mice with IgG1-cre+/− mice [20]. In this situation, GCs would form prior to ADAM10 deletion. Here we demonstrate that these recently generated mice showed no alteration in lymphoid architecture and/or GC development. Intriguingly, humoral responses to T-dependent and T-independent antigens were still clearly impaired in ADAM10Δ/ΔIgG1cre+/− mice, implicating ADAM10 in B cell terminal differentiation. Furthermore, we show that in spite of normal PC numbers, mRNA expression levels of transcription factors important for PC development, Prdm1, xbp1 and Irf4 were altered in PCs isolated from ADAM10Δ/ΔIgG1cre+/− mice. In addition, the GC transcription factor Bcl6 was elevated at both the message and protein level. These results demonstrate that ADAM10 is required for proper PC function.
Results
Generation of ADAM10Δ/ΔIgG1+/− mice
Members of the ADAM family regulate a variety of functions, including, but not limited to, cell migration, proliferation and adhesion [13]. We previously generated mice that lacked ADAM10 in all peripheral B cells (ADAM10B−/− mice) by crossing a transgenic mouse strain containing loxP sites surrounding exon 9 of adam10 (ADAM10Δ/Δ) allele with mice expressing Cre recombinase under the control of the CD19 promoter [17]. Study of these mice demonstrated a severe defect in GC formation and changes in lymphoid architecture [19]. In order to determine whether ADAM10 plays a role in PC development or if the impairment in antibody production observed in ADAM10B−/− mice was secondary to changes in architecture, we crossed ADAM10Δ/Δ mice with IgG1-cre transgenic mice and generated ADAM10Δ/ΔIgG1-cre+/−mice [20]. Previous studies have demonstrated that the IgG1-cre transgene shows specificity for GC B cells, with approximately 75% of GC B cells expressing Cre recombinase. Introduction of the IgG1-cre transgene has been shown to affect IgG1 production; therefore, we also generated controls that have Cre expression but lack ADAM10-floxxed alleles (ADAM10+/+IgG1-cre+/− mice). In order to track cells that had undergone cre-mediated recombination, we crossed ADAM10Δ/ΔIgG1-cre+/− and ADAM10+/+IgG1-cre+/− mice with R26R-EYFP+ mice, thus, generating ADAM10Δ/ΔIgG1-cre+/−R26R-EYFP+ (ADAM10Δ/ΔIgG1-cre+/−YFP+) and ADAM10+/+IgG1-cre+/−YFP+ (control) mice that express EYFP transgene following cre-mediated recombination [17].
Antibody production and GC formation in ADAM10Δ/ΔIgG1-cre+/−YFP+ mice
Our previous study demonstrated that ADAM10-deletion in all peripheral B cells (ADAM10B−/− mice) led to severe impairments in antibody production, as evidenced from decreased antibody responses to T-dependent antigens. Moreover, GC formation was severely affected in these mice. Furthermore, follicular helper T cell (TFH) numbers were also diminished in ADAM10B−/− mice [19]. In order to determine if deletion of ADAM10 in class-switched cells affected basal antibody levels, mice were bled and IgM, IgG1 and IgE levels were measured by ELISA. As depicted in Figure 1 , no differences were seen between ADAM10Δ/ΔIgG1-cre+/−YFP+ and controls.
Figure 1. ADAM10Δ/ΔIgG1-cre+/− mice have normal basal antibody levels.
Serum (A) IgM, (B) IgG1, and (C) IgE were measured by capture ELISA from 8- to 12-wk-old mice. ns: non significant.
In order to determine whether deletion of ADAM10 within GCs affected GC maintenance and the production of antigen-specific antibodies, ADAM10Δ/ΔIgG1-cre+/− mice and controls were immunized with 10 µg of NP-KLH emulsified in alum. The proportion of GC B cells, defined as B220+GL7+Fashi, was quantified by flow cytometry 14 days post-immunization. The percentage of GC B cells was comparable between ADAM10Δ/ΔIgG1-cre+/−YFP+ and controls ( Figure 2A,B ). Immunohistochemistry analysis also showed normal GCs, defined as clusters of GL7+ within B cell follicles ( Figure 2C ). Moreover, the appearance of B cell follicles was comparable to that of controls ( Figure 2C ).
Figure 2. ADAM10Δ/ΔIgG1-cre+/− mice have normal germinal center formation.
ADAM10Δ/ΔIgG1-cre+/− (▪) and controls (□) were immunized with NP-KLH emulsified in alum. Fourteen days post-immunization GC formation was assessed by flow cytometry and immunohistochemistry. (A) Representative dot plot (gated on B220+ cells). (B) Frequency of GC B cells of total B cells, representative of 5 mice per group from at least two independent studies. (C) Representative splenic sections stained with GL7 (green) and IgD (red). ns: non significant.
Interestingly, despite normal GC B cell numbers, while antigen-specific IgM levels were comparable to control mice ( Figure 3A ), antigen-specific IgG1 was significantly reduced ( Figure 3B ). Moreover, mice were also immunized with 100 µg of NP-LPS, a T-independent antigen. Antibody responses to this antigen were also impaired ( Figure 3C–D ). These results demonstrate a defect in class-switched antibody production in ADAM10Δ/ΔIgG1-cre+/− mice.
Figure 3. ADAM10Δ/ΔIgG1-cre+/− mice show impaired primary antibody responses.
ADAM10Δ/ΔIgG1-cre+/− (▪) and controls (□) were immunized with a T-dependent antigen, NP-KLH, emulsified in alum (A–B) or a T-independent antigen, NP-LPS (C–D). At the indicated times, serum samples were collected and NP-specific antibodies were measured by capture ELISA. Bars represent the mean ± SE of 5–9 mice per group (*p<0.05, **p<0.01). Data represent results obtained in at least two independent experiments.
Memory B cell development and recall antibody responses in ADAM10Δ/ΔIgG1-cre+/−YFP+ mice
Given that ADAM10Δ/ΔIgG1-cre+/− mice showed abnormal primary antibody responses, we sought to determine whether recall antibody responses were also impaired. To this end, mice were immunized with 10 µg of NP-KLH emulsified in alum and boosted 6 weeks later. As demonstrated in Figure 3 , ADAM10Δ/ΔIgG1+/− mice showed decreased production of antigen specific IgG1-antibodies following primary immunization. Moreover, recall responses were also dramatically impaired ( Figure 4A ). Interestingly, when the frequency of memory B cells was analyzed by flow cytometry 14 days following immunization, ADAM10Δ/ΔIgG1+/− mice had memory B cells percentages comparable to that of controls ( Figure 4B,C ). These data also suggest that the defects seen in secondary antibody responses result from impaired PC differentiation and/or function.
Figure 4. ADAM10Δ/ΔIgG1-cre+/− mice show impaired recall antibody responses but normal memory B cell development.
(A) ADAM10Δ/ΔIgG1-cre+/− (▪) and controls (□) were immunized with NP-KLH emulsified in alum. At the indicated times serum samples were collected and NP-specific antibodies were measured by capture ELISA. Mice were challenged with NP-KLH in alum 6 weeks following primary immunization. Bars represent the mean ± SE of 5–9 mice per group (*p<0.05, **p<0.01). Data represent results obtained in at least two independent experiments. (B–C) ADAM10Δ/ΔIgG1-cre+/− (▪) and controls (□) were immunized with NP-KLH emulsified in alum. Fourteen days following immunization, spleens were harvested and memory B cell numbers were analyzed. Memory B cells were defined as B220+IgMlo/−IgG1+CD38+ [44].(B) Representative dot plot (gated on B220+IgMlo/− cells). (C) Frequency of memory B cells of total B220+IgMlo/− cells, representative of 5 mice from at least two independent studies. ns: non significant.
Plasma cell development in ADAM10Δ/ΔIgG1-cre+/−YFP+ mice
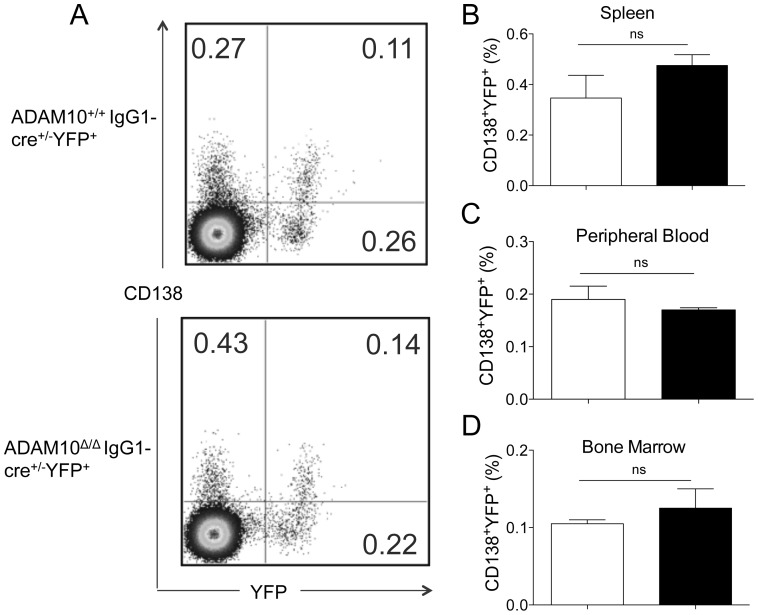
Given that ADAM10Δ/ΔIgG1-cre+/−YFP+ mice had impaired antibody responses but normal GCs, we hypothesized that the defect in antibody production resulted from aberrant PC differentiation. CD138 is marker for antibody-secreting PCs (B220lo/−CD138+ cells). Accordingly, ADAM10Δ/ΔIgG1-cre+/−YFP+ and control mice were immunized with 10 µg of NP-KLH and the percentage of Cre-expressing PCs (B220lo/−CD138+YFP+ cells) in the spleen, peripheral blood and bone marrow was determined ( Figure 5 ) [7]. The gating protocol is depicted in Figure 5A . Surprisingly, although ADAM10Δ/ΔIgG1-cre+/−YFP+ mice had markedly impaired antigen-specific IgG1 responses, they had PC percentages comparable to that of controls ( Figure 5B–D ). Previous studies revealed a similar phenotype when humoral responses were studied in B-cell specific XBP1-deficient mice [21]. XBP-1 is a protein involved in ER-stressed and is required for antibody-secretion [21]. These data suggested that ADAM10 might regulate antibody production by modulating XBP-1 expression in PCs.
Figure 5. ADAM10Δ/ΔIgG1-cre+/− mice have normal plasma cell numbers.
ADAM10Δ/ΔIgG1-cre+/− (▪) and controls (□) were immunized with NP-KLH emulsified in alum. Twenty-one days following immunization, tissues were harvested and PC numbers were analyzed. Cre-expressing plasma cells were defined as B220lo/−CD138+YFP+. (A) Representative FACS staining of ADAM10Δ/ΔIgG1-cre+/− and controls. Frequency of plasma cells of total B220lo/− cells from (B) spleen, (C) peripheral blood and (D) bone marrow. Bars represent the mean ± SE of 4–5 mice per group. Data represent results obtained in at least two independent experiments.
Abnormal gene expression in plasma cells isolated from ADAM10Δ/ΔIgG1-cre+/−YFP+ mice
Given that the phenotype observed in ADAM10Δ/ΔIgG1-cre+/−YFP+ mice resembled that of B-cell specific XBP1-deficient mice, Xbp1 levels were determined in PCs isolated from ADAM10Δ/ΔIgG1-cre+/−YFP+ and control mice. Interestingly, Xbp1 message levels were significantly reduced when compared to controls ( Figure 6A ). Studies have demonstrated that Xbp1 expression is preceded by the downregulation of Bcl6 and the increased expression of Blimp1 and IRF4 [21]. Thus, the expression of these genes was examined. PCs isolated from ADAM10Δ/ΔIgG1-cre+/−YFP+ mice also showed a reduction in message levels for Prdm1 (the gene encoding for Blmp1) ( Figure 6B ) and Irf4 ( Figure 6C ). Moreover, Bcl6 levels were significantly higher than in control PCs ( Figure 6D ). Even more striking, while PC isolated from controls had ∼60 fold more Prdm1 message than Bcl6, PCs isolated from ADAM10Δ/ΔIgG1-cre+/−YFP+ mice showed only ∼3 fold more Prdm1 than Bcl6 ( Figure 6E ). These results demonstrate that ADAM10 is required for the proper downregulation of Bcl6 and upregulation of Prdm1, Irf4 and Xbp1, and thus for optimal PC function and production of class-switched antibodies.
Figure 6. Plasma Cells from ADAM10Δ/ΔIgG1-cre+/− mice have altered gene expression.
ADAM10Δ/ΔIgG1-cre+/− and controls were immunized with NP-KLH emulsified in alum. Twenty-one days following immunization, splenic PCs were isolated via magnetic bead isolation. mRNA was isolated and (A) Xbp1 (B) Prdm1, (C) Irf4 and (D) Bcl6 message levels were determined by qPCR. (E) The ratio of Prdm1 to Bcl6 was calculated. Bars represent the mean ± SE of 3 independent studies; cells from 3 mice from each genotype pooled in each study. (*p<0.05, **p<0.01, ***p<0.001).
Consistent with the gene expression results, flow cytometry analysis revealed the presence of a B220lo/−CD138+Bcl6+ population in the spleens of ADAM10Δ/ΔIgG1-cre+/− mice following immunization. Splenocytes were isolated and stained for B220 and CD138 ( Figure 7A ). B220lo/−CD138+ cells were analyzed for non-specific (isotype) or Bcl6-specific staining ( Figure 7B,C ). Quantified results are displayed in Figure 7D . These results demonstrate that in the absence of ADAM10, Bcl6 is overexpressed at both the message and protein level.
Figure 7. Plasma cells isolated from ADAM10Δ/ΔIgG1-cre+/− mice express Bcl6.
ADAM10Δ/ΔIgG1-cre+/− and controls were immunized with NP-KLH emulsified in alum. Twenty-one days following immunization, splenic plasma cells were analyzed by Bcl6 protein expression via flow cytometry. (A) Plasma cells were defined as CD138+B220lo/−. After gating on these cells, they were analyzed for (B) non-specific (isotype) or (C) Bcl6-specific staining. (D) Frequency of Bcl6+ cells of plasma cells. Bars represent the mean ± SE of 4–5 mice. (**p<0.01).
Discussion
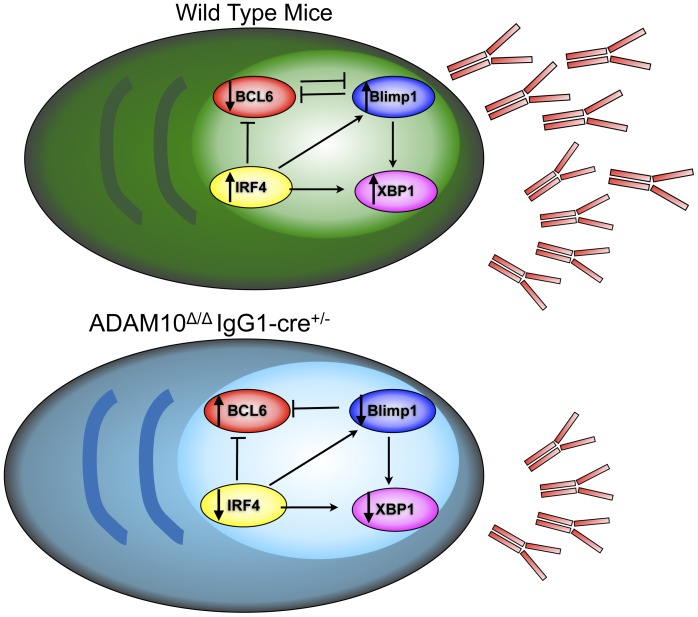
Members of the ADAM family regulate a wide range of functions, including cell migration, proliferation and adhesion [22]. ADAM10, in particular, has been recently shown to be critical for lymphocyte development through initiation of the canonical Notch signaling pathway [17], [18]. We recently published that ADAM10 is highly expressed in GC B cells. Interestingly, mice that lacked ADAM10 in all peripheral B cells fail to generate GCs and have severely impaired humoral responses. Furthermore, defects in antibody production are accompanied by changes in lymphoid architecture [19]. Here we demonstrate that mice with ADAM10 deletion in class-switched B cells (ADAM10Δ/ΔIgG1-cre+/− mice) have normal GC formation and show no changes in splenic architecture. These mice, however, showed reduced production of class-switched antibodies. Given that Cre under the control of the IgG1 promoter is preferentially expressed in class-switched cells, the current study does not allow us to determine the role of ADAM10 in IgM-producing cells. As summarized in Figure 8 , we have demonstrated that in spite of normal PC percentages, expression levels of proteins important for PC development, Prdm1, xbp1 and Irf4 were diminished compared to controls. In addition, the GC transcription factor Bcl6 was not properly downregulated in ADAM10Δ/ΔIgG1cre+/− mice. These results demonstrate that ADAM10 is required for the appropriate downregulation of Bcl6 and upregulation of Prdm1, Irf4 and Xbp1. Subsequently, antibody responses are abnormal in ADAM10Δ/ΔIgG1cre+/− mice. Thus, ADAM10 is important for proper PC function.
Figure 8. Model.
Wild type plasma cells express higher levels of Blimp1, IRF4 and XBP1, while Bcl6 is repressed. This allows for antibody secretion. In the case of ADAM10Δ/ΔIgG1-cre+/− mice, Bcl6 levels are higher than seen in wild type. Moreover, Blimp1, IRF4 and XBP1 expression are decreased, leading to impaired antibody secretion.
The transcription factor Bcl6 is necessary for GC formation and B cell proliferation. While it is well established that Bcl6 must be downregulated for class-switched PC differentiation to occur, the factors mediating this event are not fully understood [12]. It has been previously demonstrated that Stat3 activated by IL-21 can trigger Blimp1 expression by competing with Bcl6 for DNA binding sites [9]. Moreover, it has been proposed that B cell receptor (BCR) and CD40 signaling lead to Bcl6 degradation [23]. Recent studies demonstrated that ectopic Stat3 signaling could induce Blimp1 expression, even in the presence of high Bcl6 levels; however, PC differentiation did not proceed until Bcl6 levels were reduced [9]. Here we demonstrate that ADAM10 is important for Bcl6 downregulation and ADAM10 deficiency leads to impaired antibody responses.
Along with defective Bcl6 downregulation observed in ADAM10-deficient PCs, ADAM10 deletion also resulted in decreased levels of Prdm1, Irf4 and Xbp1. Blimp1 and IRF4 lead to cell cycle arrest and promote PC differentiation [24], while XBP1 is required for endoplasmic reticulum (ER) expansion and immunoglobulin production and secretion [25]. It has been very well documented that Bcl6 can inhibit Blimp1 [26]. Moreover, Blimp1 and IRF4 permit XPB1 expression [7], [25]. It is possible that ADAM10 is involved in a pathway that controls both Blimp1 and IRF4 expression, and decreased Blimp1 and IRF4 then leads to decreased XBP1 expression.
Bcl6−/− mice respond normally to T-independent antigens, demonstrating that Bcl6 is dispensable for T-independent antibody responses. On the other hand, studies have demonstrated that PC differentiation following T-independent immunizations is dependent on Blimp1 expression [4], [27], [28]. Recent studies have demonstrated that through dual BCR and toll like receptor (TLR) engagement, NP-LPS induces T-independent isotype switching [29]. Interestingly, when ADAM10Δ/ΔIgG1cre+/− mice where immunized with NP-LPS, a T-independent antigen that activated B cells through Toll like receptor 4 (TLR4) signaling, impaired antibody production was also evident. It is therefore possible that ADAM10 deletion leads to decreased Blimp1, Xbp1 and Irf4 expression in a Bcl6-independent manner. Remarkably, studies of mice deficient in a TLR4 downstream signaling molecule, MyD88, demonstrated that MyD88-deficient B cells also exhibited enhanced Bcl6 expression and diminished Blimp1 expression [30], suggesting that ADAM10 might be involved in TLR4-signaling.
ADAM10 is critical for Notch1 and Notch2 cleavage and the initiation of the canonical Notch signaling pathway [17], [18]. The role of Notch signaling in GC formation and PC differentiation, however, remains controversial. In vivo studies of B cell specific RBP-Jκ-deficient mice, a key mediator of Notch signaling, failed to reveal a defect in antibody production [31]. Consistent with this finding, a recently published report demonstrated that B cell-specific Notch2-deficient mice have normal GC formation and normal numbers of splenic PCs [32], [33]. Moreover, mice with constitutively active Notch2 intracellular domain exhibited diminished antibody responses to T-dependent and T-independent antigens and impaired GC formation [34]. On the other hand, in vitro studies have demonstrated that Notch signaling enhances B cell activation and supports B cell survival [35]. Moreover, recent studies have shown that Notch signaling synergizes with BCR and CD40 signaling to enhance murine B cell activation [36]. Further studies will be needed, however, in order to elucidate Notch1 and Notch2's role in PC formation and whether the phenotype observed in ADAM10Δ/ΔIgG1-cre+/−YFP+ mice results from impaired Notch signaling.
Another ADAM10 substrate is the IL-6 receptor [37]. Studies have demonstrated that IL-6 signaling can induce Bcl6 expression [38], [39]. In the context of a GC, IL-6 is produced by activated FDCs and promotes SHM, affinity maturation, and CSR [40]. Indeed, IL-6-deficient mice showed decreased SHM following T-dependent immunization [40]. Given that ADAM10 is responsible for the cleavage of the IL-6 receptor (IL-6R) [37], one could speculate that increased IL-6R expression could lead to increased IL-6 signaling, thus, leading to Bcl6 overexpression. Moreover, it is possible that by modulating IL-6R expression levels, ADAM10 can influence SHM and affinity maturation and CSR.
Regulation of Notch signaling is not the only way that ADAM10 is capable of modulating gene expression. Recent experiments have demonstrated that ADAM10 not only mediates RIP, but it is also subject to RIP. ADAM9 and ADAM15 have been identified as the proteases responsible for releasing the ADAM10 ectodomain, while gamma-secretase mediates the release of the ADAM10 intracellular domain (ICD). ADAM10-ICD then translocates to the nucleus and modulates gene expression [15]. Studies have demonstrated that nuclear ADAM10 can interact with androgen receptors and act like a transcription factor [41]. It is thus possible that ADAM10 regulates gene expression and PC differentiation, not by shedding of membrane proteins but through its involvement in gene regulation. ADAM10-ICD regulation of gene expression will require further study.
In conclusion, here we demonstrate that deletion of ADAM10 in class-switched cells does not impair GC formation. However, despite normal PC frequencies, ADAM10Δ/ΔIgG1-cre+/−YFP+ mice showed impaired antibody responses to T-dependent and T-independent antigens. Gene expression analysis demonstrated that PCs isolated from ADAM10Δ/ΔIgG1-cre+/−YFP+ mice expressed lower levels of Prdm1, Irf4 and Xbp1. Intriguingly, ADAM10Δ/ΔIgG1-cre+/− PCs also expressed 3-fold higher levels of Bcl6. These results demonstrate that ADAM10 is important for the expression of transcription factors that are required for PC differentiation and thus, for optimal antibody production.
Materials and Methods
Mice and immunizations
To generate ADAM10Δ/ΔIgG1-cre+/−YFP+ mice, ADAM10Δ/ΔYFP+ mice were crossed with IgG1-cre+/− mice (Jackson Mice) [19], [42]. ADAM10+/+IgG1-cre+/−YFP+ mice were used as controls. All mouse protocols were approved by the Virginia Commonwealth University Institutional Animal Care and Use Committee. Immunizations comprised of injections of 10 µg 4-Hydroxy-3-nitrophenylacetyl coupled to keyhole limpet hemocyanin at a ratio of 27∶1 (NP27KLH, referred to as NP-KLH) in 4 mg of alum or 100 µg of NP-LPS in PBS (Biosearch Technologies). For recall responses to NP-KLH, mice were boosted 6 weeks post-primary immunization with the same dose of antigen as primary immunization.
Flow cytometry and immunohistochemistry
Single cell suspension of splenocytes were stained as described previously [19]. Abs included anti-mouse unlabeled 2.4G2, FITC conjugated GL7 (GL7); PE-Cy7 conjugated B220 (RA3-6B2) and CD38 (90); allophycocyanin-conjugated GL7 (GL7) and CD138 (281-2); PE-conjugated IgD (11-26c.2a), Fas (Jo2) and PerCP-Cy5.5 conjugated IgM. Flow cytometry analysis was performed using a Canto or Aria II (BD Biosciences), and data analysis was conducted with FlowJo v8.8.7 (Tree Star). Lymphocytes were gated based on FSC vs. SSC. B cell subsets were defined as follows: Plasma cells: B220lo/−CD138+ [21]; germinal center B cells: B220+GL7+Fashi [43]; and memory B cells: B220+IgMlo/−IgG1+CD38+ [44]. For immunohistochemistry, spleens were prepared as previously described [19]. Digital images were captured, overlaid, and processed with the Confocal and LCS Lite programs (Leica).
Cell isolation and Quantitative PCR
Plasma cells were isolated via negative selection (B220−DX5− cells) and subsequent positive selection (CD138+ cells) using magnetic beads and following manufacturer's instructions (Miltenyi). RNA was extracted and cDNA was generated as previously described [19]. Primers and probes for running a TaqMan quantitative PCR (qPCR) assay were purchased from Applied Biosystems. TaqMan gene expression assays included Xbp1: Mm00457357_m1, Prdm1: Mm00476128_m1, Bcl6: Mm00477633_m1; and Irf4: Mm00516431_m1. Reaction parameters were as previously described [17]. Results were analyzed with iQ5 real-time PCR software (version 2.0).
ELISA
For total IgM and IgG1 ELISAs, samples were serially diluted and added to 96-well plates (50 µL/well) pre-coated with 5 µg/mL of goat-anti IgM and IgG, respectively (Southern Biotech). For standard curve, normal mouse IgM and IgG1 (Southern Biotech) were used. After incubation at 37°C for 1 h, bound Abs were revealed by goat-anti-IgM-AP and goat-anti-IgG1-AP, respectively (Southern Biotech). Total IgE ELISA was carried out as previously described [45]. For antigen-specific ELISAs, ELISA was carried out as described with minor modifications. Plates were coated with NP14BSA (Biosearch Technologies)(15 µg/mL in PBS) for samples and with 5 µg/mL of goat-anti Ig (Southern Biotech) in BBS for standard. Standard curves were performed by coating with anti- IgM, anti-IgG1 or IgG3 and adding known amounts of IgM, IgG1 or IgG3, respectively. The values for experimental samples are reflected as relative units (RU) because serum antigen-specific Abs were captured with antigen. The remaining steps were carried out as described.
Statistical analysis
p-values were calculated using unpaired two-tailed Student's t tests in Graphpad Prism v5. Error bars represent the SEM between samples. p<0.05 is considered statistically significant.
Acknowledgments
We thank J. Tew and S. Barbour for suggestions on the manuscript. Expert technical assistance by Hannah Zellner, Danjing Zhao and Lauren Folgosa is also acknowledged.
Funding Statement
This work was funded by the National Heart Lung Blood Institute (NHLBI) and the National Institute of Allergy and Infections disease (NIAID) of the National Institutes of Health (NIH). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Smith KGC, Hewitson TD, Nossal GJV, Tarlinton DM (1996) The phenotype and fate of the antibody-forming cells of the splenic foci. Eur J Immunol 26: 444–448. [DOI] [PubMed] [Google Scholar]
- 2. Erickson LD, Durell BG, Vogel LA, O'Connor BP, Cascalho M, et al. (2002) Short-circuiting long-lived humoral immunity by the heightened engagement of CD40. J Clin Invest 109: 613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Cozine CL, Wolniak KL, Waldschmidt TJ (2005) The primary germinal center response in mice. Curr Opin Immunol 17: 298–302 doi:10.1016/j.coi.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 4. Shapiro-Shelef M, Lin K-I, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, et al. (2003) Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 19: 607–620. [DOI] [PubMed] [Google Scholar]
- 5. Shaffer AL, Lin K-I, Kuo TC, Yu X, Hurt EM, et al. (2002) Blimp-1 Orchestrates Plasma Cell Differentiation by Extinguishing the Mature B Cell Gene Expression Program. Immunity 17: 51–62. [DOI] [PubMed] [Google Scholar]
- 6. Reimold AM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, et al. (2001) Plasma cell differentiation requires the transcription factor XBP-1. Nature 412: 300–307 doi:10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 7. Klein U, Casola S, Cattoretti G, Shen Q, Lia M, et al. (2006) Transcription factor IRF4 controls plasma cell differentiation and class-switch recombination. Nat Immunol 7: 773–782 doi:10.1038/ni1357. [DOI] [PubMed] [Google Scholar]
- 8. Alinikula J, Nera K-P, Junttila S, Lassila O (2011) Alternate pathways for Bcl6-mediated regulation of B cell to plasma cell differentiation. Eur J Immunol doi:10.1002/eji.201141553. [DOI] [PubMed] [Google Scholar]
- 9. Diehl SA, Schmidlin H, Nagasawa M, van Haren SD, Kwakkenbos MJ, et al. (2008) STAT3-mediated up-regulation of BLIMP1 Is coordinated with BCL6 down-regulation to control human plasma cell differentiation. J Immunol 180: 4805–4815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cattoretti G, Shaknovich R, Smith PM, Jäck H-M, Murty VV, et al. (2006) Stages of germinal center transit are defined by B cell transcription factor coexpression and relative abundance. 177: 6930–6939. [DOI] [PubMed] [Google Scholar]
- 11. Oracki SA, Walker JA, Hibbs ML, Corcoran LM, Tarlinton DM (2010) Plasma cell development and survival. Immunol Rev 237: 140–159 doi:10.1111/j.1600-065X.2010.00940.x. [DOI] [PubMed] [Google Scholar]
- 12. Cattoretti G, Chang CC, Cechova K, Zhang J, Ye BH, et al. (1995) BCL-6 protein is expressed in germinal-center B cells. Blood 86: 45–53. [PubMed] [Google Scholar]
- 13. Blobel CP (2005) ADAMs: key components in EGFR signalling and development. Nat Rev Mol Cell Biol 6: 32–43 doi:10.1038/nrm1548. [DOI] [PubMed] [Google Scholar]
- 14. Crawford HC, Dempsey PJ, Brown G, Adam L, Moss ML (2009) ADAM10 as a therapeutic target for cancer and inflammation. Curr Pharm Des 15: 2288–2299. [DOI] [PubMed] [Google Scholar]
- 15. Tousseyn T, Thathiah A, Jorissen E, Raemaekers T, Konietzko U, et al. (2008) ADAM10, the Rate-limiting Protease of Regulated Intramembrane Proteolysis of Notch and Other Proteins, Is Processed by ADAMS-9, ADAMS-15, and the -Secretase. Journal of Biological Chemistry 284: 11738–11747 doi:10.1074/jbc.M805894200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gibb DR, Saleem SJ, Chaimowitz NS, Mathews JA, Conrad DH (2011) The emergence of ADAM10 as a regulator of lymphocyte development and autoimmunity. Mol Immunol doi:10.1016/j.molimm.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gibb DR, Shikh el MEM, Kang D-J, Rowe WJ, Sayed el RM, et al. (2010) ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. Journal of Experimental Medicine 207: 623–635 doi:10.1084/jem.20091990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Tian L, Wu X, Chi C, Han M, Xu T, et al. (2008) ADAM10 is essential for proteolytic activation of Notch during thymocyte development. Int Immunol 20: 1181–1187 doi:10.1093/intimm/dxn076. [DOI] [PubMed] [Google Scholar]
- 19. Chaimowitz NS, Martin RK, Cichy J, Gibb DR, Patil P, et al. (2011) A Disintegrin and Metalloproteinase 10 Regulates Antibody Production and Maintenance of Lymphoid Architecture. J Immunol doi:10.4049/jimmunol.1102172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Casola S, Cattoretti G, Uyttersprot N, Koralov SB, Seagal J, et al. (2006) Tracking germinal center B cells expressing germ-line immunoglobulin gamma1 transcripts by conditional gene targeting. Proc Natl Acad Sci USA 103: 7396–7401 doi:10.1073/pnas.0602353103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Todd DJ, McHeyzer-Williams LJ, Kowal C, Lee A-H, Volpe BT, et al. (2009) XBP1 governs late events in plasma cell differentiation and is not required for antigen-specific memory B cell development. Journal of Experimental Medicine 206: 2151–2159 doi:10.1084/jem.20090738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Overall CM, Blobel CP (2007) In search of partners: linking extracellular proteases to substrates. Nat Rev Mol Cell Biol 8: 245–257 doi:10.1038/nrm2120. [DOI] [PubMed] [Google Scholar]
- 23. Ozaki K, Spolski R, Ettinger R, Kim H-P, Wang G, et al. (2004) Regulation of B cell differentiation and plasma cell generation by IL-21, a novel inducer of Blimp-1 and Bcl-6. J Immunol 173: 5361–5371. [DOI] [PubMed] [Google Scholar]
- 24. Calame KL, Lin K-I, Tunyaplin C (2003) Regulatory mechanisms that determine the development and function of plasma cells. Annu Rev Immunol 21: 205–230 doi:10.1146/annurev.immunol.21.120601.141138. [DOI] [PubMed] [Google Scholar]
- 25. Shaffer AL, Shapiro-Shelef M, Iwakoshi NN, Lee A-H, Qian S-B, et al. (2004) XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21: 81–93 doi:10.1016/j.immuni.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 26. Johnston RJ, Poholek AC, DiToro D, Yusuf I, Eto D, et al. (2009) Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science 325: 1006–1010 doi:10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Soro PG, Morales-A P, Martínez-M JA, Morales-A S, Copín SG, et al. (1999) Differential involvement of the transcription factor Blimp-1 in T cell-independent and -dependent B cell differentiation to plasma cells. J Immunol 163: 611–617. [PubMed] [Google Scholar]
- 28. Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM (1997) Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 276: 589–592. [DOI] [PubMed] [Google Scholar]
- 29. Pone EJ, Zhang J, Mai T, White CA, Li G, et al. (2012) BCR-signalling synergizes with TLR-signalling for induction of AID and immunoglobulin class-switching through the non-canonical NF-κB pathway. Nat Comms 3: 767 doi:10.1038/ncomms1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Pasare C, Medzhitov R (2005) Control of B-cell responses by Toll-like receptors. Nature 438: 364–368 doi:10.1038/nature04267. [DOI] [PubMed] [Google Scholar]
- 31. Tanigaki K, Han H, Yamamoto N, Tashiro K, Ikegawa M, et al. (2002) Notch|[ndash]|RBP-J signaling is involved in cell fate determination of marginal zone B cells. Nat Immunol 3: 443–450 doi:doi:10.1038/ni793. [DOI] [PubMed] [Google Scholar]
- 32. Saito T, Chiba S, Ichikawa M, Kunisato A, Asai T, et al. (2003) Notch2 is preferentially expressed in mature B cells and indispensable for marginal zone B lineage development. Immunity 18: 675–685. [DOI] [PubMed] [Google Scholar]
- 33. Sakurai N, Maeda M, Lee S-U, Ishikawa Y, Li M, et al. (2011) The LRF transcription factor regulates mature B cell development and the germinal center response in mice. J Clin Invest doi:10.1172/JCI45682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hampel F, Ehrenberg S, Hojer C, Draeseke A, Marschall-Schröter G, et al. (2011) CD19-independent instruction of murine marginal zone B-cell development by constitutive Notch2 signaling. Blood doi:10.1182/blood-2010-12-325944. [DOI] [PubMed] [Google Scholar]
- 35. Santos MA, Sarmento LM, Rebelo M, Doce AA, Maillard I, et al. (2007) Notch1 engagement by Delta-like-1 promotes differentiation of B lymphocytes to antibody-secreting cells. Proc Natl Acad Sci USA 104: 15454–15459 doi:10.1073/pnas.0702891104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Thomas M, Calamito M, Srivastava B, Maillard I, Pear WS, et al. (2007) Notch activity synergizes with B-cell-receptor and CD40 signaling to enhance B-cell activation. Blood 109: 3342–3350 doi:10.1182/blood-2006-09-046698. [DOI] [PubMed] [Google Scholar]
- 37. Matthews V, Schuster B, Schütze S, Bussmeyer I, Ludwig A, et al. (2003) Cellular cholesterol depletion triggers shedding of the human interleukin-6 receptor by ADAM10 and ADAM17 (TACE). J Biol Chem 278: 38829–38839 doi:10.1074/jbc.M210584200. [DOI] [PubMed] [Google Scholar]
- 38. Nurieva RI, Chung Y, Martinez GJ, Yang XO, Tanaka S, et al. (2009) Bcl6 mediates the development of T follicular helper cells. Science 325: 1001–1005 doi:10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tsuyama N, Danjoh I, Otsuyama K-I, Obata M, Tahara H, et al. (2005) IL-6-induced Bcl6 variant 2 supports IL-6-dependent myeloma cell proliferation and survival through STAT3. Biochem Biophys Res Commun 337: 201–208 doi:10.1016/j.bbrc.2005.09.036. [DOI] [PubMed] [Google Scholar]
- 40. Wu Y, Shikh el MEM, Sayed el RM, Best AM, Szakal AK, et al. (2009) IL-6 produced by immune complex-activated follicular dendritic cells promotes germinal center reactions, IgG responses and somatic hypermutation. Int Immunol 21: 745–756 doi:10.1093/intimm/dxp041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Arima T, Enokida H, Kubo H, Kagara I, Matsuda R, et al. (2007) Nuclear translocation of ADAM-10 contributes to the pathogenesis and progression of human prostate cancer. Cancer Science 98: 1720–1726 doi:10.1111/j.1349-7006.2007.00601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hobeika E, Thiemann S, Storch B, Jumaa H, Nielsen PJ, et al. (2006) Testing gene function early in the B cell lineage in mb1-cre mice. Proc Natl Acad Sci USA 103: 13789–13794 doi:10.1073/pnas.0605944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vu F, Dianzani U, Ware CF, Mak T, Gommerman JL (2008) ICOS, CD40, and lymphotoxin beta receptors signal sequentially and interdependently to initiate a germinal center reaction. J Immunol 180: 2284–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Aiba Y, Kometani K, Hamadate M, Moriyama S, Sakaue-Sawano A, et al. (2010) Preferential localization of IgG memory B cells adjacent to contracted germinal centers. Proc Natl Acad Sci USA 107: 12192–12197 doi:10.1073/pnas.1005443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Caven TH, Shelburne A, Sato J, Chan-Li Y, Becker S, et al. (2005) IL-21 dependent IgE production in human and mouse in vitro culture systems is cell density and cell division dependent and is augmented by IL-10. Cell Immunol 238: 123–134 doi:10.1016/j.cellimm.2006.03.001. [DOI] [PubMed] [Google Scholar]