Abstract
Manipulation of crops to improve their nutritional value (biofortification) and optimisation of plants for removal of toxic metals from contaminated soils (phytoremediation) are major goals. Identification of membrane transporters with roles in zinc and cadmium transport would be useful for both aspects. The P1B-ATPases play important roles in heavy metal allocation and detoxification in Arabidopsis and it is now important to elucidate their roles in monocots. We identified nine P1B-ATPases in barley and this study focuses on the functional characterization of HvHMA2, providing evidence for its role in heavy metal transport. HvHMA2 was cloned using information from EST analysis and 5′ RACE. It possesses the conserved aspartate that is phosphorylated during the reaction cycle of P-type pumps and has motifs and key residues characteristic of P1B-ATPases, falling into the P1B-2 subclass. Homologous sequences occur in three major sub-families of the Poaceae (Gramineae). Heterologous expression in Saccharomyces cerevisiae demonstrates that HvHMA2 functions as a Zn and Cd pump. Mutagenesis studies show that proposed cation coordination sites of the P1B-2 pumps are crucial for the metal responses conferred by HvHMA2 in yeast. HvHMA2 expression suppresses the Zn-deficient phenotype of the Arabidopsis hma2hma4 mutant indicating that HvHMA2 functions as a Zn pump in planta and could play a role in root to shoot Zn transport. When expressed in Arabidopsis, HvHMA2 localises predominantly to the plasma membrane.
Introduction
Plants require a range of metals in trace amounts for growth and development. These metal micronutrients include Fe, Cu, Co, Zn, Mn and Ni [1]. They can play critical structural roles in many proteins; act as catalytic components in enzymes, and function in redox reactions. The correct balance of these micronutrients is required for optimum growth and development and complex mechanisms have evolved to ensure that proteins are supplied with adequate levels of the required metal and also to cope with fluctuations in the environment [2], [3].
Zn is required for all organisms, including plants. Zn deficiency is one of the most common micronutrient deficiencies in agricultural soils and thus can lead to reductions in crop yield. Zn is also essential in human nutrition and Zn deficiency is estimated to affect more than 25% of the world's population causing impaired growth and increased susceptibility to disease. Plants at the base of the food chain are an important source of dietary Zn. Zn tends to be at a relatively low level in staple foods, and cereals such as barley, wheat and rice have relatively low levels in the grain. Therefore Zn biofortification of food crops which could lead to increased bioavailable Zn would be an important sustainable solution to address Zn malnutrition [4]. Understanding the processes that contribute to Zn uptake from the soil, translocation to the shoot and partitioning in the grain are integral to developing strategies to improve the Zn content of grain. Cadmium is a non-essential metal that can contaminate soils and is toxic to both plants and animals. It can be taken up by transporters of essential micronutrients such as Zn and Fe [5]; therefore when considering mechanisms to increase the Zn content of food it is also necessary to consider their potential to accumulate Cd [4].
The P1B-ATPases (also known as Heavy Metal ATPases or HMAs) are one of several transporter families involved in Zn transport [6], [7]. P1B-ATPases are classified into six subgroups (P1B1-6) which are proposed to have distinct metal binding and transport specificities [8]. There are eight P1B-ATPases in Arabidopsis thaliana and four have some role in Zn transport. AtHMA1 is found at the inner envelope of the chloroplast and contributes to Zn(II) detoxification by reducing the Zn content of plastids [9]. It is also reported to load Cu into the stroma, supply Cu to chloroplast Cu/Zn superoxide dismutase [10], and transport Ca [11]. AtHMA2 and AtHMA4 play key roles in translocation of Zn from root to shoot with the hma2hma4 double mutant exhibiting a strong Zn nutritional deficiency [12], [13]. They are also the main route by which Cd is transferred to the shoot [14]. AtHMA4 also plays a role in Cd detoxification at elevated Cd levels [15]. AtHMA3 is proposed to function in vacuolar sequestration of Zn, Cd, Co and Pb, suggesting a detoxification role [16].
As many of our staple food sources such as the cereals rice and wheat are monocots, it is important to understand the function of P1B-ATPases in these species. From genome sequence analysis, nine HMA genes have been identified in rice (OsHMA1-9). The first to be characterised was OsHMA9 [17]. Phylogenetically OsHMA9 clusters with the Arabidopsis Cu pumps AtHMA5-8 [18]; however, phenotypic analysis of rice oshma9 mutants suggested a broader role as mutants were sensitive to high Cu, Zn, Cd and Pb [17]. Subsequently OsHMA3, which clusters with AtHMA2, 3 and 4 in the Zn/Cd/Pb (P1B-2) sub-group, was shown to be a vacuolar Cd uptake transporter in roots, reducing cytoplasmic Cd levels and consequently transport of Cd to the shoot [19], [20], [21]. It remains to be shown whether OsHMA3 also transports other metals. Recently OsHMA2 was shown to mediate Cd efflux when expressed in yeast [22] and mutant analysis in rice suggests that OsHMA2 is a major transporter of Zn and Cd from roots to shoots [23]. Despite the importance of this family of transporters, we know virtually nothing in the major temperate cereals such as wheat and barley. To address this we describe the cloning and functional analysis of HvHMA2, a barley P1B-ATPase from the Zn/Cd/Pb (P1B-2) sub-group. Using a variety of approaches we show that it can transport the essential micronutrient Zn; however it can also transport the toxic contaminant Cd. We also show that key residues postulated to form part of the cation-binding site in P1B-2-ATPases are crucial for the metal responses conferred by HvHMA2 in yeast.
Results
Primary structure of HvHMA2
HvHMA2 was amplified by RT-PCR using sequence information from EST analysis and 5′ RACE (figure S1). HvHMA2 contains an open reading frame of 3027 bp, encoding a protein of 1009 amino acids and 108,456 molecular mass. Proteins showing high similarity in the P1B-2 subclass include putative heavy-metal transporting P-type ATPases from plants and bacteria (http://www.ncbi.nlm.nih.gov/BLAST/). Highest homology (91% identity) is to a wheat sequence TaHMA2, which has not yet been functionally characterized. Full-length homologues of HvHMA2 were also identified in other members of the Poaceae: two in brachypodium and rice, three in sorghum and four in maize. A barley OsHMA3 homologue (HvHMA3) was also recently submitted to NCBI and has 52% identity to HvHMA2. Percentage identities and alignments of HvHMA2 to homologues in other plants are shown in Table S1 and Figure 1. Hydropathy analysis suggests HvHMA2 has 8 transmembrane domains (TMs) (figure S2).
Figure 1. Alignment of HvHMA2 with various plant P1B-ATPases showing the predicted TMs.
Swissprot TM predictions for AtHMA2 are indicated, except TM6 was extended according to the SOSUI prediction. Shaded boxes indicate motifs conserved in P-type ATPases, other boxes indicate motifs in P1B-ATPase subgroup, and asterisks indicate residues conserved in P1B-2-ATPases that may co-ordinate the metal ion during transmembrane transport and contribute to ion specificity. Conserved (upper-case) and semi-conserved (lower-case) amino acids are indicated beneath the alignment.
HvHMA2 and the wheat homologue TaHMA2 contain motifs found in all P-type ATPases including the conserved aspartate (D400 in HvHMA2, D399 in TaHMA2) that is phosphorylated during the reaction cycle (Figure 1 and 2a). Both also have motifs characteristic of P1B-ATPases [18] including the HP locus in the predicted large cytoplasmic loop (present in most P1B-ATPases but not in other P-types) (Figures 1 and 2a). P1B-ATPases usually have putative heavy Metal-Binding Domains (MBDs) in the N or C termini and the CPx/SPC motif in TM6 [18], [13]. HvHMA2 and TaHMA2 contain the CPC motif in the predicted TM6 (C356 PC in HvHMA2). A “heavy-metal-associated domain” in the HvHMA2 and TaHMA2 N-termini is recognized by the pfam and PROSITE databases (http://ca.expasy.org/prosite; www.sanger.ac.uk/software/pfam). Within this domain the motif GxCCxxE occurs in all the plant P1B-2 sub-class, whereas one or more copies of the motif GMxCxxC occur in P1B-2 ATPases from other organisms.
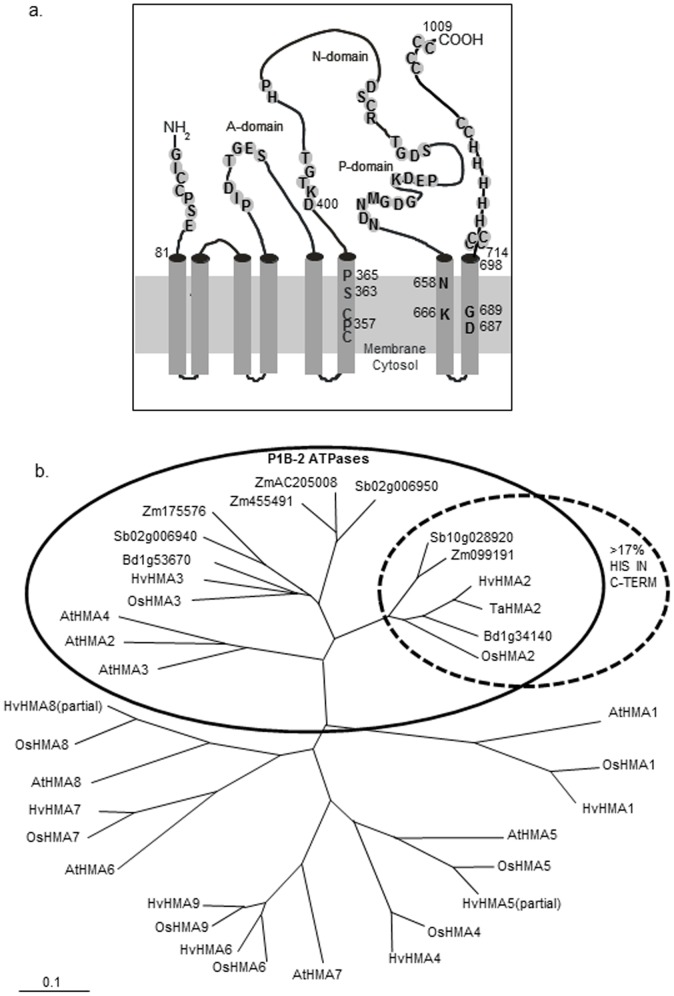
Figure 2. a. Prediction model for the transmembrane topology of HvHMA2.
Schematic diagram illustrating predicted TMs and key motifs. Residues shown in TMs are postulated to coordinate metals during transport. For HvHMA2 the putative cytoplasmic C-terminal metal-binding domain is 307 aa including 56 His and 18 Cys residues plus 4 Cys pairs. Amino acid numbers relevant to the deletion and substitution mutants are included. b. Dendrogram of P1B-ATPases. Includes: all rice and Arabidopsis P1B-ATPases, best available barley sequences and predicted P1B-2-ATPases identified through Aramemnon for maize, Brachypodium and sorghum. The P1B-2 (Zn/Cd) ATPase subgroup is circled, and the sub-set with >17% His residues in the predicted C-termini is indicated. Those P1B-2-ATPases that do not fall within this subset feature <9% His residues in the predicted C-termini. Partial sequences are indicated. Scale bar indicates amino acid substitutions per site.
PIB-ATPases are classified into subsets 1–6 depending on potential cation coordinating residues present in the 6th, 7th and 8th TMs [8], modified by [18]. For the P1B-2 Zn/Cd/Pb transporting sub-group these are TM6: CPCx4SxP; TM7: Nx7K; TM8: DxG (shown for HvHMA2 in Figure 2a, Table S2). Numbered for HvHMA2 (subtract 1 for TaHMA2 numbers) they are: C356, P357, C358, S363 and P365 in TM6, N658 and K666 in TM7 and D687 and G689 in TM8 (Figure 1, Figure 2a).
Phylogenetic analysis of barley HMAs
The phylogenetic tree (Figure 2b) relates HvHMA2 to rice P1B-type ATPase sequences, their barley homologues, Arabidopsis P1B-types, and HvHMA2 (P1B-2) homologues from other monocots. Separate sequences previously classified as distinct HMAs, HvHMA1 and HvHMA10, are here combined as HvHMA1 in line with rice genes and recent barley EST data.
The monocot P1B-2 ATPases identified here fall into two subgroups that differ notably in the composition of their predicted cytoplasmic C-termini; those of HvHMA2, TaHMA2, OsHMA2, maize, sorghum and brachypodium homologues contain high percentages of His residues (>17%) as well as up to 6 CC pairs distributed throughout (Figures 1 and 2b). Those that lack the His-rich C-termini (including OsHMA3 and HvHMA3) have a conserved W instead of the conserved G between TM7 and TM8 (G676 in HvHMA2).
Functional analysis of HvHMA2 in Saccharomyces cerevisiae
HvHMA2 transports Zn and Cd in yeast
HvHMA2 conferred Cd sensitivity to wild-type (wt) yeast (Figure 3). A transport deficient mutant was produced in which the conserved aspartate residue of HvHMA2 was mutated to alanine, hvhma2(D400A). Cd sensitivity was abolished in this mutant, indicating that HvHMA2-dependent Cd sensitivity was due to transport activity. A similar response was also seen in the Cd2+-sensitive ycf1 yeast mutant (results not shown). Previously we have shown that AtHMA4 confers Cd resistance to wt yeast when expressed in the p426 vector [15], [13]. To demonstrate that the effects observed were not due to different vectors, we expressed AtHMA4 in the pYTV vector used in this study for HvHMA2. AtHMA4 still confers resistance to Cd in this vector (Figure 4).
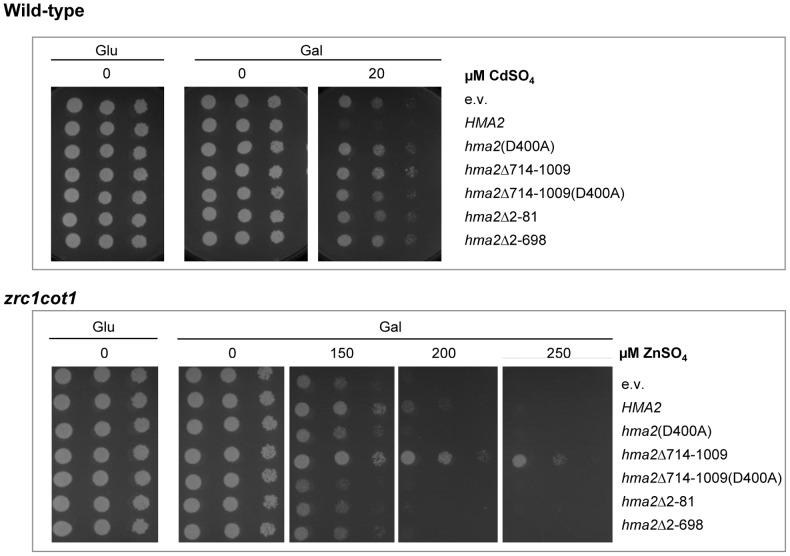
Figure 3. Heterologous expression of HvHMA2 in yeast.
HvHMA2 expression under a Gal-inducible promoter confers Cd sensitivity to wild-type yeast (top) and Zn resistance to zrc1cot1 mutant yeast (bottom) compared to empty vector (e.v) transformed control yeast. Mutant forms of the pump are: C-terminally deleted, hma2Δ714-1009; N-terminally deleted, hma2Δ2-81; C-terminal region alone, hma2Δ2-698; mutant with critical aspartate mutated, hma2(D400A). Photographs show undiluted, 1/10 and 1/100 dilutions of aliquots on agar containing either glucose or galactose as the carbon source, and varying concentrations of CdSO4 or ZnSO4 as indicated.
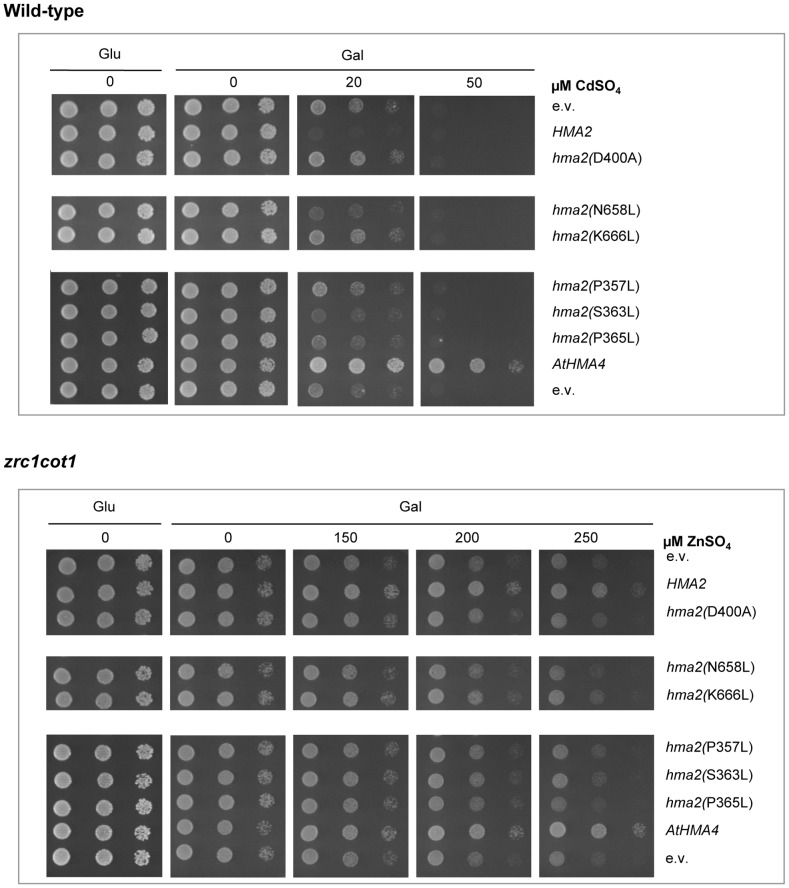
Figure 4. Mutating key residues in HvHMA2 alters metal responses.
Mutation of predicted critical residues decreases the Cd sensitivity conferred by HvHMA2 on wt yeast (top) and the Zn resistance conferred on zrc1cot1 mutant yeast (bottom). Yeast are transformed with galactose-inducible HvHMA2, HvHMA2 with mutation of N658, S363, P365, K666 or P357 to leucine or D400 to alanine, AtHMA4 or empty vector (e.v). Photographs show undiluted, 1/10 and 1/100 dilutions of aliquots on agar containing either glucose or galactose as the carbon source, with CdSO4 or ZnSO4 concentrations as indicated.
Expression of HvHMA2 partially alleviated the Zn sensitivity of the zrc1cot1 mutant (Figure 3) and this effect was lost in the D400A mutant indicating that HvHMA2 can transport Zn. HvHMA2 had no marked effect on the Cu, Co, Ni or Mn sensitivity of wt yeast (data not shown).
Role of HvHMA2 N and C termini
The C-terminally truncated version (HvHMA2Δ714-1009) which has the last 296 residues deleted confers marked Zn resistance on the zrc1cot1 mutant; this is due to transport function as it is eliminated in the corresponding D400A mutant (HvHMA2Δ714-1009(D400A)) (Figure 3). C-terminal truncation of HvHMA2 completely abolished its ability to confer Cd sensitivity (Figure 3). This truncated version, like the full-length HvHMA2, had no effect on the Cu, Co, Ni or Mn sensitivity of wt yeast (data not shown). Deletion of the N-terminus from HvHMA2 (HvHMA2Δ2-81) eliminated Cd and Zn transport (Figure 3). No effect of expression of HvHMA2Δ2-81 was observed on Cu, Co, or Ni sensitivity of wt yeast (data not shown). Expression of just the C-terminus part of HvHMA2 (HvHMA2Δ2-698) has no marked effect on Cd sensitivity in wt yeast, Zn sensitivity of zrc1cot1 (Figure 3), or on Cu, Co, Ni or Mn sensitivity of wt yeast (data not shown).
Effect of mutations in putative metal coordination sites in HvHMA2
To investigate the functional significance of some of the invariant residues in TM6 and 7 for P1B-2 ATPases (see above) we generated the HvHMA2 mutants, P357L, S363L, P365L, N658L and K666L, and expressed them in yeast. The HvHMA2 mutation P357L alters a predicted critical residue/ion specificity determinant in TM6. This proline is part of the CPC motif, a characteristic motif found in P1B-ATPases. As seen in Figure 4, the HvHMA2(P357L) mutant no longer conferred Cd sensitivity to wt yeast and restored growth to control (e.v.) levels. It also abolished the slight Zn resistance conferred on zrc1cot1 yeast mutant by HvHMA2. Two TM6 mutations, HvHMA2(S363L) and HvHMA2(P365L), also decreased the ability of HvHMA2 to confer Cd sensitivity to wt yeast and Zn resistance to zrc1cot1 mutant yeast, although not quite to the same extent as the D400A or P357L mutations (Figure 4). The TM7 mutant HvHMA2(N658L) reduced but did not eliminate the Cd sensitivity conferred to wt yeast compared to the HvHMA2 construct whereas Cd sensitivity was abolished in the (TM7) HvHMA2(K666L) mutant (Figure 4). In the zrc1cot1 yeast mutant both the N658L and K666L mutations abolished the Zn resistance conferred by HvHMA2 (Figure 4).
Tissue and membrane distribution of HvHMA2
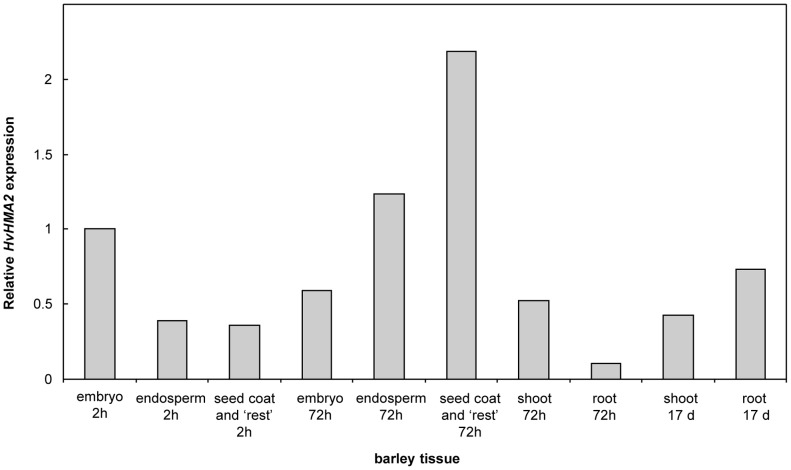
Analysis of microarray data sets shows that HvHMA2 and TaHMA2 have similar expression patterns (Figure S3). We used real-time PCR to show that HvHMA2 expression occurs in all tissues of the germinating grain (Figure S4, Figure 5). HvHMA2 expression decreases in embryo and increases in endosperm tissue between 2 and 72 h, while highest expression is seen in seed coat and other tissue remaining after embryo and endosperm removal (Figure 5, figure S4). HvHMA2 was also expressed in more mature tissues, being found in both root and shoot tissues of 17 day old plants (Figure 5). To determine the membrane localisation of HvHMA2, it was expressed with a GFP tag in Arabidopsis. This showed that HvHMA2 was predominantly localised in the plasma membrane (PM) in root and cotyledon cells (Figure 6 a, b); in addition in some cotyledon cells it was detected in the chloroplasts (Figure 6 c–f).
Figure 5. Expression pattern of HvHMA2 in different barley tissues.
Relative HvHMA2 gene expression levels determined using real-time PCR (average of 2 biological repeats each repeated in triplicate). Time after imbibition, tissues as illustrated in supplementary figure 4.
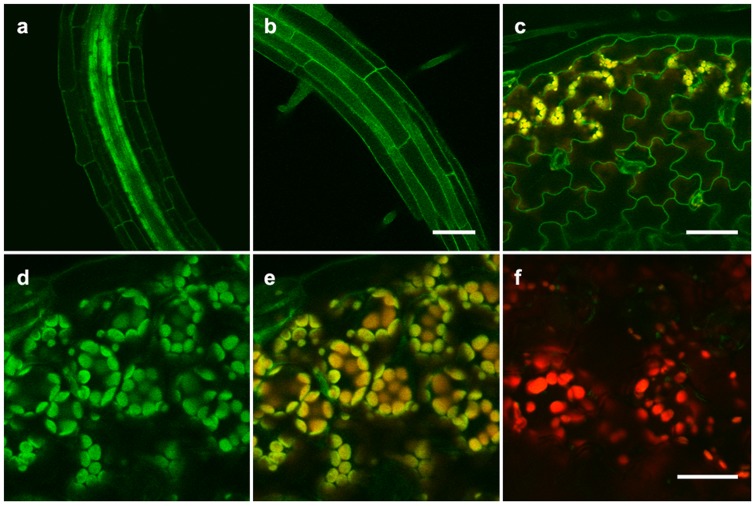
Figure 6. GFP-HvHMA2 localizes to the plasma membrane (PM) and chloroplasts of Arabidopsis.
(a) The PM is marked in all root cells within the root-hair initiation zone. Cells of the stele appear very bright at this stage. (b) In more mature root regions, the PM is very bright. Scale bar (a and b) = 50 µm. (c) In cotyledons, GFP-HvHMA2 marks the PM of epidermal cells (green) and the chloroplasts of mesophyll cells (orange). Orange colouring in chloroplasts results from overlay of the GFP and chlorophyll autofluorescence signals. Scale bar = 50 µm. (d–e) Higher magnification of chloroplast localization in cotyledons. GFP and chlorophyll autofluorescence are overlain in (e). (f) Chlorophyll autofluorescence (red) from cotyledon mesophyll cells of a untransformed control plant overlain with green channel emission collected as in (e). Note that very little green signal appears in wt chloroplasts. Scale bar (d–f) = 25 µm.
HvHMA2 expression rescues the Zn-dependent growth phenotype of the Arabidopsis hma2hma4 mutant
The Arabidopsis hma2hma4 mutant is severely stunted due to the lack of Zn translocation from root to shoot, a process dependent on AtHMA2 and AtHMA4 [12]. This stunted phenotype was clearly seen under the conditions used in this study (Figure 7, figure S5) although we did not observe chlorosis [12]. The wt phenotype was restored to the mutant by supplying additional Zn to the plants [12; ] figure S5. To test whether HvHMA2 functions in Zn transport in planta, it was expressed in the hma2hma4 mutant. Several independent lines were generated and expression of HvHMA2 in these plants was confirmed using RT-PCR (figure S6). When grown on soil alongside wt and hma2hma4 mutants, T2 plants of these lines segregated with an approximate 3∶1 distribution of wt∶stunted (hma2hma4-type) phenotype. PCR on genomic DNA isolated from T2 plants confirmed that hma2hma4 mutant plants transformed with HvHMA2 had a wt phenotype, whereas HvHMA2 was not detected in those T2 transformant plants that had a stunted phenotype (data not shown). Suppression of the hma2hma4 stunted phenotype by HvHMA2 is shown in Figure 7. Rosette diameter and bolt height were determined as a measure of rescue, and shows that these lines had a significantly greater average rosette diameter and bolt height than hma2hma4 mutants (Figure 7). The T2 population was analysed because suppression of the stunted phenotype was markedly reduced in homozygous HvHMA2-transformant T3 plants, possibly due to silencing. To determine the effect of HvHMA2 expression on the ionomic profile, plants were grown on soil supplemented with Cd as well as essential micronutrients; under these conditions the Arabidopsis hma2hma4 mutant has low shoot Zn and Cd concentrations and a high shoot Cu concentration compared to wt Arabidopsis (Figure 8; [13]). In hma2hma4 mutant lines expressing HvHMA2 (T2) the average shoot Zn, Cd and Cu levels were partially restored to the levels observed in wt Arabidopsis: Zn and Cd levels were increased compared to hma2hma4, while Cu levels were decreased (Figure 8).
Figure 7. Expression of HvHMA2 rescues the Zn-deficiency phenotype of the hma2hma4 mutant.
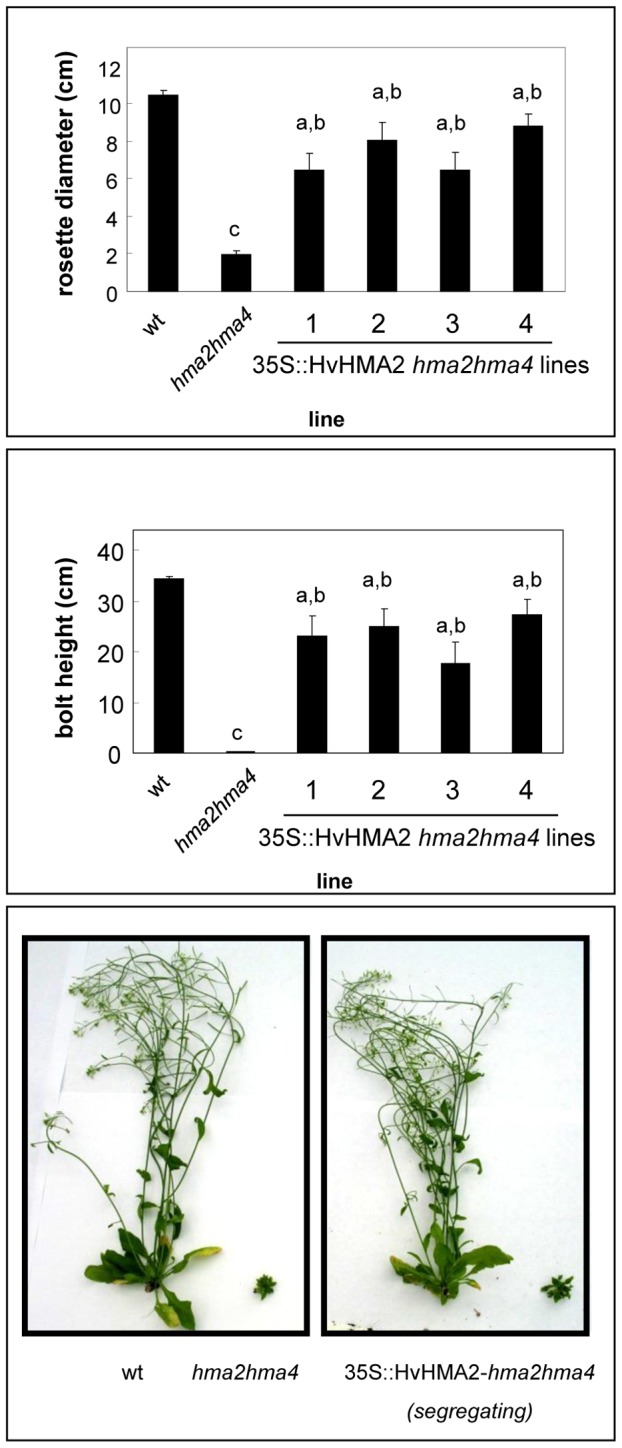
Growth of 35S::HvHMA2 hma2hma4 plants compared to untransformed hma2hma4 and wild-type (wt) plants. These were soil-grown plants not supplemented with nutrient solution. Top, rosette diameter and Middle, bolt height (41 days). Values are means+/−S.E (n = 12, T2 plants). Student's t-test was used to determine significance levels: a, significant difference between HvHMA2-expressing line and hma2hma4 mutant (P<0.05); b, significant difference between HvHMA2-expressing line and wt (P<0.05); c, significant difference between wt and hma2hma4 (P<0.05). Bottom, representative plants are shown.
Figure 8. HvHMA2 partially restores the Zn, Cd and Cu shoot levels when expressed in hma2hma4 mutants.
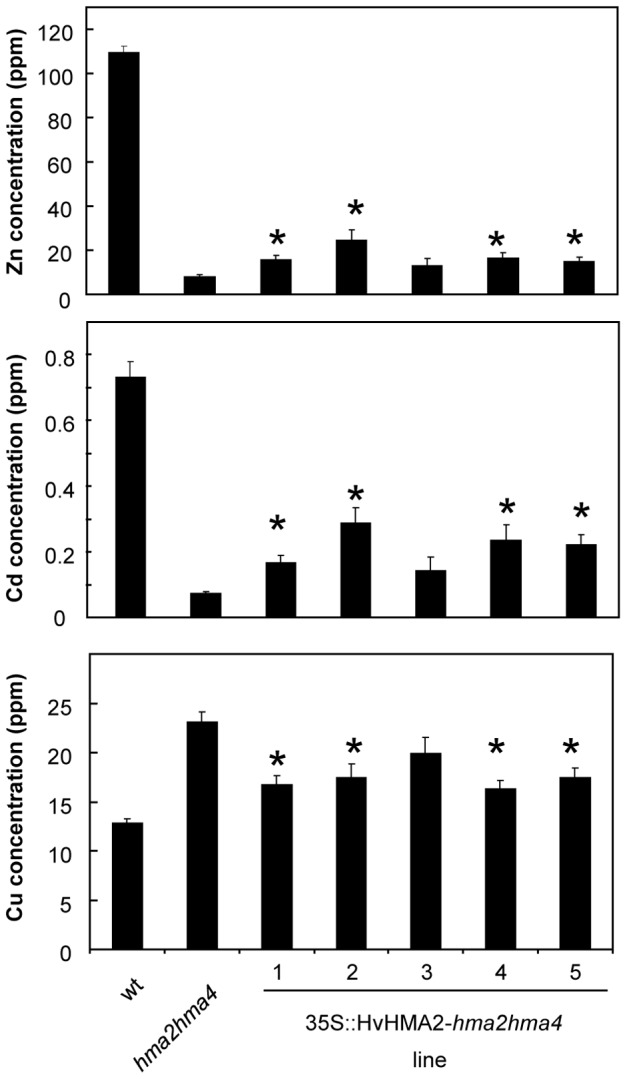
Levels of Zn (top), Cd (middle) and Cu (bottom) in shoots of soil-grown plants. These were soil-grown plants supplemented with nutrient solution according to [42]. Wild-type (wt), hma2hma4 mutant and 35S::HvHMA2-hma2hma4 T2 plants are shown. Values are means+/−S.E (n = 12 plants). Student's t-test was used to determine significance levels: *, significant difference between HvHMA2-expressing line and hma2hma4 mutant (P<0.05).
Silique growth is not fully rescued in the 35S::HvHMA2-hma2hma4 lines and this correlates with reduced Zn content
When grown in soil with no nutrient supplementation, the HvHMA2-hma2hma4 Arabidopsis plants flowered and formed siliques although these tended to be shorter than wt siliques (Figure 9a and b). This was also observed in AtHMA4-hma2hma4 plants [13]. Viable seed were produced from these plants. Elemental analysis carried out on siliques showed that only Zn levels were significantly different in HvHMA2-hma2hma4 plants compared to wt (Figure 9c).
Figure 9. HvHMA2-expressing hma2hma4 plants have shorter siliques than wild-type and are reduced in Zn content.
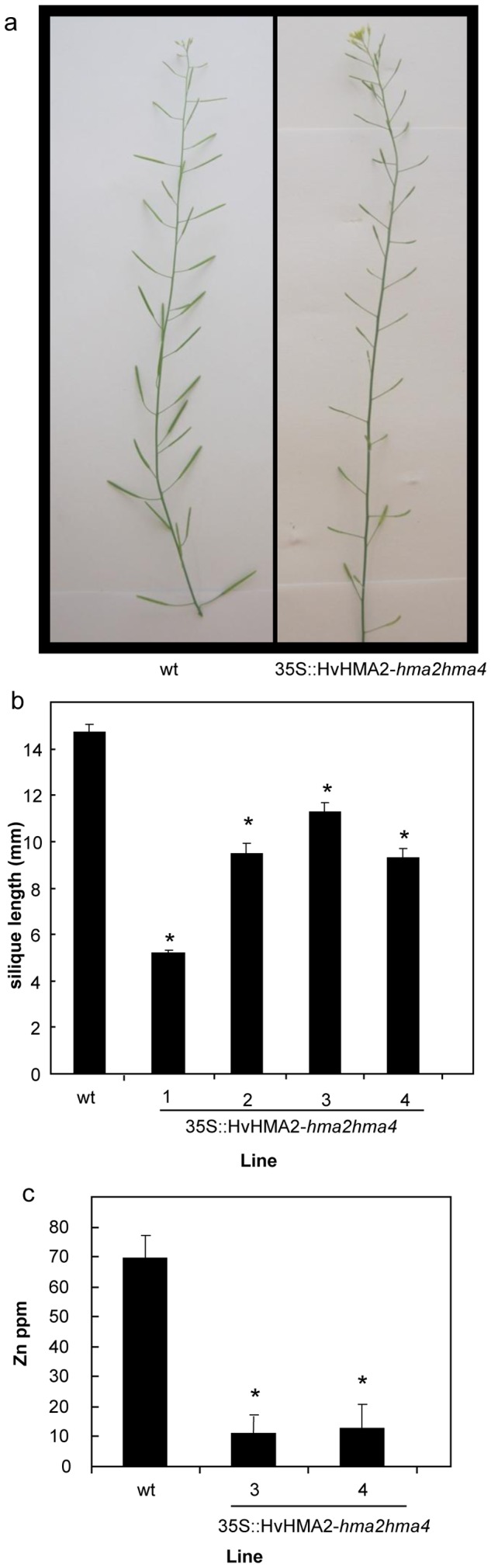
(a). Images of siliques from wt and HvHMA2-expressing hma2hma4 line. (b). Silique length. Values are means+/−S.E (n = 40 siliques from 5 plants producing bolts for each line). (c). Zn levels in siliques of wt and 35S::HvHMA2-hma2hma4 lines. Results represent the mean Zn concentration determined from approximately 20 siliques per plant using three plants per line. For a, b and c plants were grown in soil for 49 days and had no additional nutrient supplementation. Student's t-test was used to determine significance levels: *, significant difference between transgenic lines and wt (P≤0.05).
Discussion
Barley is a major crop and represents a good model for studying metal transport in cereals. We are focussing on the P1B-ATPase family of ion transporters to understand the role they play in metal transport and homeostasis. HMA2 homologues appear to be highly conserved in Poaceae and they all contain the residues that put them into the P1B-2 subclass [8]. Functional characterisation of Arabidopsis P1B-2 ATPases AtHMA2, 3 and 4 indicates they are Zn/Cd pumps [24], [15], [25], [26], [16]. AtHMA2 is at the PM of Arabidopsis root pericycle cells [12], [27], [28] while AtHMA3 is vacuolar [16]. Rice has two P1B-2 ATPases and evidence indicates OsHMA3 is a vacuolar Cd pump in roots (as yet there is no evidence that it can transport Zn; [19]) while OsHMA2 functions in Cd and Zn transport at the plasma membrane [23]. We provide evidence that barley possesses two P1B-2 ATPases, HvHMA2 and 3, and importantly we demonstrate that HvHMA2 is able to transport both Zn and Cd. Results indicate that HvHMA2 localises predominantly to the PM although we also detected some chloroplast expression. The PM localisation is consistent with data showing that HvHMA2 may function in root to shoot translocation of Zn while a function in the chloroplast would require further investigation.
HvHMA2 functions as a Zn/Cd pump in yeast
Studies in yeast show that HvHMA2 can transport Zn and Cd. Heterologous expression in yeast has been used previously to indicate potential substrates for HMAs and there are reports of expression resulting in both metal sensitivity and resistance [24], [15], [29], [30], [13]. This probably relates to their predominant membrane location when expressed in yeast. For example, the ER is thought to be a Cd-sensitive compartment in yeast; if pumps expressed here transport Cd into the ER lumen then Cd sensitivity may be observed [30], [31], [32], [33]. PM expression could alternatively result in Cd resistance for an efflux pump. HvHMA2 confers Cd sensitivity to wt yeast and this is eliminated in a transport null mutant where the conserved aspartate necessary for phosphorylation and functioning of these pumps was mutated to alanine. This suggests that HvHMA2 is transporting Cd into a sensitive compartment. HvHMA2 can also function in Zn transport as indicated by the Zn resistance observed when expressing this pump in the Zn-sensitive zrc1cot1 mutant. Similarly, this was eliminated in the transport null version of the pump. This suggests that a proportion of HvHMA2 may be expressed at the PM and function in efflux out of the cell and/or that transport of Zn into the compartment where HvHMA2 is localised confers Zn resistance to this yeast mutant. Dual locations for related pumps have been observed when expressed in yeast [33]. We were not able to observe a signal when HvHMA2-GFP was expressed in yeast, possibly due to low levels of expression or instability caused by the tag, and so we cannot comment on its localisation, but the ability of HvHMA2 to confer Cd sensitivity and Zn resistance in yeast facilitated further structure/function analysis. Future studies using epitope tags or an antibody to HvHMA2 could help elucidate the localisation of HvHMA2 in yeast.
Functional significance of putative metal-binding domains and metal coordination sites in HvHMA2
We used the yeast system to study the functional significance of key residues in HvHMA2 and the relative importance of particular regions of the protein. The N-terminal domain contains the GxCCxxE motif that appears to be conserved in the N termini of all plant P1B-2 ATPases; like the N-terminal GxxCxxC motifs of Cu-transporting ATPases these motifs may bind and also regulate metal transport [34]. Mutation of either of the cysteine residues in AtHMA4 abolished its ability to complement the Zn hypersensitivity of zrc1 and the Cd hypersensitivity of the ycf1 mutant yeast strains [26]. Mutagenesis studies indicate the CCxxE motif binds Zn and Cd with greater affinity than other metals, and mutation of these residues alters the metal-binding affinity of the N-terminal domain and reduces the ATPase activity but not the metal dependence of the pump [35]. In addition, the N-terminally deleted AtHMA2 and a mutant in which the cysteine residues in the GICCTSE motif were mutated failed to restore the growth of the Arabidopsis hma2hma4 mutant to wt levels as seen for plants transformed with a non-mutated version of AtHMA2 [28]. This suggests that the N-terminal region is crucial for function and this is supported by the results shown here for HvHMA2, with deletion of this region eliminating Cd sensitivity conferred to wt yeast and Zn resistance conferred to the zrc1cot1 mutant. We cannot rule out effects on targeting and expression levels at this stage and further studies are necessary to determine whether it is the vicinal cysteines and the subsequent glutamate which have a crucial role as seen for AtHMA2.
The most marked effect of deleting the C-terminus of HvHMA2 was seen in the zrc1cot1 yeast mutant as this conferred greater Zn resistance than the full length version. Whether this is due to transport differences, or to targeting differences with more of the mutant form being present at the PM pumping Zn out of the cell is not known. The C-terminal region may also function as an autoregulatory domain as has been suggested for AtHMA4 [15], [30]. The C-terminal domain of AtHMA2 is not essential for function in planta as deleting it seemed to have only a minor effect on the ability of this pump to restore the growth of the hma2hma4 mutant [28]. In contrast, deletion of this domain in AtHMA4 suppressed its rescue of this mutant suggesting an important role in planta [13]. Results expressing AtHMA4 constructs in tobacco suggest that the full-length pump is required for enhanced transfer of Zn from root to shoot [36]. Deletion of the rice OsHMA2 C-terminus reduces its ability to translocate Zn and Cd from root to shoot [23]. Expression of the AtHMA4 C-terminal region alone in yeast confers strong Cd resistance to wt yeast [29], [13] and also Zn resistance to the zrc1cot1 mutant [13]. Studies indicate that this is due to binding of Zn and Cd [30]. In contrast, expression of the C-terminal region from HvHMA2 has little effect on yeast metal tolerance suggesting that this may not show strong binding of these metals in yeast.
Analysis of SERCA pumps and sequence comparisons between different classes of P1B-ATPases has allowed key residues to be identified that may be important in metal coordination and transport [8], [18], [37], [38]. We identified invariant residues and key putative metal coordination residues in TMs 6 and 7 of HvHMA2, and tested the effect of these mutations on the yeast Cd response. Following expression in wt yeast, mutants P357L (in the CPC motif), S363L, P365L, K666L and N658L all decreased the Cd sensitivity compared to non-mutated HvHMA2 with P357L and K666L being the most effective. All mutation also reduced Zn resistance conferred by HvHMA2 to zrc1cot1.This suggests that these residues are important and in some cases crucial for transport function, although we cannot rule out effects on expression levels or targeting as being influencing factors. Further studies are now required to determine the exact functional significance of these mutations in HvHMA2 and epitope tagging could help answer the question of localisation. Few studies have tested the functional significance of different residues in plant sequences. Mutating the CPC motif to GPC eliminated the Cd and Zn resistance conferred on yeast by AtHMA4 [15], and CPC to SPC in AtHMA4 abolished its ability to rescue the ycf1 mutant on elevated Cd and the zrc1 mutant on high Zn [26]. Interestingly, substitutions in the P1B-1 ATPase AtHMA5 of both the latter proline in CPC(x)6P motif of TM6 in the Chisdra-2 ecotype and of N923 in the Cape Verde Island ecotype (TM7, equivalent to HvHMA2 N658), were both associated with Cu sensitivity and low capacity of Cu translocation from roots to shoots, indicating that these are also important residues in the P1B-1 subclass of pumps [39].
Some of these residues have been investigated in P1B-2 ATPases from other organisms. For HvHMA2 we observed the greatest decrease in HvHMA2-conferred Zn resistance with the mutants K666L, P357L (within the CPC motif) and D400A (the phosphorylated aspartate). Similarly in ZntA of Escherichia coli the mutation K693N (TM7, equivalent to HvHMA2(K666)) abolished Zn-stimulated ATPase activity completely, although Zn-dependent phosphorylation by ATP still occurred [40]. Also in ZntA, mutants in the CPC motif have been investigated: C392A, P393A, and C394A lost the ability to bind a metal ion with high affinity in the transmembrane domain, while histidine and serine substitutions at C392 and C394 abolished binding of Pb2+ but not other divalent metal ions [37]. Our data support a model whereby the CPC motif of TM6 and the conserved lysine in TM7 are parts of the transmembrane metal-binding site.
HvHMA2 suppresses the stunted phenotype of the hma2hma4 mutant by partially restoring the elemental balance
Root to shoot Zn transfer is obviously an important step in crops and this would be a key process in barley to ensure that Zn is moved to the shoot where it could be available for transport to the grain; HvHMA2 could potentially play a role in root to shoot transport. The Arabidopsis hma2hma4 mutant is defective in root to shoot translocation of Zn, and can be rescued by application of high levels of Zn to the soil [12]. This is a useful system for investigating the function of Zn/Cd P1B-ATPases and exploring the ability of related pumps to transport Zn [28], [13]. We showed that HvHMA2 can function in root to shoot transfer as when expressed in the hma2hma4 mutant it suppressed the stunted phenotype, restoring growth to wt levels. HvHMA2 expression also resulted in a small but significant increase in shoot Zn concentration (determined in leaves before bolting). It has previously been noted that partial rescue of the hma2hma4 shoot Zn concentration to around 30% of wt levels was seen in AtHMA4-hma2hma4 lines [13] and this was sufficient to fully rescue the stunted phenotype of the hma2hma4 double mutant. A similar rescue is seen here when expressing HvHMA2 indicating that the barley transporter, HvHMA2 enhances root to shoot transport of Zn when expressed in Arabidopsis.
HvHMA2-hma2hma4 plants also flowered and produced siliques but these were generally shorter than wt. We have previously observed this for AtHMA4-hma2hma4 plants [13]. The only element that was significantly reduced in siliques of HvHMA2-expressing lines was Zn (Figure 9), suggesting that the reduced Zn content may lead to the shorter siliques observed here. Indeed reduced silique length has been observed previously in Arabidopsis wt plants when Zn supply was reduced [41]. It would be interesting to express HvHMA2 under the AtHMA2 or AtHMA4 promoter to determine whether the partial rescue of the siliques was a consequence of expression under the 35S promoter rather than a difference in activity of AtHMA2/4 and HvHMA2.
Interestingly, the shoot Cu concentration of the hma2hma4 mutant is higher than the wt [13]. When HvHMA2 is expressed in the hma2hma4 mutant, Cu levels in shoots are reduced towards wt as seen when expressing AtHMA4 in this mutant [13]. It could be that this is due to these pumps partially restoring the Zn balance which then has the indirect effect of restoring the Cu balance, as no direct transport of Cu by HvHMA2 or AtHMA4 has been shown.
Role of HvHMA2 in Zn transport in grain
We analysed HvHMA2 expression in a number of published microarray datasets. A time course of gene expression in developing barley grain indicated highest expression of HvHMA2 in ‘endosperm plus aleurone’ at 16 and 25 days after flowering. Expression in these tissues also increased following imbibition (figure S3b). In comparison, expression in ‘embryo plus scutellum’ increased during grain development and was maximal in the mature grain; it decreased during imbibition (figure S3b). Laser capture microdissection was used to study expression of potential Zn transporter genes in the different cell layers of barley grain at 20 days after pollination and results indicated that HvHMA2 is most highly expressed in transfer cells with lower levels in the aleurone layer, endosperm and embryo [42]. To extend this data we used real-time PCR to investigate expression of HvHMA2 in germinating grain. HvHMA2 expression is slightly reduced in embryo tissue between 2 and 72 hr imbibition while it is increased in the endosperm cells. Interestingly the remaining tissue left after dissecting out the embryo and endosperm shows the highest levels of HvHMA2 expression indicating that HvHMA2 may function in pumping Zn in this tissue to the endosperm and embryo.
In summary, this study provides evidence that HvHMA2 functions in Zn and Cd transport and may play a similar role to AtHMA2 and 4 in Arabidopsis in transferring these ions from roots to shoots. Further work is required to determine the exact physiological role of this family of pumps in the grain and whether the manipulation of expression levels of HvHMA2 in barley can be used to alter Zn content.
Experimental Procedures
Plant materials
For growth and elemental analyses of Arabidopsis plants (wt, hma2hma4 mutants and HvHMA2-transformants), plants were grown as described previously [13]. For elemental analysis of leaf material collected before bolting, plants were grown in soil supplemented with sub-toxic concentrations of various elements including 0.09 ppm Cd and were regularly watered with Fe-HBED and 0.25× Hoagland's solution [43]. Grain from Hordeum vulgare L. cultivar Golden Promise was heat treated at 45°C for 48 hours and then imbibed on water soaked absorbent paper in sealed petri dishes at 20°C to initiate germination. Grain tissues were separated and used to prepare RNA. To isolate leaf and root material from more mature plants, barley plants were grown on vermiculite.
DNA and RNA isolation and cDNA synthesis
Genomic DNA was prepared using the DNAmite kit (Microzone Ltd, UK). RNA was prepared using a phenol-SDS extraction and LiCl precipitation method based on [44], except for barely mature root and shoot material which was isolated using TRIzol Reagent (Invitrogen Life Technologies). cDNA was produced using the Superscript III kit (Invitrogen, UK).
RT-PCR to detect expression of HvHMA2 in hma2hma4 mutants
RT-PCR was performed with Biomix taq (Bioline, UK). All primers used in this study are given in Table S3. Actin 2, used as the control, was amplified using primers spanning an intron (Actin2.f = 5′-ggtaacattgtgctcagtggtgg-3′, Actin2.R = 5′-ctcggccttggagatccacatc-3′, 28 cycles) while the transgene, HvHMA2 was detected using primers HvHMA2rt.F (5′-tcaatgcagcacagaacaca-3′) and HvHMA2rt.R (5′-ggccagcttgaacaaacatt-3′) (30 cycles). Real-Time PCR reactions were carried out as previously described [45] using the above primers for HvHMA2. RNABP was the control gene with primers RNABP-F (5′- cgcccagttatccatccatcta-3′) and RNABP-R (5′- aaaaacaccacaggaccggac-3′).
Cloning of HvHMA2 and creation of Entry clones for Gateway-cloning
Partial sequence for HvHMA2 (from 3rd TM into 3′ UTR) was obtained by alignment of AtHMA2, 3 and 4 sequences with barley EST sequences (http://harvest.ucr.edu/, http://www.plantgdb.org/, http://www.scri.ac.uk/, http://earth.lab.nig.ac.jp/ and http://www.ncbi.nlm.nih.gov/) and primers HvHMA2rB, HvHMA2fC(EcoRV), HvHMA2hingeR, and HvHMA2u3R(EcoRI) were designed. HvHMA2 N-terminal sequence was obtained by 5′ RACE (Generacer kit, Invitrogen) using the RACE forward primer and reverse primer HvHMA2rB on leaf cDNA. The PCR product was re-amplified using the nested RACE forward primer and HvHMA2rB. The resultant sequence was used to design the primer HvHMA2atgF(EcoRV) spanning the HvHMA2 translational start. N-terminal and C-terminal halves of the protein were amplified from barley leaf cDNA using primers HvHMA2atgF(EcoRV) and HvHMA2hingeR or HvHMA2fC(EcoRV) and HvHMA2u3R(EcoRI), respectively. Full length HvHMA2 was amplified using primers HvHMA2atgF(EcoRV) and HvHMA2u3R(EcoRI) with Pfu DNA polymerase (Promega) (Ta 60°C), then the 3 kb product was re-amplified. 5′ A overhangs were added and the product was AT-cloned into pGEM-T easy (Invitrogen) to create pGEMTe.HvHMA2.FL. Sequencing confirmed EST and RACE data. Full length HvHMA2 sequence was amplified from this using a topo-adapted forward primer (HvHMA2topoF) and the reverse primer HvHMA2with-stop. The resultant PCR product was topoisomerase-cloned into pENTR/D-TOPO (Invitrogen) and transformed into E.coli to create pENTR:HvHMA2(with-stop). The HvHMA2 insert was fully sequenced.
Sequence analysis
Hydropathy analysis was performed with Expasy protscale (http://www.expasy.ch/tools/protscale.html).
The sequence alignment was prepared using ClustalW2 [46] and annotated with GeneDoc (www.psc.edu/biomed/genedoc) using information from Swissprot (http://www.expasy.ch/) and SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/; [47]. Brachypodium, sorghum and maize sequences from http://aramemnon.uni-koeln.de/ are: Bd1g34140, Bd1g53670, Sb02g006940, Sb02g006950, Sb10g028920, Zm455491, ZmAC205008_FGT002, Zm175576 and Zm099191. NCBI accession numbers are: TaHMA2: DQ490135; OsHMA2 and OsHMA3: ADU53143 and BAJ25745 respectively (both from cv japonica). Barley sequences are BAK06002 (HvHMA1), BAK00726, BAJ93769, BAJ93251, BAK07450, BAJ87066 (HvHMA3 to 7 respectively) and BAJ96159 (HvHMA9). For HvHMA8 and other rice sequence details see [18]. The dendrogram was constructed using ClustalW2 and Treeview [48]. Sequence similarity and identity was calculated using MatGAT 2.02 [49]. Barley and wheat HMA2 expression data were retrieved using Webcomparator (http://contigcomp.acpfg.com.au/) [50].
Generation of HvHMA2 transformed Arabidopsis plants
HvHMA2 plant expression constructs were created by Gateway (Invitrogen) recombination of pENTR:HvHMA2(with-stop) into pEarleyGate100 [51] to create the non-tagged construct pEG100 35S::HvHMA2, and into pMDC43 [52] to create pMDC43 35S::GFP-HvHMA2 with GFP fused to the N-terminus of HvHMA2. Constructs were electroporated into Agrobacterium tumefaciens GV3101 and transformed using the floral dip method [53] into Arabidopsis wt or hma2hma4 plants (grown with Zn) [12], [13].
Preparation of HvHMA2 constructs for expression in yeast
HvHMA2 and mutants (below) were recombined into pYTV [54], under the galactose-inducible Gal1 promoter.
Deletion mutants
HvHMA2 deletion mutations generated by PCR and topo-cloned into pENTR/D-TOPO were: HvHMA2Δ714-1009 (deletion of predicted cytoplasmic C-terminus); primers HvHMA2topoF and HvHMA2Δ714-1009R. HvHMA2Δ2-81 (deletion of predicted cytoplasmic N-terminus); primers HvHMA2Δ2-81F and HvHMA2with-stop. HvHMA2Δ2-698 (deletion of all except predicted cytoplasmic C-terminus); primers HvHMA2Δ2-698F and HvHMA2with-stop.
Transport-null substitution mutants HvHMA2(D400A) and HvHMA2Δ714-1009 (D400A)
Site-directed mutagenesis (Quikchange XL, Stratagene) (primers HvHMA2D400A and HvHMA2D400Arc) was performed on pENTR:HvHMA2(with-stop) and pENTR:HvHMA2Δ714-1009 to mutate the critical phosphorylated aspartate of the D400KTGT motif to alanine.
HvHMA2 substitution mutants of proposed ion-specificity determinant residues
HvHMA2(P357L), HvHMA2(S363L) and HvHMA2(P365L) were created by PCR with primers HvHMA2Δ2-81F and either HvHMA2-P357L.R, HvHMA2-S363L.R or HvHMA2-P365L.R. Residues were mutated to leucine as this is a hydrophobic amino acid suggested to cause minimal disruption to helix formation while still having a relatively small side chain [55]. Products were AatII/FspI-digested and inserted into AatII/FspI pENTR:HvHMA2(with-stop). Primer HvHMA2-P357L.R also includes a silent mutation which abolishes a SalI site. HvHMA2(N658L) and HvHMA2(K666L) were made similarly with primers HvHMA2fC(EcoRV)and HvHMA2-N658L.R or HvHMA2-K666L.R. The product was FspI/MfeI-digested and inserted into FspI/MfeI pENTR:HvHMA2(with-stop).
Yeast strains and heterologous expression of various HvHMA2 constructs
Wt S. cerevisiae BY4741, the Cd-sensitive ycf1 mutant, and yeast transformation were as previously described [15], [45], [13]. The Zn-hypersensitive zrc1cot1 mutant (MATa;his3Δ1;leu2Δ0; met15Δ0;ura3Δ0; zrc1::natMX cot1::kanMX4) was obtained from Dr. U. Kramer (Heidleberg, Germany).
Metal sensitivity tests of HvHMA2 transformed yeast
Tests were performed as described previously [13] except plates were incubated for 2 days.
Microscopy
Seedlings of both 35S::GFP-HMA2 expressing plants and wt Col0 were grown as described in [56]. Plants were mounted on microscope slides in water for imaging using a Zeiss LSM 510META confocal system (Carl Zeiss Ltd., Welwyn Garden City, UK). GFP and chlorophyll were excited using the 488 nm line of an argon ion laser. GFP emission was detected between 505–530 nm and chlorophyll autofluorescence was detected using a LP580 filter.
Ionomic analysis
Elemental analysis of leaf material before bolting was carried out using ICP-MS [43]. Elemental analysis of silique material was carried out by ICP-OES.
Accession numbers
The Genbank accession number for HvHMA2 cDNA cloned in this study is GU177852.
Supporting Information
RT-PCR amplification of HvHMA2 . The N-terminal and C-terminal parts and the full length HvHMA2 sequence were amplified from barley leaf cDNA (lanes 1–3 respectively) using information from EST analysis and 5′ RACE. Full-length HvHMA2 PCR product was re-amplified from the ∼3 kB product (lane 4).
(TIF)
Hydropathy analysis of P1B-2–ATPases. Hydropathy analyses indicate that locations of predicted TM domains are highly conserved in the primary structure of P1B-2 -ATPases.
(TIF)
Tissue expression pattern of HvHMA2 and TaHMA2 . a. Microarray expression data for barley (solid line) and wheat (broken line) indicates HMA2 is expressed in all tissues, with highest expression in anthers. Unbroken line, HvHMA2; broken line, TaHMA2. Tissue key: gem: germinating seed embryo; rad: germinating seed radicle; roo: germinating seed root; col: germinating seed coleoptile; cro: seedling crown; lea: seedling leaf; brc: floral bracts before anthesis; inf: immature inflorescence; ant: anthers before anthesis; pst: pistil before anthesis; car5: caryopsis 5 DAP (days after pollination); en22: endosperm 22 DAP; em22: embryo 22 DAP; car10: caryopsis 10 DAP; car16: caryopsis 16 DAP. b. Normalized expression values for two replicate experimental series based on independently grown plant material indicates HvHMA2 expression in grain tissues varies during grain maturation and germination. Time: daf, days after flowering; hai, hours after imbibition. Data extracted from supplementary data of [57].
(TIF)
Tissues used for RT-PCR. Top: Preparation of tissues for RNA extraction after 2 h imbibition. Bottom: Preparation of tissues for RNA extraction after 72 h imbibition or longer.
(TIF)
Zn restores growth of the Arabidopsis hma2hma4 mutant to wild-type levels. Top, Rosette diameter measured in plants with or without Zn (3 mM) supplied throughout the growth period. Mean (±S.E) is shown from a representative experiment (n = 12 plants). Bottom, representative plants showing the effect of Zn on the growth of hma2hma4 mutant.
(TIF)
Arabidopsis hma2hma4 plants are expressing HvHMA2 . RT-PCR shows expression of HvHMA2 (top) in four independent lines of the Arabidopsis hma2hma4 mutant transformed with HvHMA2 under the 35S-promoter (35S::HvHMA2 hma2hma4 lines). Wild-type (wt) and hma2hma4 mutant are shown as controls. Actin control levels are similar for all lines (bottom).
(TIF)
Analysis of protein sequence homology between HvHMA2 and P1B-2 P-types from Arabidopsis, wheat, rice, sorghum and brachypodium.
(DOC)
nvariant amino acids in TMs 6, 7 and 8 ofI P1B-2 P-type ATPases that may be involved in coordinating metals during transport.
(DOC)
Primers used for the cloning of HvHMA2 and the generation of mutants.
(DOC)
Acknowledgments
We are grateful to Rachel Gibson and Adrienne Payne for technical assistance, Professor Christopher Cobbett (Melbourne, Australia) for providing hma2hma4 mutant seed and to Professor Ute Krämer, (Bochum, Germany) for the zrc1cot1 mutant.
Funding Statement
This work was supported by the European Union Framework Programme 6 as part of the Integrated Project Public health impact of long-term, lowlevel mixed element exposure in susceptible population strata (PHIME) (contract no FOOD-CT-2006- 016253); it reflects only the authors' views. The Community is not liable for any use that may be made of the information contained therein. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Williams L, Salt DE (2009) The plant ionome coming into focus. CurrOpin Plant Biol 12: 247–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Puig S, Penarrubia L (2009) Placing metal micronutrients in context: transport and distribution in plants. Curr Opin Plant Biol 12: 299–306. [DOI] [PubMed] [Google Scholar]
- 3.Williams LE, Pittman JK (2010) Dissecting pathways involved in manganese homeostasis and stress in higher plant cells. In Cell Biology of Metals and Nutrients; Plant Cell Monographs 17, 95–117. Cell Biology of Metals and Nutrients; Plant Cell Monographs 17, 95–117. Hell R, Mendel RR, editors. Springer-Verlag Berlin Heidelberg. [Google Scholar]
- 4. Palmgren MG, Clemens S, Williams LE, Kraemer U, Borg S, et al. (2008) Zinc biofortification of cereals: problems and solutions. Trends Plant Sci 13: 464–473. [DOI] [PubMed] [Google Scholar]
- 5. Verbruggen N, Hermans C, Schat H (2009) Mechanisms to cope with arsenic or cadmium excess in plants. Curr Opin Plant Biol 12: 364–372. [DOI] [PubMed] [Google Scholar]
- 6. Williams LE, Pittman JK, Hall JL (2000) Emerging mechanisms for heavy metal transport in plants. Biochim Biophys Acta 1465: 104–126. [DOI] [PubMed] [Google Scholar]
- 7. Hall JL, Williams LE (2003) Transition metal transporters in plants. J Exp Bot 54: 2601–2613. [DOI] [PubMed] [Google Scholar]
- 8. Argüello JM (2003) Identification of ion-selectivity determinants in heavy-metal transport P1B-type ATPases. J Membr Biol 195: 93–108. [DOI] [PubMed] [Google Scholar]
- 9. Kim YY, Choi H, Segami S, Cho HT, Martinoia E, et al. (2009) AtHMA1 contributes to the detoxification of excess Zn(II) in Arabidopsis. Plant J 58: 737–753. [DOI] [PubMed] [Google Scholar]
- 10. Seigneurin-Berny D, Gravot A, Auroy P, Mazard C, Kraut A, et al. (2006) HMA1, a new Cu-ATPase of the chloroplast envelope, is essential for growth under adverse light conditions. J Biol Chem 281: 2882–2892. [DOI] [PubMed] [Google Scholar]
- 11. Moreno I, Norambuena L, Maturana D, Toro M, Vergara C, et al. (2008) AtHMA1 is a thapsigargin-sensitive Ca2+/Heavy metal pump. J Biol Chem 283: 9633–9641. [DOI] [PubMed] [Google Scholar]
- 12. Hussain D, Haydon MJ, Wang Y, Wong E, Sherson SM, et al. (2004) P-type ATPase heavy metal transporters with roles in essential zinc homeostasis in Arabidopsis. Plant Cell 16: 1327–1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mills RF, Valdes B, Duke M, Peaston KA, Lahner B, et al. (2010) Functional significance of AtHMA4 C-terminal domain in planta . PloS ONE 5: e13388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wong CKE, Cobbett CS (2009) HMA P-type ATPases are the major mechanism for root-to-shoot Cd translocation in Arabidopsis thaliana . New Phytol 181: 71–78. [DOI] [PubMed] [Google Scholar]
- 15. Mills RF, Francini A, Ferreira da Rocha PSC, Baccarini PJ, Aylett M, et al. (2005) The plant P1B-type ATPase AtHMA4 transports Zn and Cd plays a role in detoxification of transition metals supplied at elevated levels. FEBS Lett 579: 783–791. [DOI] [PubMed] [Google Scholar]
- 16. Morel M, Crouzet J, Gravot A, Auroy P, Leonhardt N, et al. (2009) AtHMA3, a P1B-ATPase allowing Cd/Zn/Co/Pb vacuolar storage in Arabidopsis. Plant Physiol 149: 894–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee S, Kim YY, Lee Y, An G (2007) Rice P-1B-type heavy-metal ATPase, OsHMA9, is a metal efflux protein. Plant Physiol 145: 831–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Williams LE, Mills RF (2005) P1B-ATPases – an ancient family of transition metal pumps with diverse functions in plants. Trends Plant Sci 10: 491–502. [DOI] [PubMed] [Google Scholar]
- 19. Miyadate H, Adachi S, Hiraizumi A, Tezuka K, Nakazawa N, et al. (2011) OsHMA3, a P-1B-type of ATPase affects root-to-shoot cadmium translocation in rice by mediating efflux into vacuoles. New Phytol 189: 190–199. [DOI] [PubMed] [Google Scholar]
- 20. Ueno D, Yamaji N, Kono I, Huang CF, Ando T, et al. (2010) Gene limiting cadmium accumulation in rice. PNAS 107: 16500–16505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ueno D, Koyama E, Yamaji N, Ma JF (2011) Physiological, genetic, and molecular characterization of a high-Cd-accumulating rice cultivar, Jarjan. J Exp Bot 62: 2265–2272. [DOI] [PubMed] [Google Scholar]
- 22. Nocito FF, Lancilli C, Dendena B, Lucchini G, Sacchi GA (2011) Cadmium retention in rice roots is influenced by cadmium availability, chelation and translocation. Plant Cell Environ 34: 994–1008. [DOI] [PubMed] [Google Scholar]
- 23. Sato-Nagasawa N, Mori M, Nakazawa N, Kawamoto T, Nagato Y, et al. (2012) Mutations in rice (Oryza sativa) heavy metal ATPase2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol 53: 213–224. [DOI] [PubMed] [Google Scholar]
- 24. Mills RF, Krijger GC, Baccarini PJ, Hall JL, Williams LE (2003) Functional expression of AtHMA4, a P1B-type ATPase in the Zn/Co/Cd/Pb subclass. Plant J 35: 164–176. [DOI] [PubMed] [Google Scholar]
- 25. Eren E, Argüello JM (2004) Arabidopsis HMA2, a divalent heavy-transporting PIB-ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol 136: 3712–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Verret F, Gravot A, Auroy P, Preveral S, Forrestier C, et al. (2005) Heavy metal transport by AtHMA4 involves the N-terminal degenerated metal binding domain and the C-terminal His(11) stretch. FEBS Lett 579: 1515–1522. [DOI] [PubMed] [Google Scholar]
- 27. Sinclair SA, Sherson SM, Jarvis R, Camakaris J, Cobbett CS (2007) The use of the zinc-fluorophore, Zinpyr-1, in the study of zinc homeostasis in Arabidopsis roots. New Phytol 174: 39–45. [DOI] [PubMed] [Google Scholar]
- 28. Wong CKE, Jarvis RS, Sherson SM, Cobbett CS (2009) Functional analysis of the heavy metal binding domains of the Zn/Cd-transporting ATPase, HMA2, in Arabidopsis thaliana . New Phytol 181: 79–88. [DOI] [PubMed] [Google Scholar]
- 29. Bernard C, Roosens N, Czernic P, Lebrun M, Verbruggen N (2004) A novel CPx-ATPase from the cadmium hyperaccumulator Thlaspi caerulescens . FEBS LETT 569: 140–148. [DOI] [PubMed] [Google Scholar]
- 30. Baekgaard L, Mikkelsen MD, Sorensen DM, Hegelund JN, Persson DP, et al. (2010) A combined zinc/cadmium sensor and zinc/cadmium export regulator in a heavy metal pump. J Biol Chem 285: 31243–31252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Clemens S, Bloss T, Vess C, Neumann D, Nies DH, et al. (2002) A transporter in the endoplasmic reticulum of Schizosaccharomyces pombe cells mediates zinc storage and different. J Biol Chem 277: 18215–18221. [DOI] [PubMed] [Google Scholar]
- 32. Wu C-C, Bal N, Perard J, Lowe J, Boscheron C, et al. (2004) A cloned prokaryotic Cd2+ P-type ATPase increases yeast sensitivity to Cd2+ . Biochem Biophys Res Commun 324: 1034–1040. [DOI] [PubMed] [Google Scholar]
- 33. Courbot M, Willems G, Motte P, Arvidsson S, Roosens N, et al. (2007) A major quantitative trait locus for cadmium tolerance in Arabidopsis halleri colocalizes with HMA4, a gene encoding a heavy metal ATPase. Plant Physiol 144: 1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. LeShane ES, Shinde U, Walker JM, Barry AN, Blackburn NJ, et al. (2010) Interactions between Copper-binding Sites Determine the Redox Status and Conformation of the Regulatory N-terminal Domain of ATP7B. J Biol Chem 285: 6327–6336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eren E, Gonzalez-Guerrero M, Kaufman BM, Argüello JM (2007) Novel Zn2+ coordination by the regulatory N-terminus metal binding domain of Arabidopsis thaliana Zn2+-ATPase HMA2. Biochem 46: 754–7764. [DOI] [PubMed] [Google Scholar]
- 36. Siemianowski O, Mills RF, Williams LE, Antosiewicz DM (2011) Expression of the P1B-type ATPase AtHMA4 in tobacco modifies Zn and Cd root to shoot partitioning and metal tolerance. Plant Biotechnol J 9: 64–74. [DOI] [PubMed] [Google Scholar]
- 37. Dutta SJ, Junbo L, Hou Z, Mitra B (2006) Conserved aspartic acid 714 in transmembrane segment 8 of the ZntA subgroup of P1B-type ATPases is a metal binding residue. Biochem 45: 5923–5931. [DOI] [PubMed] [Google Scholar]
- 38. Dutta SJ, Junbo L, Stemmler AJ, Mitra B (2007) Conservative and nonconservative mutations of the transmembrane CPC motif in ZntA: effect on metal selectivity and activity. Biochem 46: 3692–3703. [DOI] [PubMed] [Google Scholar]
- 39. Kobayashi Y, Kuroda K, Kimura K, Southron-Francis JL, Furuzawa A, et al. (2008) Amino acid polymorphisms in strictly conserved domains of a P-type ATPase HMA5 are involved in the mechanism of copper tolerance variation in Arabidopsis. Plant Physiol 148: 969–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Okkeri J, Haltia T (2006) The metal-binding sites of the zinc transporting P-type ATPase of E. coli Lys693 and Asp714 in the seventh and eighth transmembrane segments of ZntA contribute to the coupling of metal binding and ATPase activity. Biochem Biophy Acta 1575: 1485–1495. [DOI] [PubMed] [Google Scholar]
- 41. Talukdar S, Aarts MGM (2008) Arabidopsis thaliana and Thlaspi caerulescens respond comparably to low zinc supply. Plant and Soil 306: 85–94. [Google Scholar]
- 42. Tauris B, Borg S, Gregersen PL, Holm PB (2009) A roadmap for zinc trafficking in the developing barley grain based on laser capture microdissection and gene expression profiling. J Exp Bot 60: 1333–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lahner B, Gong JM, Mahmoudian M, Smith EL, Abid KB, et al. (2003) Genomic scale profiling of nutrient and trace elements in Arabidopsis thaliana . Nature Biotechnol 21: 1215–1221. [DOI] [PubMed] [Google Scholar]
- 44. Verwoerd TC, Dekker BMM, Hoekema A (1989) A small-scale procedure for the rapid isolation of plant RNAs. Nucleic Acids Res 17: 2362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Mills RF, Doherty ML, Lopez-Marques RL, Weimar T, Dupree P, et al. (2008) ECA3, a Golgi-localized P-2A-type ATPase, plays a crucial role in manganese nutrition in Arabidopsis. Plant Physiol 146: 116–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. (2007) ClustalW and ClustalX version 2. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 47. Hirokawa T, Boon-Chieng S, Mitaku S (1998) SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14: 378–379. [DOI] [PubMed] [Google Scholar]
- 48. Page RDM (1996) TREEVIEW: An application to display phylogenetic trees on personal computers. Comp Appl Biosci 12: 357–358. [DOI] [PubMed] [Google Scholar]
- 49. Campanella JJ, Bitincka L, Smalley J (2003) MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4: 29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Schreiber AW, Sutton T, Caldo RA, Kalashyan E, Lovell B, et al. (2009) Comparative transcriptomics in the Triticeae. BMC Genomics 10: 285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Earley KW, Haag JR, Pontes O, Opper K, Juehne T, et al. (2006) Gateway-compatible vectors for plant functional genomics and proteomics. Plant J 45: 616–629. [DOI] [PubMed] [Google Scholar]
- 52. Curtis MD, Grossniklaus U (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta . Plant Physiol 133: 462–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana . Plant J 16: 735–743. [DOI] [PubMed] [Google Scholar]
- 54. Gong W, Shen YP, Ma LG, Pan Y, Du YL, et al. (2004) Genome-wide ORFeome cloning and analysis of Arabidopsis transcription factor genes. Plant Physiol 135: 773–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Lyu PC, Sherman JC, Chen A, Kallenbach NR (1991) α-helix stabilization by natural and unnatural amino acids with alkyl side chains. Proc Natl Acad Sci USA 88: 5317–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kurup S, Runions J, Kohler U, Laplaze L, Hodge S, et al. (2005) Marking cell lineages in living tissues. Plant J 42: 444–453. [DOI] [PubMed] [Google Scholar]
- 57. Sreenivasulu N, Usadel B, Winter A, Radchuk V, Scholz U, et al. (2008) Barley grain maturation and germination: Metabolic pathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan profiling tools. Plant Physiol 146: 1738–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
RT-PCR amplification of HvHMA2 . The N-terminal and C-terminal parts and the full length HvHMA2 sequence were amplified from barley leaf cDNA (lanes 1–3 respectively) using information from EST analysis and 5′ RACE. Full-length HvHMA2 PCR product was re-amplified from the ∼3 kB product (lane 4).
(TIF)
Hydropathy analysis of P1B-2–ATPases. Hydropathy analyses indicate that locations of predicted TM domains are highly conserved in the primary structure of P1B-2 -ATPases.
(TIF)
Tissue expression pattern of HvHMA2 and TaHMA2 . a. Microarray expression data for barley (solid line) and wheat (broken line) indicates HMA2 is expressed in all tissues, with highest expression in anthers. Unbroken line, HvHMA2; broken line, TaHMA2. Tissue key: gem: germinating seed embryo; rad: germinating seed radicle; roo: germinating seed root; col: germinating seed coleoptile; cro: seedling crown; lea: seedling leaf; brc: floral bracts before anthesis; inf: immature inflorescence; ant: anthers before anthesis; pst: pistil before anthesis; car5: caryopsis 5 DAP (days after pollination); en22: endosperm 22 DAP; em22: embryo 22 DAP; car10: caryopsis 10 DAP; car16: caryopsis 16 DAP. b. Normalized expression values for two replicate experimental series based on independently grown plant material indicates HvHMA2 expression in grain tissues varies during grain maturation and germination. Time: daf, days after flowering; hai, hours after imbibition. Data extracted from supplementary data of [57].
(TIF)
Tissues used for RT-PCR. Top: Preparation of tissues for RNA extraction after 2 h imbibition. Bottom: Preparation of tissues for RNA extraction after 72 h imbibition or longer.
(TIF)
Zn restores growth of the Arabidopsis hma2hma4 mutant to wild-type levels. Top, Rosette diameter measured in plants with or without Zn (3 mM) supplied throughout the growth period. Mean (±S.E) is shown from a representative experiment (n = 12 plants). Bottom, representative plants showing the effect of Zn on the growth of hma2hma4 mutant.
(TIF)
Arabidopsis hma2hma4 plants are expressing HvHMA2 . RT-PCR shows expression of HvHMA2 (top) in four independent lines of the Arabidopsis hma2hma4 mutant transformed with HvHMA2 under the 35S-promoter (35S::HvHMA2 hma2hma4 lines). Wild-type (wt) and hma2hma4 mutant are shown as controls. Actin control levels are similar for all lines (bottom).
(TIF)
Analysis of protein sequence homology between HvHMA2 and P1B-2 P-types from Arabidopsis, wheat, rice, sorghum and brachypodium.
(DOC)
nvariant amino acids in TMs 6, 7 and 8 ofI P1B-2 P-type ATPases that may be involved in coordinating metals during transport.
(DOC)
Primers used for the cloning of HvHMA2 and the generation of mutants.
(DOC)