Abstract
Consequences of expression of the protein tyrosine phosphatase nonreceptor 22 (PTPN22) gain-of-function variant were evaluated in leukocytes from patients with anti-neutrophil cytoplasmic autoantibody (ANCA) disease. The frequency of the gain-of-function allele within the Caucasian patient cohort was 22% (OR 1.45), compared to general American Caucasian population (16.5%, p = 0.03). Examination of the basal phosphatase activity of PTPN22 gain-of-function protein indicated persistently elevated activity in un-stimulated peripheral leukocytes, while basal activity was undetectable in leukocytes from patients without the gain-of-function variant. To examine consequences of persistently high PTPN22 activity, the activation status of ERK and p38 MAPK were analyzed. While moderate levels of activated ERK were observed in controls, it was undetectable in leukocytes expressing PTPN22 gain-of-function protein and instead p38MAPK was up-regulated. IL-10 transcription, reliant on the ERK pathway, was negatively affected. Over the course of disease, patients expressing variant PTPN22 did not show a spike in IL-10 transcription as they entered remission in contrast to controls, implying that environmentally triggered signals were blunted. Sustained activity of PTPN22, due to the gain-of-function mutation, acts as a dominant negative regulator of ERK activity leading to blunted cellular responsiveness to environmental stimuli and expression of protective cytokines.
Introduction
Anti-neutrophil Cytoplasmic Autoantibody (ANCA) disease is multifactorial in origin, as with many autoimmune diseases, involving complex interactions of genetic polymorphisms, epigenetic changes and environmental influences [1]–[5]. The list of genes associated with ANCA disease includes one generalized to autoimmune propensity, the protein tyrosine phosphatase non-receptor 22 (PTPN22) [6]–[24]. In 2004, a single nucleotide polymorphism (SNP) in the PTPN22 gene was identified that resulted in a protein modification, which disrupted the regulatory domain of the phosphatase conferring a gain-of-function phenotype [25], [26]. The following year this genetic variant was linked with proteinase 3(PR3)-ANCA disease in a cohort of patients from Germany [24] and then in 2009 a similar association was made in a study of a cohort from Great Britain [27]. Now considered an autoimmunity-predisposing allele, this polymorphism strongly correlates with numerous other autoimmune diseases including type 1 diabetes (T1D) [28]–[32], rheumatoid arthritis (RA) [25], [33]–[38], systemic lupus erythematosus (SLE) [26], [39]–[41], Graves' disease [42], [43], and generalized vitiligo [44].
The aim of the studies presented here is to investigate effects of the gain-of-function variant on signaling responses in leukocytes from patients with ANCA disease and how these influence immunological events. ANCA have two primary targets, PR3 and myeloperoxidase (MPO), which are expressed solely on the surface of neutrophils and monocytes. Because PTPN22 is uniquely expressed in hematopoietic cell types, studies of the gain-of-function polymorphism are fundamentally important in this disease [25]. Binding of ANCA to its antigens stimulates cellular signal transduction pathways causing changes in gene transcription, cell activation status, and ultimately, neutrophil degranulation [45]–[49]. It is the aberrant release of neutrophils' noxious constituents that causes inflammation of vessel walls and injury of highly vascularized organs such as the kidney and lung [50]–[53].
What makes a gain-of-function variant of PTPN22 particularly interesting, especially in the framework of multifactorial systemic autoimmune diseases like ANCA disease, is that protein tyrosine phosphatases (PTPs) serve as “sensors” and “transmitters” for environmental signals [54]. Alterations in genes encoding protein tyrosine phosphatases broadly affect kinase-phosphatase systems with deleterious effects on cellular equilibrium. PTPN22 is known to modulate the activity of the RAS and SRC-family signaling pathways, both of which are major pathways involved in immune modulation [55], [56]. Due to the proximal position of the RAS and SRC-family in numerous signal transduction cascades, including extracellular signal-regulated kinase (ERK), JNK, and p38 MAPK [57], inappropriate regulation would impact immune cell functions, including those emanating from integrins, Fc receptors, growth factor receptors, and cytokine receptors [58]–[61].
Intuitively, a function-altering, genetic polymorphism in PTPN22 coupled with environmental exposures would place an individual at a higher risk for developing autoimmune disease. Environmental factors known to impact ANCA disease, at both disease onset and relapse, include bacterial and viral infections [62]–[64], aging [2], [65], seasonal changes [66] and silica exposure [3]. We have evidence that one manifestation of these factors is perturbation of epigenetic regulation of gene transcription. We found that gene silencing marks were altered in leukocytes of patients with ANCA disease resulting in aberrant transcription at the gene locus for PR3 and MPO [4], [67]. The gain-of-function variant could also be deviant in transmission of environmentally-induced epigenetic signals. For example, the JmjC-domain containing histone demethylase, JMJD3, is “induced” when the cell “senses” bacterial products and inflammatory cytokines within the microenvironment [68], [69]. We draw particular attention to JMJD3 because mRNA levels were abnormally high in ANCA disease patients concurrent with loss of epigenetic methylation marks at PRTN3 and MPO loci [4].
Here we describe a study demonstrating how a genetic polymorphism can disrupt “sensors” of the signaling milieu. The data indicate that the PTPN22 gain-of-function variant confers abnormally high basal phosphatase activity perturbing proper responses to external stimuli in circulating neutrophils and lymphocytes of patients with ANCA disease.
Materials and Methods
Patients and clinical analysis
Patients with biopsy-proven ANCA disease enrolled in this study were diagnosed between 1985 and 2009, and followed in a life-long registry by physicians in the Glomerular Disease Collaborative Network (GDCN). Methods of identifying and enrolling patients in the GDCN have been described [65], [70], [71]. All study materials were given Institutional Review Board approval for human subjects' research (IRB study #97-0523) by the UNC-CH Office of Human Research Ethics. Study subjects gave informed, written consent and participated according to UNC Institutional Review Board guidelines. A total of 230 Caucasian patients with ANCA disease participated in the PTPN22 genotyping study. Patients were categorized by diagnosis: granulomatosis with polyangiitis (GPA) [72]–[74], microscopic polyangiitis (MPA), Churg-Strauss syndrome (CSS), and renal-limited disease (Lim) [75], [76]. ANCA serotypes were determined by indirect immunofluorescence and/or antigen-specific PR3 and MPO enzyme-linked immune-absorbent assays (ELISA) (Invitrogen, Carlsbad, CA, USA) [77], [78]. Of 230 Caucasian ANCA-patients, 107 were PR3-ANCA and 109 were MPO-ANCA patients; 74 patients were diagnosed with GPA, 110 with MPA, 40 with renal limited, three with CSS, two with pulmonary capillaritis and one with neuro-limited disease. The Birmingham Vasculitis Activity Score (BVAS) 2003 version was used to rank disease severity activity: remission (BVAS = 0), active+ (BVAS 1–4), active++ (BVAS 5–9) and active+++ (BVAS≥10).
PTPN22 genotyping
Genomic DNA was extracted from leukocytes in EDTA-treated blood using the Puregene DNA Purification System (Puregene, Minneapolis, MN, USA). DNA quality was spectrophotometrically determined by OD 260/280 nm ratios and by agarose gel visualization. Genotyping for SNP C1858T (rs2476601) and G788A (rs33996649) was performed using TaqMan-SNP-Genotyping Assay (Applied Biosystems, Foster City, CA). The primer sequences for G788A were: forward 5′ TTTGAACTAATGAAGGCCTCTGTGT 3′ and reverse 5′ ATTCCTGAGAACTTCAGTGTTTTCAGT 3′. The specific minor groove binder probe sequences were 5′ TTGATCCGGGAAATG 3′ (FAM) and 5′ TTGATCCAGGAAATG 3′ (VIC). The primer and the specific minor groove binder probe sequences for C1858T were commercially available and pre-designed by Applied Biosystems. TaqMan was performed by ABI PRISM 7900HT sequence detection system (Applied Biosystems).
PTPN22 (lymphoid tyrosine phosphatase) activity assay
For phosphatase activity, total leukocytes were obtained after lysis of erythrocytes using RBC Lysis Buffer (NH4Cl) [79]. Patients analyzed included those with the gain-of-function variant (n = 12), non-variant (n = 12) and loss-of-function (n = 3). For analysis of specific cell types, neutrophils and lymphocytes&monocytes were separated from blood by Plasmagel (ZeptoMetrix, Buffalo, NY, USA) and Histopaque 1077 (Sigma, St. Louis, MO, USA).
Microtiter plate wells were coated in duplicate with mouse anti-human PTPN22 antibody (Abnova, Taipei, Taiwan) at a 1∶100 dilution and incubated overnight at 4°C. Normal mouse IgG served as a mock control. Leukocytes were lysed in lysis buffer (20 mM Tris-HCl, 150 mM NaCl, and 1 mM EDTA, pH 7.4, with 1 mM of phenylmethanesulphonylfluoride, 10 mg/ml of aprotinin, 10 mg/ml of leupeptin, 10 mg/ml of soybean trypsin inhibitor) at a concentration of 5×106 cells/ml. Lysate was added into each pre-coated well (100 µl) and incubated for 3 hrs at room temperature (RT). After washing with lysis buffer without protein inhibitor, 100 µl of phosphatase substrate (p-NPP, Bio-Rad, Hercules, CA, USA) (in 100 mM Bis-Tris, pH 6.0, 5 mM DTT buffer) was added to each well [80]. Phosphatase activity was detected by a VersaMax Microplate Reader (Molecular Devices, Sunnyvale, CA, USA) at 405 nm.
PTPN22 protein was quantitated by capture enzyme-linked immunosorbant assay. Wells were coated with mouse anti-human PTPN22 antibody, or normal mouse IgG as a mock control. An aliquot of fresh cell lysate was added and PTPN22 protein detected with rabbit anti-PTPN22 (1∶200, Lifespan, Providence, RI, USA) and secondary antibody AKP-conjugated goat anti rabbit IgG (H+L) (1∶5000, Pierce, Rockford, IL, USA). For PTPN22-responsiveness studies, total leukocytes were pre-treated with PMA (100 ng/µl) as described (Sigma) for 10 mins at 37°C.
Western blot method for ERK/pERK and p38/pp38 detection
Samples were analyzed from non-variant (n = 3), loss-of-function (n = 2) and gain-of-function (n = 4) ANCA patients. Fresh cell pellets were lysed in SDS sample buffer. Denatured protein was run on 8% Tris-HCl gel and transferred to nitrocellulose membrane (Schleicher and Schuell, Keene, NH, USA). After blocking, the membranes were incubated overnight at 4°C with appropriate dilutions of unconjugated primary antibodies, including mouse anti-human PTPN22 (Abnova), polyclonal anti-ERK (Abcam, Cambridge, MA, USA), polyclonal anti-phosphor-P44/42 MAP kinase (Cell Signaling, Danvers, MA, USA), polyclonal anti-P38 and polyclonal phosphor-P38 antibodies (Abcam). After washing, the membranes were incubated with horseradish peroxidase-conjugated goat anti-mouse or -rabbit IgG (H+L) (Jackson ImmunoResearch, West Grove, PA) for 1 h. Proteins were detected with Super-Signal West Pico Chemiluminescent Substrate (Pierce).
Densitometric scanning analysis of the ratio of intensity pERK/ERK and pp38/p38 was performed by ImageMaster VDS software.
Analysis of microarray data
RNA was isolated from circulating leukocytes of gain-of-function (n = 4) and non-variant (n = 12) patients with ANCA disease [67], [79], [81]. The Affymetrix microarray gene chip was used for identification of gene expression levels, as previously described [67], [79], [81]. The data were then imported into the Partek Genomics Suite 6.4 program (Partek, St Louis, CA, USA) for an ANOVA statistical analysis and differentially expressed genes (≥2.0-fold, p-value <0.05) within the gain-of-function group were compared to non-variant group. The molecular network analysis was performed using Ingenuity Pathways Analysis (IPA) (Ingenuity Systerms, Redwood City, CA, USA) and the expression profile of genes from gain-of-function and non-variant groups was visualized using a principal components analysis (PCA) mapped scatter plot in Partek program.
Taqman PCR analyses for IL-10 gene expression
IL-10 primers and probes were purchased from Applied Biosystems. Fluorescence emission was monitored using the ABI PRISM 7900 HT sequence detection system. Relative level of total leukocyte RNA was determined by standard 2(−ΔΔCt) calculations and expressed as fold change of reference control samples. Cytochrome c oxidase subunit 5B (COX5B) was used as a RNA loading standard [67], [81], [82].
Statistical analysis
Differences in genotyping tests between ANCA patients and controls were analyzed by chi-square test. The direction and strength of these differences were assessed by calculating odds ratios. All of the alleles detected in our study were tested for the Hardy-Weinberg equilibrium. Clinical comparisons between patients with and without C1858T SNP for categorical measures were performed using chi-square tests. Continuous measures were compared using Wilcoxon rank sum test. A corrected p-value of <0.05 was considered significant. Wilcoxon Two-Sample test were used for comparisons of continuous measures and paired data were analyzed by the Signed Rank Test. A corrected p-value of <0.05 was considered significant. All statistical analyses were performed using SAS statistical program (SAS Institute, Inc., Cary, NC, USA).
Results
Identification of patients with the risk-associated allele of PTPN22
The gain-of-function allelic variant has a SNP changing a cytosine to a thymine (C1858T) which converts the codon from one coding for arginine (R) to one for tryptophan (W) (R620W), and this amino acid change confers a gain-of-function phenotype. A total of 230 Caucasian patients were genotyped for the risk-associated allele of PTPN22 (C1858T) using a TaqMan-SNP-Genotyping Assay. Different from cohorts studied in previous reports, the patient cohort studied here included both PR3- and MPO-ANCA groups. There was a significant association of the C1858T SNP allele in patients of Caucasian descent (22.2%) compared to the general American Caucasian population frequency (16.5%, p = 0.03) [32], with an odds ratio (OR) of 1.45 (95% confidence interval 1.02–2.04) (Table 1). The frequency was significantly higher in patients with a PR3-ANCA serotype (24.3%, p = 0.03, OR 1.63, 95% confidence interval 1.02–2.60) compared to American Caucasian population, but not in those with a MPO-ANCA serotype (20.2%, p = 0.32) (Table 1). Carriage of the variant allele had no influence on a diagnosis of GPA, (23.6%, p = 0.11) or with MPA (21.6%, p = 0.16) compared to the general American Caucasian population.
Table 1. Frequency of PTPN22 risk-allele (T1858) genotype in Caucasian ANCA patients.
| C/C | C/T+T/T | OR (95%CI) | p-value | |
| All Patients | 179(77.8%) | 51(22.2%) | 1.45 (1.02–2.04) | 0.03 |
| *PR3-ANCA | 81(75.7%) | 26(24.3%) | 1.63 (1.02–2.60) | 0.03 |
| *MPO-ANCA | 87(79.8%) | 22(20.2%) | 1.28 (0.78–2.10) | 0.32 |
# Zheng W, 2005 (35).
Excluded from analysis: 6 ANCA-neg; 5 PR3+MPO dual serology: 3 p-ANCA+ANA positives.
For completeness, genotypic analysis was performed to determine the frequency of the loss-of-function variant in PTPN22 (G788A, rs33996649) in the patient cohort. This polymorphism results in an amino acid change in residue 263 from arginine (R) to glutamine (Q) (R263Q) conferring a loss-of-function phenotype, and has been proposed to have a protective effect in SLE [80]. The frequency of this allele in ANCA patients was similar to the general American Caucasian population (4.8% vs 3.0%, p = 0.21) (Table 2) [80].
Table 2. Frequency of PTPN22 of protective allele (A788) in Caucasian ANCA patients.
| G/G | G/A+A/A | OR (95%CI) | p-value | |
| Reference Controls | 550(97.0%) | 17(3.0%) | ||
| Patients | 219(95.2%) | 11(4.8%) | 0.62 (0.28–1.33) | 0.21 |
Combining the reported frequencies of the risk allele, C1858T, in ANCA disease [24], [27] with the frequency observed in this USA cohort, a meta-analysis was performed. Even with the population differences, the combined odds ratio was 1.49 (95% confidence interval 1.28–1.73) (p<0.0001) (Table 3).
Table 3. Meta-analysis of the frequency of the PTPN22 C1858T SNP in ANCA Disease.
| CT+TT (%) | CC (%) | OR (95% Cl) | p-value | ||
| British cohort [27] | 1.45(1.20–1.76) | 0.0001 | |||
| ANCA | 155 (24.76%) | 471(75.24%) | |||
| Controls | 1368 (18.46%) | 6044(81.54%) | |||
| German cohort [24] | 1.71(1.15–2.54) | 0.0078 | |||
| ANCA | 57 (28.64%) | 142(71.36%) | |||
| Controls | 76(19.05%) | 323(80.95%) | |||
| USA cohort | 1.45(1.02–2.04) | 0.0368 | |||
| ANCA | 51 (22.17%) | 179(77.83%) | |||
| Controls | 194 (16.47%) | 984(83.53%) | |||
| Total | 1.49(1.28–1.73) | <0.0001 | |||
| ANCA | 263(24.93%) | 792(75.07%) | |||
| Controls | 1638(18.22%) | 7351(81.78%) |
Assessment of functional changes attributed to the gain-of-function variant of PTPN22
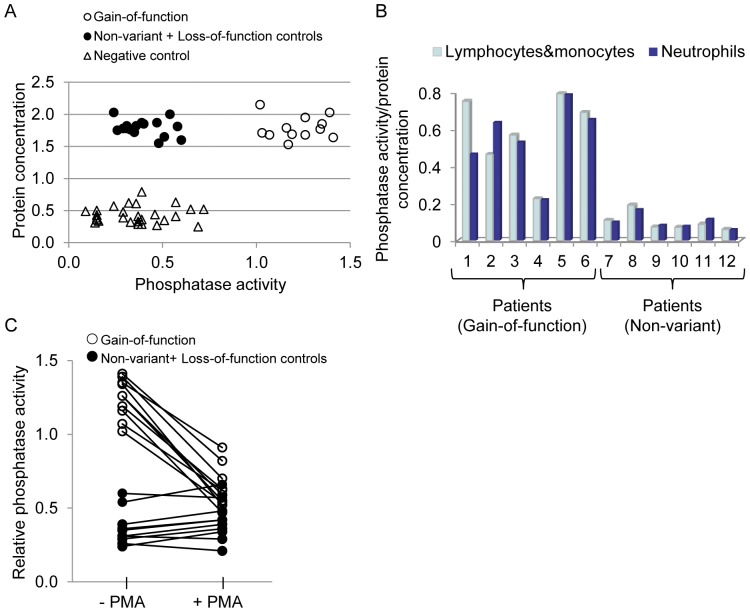
Based on the assertion that the PTPN22 variant (R620W) confers a gain-of-function phenotype, we hypothesized that unstimulated peripheral leukocytes from patients carrying this allele would have higher basal activity. Evaluations included both patients in remission and with active disease (Table 4). To determine amount of activity/protein concentration, PTPN22 protein was captured from total leukocyte lysates using an anti-PTPN22 antibody on two separate micro-titer plates. One was analyzed for total protein captured and the other for activity status of the captured protein. All patients with the gain-of-function variant (R620W) (n = 12) expressed high basal PTPN22 phosphatase activity in un-stimulated leukocytes, in stark contrast to controls with undetectable activity, including both leukocytes expressing the loss-of-function alleles (n = 3) and non-variant alleles (n = 12) (1.22±0.14 versus 0.41±0.12, p<0.0001) (Figure 1A). High basal phosphatase activity was present in R620W neutrophils (n = 6) (5.87±1.50 versus 1.04±0.38, p = 0.0004) as well as lymphocytes&monocytes (n = 6), but not in non-variant controls (n = 6) (p = 0.0003) (Figure 1B).
Table 4. Characteristics of patients with ANCA disease enrolled in the functional studies.
| Patient/ | Age | Gender | Diagnosis | ANCA | Disease |
| Genotype | subtype | activity | |||
| Gain-of-function | |||||
| P03* | 70 | F | GPA | PR3-ANCA | remission |
| P06* | 35 | M | GPA | PR3-ANCA | active+ |
| P07* | 63 | F | MPA | PR3-ANCA | remission |
| P08* | 25 | F | MPA | PR3-ANCA | remission |
| P09 | 55 | F | GPA | PR3-ANCA | active++ |
| P12 | 33 | F | MPA | MPO-ANCA | remission |
| P17 | 52 | M | Lim | MPO-ANCA | active+ |
| P19 | 58 | M | GPA | PR3-ANCA | remission |
| P20 | 61 | F | MPA | MPO-ANCA | active+ |
| P21 | 57 | F | Lim | MPO-ANCA | remission |
| P24 | 74 | F | GPA | ANCA-Neg | active++ |
| P27 | 55 | M | GPA | PR3-ANCA | remission |
| Loss-of-function | |||||
| P02* | 76 | M | GPA | PR3-ANCA | remission |
| P05* | 56 | M | GPA | PR3-ANCA | remission |
| P25 | 61 | M | GPA | PR3-ANCA | active++ |
| Non-variant | |||||
| P01* | 56 | F | MPA | MPO-ANCA | remission |
| P04* | 73 | M | GPA | MPO-ANCA | remission |
| P10* | 86 | M | MPA | MPO-ANCA | remission |
| P11 | 21 | M | GPA | PR3-ANCA | active+ |
| P13 | 54 | M | Lim | MPO+PR3 | active+ |
| P14 | 42 | F | CSS | MPO-ANCA | active++ |
| P15 | 45 | F | GPA | PR3-ANCA | active+ |
| P16 | 60 | M | GPA | PR3-ANCA | active++ |
| P18 | 34 | M | GPA | PR3-ANCA | remission |
| P22 | 51 | F | MPA | PR3-ANCA | active+ |
| P23 | 59 | F | GPA | PR3-ANCA | active+ |
| P26 | 78 | F | Lim | MPO-ANCA | active++ |
Patient's sample included in western blot analysis of signaling pathways.
Figure 1. PTPN22 phosphatase activity in leukocytes.
Basal level of PTPN22 phosphatase activity was high in leukocytes expressing the gain-of-function variant, (A) PTPN22 protein was active in all samples with the gain-of-function PTPN22 (R620W), while activity was undetectable in non-variant and loss-of-function control groups (p<0.0001). Activity values were plotted against total protein captured on ELISA plate using mouse-anti-PTPN22 antibody. For mock-controls, ELISA wells were coated with normal mouse IgG in parallel (B) High basal PTPN22 phosphatase activity was present in neutrophils (p = 0.0004) and lymphocytes&monocytes (p = 0.0003) (activity calculated as fold-increase above controls). (C) High basal phosphatase activity was significantly down-regulated after PMA treatment (p<0.0001), while there were no changes in non-variant controls (p = 0.75).
We asked if we could modulate this high basal phosphatase activity by treating the leukocytes with the powerful stimulant PMA (n = 10 with sufficient sample). PTPN22 phosphatase activity was significantly down-regulated (1.25±0.13 versus 0.64±0.14, p<0.0001) with the mean of decreases 0.61±0.13, while no change was observed in either the loss-of-function controls (n = 3) (0.27±0.04 versus 0.28±0.07, p = 0.07), with the mean of the decreases −0.01±0.08 or the non-variant controls (n = 7) (0.41±0.12 versus 0.47±0.11, p = 0.75) with the mean of the decreases −0.07±0.05 (Figure 1C). The data imply that constitutive phosphatase activity of the variant remains susceptible to pharmacological agents.
Downstream effects of PTPN22 variant high basal activity
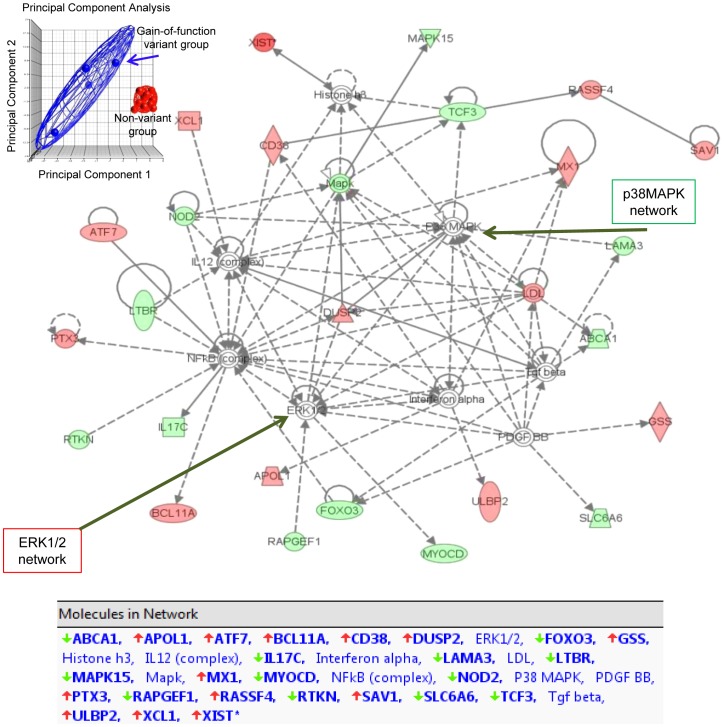
To examine whether the high basal activity of the gain-of-function PTPN22 variant, observed in un-stimulated leukocytes, was affecting primary signaling pathways, we mined our existing Affymetrix array database. Four of the patients enrolled in that study were carriers of the gain-of-function PTPN22 variant (Table 5). Comparisons between groups indicated that the high basal activity of the PTPN22 variant caused global changes in gene transcription. Analysis identified that 151 genes (98 up and 53 down) were differentially regulated (≥2.0-fold and p<0.05). Bioinformatic analysis using principal component analysis (PCA) showed remarkably different gene expression profiles comparing leukocytes with the gain-of-function genotype compared with non-variants (Figure 2). Genes with correlated expression profiles tend to cluster tightly into a small-size elliposoid by the wire mesh, while genes with less similar expression profiles form a looser cluster with a larger size of ellipsoid (Figure 2). The analysis indicates that there are dramatic intrinsic differences in signaling pathways associated with the gain-of-function polymorphism. Analysis using the Ingenuity Pathway Tools (IPA) software indicated that the primary networks affected were those involving ERK, p38MAPK and NFκB (Figure 2).
Table 5. Characteristics of patients with ANCA disease enrolled in the Affymetrix microarray study.
| Patient/ | Age | Gender | Diagnosis | ANCA | Disease |
| Genotype | subtype | activity | |||
| Gain-of-function | |||||
| 1 | 26 | F | MPA | MPO-ANCA | remission |
| 2 | 45 | F | MPA | MPO-ANCA | active+ |
| 3 | 54 | F | MPA | MPO-ANCA | active++ |
| 4 | 54 | F | MPA | MPO-ANCA | active++ |
| Loss-of-function | |||||
| 5 | 68 | M | GPA | PR3-ANCA | active++ |
| Non-variant | |||||
| 6 | 71 | M | GPA | PR3-ANCA | remission |
| 7 | 61 | F | MPA | MPO-ANCA | active+ |
| 8 | 56 | M | GPA | PR3-ANCA | active++ |
| 9 | 55 | M | GPA | PR3-ANCA | active++ |
| 10 | 38 | M | MPA | PR3-ANCA | active++ |
| 11 | 60 | F | MPA | MPO-ANCA | active++ |
| 12 | 55 | M | GPA | PR3-ANCA | active++ |
| 13 | 72 | M | Lim | PR3-ANCA | active+++ |
| 14 | 79 | M | MPA | MPO-ANCA | active+++ |
| 15 | 60 | M | MPA | PR3-ANCA | active+++ |
| 16 | 17 | F | MPA | PR3-ANCA | active+++ |
Figure 2. Bioinformatics analysis of Affymetrix microarray gene expression data, comparing leukocytes with the gain-of-function genotype to those with a non-variant genotype.
Principal Component Analysis (PCA) scatter plot using Partek analysis is shown in the upper left corner. PCA is mathematically defined as an orthogonal linear transformation that transforms the data to a new coordinate system such that the greatest variance by any projection of the data comes to lie on the first coordinate (called the first principal component), the second greatest variance on the second coordinate, and so on. Each dot represents a patient's expression profile; the blue color dots represent gain-of-function and red show non-variant genotypes. Analysis using the Ingenuity Pathway Tools (IPA) software utilizes a repository of biological interactions and functional annotations created from millions of individually modeled relationships. The genes in red indicate increased expression and blue represents decreased expression, comparing gain-of-function with non-variant individuals. Primary networks identified were ERK1/2, p38MAPK, and NFκB networks.
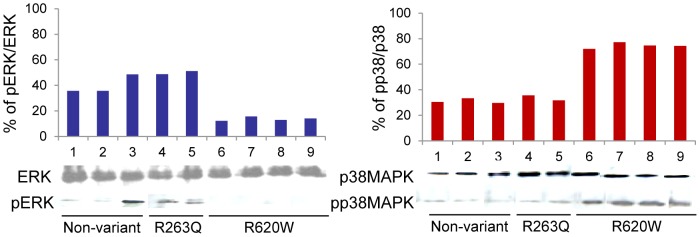
These data are consistent with reports that PTPN22 function regulates signaling molecules leading to activation of ERK1,2 [83], [84]. We examined the phosphorylation status of ERK1,2 in four patients with PTPN22 (R620W), two patients with PTPN22 loss-of-function allele (R263Q) and three patients with the normal allele (Table 4). Phosphorylated/active ERK was undetectable with PTPN22 gain-of-function activity, in contrast to controls. Instead, the phosphorylated/active p38 mitogen-activated protein kinase (p38 MAPK) form was elevated (Figure 3).
Figure 3. Analysis of ERK1,2 and p38MAPK phosphorylation status.
Bar graphs represent the ratio of intensity pERK/ERK and pp38/p38MAPK as quantitated by densitometric scanning analysis using ImageMaster VDS software. Western blot analysis for ERK1,2 and p38MAPK activation demonstrates PTPN22 gain-of-function (R620) exerts a negative effect on the ERK signaling pathway, compared to loss-of-function (R263Q) and non-variant controls. In contrast p38 mitogen-activated protein kinase (p38 MAPK) was increased with the gain-of-function (R620) phenotype.
IL-10 gene expression is down regulated in leukocytes with the PTPN22 gain-of-function (R620W) variant
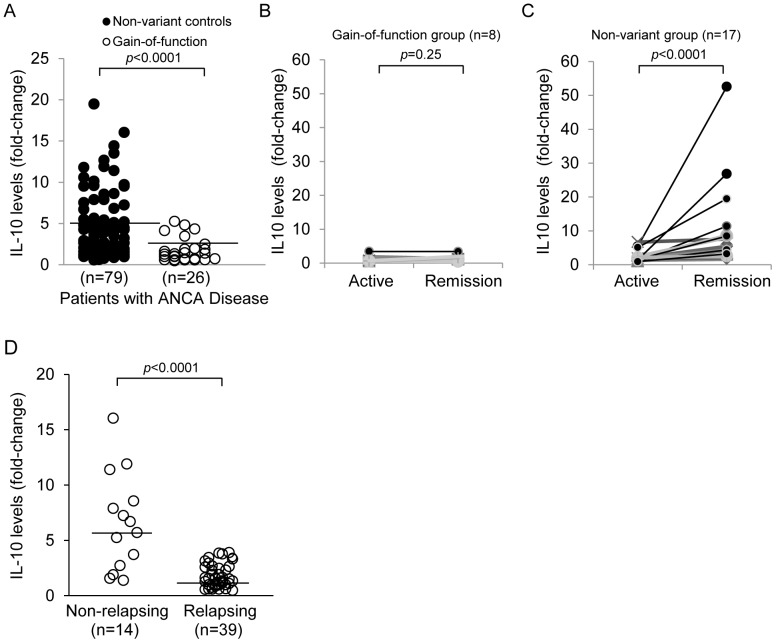
Maximal IL-10 production requires signaling through activated ERK and the downstream phosphorylation of the Sp1 transcription factor [85], [86]. Based on decreased activity of ERK with the gain-of-function (R620W) variant, we hypothesized the IL-10 gene expression would be negatively affected. IL-10 mRNA levels were significantly lower in patients with the gain-of-function variant (n = 26), as compared to non-variant controls (n = 79)(1.8±1.4 versus 5.0±4.1, p<0.0001) (Figure 4A). Longitudinally, baseline IL-10 transcripts did not increase in patients having the gain-of-function genotype as their disease progressed from active disease (BVAS≥1) to remission (BVAS = 0) (n = 8) (1.17±1.01 versus 1.50±0.88, p = 0.25) with the mean of the increase 0.33±0.61 (Figure 4B). In contrast, patients without the SNP showed a robust increase in IL-10 transcripts as remission was achieved (n = 17) (2.37±1.69 versus 10.19±12.78, p<0.0001) with the mean of the increase 7.82±12.01 (Figure 4C). Epidemiological analyses indicated that patients having gain-of-function (R620W) variant progressed to end-stage kidney disease (ESKD) on average 20 months faster (18% vs. 9%, p = 0.04). No substantial differences were found in regard to gender, ANCA serotype, disease diagnosis, treatment resistance, or organ involvement.
Figure 4. IL-10 mRNA expression.
(A) IL-10 transcript levels are reduced in leukocytes from PTPN22 (R620W) positive patients. (A) IL-10 mRNA expression, which is mediated through the ERK pathway, was significantly lower in gain-of-function patients (p<0.0001) by quantitative TaqMan PCR. (B) Longitudinally, the baseline level of IL-10 message in patients with the gain-of-function variant did not increase as they transitioned from active disease to remission (p = 0.25). (C) In contrast, patients with normal PTPN22 showed a robust increase in IL-10 as they entered remission (p<0.0001). (D) Decreased IL-10 levels were associated with the relapsing group (n = 39, 1.8±1.15), and higher level in the non-relapse patient group (n = 14, 6.6±4.4, p<0.0001).
It was reported that lower IL-10 levels in remission are associated with a higher relapse rate in long-term follow-up [87]. Analysis of our cohort indicated that higher IL-10 transcript levels were associated with non-relapsing disease (n = 14, 6.6±4.4, p<0.0001), while lower levels were associated with a relapsing-disease history (n = 39, 1.8±1.15) (Figure 4D). There were 14 patients in this study that had the gain-of-function SNP. Three (21%) were in the non-relapsing group while 11(79%) were in the relapsing-disease group.
Discussion
This is the first report of studies on the basal activity of the PTPN22-gain-of-function protein in non-stimulated leukocytes, immediately following blood draw. High basal PTPN22 phosphatase activity was detected in leukocytes of every patient tested who had the PTPN22 SNP (C1858T) genotype, while their non-variant counterparts had undetectable activity. We were intrigued that high basal PTPN22 phosphatase activity was present in neutrophils expressing the gain-of-function variant. Interestingly, one consequence was activation of the p38 MAPK pathway. P38 MAPK regulates macrophage and neutrophil functional responses, including respiratory burst activity, and chemotaxis. High activity was also present in the lymphocyte&monocyte pool which would support the findings of altered T cell function in Type-1 diabetes and Jurkat T leukemia cells overexpressing the transfected gain-of-function variant [80], [88]–[90].
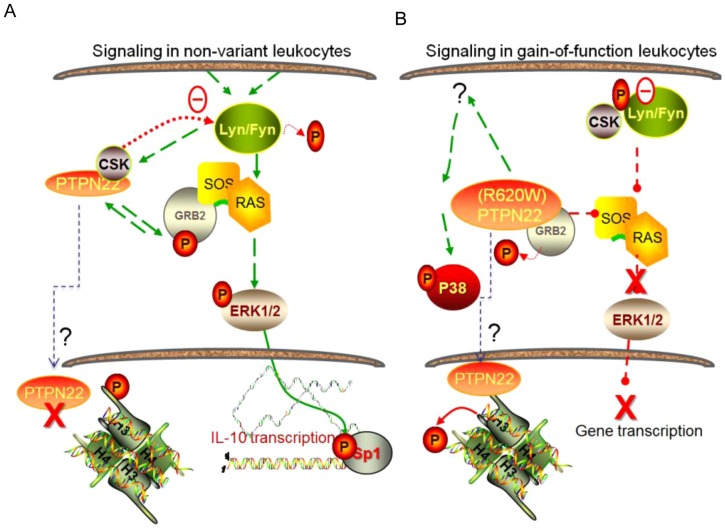
Constitutive activity of PTPN22 gain-of-function variant was associated with global changes in the transcriptome, inasmuch as the assessment of the small cohort studied here. Activity of normal PTPN22 is under regulatory constraints, one of which is inhibition by phosphorylation on Ser-35 by protein kinase C (PKC) [91]. The mechanistic pathway of PTPN22 is under study by many groups and to date, functional partners of normal PTPN22 include growth factor receptor-bound protein 2 (GRB-2), and C-SRC kinase (CSK) [89]. The schematic in Figure 5A illustrates some of the signaling pathways reportedly linked with PTPN22 function, and in Figure 5B a prediction – based on the data presented here – of how they may be perturbed by constitutively high PTPN22 activity.
Figure 5. Signaling pathways disrupted by the gain-of-function variant of PTPN22.
(A) Signaling pathways affected by PTPN22 include SRC-family kinases (Lyn/Fyn) and RAS pathways [83], [84]. It can affect the activity of SRC-family kinases through regulation of CSK (cSRC Kinase) [96], [97]. PTPN22 can affect RAS activity through binding to GRB-2 (Growth factor receptor-bound protein 2). ERK1,2 phosphorylates and activates many transcription factors, including the transcription factor Sp1 depicted here, which regulates the transcription of IL-10 [86]. (B) Changes in PTPN22 function due to the gain-of-function phenotype. PTPN22 (R620W) amino acid change lies within a domain that binds CSK, resulting in a greatly reduced binding [84]. Thus CSK is available for binding and inhibiting SRC. Also, a gain-of function alteration could act as a super-antagonist of epigenetic nucleosome remodeling, based on reports that PTPs directly dephosphorylate histone tails [98]. The gain-of-function phosphatase activity also affects the function of GRB2 restraint of RAS signaling and a loss of ERK1/2 phosphorylation/activation and loss of Sp1 transcriptional activity.
It is reported that decreased IL-10 production during remission is a predictor of relapse in ANCA disease [87]. Results in our study are in agreement. Moreover, we found that IL-10 message was lower in patients expressing the PTPN22 gain-of-function variant and that a high proportion of these fell into the relapsing-disease category. Reduced IL-10 responses in patients with the gain-of-function variant can be considered as a deviation in transmission of signals within the microenvironment of the body. We examined gene expression of the anti-inflammatory cytokine IL-10, because it is known to be responsive to environmental triggers [86], [92]; for example, neutrophils secrete high amounts of IL-10 in response to bacterial products [93]. Relevant to this discussion, expression of IL-10 is regulated through epigenetic mechanisms at the IL-10 locus through signals transmitted by the ERK signaling pathway, which is responsive to many cytokines, growth factors, and importantly environmental stress [85], [86], [94].
Although the allelic variant of PTPN22 has been reported as a predisposing factor in ANCA disease [95], this is the first report of its frequency in a USA patient cohort. In this patient cohort, association of the disease-associated allele was skewed toward PR3-ANCA disease. In concordance, the geographic distribution of this variant allele is highest in countries where PR3-ANCA predominates with the highest in Finland (15.5%), Sweden (12%) and then UK (8%), decreasing southward to Spain (6%) and Italy (2%). Combined, these studies provide statistical power to support an association between the PTPN22 variant and ANCA disease (OR 1.49, p = 4.15×10−6). The allele is nearly absent in African American and Asian populations [90].
PTPN22 integrates and transmits environmental changes through dynamic signalling molecules. Studies of the gain-of-function variant in multifactorial autoimmune diseases, such as ANCA disease, provides the opportunity to understand how disease outcome can be influenced by a complex interplay of genetic regulation and environmental influences.
Funding Statement
This work was supported by the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK)/National Institutes of Health (NIH) Grant 2PO1DK 58335. Yali Cao would like to thank the China Scholarship Council for the financial assistance. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Falk RJ, Jennette JC (1997) ANCA small-vessel vasculitis. J Am Soc Nephrol 8: 314–22. [DOI] [PubMed] [Google Scholar]
- 2. Hogan SL, Cooper GS, Savitz DA, Nylander-French LA, Parks CG, et al. (2007) Association of silica exposure with anti-neutrophil cytoplasmic autoantibody small-vessel vasculitis: a population-based, case-control study. Clin J Am Soc Nephrol 2: 290–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hogan SL, Satterly KK, Dooley MA, Nachman PH, Jennette JC, et al. (2001) Silica exposure in anti-neutrophil cytoplasmic autoantibody-associated glomerulonephritis and lupus nephritis. J Am Soc Nephrol 12: 134–42. [DOI] [PubMed] [Google Scholar]
- 4. Ciavatta DJ, Yang J, Preston GA, Badhwar AK, Xiao H, et al. Epigenetic basis for aberrant upregulation of autoantigen genes in humans with ANCA vasculitis. J Clin Invest 120: 3209–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Willcocks LC, Lyons PA, Rees AJ, Smith KG (2010) The contribution of genetic variation and infection to the pathogenesis of ANCA-associated systemic vasculitis. Arthritis Res Ther 12: 202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Huang D, Zhou Y, Hoffman GS (2001) Pathogenesis: immunogenetic factors. Best Pract Res Clin Rheumatol 15: 239–58. [DOI] [PubMed] [Google Scholar]
- 7. Giscombe R, Wang X, Huang D, Lefvert AK (2002) Coding sequence 1 and promoter single nucleotide polymorphisms in the CTLA-4 gene in Wegener's granulomatosis. J Rheumatol 29: 950–3. [PubMed] [Google Scholar]
- 8. Griffith ME, Lovegrove JU, Gaskin G, Whitehouse DB, Pusey CD (1996) C-antineutrophil cytoplasmic antibody positivity in vasculitis patients is associated with the Z allele of alpha-1-antitrypsin, and P-antineutrophil cytoplasmic antibody positivity with the S allele. Nephrol Dial Transplant 11: 438–43. [PubMed] [Google Scholar]
- 9. Segelmark M, Elzouki AN, Wieslander J, Eriksson S (1995) The PiZ gene of alpha 1-antitrypsin as a determinant of outcome in PR3- ANCA-positive vasculitis. Kidney Int 48: 844–50. [DOI] [PubMed] [Google Scholar]
- 10. Gencik M, Meller S, Borgmann S, Sitter T, Menezes Saecker AM, et al. (2000) The association of CD18 alleles with anti-myeloperoxidase subtypes of ANCA-associated systemic vasculitides. Clin Immunol 94: 9–12. [DOI] [PubMed] [Google Scholar]
- 11. Meller S, Jagiello P, Borgmann S, Fricke H, Epplen JT, et al. (2001) Novel SNPs in the CD18 gene validate the association with MPO-ANCA+ vasculitis. Genes Immun 2: 269–72. [DOI] [PubMed] [Google Scholar]
- 12. Jagiello P, Gencik M, Arning L, Wieczorek S, Kunstmann E, et al. (2004) New genomic region for Wegener's granulomatosis as revealed by an extended association screen with 202 apoptosis-related genes. Hum Genet 114: 468–77. [DOI] [PubMed] [Google Scholar]
- 13. Zhou Y, Huang D, Farver C, Hoffman GS (2003) Relative importance of CCR5 and antineutrophil cytoplasmic antibodies in patients with Wegener's granulomatosis. J Rheumatol 30: 1541–7. [PubMed] [Google Scholar]
- 14. Borgmann S, Endisch G, Hacker UT, Song BS, Fricke H (2003) Proinflammatory genotype of interleukin-1 and interleukin-1 receptor antagonist is associated with ESRD in proteinase 3-ANCA vasculitis patients. Am J Kidney Dis 41: 933–42. [DOI] [PubMed] [Google Scholar]
- 15. Bartfai Z, Gaede KI, Russell KA, Murakozy G, Muller-Quernheim J, et al. (2003) Different gender-associated genotype risks of Wegener's granulomatosis and microscopic polyangiitis. Clin Immunol 109: 330–7. [DOI] [PubMed] [Google Scholar]
- 16. Zhou Y, Giscombe R, Huang D, Lefvert AK (2002) Novel genetic association of Wegener's granulomatosis with the interleukin 10 gene. J Rheumatol 29: 317–20. [PubMed] [Google Scholar]
- 17. Gencik M, Meller S, Borgmann S, Fricke H (2000) Proteinase 3 gene polymorphisms and Wegener's granulomatosis. Kidney Int 58: 2473–7. [DOI] [PubMed] [Google Scholar]
- 18. Tsuchiya N, Kobayashi S, Kawasaki A, Kyogoku C, Arimura Y, et al. (2003) Genetic background of Japanese patients with antineutrophil cytoplasmic antibody-associated vasculitis: association of HLA-DRB1*0901 with microscopic polyangiitis. J Rheumatol 30: 1534–40. [PubMed] [Google Scholar]
- 19. Szyld P, Jagiello P, Csernok E, Gross WL, Epplen JT (2006) On the Wegener granulomatosis associated region on chromosome 6p21.3. BMC Med Genet 7: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dijstelbloem HM, Scheepers RH, Oost WW, Stegeman CA, Van Der Pol WL, et al. (1999) Fcgamma receptor polymorphisms in Wegener's granulomatosis: risk factors for disease relapse. Arthritis Rheum 42: 1823–7. [DOI] [PubMed] [Google Scholar]
- 21. Edberg JC, Wainstein E, Wu J, Csernok E, Sneller MC, et al. (1997) Analysis of FcgammaRII gene polymorphisms in Wegener's granulomatosis. Exp Clin Immunogenet 14: 183–95. [PubMed] [Google Scholar]
- 22. Tse WY, Abadeh S, Jefferis R, Savage CO, Adu D (2000) Neutrophil FcgammaRIIIb allelic polymorphism in anti-neutrophil cytoplasmic antibody (ANCA)-positive systemic vasculitis. Clin Exp Immunol 119: 574–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, et al. (2007) FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet 39: 721–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Jagiello P, Aries P, Arning L, Wagenleiter SE, Csernok E, et al. (2005) The PTPN22 620W allele is a risk factor for Wegener's granulomatosis. Arthritis Rheum 52: 4039–43. [DOI] [PubMed] [Google Scholar]
- 25. Begovich AB, Carlton VE, Honigberg LA, Schrodi SJ, Chokkalingam AP, et al. (2004) A missense single-nucleotide polymorphism in a gene encoding a protein tyrosine phosphatase (PTPN22) is associated with rheumatoid arthritis. Am J Hum Genet 75: 330–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kyogoku C, Langefeld CD, Ortmann WA, Lee A, Selby S, et al. (2004) Genetic association of the R620W polymorphism of protein tyrosine phosphatase PTPN22 with human SLE. Am J Hum Genet 75: 504–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Carr EJ, Niederer HA, Williams J, Harper L, Watts RA, et al. (2009) Confirmation of the genetic association of CTLA4 and PTPN22 with ANCA-associated vasculitis. BMC Med Genet 10: 121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bottini N, Musumeci L, Alonso A, Rahmouni S, Nika K, et al. (2004) A functional variant of lymphoid tyrosine phosphatase is associated with type I diabetes. Nat Genet 36: 337–8. [DOI] [PubMed] [Google Scholar]
- 29. Smyth D, Cooper JD, Collins JE, Heward JM, Franklyn JA, et al. (2004) Replication of an association between the lymphoid tyrosine phosphatase locus (LYP/PTPN22) with type 1 diabetes, and evidence for its role as a general autoimmunity locus. Diabetes 53: 3020–3. [DOI] [PubMed] [Google Scholar]
- 30. Ladner MB, Bottini N, Valdes AM, Noble JA (2005) Association of the single nucleotide polymorphism C1858T of the PTPN22 gene with type 1 diabetes. Hum Immunol 66: 60–4. [DOI] [PubMed] [Google Scholar]
- 31. Begovich AB, Caillier SJ, Alexander HC, Penko JM, Hauser SL, et al. (2005) The R620W polymorphism of the protein tyrosine phosphatase PTPN22 is not associated with multiple sclerosis. Am J Hum Genet 76: 184–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zheng W, She JX (2005) Genetic association between a lymphoid tyrosine phosphatase (PTPN22) and type 1 diabetes. Diabetes 54: 906–8. [DOI] [PubMed] [Google Scholar]
- 33. Van Oene M, Wintle RF, Liu X, Yazdanpanah M, Gu X, et al. (2005) Association of the lymphoid tyrosine phosphatase R620W variant with rheumatoid arthritis, but not Crohn's disease, in Canadian populations. Arthritis Rheum 52: 1993–8. [DOI] [PubMed] [Google Scholar]
- 34. Orozco G, Sanchez E, Gonzalez-Gay MA, Lopez-Nevot MA, Torres B, et al. (2005) Association of a functional single-nucleotide polymorphism of PTPN22, encoding lymphoid protein phosphatase, with rheumatoid arthritis and systemic lupus erythematosus. Arthritis Rheum 52: 219–24. [DOI] [PubMed] [Google Scholar]
- 35. Hinks A, Barton A, John S, Bruce I, Hawkins C, et al. (2005) Association between the PTPN22 gene and rheumatoid arthritis and juvenile idiopathic arthritis in a UK population: further support that PTPN22 is an autoimmunity gene. Arthritis Rheum 52: 1694–9. [DOI] [PubMed] [Google Scholar]
- 36. Simkins HM, Merriman ME, Highton J, Chapman PT, O'donnell JL, et al. (2005) Association of the PTPN22 locus with rheumatoid arthritis in a New Zealand Caucasian cohort. Arthritis Rheum 52: 2222–5. [DOI] [PubMed] [Google Scholar]
- 37. Criswell LA, Pfeiffer KA, Lum RF, Gonzales B, Novitzke J, et al. (2005) Analysis of families in the multiple autoimmune disease genetics consortium (MADGC) collection: the PTPN22 620W allele associates with multiple autoimmune phenotypes. Am J Hum Genet 76: 561–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lee AT, Li W, Liew A, Bombardier C, Weisman M, et al. (2005) The PTPN22 R620W polymorphism associates with RF positive rheumatoid arthritis in a dose-dependent manner but not with HLA-SE status. Genes Immun 6: 129–33. [DOI] [PubMed] [Google Scholar]
- 39. Robertson J, Wu J, Arends J, Glass W 2nd, Southwood S, et al. (2005) Characterization of the T-cell epitope that causes anti-GBM glomerulonephritis. Kidney Int 68: 1061–70. [DOI] [PubMed] [Google Scholar]
- 40. Reddy MV, Johansson M, Sturfelt G, Jonsen A, Gunnarsson I, et al. (2005) The R620W C/T polymorphism of the gene PTPN22 is associated with SLE independently of the association of PDCD1. Genes Immun 6: 658–62. [DOI] [PubMed] [Google Scholar]
- 41. Behrens TW, Graham RR, Kyogoku C, Baechler EC, Ramos PS, et al. (2005) Progress towards understanding the genetic pathogenesis of systemic lupus erythematosus. Novartis Found Symp 267: 145–60; discussion 60–4. [DOI] [PubMed] [Google Scholar]
- 42. Velaga MR, Wilson V, Jennings CE, Owen CJ, Herington S, et al. (2004) The codon 620 tryptophan allele of the lymphoid tyrosine phosphatase (LYP) gene is a major determinant of Graves' disease. J Clin Endocrinol Metab 89: 5862–5. [DOI] [PubMed] [Google Scholar]
- 43. Skorka A, Bednarczuk T, Bar-Andziak E, Nauman J, Ploski R (2005) Lymphoid tyrosine phosphatase (PTPN22/LYP) variant and Graves' disease in a Polish population: association and gene dose-dependent correlation with age of onset. Clin Endocrinol (Oxf) 62: 679–82. [DOI] [PubMed] [Google Scholar]
- 44. Canton I, Akhtar S, Gavalas NG, Gawkrodger DJ, Blomhoff A, et al. (2005) A single-nucleotide polymorphism in the gene encoding lymphoid protein tyrosine phosphatase (PTPN22) confers susceptibility to generalised vitiligo. Genes Immun 6: 584–7. [DOI] [PubMed] [Google Scholar]
- 45. Lai KN, Leung JC, Rifkin I, Lockwood CM (1994) Effect of anti-neutrophil cytoplasm autoantibodies on the intracellular calcium concentration of human neutrophils [see comments]. Lab Invest 70: 152–62. [PubMed] [Google Scholar]
- 46. Lai KN, Lockwood CM (1991) The effect of anti-neutrophil cytoplasm autoantibodies on the signal transduction in human neutrophils. Clin Exp Immunol 85: 396–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Porges AJ, Redecha PB, Kimberly WT, Csernok E, Gross WL, et al. (1994) Anti-neutrophil cytoplasmic antibodies engage and activate human neutrophils via Fc gamma RIIa. J Immunol 153: 1271–80. [PubMed] [Google Scholar]
- 48. Reumaux D, Vossebeld PJ, Roos D, Verhoeven AJ (1995) Effect of tumor necrosis factor-induced integrin activation on Fc gamma receptor II-mediated signal transduction: relevance for activation of neutrophils by anti-proteinase 3 or anti-myeloperoxidase antibodies. Blood 86: 3189–95. [PubMed] [Google Scholar]
- 49. Calderwood JW, Williams JM, Morgan MD, Nash GB, Savage CO (2005) ANCA induces beta2 integrin and CXC chemokine-dependent neutrophil-endothelial cell interactions that mimic those of highly cytokine-activated endothelium. J Leukoc Biol 77: 33–43. [DOI] [PubMed] [Google Scholar]
- 50. Falk RJ, Terrell RS, Charles LA, Jennette JC (1990) Anti-neutrophil cytoplasmic autoantibodies induce neutrophils to degranulate and produce oxygen radicals in vitro. Proc Natl Acad Sci U S A 87: 4115–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Jennette JC, Wilkman AS, Falk RJ (1989) Anti-neutrophil cytoplasmic autoantibody-associated glomerulonephritis and vasculitis. Am J Pathol 135: 921–30. [PMC free article] [PubMed] [Google Scholar]
- 52. Charles LA, Falk RJ, Jennette JC (1992) Reactivity of antineutrophil cytoplasmic autoantibodies with mononuclear phagocytes. J Leukoc Biol 51: 65–8. [DOI] [PubMed] [Google Scholar]
- 53. Deremee RA (1991) Antineutrophil cytoplasmic autoantibody-associated diseases: a pulmonologist's perspective. Am J Kidney Dis 18: 180–3. [DOI] [PubMed] [Google Scholar]
- 54. Samet JM, Bell ML (2004) Commentary: nitrogen dioxide and asthma redux. Int J Epidemiol 33: 215–6. [DOI] [PubMed] [Google Scholar]
- 55. Hill RJ, Zozulya S, Lu YL, Ward K, Gishizky M, et al. (2002) The lymphoid protein tyrosine phosphatase Lyp interacts with the adaptor molecule Grb2 and functions as a negative regulator of T-cell activation. Exp Hematol 30: 237–44. [DOI] [PubMed] [Google Scholar]
- 56. Gjorloff-Wingren A, Saxena M, Williams S, Hammi D, Mustelin T (1999) Characterization of TCR-induced receptor-proximal signaling events negatively regulated by the protein tyrosine phosphatase PEP. Eur J Immunol 29: 3845–54. [DOI] [PubMed] [Google Scholar]
- 57. Van Vliet C, Bukczynska PE, Puryer MA, Sadek CM, Shields BJ, et al. (2005) Selective regulation of tumor necrosis factor-induced Erk signaling by Src family kinases and the T cell protein tyrosine phosphatase. Nat Immunol 6: 253–60. [DOI] [PubMed] [Google Scholar]
- 58. Hermiston ML, Zikherman J, Zhu JW (2009) CD45, CD148, and Lyp/Pep: critical phosphatases regulating Src family kinase signaling networks in immune cells. Immunol Rev 228: 288–311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Chow LM, Fournel M, Davidson D, Veillette A (1993) Negative regulation of T-cell receptor signalling by tyrosine protein kinase p50csk. Nature 365: 156–60. [DOI] [PubMed] [Google Scholar]
- 60. Fernandez R, Suchard SJ (1998) Syk activation is required for spreading and H2O2 release in adherent human neutrophils. J Immunol 160: 5154–62. [PubMed] [Google Scholar]
- 61. Mocsai A, Ligeti E, Lowell CA, Berton G (1999) Adhesion-dependent degranulation of neutrophils requires the Src family kinases Fgr and Hck. J Immunol 162: 1120–6. [PubMed] [Google Scholar]
- 62. Davies DJ, Moran JE, Niall JF, Ryan GB (1982) Segmental necrotising glomerulonephritis with antineutrophil antibody: possible arbovirus aetiology? Br Med J 285: 606–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stegeman CA, Tervaert JW, Sluiter WJ, Manson WL, De Jong PE, et al. (1994) Association of chronic nasal carriage of Staphylococcus aureus and higher relapse rates in Wegener granulomatosis. Ann Intern Med 120: 12–7. [DOI] [PubMed] [Google Scholar]
- 64. Pudifin DJ, Duursma J, Gathiram V, Jackson TF (1994) Invasive amoebiasis is associated with the development of anti-neutrophil cytoplasmic antibody. Clin Exp Immunol 97: 48–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hogan SL, Falk RJ, Chin H, Cai J, Jennette CE, et al. (2005) Predictors of relapse and treatment resistance in antineutrophil cytoplasmic antibody-associated small-vessel vasculitis. Ann Intern Med 143: 621–31. [DOI] [PubMed] [Google Scholar]
- 66. Falk RJ, Hogan S, Carey TS, Jennette JC (1990) Clinical course of anti-neutrophil cytoplasmic autoantibody-associated glomerulonephritis and systemic vasculitis. The Glomerular Disease Collaborative Network [see comments]. Ann Intern Med 113: 656–63. [DOI] [PubMed] [Google Scholar]
- 67. Yang JJ, Pendergraft WF, Alcorta DA, Nachman PH, Hogan SL, et al. (2004) Circumvention of normal constraints on granule protein gene expression in peripheral blood neutrophils and monocytes of patients with antineutrophil cytoplasmic autoantibody-associated glomerulonephritis. J Am Soc Nephrol 15: 2103–14. [DOI] [PubMed] [Google Scholar]
- 68. De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, et al. (2009) Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. Embo J 28: 3341–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, et al. (2007) The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 130: 1083–94. [DOI] [PubMed] [Google Scholar]
- 70. Hogan SL, Nachman PH, Wilkman AS, Jennette JC, Falk RJ (1996) Prognostic markers in patients with antineutrophil cytoplasmic autoantibody-associated microscopic polyangiitis and glomerulonephritis. J Am Soc Nephrol 7: 23–32. [DOI] [PubMed] [Google Scholar]
- 71. Pagnoux C, Hogan SL, Chin H, Jennette JC, Falk RJ, et al. (2008) Predictors of treatment resistance and relapse in antineutrophil cytoplasmic antibody-associated small-vessel vasculitis: comparison of two independent cohorts. Arthritis Rheum 58: 2908–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Falk RJ, Gross WL, Guillevin L, Hoffman G, Jayne DR, et al. Granulomatosis with Polyangiitis (Wegener's): An Alternative Name for Wegener's Granulomatosis. J Am Soc Nephrol 22: 587–8. [DOI] [PubMed] [Google Scholar]
- 73. Falk RJ, Gross WL, Guillevin L, Hoffman G, Jayne DR, et al. Granulomatosis with polyangiitis (Wegener's): an alternative name for Wegener's granulomatosis. Ann Rheum Dis 70: 704. [DOI] [PubMed] [Google Scholar]
- 74. Falk RJ, Gross WL, Guillevin L, Hoffman GS, Jayne DR, et al. Granulomatosis with polyangiitis (Wegener's): an alternative name for Wegener's granulomatosis. Arthritis Rheum 63: 863–4. [DOI] [PubMed] [Google Scholar]
- 75. Falk RJ, Jennette JC (1988) Anti-neutrophil cytoplasmic autoantibodies with specificity for myeloperoxidase in patients with systemic vasculitis and idiopathic necrotizing and crescentic glomerulonephritis. N Engl J Med 318: 1651–7. [DOI] [PubMed] [Google Scholar]
- 76. Jennette JC, Falk RJ, Andrassy K, Bacon PA, Churg J, et al. (1994) Nomenclature of systemic vasculitides. Proposal of an international consensus conference. Arthritis Rheum 37: 187–92. [DOI] [PubMed] [Google Scholar]
- 77. Falk RJ, Jennette JC (1988) Anti-neutrophil cytoplasmic autoantibodies with specificity for myloperoxidase in patients with systemic vasculitis and idiopathic necrotizing and crescentic glomerulonephritis. N Engl J Med 318: 1651–7. [DOI] [PubMed] [Google Scholar]
- 78. Hagen EC, Ballieux BE, Van Es LA, Daha MR, Van Der Woude FJ (1993) Antineutrophil cytoplasmic autoantibodies: a review of the antigens involved, the assays, and the clinical and possible pathogenetic consequences. Blood 81: 1996–2002. [PubMed] [Google Scholar]
- 79. Yang JJ, Preston GA, Alcorta DA, Waga I, Munger WE, et al. (2002) Expression profile of leukocyte genes activated by anti-neutrophil cytoplasmic autoantibodies (ANCA). Kidney Int 62: 1638–49. [DOI] [PubMed] [Google Scholar]
- 80. Orru V, Tsai SJ, Rueda B, Fiorillo E, Stanford SM, et al. (2009) A loss-of-function variant of PTPN22 is associated with reduced risk of systemic lupus erythematosus. Hum Mol Genet 18: 569–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Alcorta DA, Barnes DA, Dooley MA, Sullivan P, Jonas B, et al. (2007) Leukocyte gene expression signatures in antineutrophil cytoplasmic autoantibody and lupus glomerulonephritis. Kidney Int 72: 853–64. [DOI] [PubMed] [Google Scholar]
- 82. Alcorta D, Preston G, Munger W, Sullivan P, Yang JJ, et al. (2002) Microarray studies of gene expression in circulating leukocytes in kidney diseases. Exp Nephrol 10: 139–49. [DOI] [PubMed] [Google Scholar]
- 83. Nagao M, Yamauchi J, Kaziro Y, Itoh H (1998) Involvement of protein kinase C and Src family tyrosine kinase in Galphaq/11-induced activation of c-Jun N-terminal kinase and p38 mitogen-activated protein kinase. J Biol Chem 273: 22892–8. [DOI] [PubMed] [Google Scholar]
- 84. Fiorillo E, Orru V, Stanford SM, Liu Y, Salek M, et al. (2010) Autoimmune-associated PTPN22 R620W variation reduces phosphorylation of lymphoid phosphatase on an inhibitory tyrosine residue. J Biol Chem 285: 26506–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Slack EC, Robinson MJ, Hernanz-Falcon P, Brown GD, Williams DL, et al. (2007) Syk-dependent ERK activation regulates IL-2 and IL-10 production by DC stimulated with zymosan. Eur J Immunol 37: 1600–12. [DOI] [PubMed] [Google Scholar]
- 86. Lucas M, Zhang X, Prasanna V, Mosser DM (2005) ERK activation following macrophage FcgammaR ligation leads to chromatin modifications at the IL-10 locus. J Immunol 175: 469–77. [DOI] [PubMed] [Google Scholar]
- 87. Hruskova Z, Rihova Z, Mareckova H, Jancova E, Rysava R, et al. (2009) Intracellular cytokine production in ANCA-associated vasculitis: low levels of interleukin-10 in remission are associated with a higher relapse rate in the long-term follow-up. Archives of medical research 40: 276–84. [DOI] [PubMed] [Google Scholar]
- 88. Vang T, Congia M, Macis MD, Musumeci L, Orru V, et al. (2005) Autoimmune-associated lymphoid tyrosine phosphatase is a gain-of-function variant. Nat Genet 37: 1317–9. [DOI] [PubMed] [Google Scholar]
- 89. Wu J, Katrekar A, Honigberg LA, Smith AM, Conn MT, et al. (2006) Identification of substrates of human protein-tyrosine phosphatase PTPN22. J Biol Chem 281: 11002–10. [DOI] [PubMed] [Google Scholar]
- 90. Vang T, Miletic AV, Bottini N, Mustelin T (2007) Protein tyrosine phosphatase PTPN22 in human autoimmunity. Autoimmunity 40: 453–61. [DOI] [PubMed] [Google Scholar]
- 91. Yu X, Sun JP, He Y, Guo X, Liu S, et al. (2007) Structure, inhibitor, and regulatory mechanism of Lyp, a lymphoid-specific tyrosine phosphatase implicated in autoimmune diseases. Proc Natl Acad Sci U S A 104: 19767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Fillatreau S, Gray D, Anderton SM (2008) Not always the bad guys: B cells as regulators of autoimmune pathology. Nat Rev Immunol 8: 391–7. [DOI] [PubMed] [Google Scholar]
- 93. Zhang X, Majlessi L, Deriaud E, Leclerc C, Lo-Man R (2009) Coactivation of Syk kinase and MyD88 adaptor protein pathways by bacteria promotes regulatory properties of neutrophils. Immunity 31: 761–71. [DOI] [PubMed] [Google Scholar]
- 94. Dillon S, Agrawal S, Banerjee K, Letterio J, Denning TL, et al. (2006) Yeast zymosan, a stimulus for TLR2 and dectin-1, induces regulatory antigen-presenting cells and immunological tolerance. J Clin Invest 116: 916–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Martorana D, Maritati F, Malerba G, Bonatti F, Alberici F, et al. (2012) PTPN22 R620W polymorphism in the ANCA-associated vasculitides. Rheumatology (Oxford) [DOI] [PubMed] [Google Scholar]
- 96. Cloutier JF, Veillette A (1996) Association of inhibitory tyrosine protein kinase p50csk with protein tyrosine phosphatase PEP in T cells and other hemopoietic cells. EMBO J 15: 4909–18. [PMC free article] [PubMed] [Google Scholar]
- 97. Roskoski R Jr (2005) Src kinase regulation by phosphorylation and dephosphorylation. Biochem Biophys Res Commun 331: 1–14. [DOI] [PubMed] [Google Scholar]
- 98. Rosenfeld MG, Lunyak VV, Glass CK (2006) Sensors and signals: a coactivator/corepressor/epigenetic code for integrating signal-dependent programs of transcriptional response. Genes Dev 20: 1405–28. [DOI] [PubMed] [Google Scholar]