Abstract
Survival in the majority of high grade astrocytoma (HGA) patients is very poor, with only a rare population of long-term survivors. A better understanding of the biological factors associated with long-term survival in HGA would aid development of more effective therapy and survival prediction. Factors associated with long-term survival have not been extensively studied using unbiased genome-wide expression analyses. In the present study, gene expression microarray profiles of HGA from long-term survivors were interrogated for discovery of survival-associated biological factors. Ontology analyses revealed that increased expression of immune function-related genes was the predominant biological factor that positively correlated with longer survival. A notable T-cell signature was present within this prognostic immune gene-set. Using immune cell-specific gene classifiers, both T-cell and myeloid linage-associated genes were shown to be enriched in HGA from long versus short-term survivors. Association of immune function and cell-specific genes with survival was confirmed independently in a larger publicly available glioblastoma gene expression microarray dataset. Histology was used to validate the results of microarray analyses in a larger cohort of long-term survivors of HGA. Multivariate analyses demonstrated that increased immune cell infiltration was a significant independent variable contributing to longer survival, as was Karnofsky/Lansky performance score. These data provide evidence of a prognostic anti-tumor adaptive immune response and rationale for future development of immunotherapy in HGA.
INTRODUCTION
Median survival in high grade astrocytoma (HGA), consisting predominantly of glioblastoma (GBM) and anaplastic astrocytoma (AA) is 15 months and 3 years respectively (1, 2). Few robust prognostic factors have been identified in HGA, hindering patient care and stratification in clinical trials. Currently established clinical risk factors include Karnofsky/Lansky performance score, a measure of patient wellbeing that is widely utilized in oncology, and age (3). O-6-methylguanine-DNA methyltransferase (MGMT) promoter methylation status and isocitrate dehydrogenase 1 (IDH1) mutational status have been established as molecular prognostic factors in adult HGA (4, 5).
Gene expression microarray analyses have been employed to identify novel prognostic biomarkers in HGA. This unbiased genome-wide approach has the additional benefit of providing insight into the biological mechanisms of tumorigenesis that can be exploited for the development of more effective therapies. Despite numerous studies that have identified prognostic gene signatures in HGA using microarray technology there remains no predictor of survival that has proven robustly reproducible from study to study (6-10). This may be due to the effects of biological heterogeneity inherent in HGA combined with the typically limited duration and range of survival.
While prognosis is poor for the majority of HGA, a small but discrete subgroup of long-term survivors exists, with 3-5% of GBM patients surviving longer than 3 years. The driving hypothesis for this study is that a more pronounced and biologically informative prognostic gene signature could be obtained by gene expression microarray analysis of long-term survivors of HGA. Using this approach, the present study identified immune function as the predominant ontology associated with long-term survival in pediatric and adult HGA. Histological validation in a larger cohort of long-term survivors demonstrated that increased infiltration of immune cells was prognostically favorable.
MATERIALS AND METHODS
Patient cohort and sample collection
Both adult and pediatric HGA samples, including GBM and AA, were included in this study cohort. Using broad age and diagnostic categories substantially increased the number of long-term survivors available to the study. This inclusive approach also has the potential to identify broad prognostic factors in HGA. Multivariate analyses were employed to address the confounding effect of age, tumor grade and a number of other potentially prognostic factors in this cohort.
For the initial discovery cohort (gene-expression microarray analysis), surgical tumor samples were obtained from 26 patients who presented between 1990 and 2008 for treatment of HGA at the University Hospital or The Children’s Hospital (Denver, CO) who were diagnosed with GBM or AA according to World Health Organization (WHO) guidelines (11). Tumor was either snap frozen in liquid nitrogen or placed in RNAlater storage solution (Qiagen, Valencia, CA) at the time of resection. Survival data was available for all patients in this study, which was conducted in compliance with institutional review board regulations (COMIRB 95-500 and 05-0149). Included in this study were 3 long-term HGA survivors (2 adult GBM and 1 pediatric GBM), with a median survival of 7.0 years (range 6.0-8.5 years). Clinical details are provided in Supplemental Table IA, including previously identified survival-associated factors of age at diagnosis, Karnofsky/Lansky performance score, diagnosis, tumor location, extent of surgery and therapy received. The microarray analysis control cohort consisted of 23 standard survival HGA samples (21 GBM and 2 AA) of which 18 were from pediatric patients. The median survival for this cohort was 12 months (range 1 to 40 months).
Validation of the results of gene-expression microarray analyses was performed in a publicly available Affymetrix genechip dataset used by Phillips et. al to identify molecular subclasses of high-grade glioma (GSE4271) (10). Gene expression profiles were available for 50 primary samples of GBM from deceased patients. The median survival of this cohort was 14.7 months (range 0.7-74 months), of which 8 survived longer than 3 years.
In the secondary validation cohort formalin-fixed paraffin-embedded (FFPE) material from 33 patients was obtained from archival diagnostic specimen banks of the pathology departments of the University Hospital or The Children’s Hospital. Included in this study were 14 long-term HGA survivors (12 GBM and 2 AA) of which 4 were pediatric. For the purposes of this study long-term survivors were defined as anyone who survived longer than 5 years with a diagnosis of GBM and 15 years with a diagnosis of AA. This long-term survival cohort included the same 3 long-term survivors used in the microarray study. The median survival for the HGA long term survivor cohort was 8.2 years (range 5.5-16.6 years). The control cohort consisted of 19 standard survival HGA samples (13 GBM, 6 AA) of which 10 were pediatric. The median survival for this cohort was 10 months (range 1-31 months).
Gene expression microarray analysis
Five micrograms of RNA that had been extracted from tumor was amplified, biotin-labeled (ENZO, Farmingdale, NY) and hybridized to Affymetrix HG-U133 Plus 2 microarray chips (Affymetrix, Santa Clara, CA). Analysis of gene expression microarray data was performed using Bioconductor functions written in the R programming language (http://www.bioconductor.org). Microarray data CEL files were background corrected and normalized using the GC Robust Multi-array Average (gcRMA) algorithm (12), resulting in log 2 expression values. The Affymetrix HG-U133 plus 2 microarray contains 54,675 probe sets including multiple probe sets for the same gene. To reduce error associated with multiple testing and to avoid bias in enrichment statistics from genes containing multiple probesets, a filtered list containing a single probe set for each gene that possessed the highest gcRMA expression level across all samples used was created (20,722 genes). The microarray data discussed in this publication have been deposited in NCBI’s Gene Expression Omnibus (13) and are publicly accessible through GEO Series accession number GSE33331 (http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33331).
GBM patient samples from the Phillips cohort (GSE4271) was analyzed on both Affymetrix HG-U133A and HG-U133B platforms. CEL files specific to the 2 platforms were normalized separately using gcRMA normalization. These datasets were then combined and filtered to obtain single probesets for each gene as described above.
Gene ontology analyses
Two computer-based ontology analysis tools were used to measure enrichment of specific gene functions: DAVID (Database for Annotation, Visualization and Integrated Discovery: http://david.abcc.ncifcrf.gov) (14) and GSEA (Gene Set Enrichment Analysis: http://www.broad.mit.edu/gsea) (15). Both analyses were used to assess gene lists for enrichment of genes annotated with specific Gene Ontology Project (GO) biological process terms (http://www.geneontology.org) (16). Enrichment is defined as more genes that are associated with a particular variable than would be expected by chance. Briefly, DAVID is a web-based resource that provides GO term fold-enrichment scores and p-values for lists of genes that have already been identified by the user as significantly associated with a particular variable. GSEA creates a ranked list of genes based on user selected criteria such as signal-to-noise, t-tests, etc. For the purposes of this study, genes were ranked on their correlation with survival time. Thus, unlike DAVID, GSEA takes into account the strength of association rather than simply whether a gene is significantly associated or not. The output for both ontology analysis tools is an enrichment score with associated Student’s t-test p-values for each GO term.
Enrichment analysis of specific immune cell lineage genes
Gene expression microarray data derived from surgical brain tumor specimens contain genes expressed by non-tumor cells of the tumor microenvironment. To address this, a technique was developed to identify and predict specific expression patterns of immune-related cells, based on data from previously isolated cell populations, should they be found within the tumor microenvironment of HGA. Classifier gene lists for discrimination of specific immune cell lineages were created for this study from publicly available Affymetrix HG-U133A gene expression cell and tissue profiles at BioGPS (17). This dataset consists of previously acquired gene expression data for isolated cell types including multiple immune cell lineages and brain tissues. The full BioGPS dataset was filtered to create a dataset consisting of 2 samples each from a range of immune cell lineages -dendritic cells (BDCA4), monocytes (CD14), B-cells (CD19), myeloid cells (CD33), helperT-cells (CD4), natural killer (NK) cells (CD56) and cytotoxic T-cells (CD8). Samples from normal brain tissues were also included as controls and consisted of 2 each of the following – amygdala, caudate nucleus, cerebellum, cerebellum peduncles, cingulate cortex, fetal brain, globus pallidus, hypothalamus, medulla oblongata, occipital lobe, olfactory lobe, parietal lobe, pituitary, pons, prefrontal cortex, spinal cord, subthalamic nucleus, superior cervical ganglion, temporal lobe, thalamus, trigeminal ganglion and whole brain. Immune lineage classifier gene lists were thus created for each immune cell lineage listed above. Fold change gene expression and associated p-values (Students 2-tailed t-test) were generated for each immune cell type profile (n=2) versus all other immune cell and brain tissue profiles combined (n=58). In addition, a combined T-cell lineage classifier was created by pooling both helper (CD4) and cytotoxic (CD8) T-cell samples and a combined myeloid lineage classifier by pooling monocyte (CD14) and myeloid cell (CD33) samples. Classifier genes were selected as those with p < 0.05, an increase over controls of greater than 15-fold and filtered to remove duplicate gene symbols (Supplemental Table II). The ability of classifier gene lists to distinguish discrete immune cell lineages was strengthened by using only genes that were 15-fold higher than controls.
Enrichment of HGA survival associated genes in gene-expression microaray validation dataset
HGA gene expression microarray profiles were filtered to generate gene lists that were either positively or negatively correlated with survival (p<0.05). Genes that were postitively correlated with survival were further filtered to contain only genes that were annotated with “immune response” GOterm (6955) (Supplemental Table II). The 3 HGA survival associated genesets were combined with the specific immune cell lineage classifier gene sets described above for GSEA analysis of the Phillips validation dataset.
Immunohistochemistry
Immunohistochemistry (IHC) was performed on 5 micron FFPE tumor tissue sections. Slides were deparaffinized and then subjected to optimal antigen retrieval protocols. Subsequent steps were performed using the EnVision-HRP kit (Dako, Glostrup, Denmark) on a Dako autostainer according to standard protocol. Incubation with primary antibody was performed for 2 hours. The following dilutions of primary antibody were used, and applied to the sections for 1 hour: 1:250 allograft inhibitory factor-1 (AIF1) (Cat#01-1974) from Waco Pure Chemicals (Richmond, VA); 1:2 Pre-diluted CD4 (SP35; Cat#104R-18) from Cell Marque (Rocklin, CA) and 1:100 CD8 (C8/144B; Cat#M7103) from Dako. Each of these antibodies stained a discrete subpopulation of cells that were distributed throughout the parenchyma of the tumor. Sections were counterstained with hematoxylin. Slides were analyzed with the Olympus BX40 microscope, x40 objective lens and x10 eyepiece (Olympus, Center Valley, CA). Images were captured using an Optronics MicroFire 1600 X 1200 camera and PictureFrame 2.3 imaging software (Optronics, Goleta, CA). Slides were scored in a blinded fashion, with infiltrating cell abundances measured as the mean number of positive staining cells per multiple fields of view (number dependent on sample size) at 400x magnification.
Statistical analysis
The Kaplan–Meier method was used to estimate the probability of survival as a function of time. Survival was calculated from the date of initial diagnosis to the date of death from any cause; patients alive at time of analysis were censored. Differences between survival curves were analyzed for significance using the log-rank test. Multivariate analysis of the relative importance of factors to survival was performed using the Cox proportional hazards method.
RESULTS
Genes positively correlated with long-term survival in HGA are predominantly immune-related
In the initial gene ontology analysis, DAVID was used to identify enriched biological functions in genes associated with survival as a continuous variable. As input for DAVID ontology analysis, a list of 1106 genes that positively correlated (p<0.05 estimated by Pearson correlation test) with survival in HGA (n=26) was created from all 20,722 genes. Using the same approach, a list of 469 genes that were negatively correlated with survival was also created. DAVID showed that the predominant ontology in genes positively correlated with survival were immune-related (Table I). Cell cycle and M-phase related ontologies were found to be enriched in genes that were negatively correlated with survival. These cell cycle ontologies reached greater statistical significance than the positive survival correlate enriched ontologies. GSEA was used as a secondary analysis of functional enrichment in survival associated gene lists. Similar to the DAVID results, GSEA identified 3 of the highest 10 ontologies in genes that positively correlated with survival as immune-related (Table I). In the reverse analysis, amongst those genes that were negatively correlated with survival, the highest enriched GO terms were cell cycle-related.
Table I.
Ontologic analyses of genes associated with survival in high grade astrocytoma*
| GOTERM Annotation | GOterm ID# | Enrichment | ||
|---|---|---|---|---|
| Fold- | p-value | |||
| DAVID ontology analysis | ||||
| Positively correlated with survival | ||||
| 1 | Immune system process | 2376 | 2.69 | 2.16×10−32 |
| 2 | Immune response | 6955 | 3.02 | 1.61×10−29 |
| 3 | Regulation of immune system process | 2682 | 2.84 | 4.27×10−14 |
| 4 | Defense response | 6952 | 2.35 | 1.50×10−13 |
| 5 | Regulation of immune response | 50776 | 3.42 | 4.76×10−13 |
| 6 | Positive regulation of immune system process | 2684 | 3.32 | 8.00×10−13 |
| 7 | Cell activation | 1775 | 3.04 | 1.34×10−12 |
| 8 | Response to external stimulus | 9605 | 1.99 | 5.26×10−12 |
| 9 | Response to wounding | 9611 | 2.35 | 1.04×10−11 |
| 10 | Inflammatory response | 6954 | 2.79 | 1.51×10−11 |
| Negatively correlated with survival | ||||
| 1 | M phase | 279 | 8.54 | 1.00×10−48 |
| 2 | Cell cycle phase | 22403 | 7.23 | 3.05×10−46 |
| 3 | Cell cycle | 7049 | 5.05 | 6.54×01−46 |
| 4 | Cell cycle process | 22402 | 5.82 | 4.76×01−43 |
| 5 | M phase of mitotic cell cycle | 87 | 9.73 | 6.47×10−41 |
| 6 | Nuclear division | 280 | 9.74 | 3.17×01−40 |
| 7 | Mitosis | 7067 | 9.74 | 3.17×10−40 |
| 8 | Organelle fission | 48285 | 9.36 | 3.55×10−39 |
| 9 | Mitotic cell cycle | 278 | 6.79 | 1.03×01−36 |
| 10 | Cell division | 51301 | 6.51 | 5.58×01−27 |
| GOTERM Annotation | GOterm ID# | Enrichment | ||
|---|---|---|---|---|
| NES | p-value | |||
| GSEA ontology analysis | ||||
| Positively correlated with survival | ||||
| 1 | Generation of precursor metabolites and energy | 6091 | 1.94 | 0.0112 |
| 2 | Immune system process | 2376 | 1.92 | 0.0421 |
| 3 | Immune response | 6955 | 1.90 | 0.0521 |
| 4 | Response to drug | 42493 | 1.89 | 0.0137 |
| 5 | Protein oligomerization | 51295 | 1.88 | 0.0056 |
| 6 | Humoral immune response | 6959 | 1.84 | 0.0264 |
| 7 | Regulation of protein secretion | 50708 | 1.83 | 0.0041 |
| 8 | Positive regulation of MAP kinase activity | 43407 | 1.78 | 0.0161 |
| 9 | Monocarboxylic acid metabolic process | 32787 | 1.74 | 0.0372 |
| 10 | Response to chemical stimulus | 42221 | 1.73 | 0.0609 |
| Negatively correlated with survival | ||||
| 1 | Protein amino acid autophosphorylation | 46777 | 2.07 | 0.0020 |
| 2 | Protein autoprocessing | 16540 | 1.99 | 0.0040 |
| 3 | Establishment and/or maintenance of chromatin architecture |
6325 | 1.75 | 0.0565 |
| 4 | Microtubule cytoskeleton organization and biogenesis |
226 | 1.72 | 0.0362 |
| 5 | Cell cycle process | 22402 | 1.68 | 0.0971 |
| 6 | Cell cycle phase | 22403 | 1.67 | 0.0937 |
| 7 | Chromosome organization and biogenesis | 51276 | 1.65 | 0.0917 |
| 8 | mRNA processing | 6397 | 1.65 | 0.0891 |
| 9 | DNA replication | 6260 | 1.63 | 0.0886 |
| 10 | DNA recombination | 6310 | 1.62 | 0.0779 |
The top 10 enriched ontologies for positive and negative correlates of survival using DAVID and GSEA ranked according to p-value and NES, respectively.
Abbreviations: GSEA, Gene Set Enrichment Analysis; DAVID, Database for Annotation, Visualization and Integrated Discovery; NES, normalized enrichment score.
Ontological analysis using GO is dependent on curator driven annotation of gene functions based on traceable author statements and/or inferences from electronic annotations. As a consequence, genes that have not yet been annotated are ignored by the computer based ontological analyses described above. Therefore, the 1106 genes positively correlated with survival genes used above were manually reviewed to identify those with a documented predominant role in specific immune mechanisms. This process identified 19% (205/1106) genes that were related to immune function. The results of this analysis, with genes listed and categorized into subgroups according to their documented role in specific immune mechanisms, are provided in Supplemental Table IB. Notably, 99 immune-related genes (48%) were found that did not have an annotation of “immune response” (GOterm 6955) , including a number of genes of key CD immune markers such as CD2, CD3, CD33 and CD40, underscoring the importance of manual review of gene lists as performed here.
A number of notable genes associated with the adaptive immune system were identified in those genes positively correlated with longer survival in HGA. In particular a large number of genes known to be expressed by T-cells (CD3D, CD3E, CD3G, CD8B, TRAC, TRAT1, VAV1 and ZAP70) were shown to be associated with long-term survival. Additionally, multiple components of the innate immune system, including genes associated with microglia/macrophages (AIF1, CD68, CD86, CIITA, HLA-DOA, HLA-DQB2, HLA-DRB1, HLA-DRB6, NOD2) were present in this prognostic immune gene-set. Several toll-like receptors were also associated with long-term survival (TLR2, 3, 5, 6, 7 and 8).
Known negative regulators of immune function in gliomas were examined to identify any association with shorter survival in HGA. Interleukin-10 (IL-10) was found to be associated with longer survival (p=0.0127), and no significant survival association was observed for IL-13, FOXP3, STAT3, TGFB1 or TGFB2 gene expression.
Lymphoid lineage specific genes are enriched in HGA long-term survivors
To clarify which immune cell types might account for the immune function enrichment identified above, genes uniquely specific to each immune cell lineage were identified using existing publicly available gene expression data. Gene expression microarray data obtained from purified immune cell lineages were used to created classifier gene sets. These classifier genesets were then used in combination with GSEA to indicate the presence of specific types of infiltrating immune cells in HGA from long term survivors. This analysis revealed that lymphoid cell lineage classifiers helper T-cell (CD4), combined T-cell (CD4 and CD8) and natural killer cell (CD56) were statistically significantly enriched (p<0.05) in the long-term survivor associated genes (Table II). Cytotoxic T-cells (CD8) and myeloid lineages (combined myeloid lineage (CD14 and CD33), myeloid cell (CD33) and monocyte (CD14)) approached significance (p<0.1). B-cells (CD19) and dendritic cells (BDCA4) were the least enriched cell types. Conversely, none of these immune cell lineage classifiers were enriched in the genes associated with shorter survival in HGA.
Table II.
Immune cell lineage enrichment analysis of genes correlated with survival in high grade astrocytoma.
| Immune cell lineage | Enrichment | ||
|---|---|---|---|
| NES | p-value | ||
| Positively correlated with survival | |||
| 1 | CD4 helper T-cell | 2.02 | 0.0197 |
| 2 | T-cell lineage (CD4 and CD8 combined) | 1.96 | 0.0430 |
| 3 | CD56 natural killer cell | 1.95 | 0.0406 |
| 4 | Myeloid lineage (CD14 and CD33 combined) | 1.74 | 0.0559 |
| 5 | CD8 cytotoxic T-cell | 1.65 | 0.0986 |
| 6 | CD33 myeloid cell | 1.65 | 0.0824 |
| 7 | CD14 monocyte | 1.61 | 0.0936 |
| 8 | CD19 B-cell | 1.41 | 0.1684 |
| 9 | BDCA4 dendritic cell | 1.32 | 0.2119 |
| Negatively correlated with survival | |||
| No genesets were enriched | |||
Validation of survival associated immune response gene expression signature
Following primary analysis of the in-house HGA dataset a publicly available gene expression microarray dataset (Phillips; GSE4271) was used to confirm our findings (10). The patient cohort represented by this dataset contained 50 primary GBM that were from deceased patients of which 8 survived longer than 3 years (range 3.5-6.2 years).
This dataset was subject to the same ontology and immune cell lineage analyses as described above. Expression of all 19110 HG-U133A and U133B best expressed probesets in the Phillips dataset were correlated (Pearson’s) with survival as a continual variable. This approach identified 1391 genes that were positively correlated with survival (p<0.05). Ontology analysis of these genes using DAVID identified immune response as the most highly enriched biological process associated with long-term survivor GBM samples (Table III). Similar to the DAVID results, GSEA identified 2 of the highest 10 ontologies in genes that positively correlated with survival as inflammation and immune-related (Table III). These results match the results of the in-house analysis thus supporting the hypothesis that host immunity contributes significantly to long-term survival in HGA. In the converse analysis, 1181 genes were significantly correlated with shorter survival. DAVID and GSEA analyses demonstrated that the predominant ontologies associated with short-term survival were mitosis-related, again matching the results of the in-house dataset analysis (Table III). Enrichment of genes associated with specific immune cell lineages in the Phillips dataset was measured using GSEA as described above, but with the addition of HGA positive and negative correlate genesets generated from our in-house dataset (n=1106 and n=469 respectively). A third geneset was created by using only “immune response” (GOterm 6955) annotated genes that were significantly positively correlated survival (n=117). Seven of the 9 immune cell lineages were shown to be associated with long-term survival (Supplemental table IC). In contrast to the HGA in-house dataset, monocyte and myeloid linage-associated genes were significantly enriched in long-terms survivors in the Phillips dataset. HGA positive and negative survival-correlate genes were significantly enriched in the corresponding Phillips dataset survival correlated genes. Of note, a higher enrichment was seen in “immune response” annotated HGA positive correlates than in the full HGA positive correlate geneset.
Table III.
Ontology analyses of genes associated with survival in glioblastoma in the Phillips (GSE4271) dataset*
| GOTERM Annotation | GOterm ID# | Enrichment | ||
|---|---|---|---|---|
| Fold- | p-value | |||
| DAVID ontology analysis | ||||
| Positively correlated with survival | ||||
| 1 | Immune response | 6955 | 1.57 | 2.45 ×10−4 |
| 2 | Defense response | 6952 | 1.58 | 4.79 ×10−4 |
| 3 | Regulation of transcription from RNA polymerase II promoter |
6357 | 1.51 | 6.12 ×10−4 |
| 4 | Enzyme linked receptor protein signaling pathway | 7167 | 1.70 | 0.00230 |
| 5 | Vascular development | 1944 | 1.80 | 0.00352 |
| 6 | Cellular defense response | 6968 | 2.91 | 0.00392 |
| 7 | Taxis | 42330 | 2.02 | 0.00464 |
| 8 | chemotaxis | 6935 | 2.02 | 0.00464 |
| 9 | Blood vessel development | 1568 | 1.78 | 0.00494 |
| 10 | Positive regulation of protein kinase activity | 45860 | 1.81 | 0.00569 |
| Negatively correlated with survival | ||||
| 1 | DNA metabolic process | 6259 | 3.10 | 6.67 ×10−23 |
| 2 | Cell cycle | 7049 | 2.43 | 6.14 ×10−19 |
| 3 | DNA replication | 6260 | 4.17 | 7.75 ×10−17 |
| 4 | Cell cycle process | 22402 | 2.59 | 2.95 ×10−16 |
| 5 | Response to DNA damage stimulus | 6974 | 2.98 | 2.14 ×10−15 |
| 6 | Cell cycle phase | 22403 | 2.77 | 3.34 ×10−14 |
| 7 | DNA repair | 6281 | 3.20 | 5.34 ×10−13 |
| 8 | M phase | 279 | 2.92 | 5.66 ×10−13 |
| 9 | Mitotic cell cycle | 278 | 2.73 | 2.33 ×10−12 |
| 10 | Cell division | 51301 | 2.86 | 4.11 ×10−11 |
| GOTERM Annotation | GOterm ID# | Enrichment | ||
|---|---|---|---|---|
| NES | p-value | |||
| GSEA ontology analysis | ||||
| Positively correlated with survival | ||||
| 1 | Peptidyl tyrosine modification | 18212 | 2.20 | < 0.001 |
| 2 | Peptidyl tyrosine phosphorylation | 18108 | 2.11 | < 0.001 |
| 3 | Regulation of peptidyl tyrosine phosphorylation | 50730 | 2.08 | < 0.001 |
| 4 | Regulation of protein amino acid phosphorylation | 1932 | 1.98 | < 0.001 |
| 5 | Inflammatory response | 6954 | 1.93 | < 0.001 |
| 6 | Positive regulation of protein amino acid phosphorylation |
1934 | 1.86 | < 0.001 |
| 7 | JAK-STAT cascade | 7259 | 1.81 | 0.00361 |
| 8 | Defense response | 6952 | 1.80 | < 0.001 |
| 9 | Small GTPase mediated signal transduction | 7264 | 1.78 | < 0.001 |
| 10 | Ras protein signal transduction | 7265 | 1.77 | 0.00167 |
| Negatively correlated with survival | ||||
| 1 | Cell cycle process | 22402 | 2.75 | < 0.001 |
| 2 | Cell cycle phase | 22403 | 2.74 | < 0.001 |
| 3 | M phase | 279 | 2.64 | < 0.001 |
| 4 | Mitotic cell cycle | 278 | 2.60 | < 0.001 |
| 5 | DNA-dependent DNA replication | 6261 | 2.59 | < 0.001 |
| 6 | Cell cycle checkpoint | 75 | 2.47 | < 0.001 |
| 7 | Cell cycle | 7049 | 2.46 | < 0.001 |
| 8 | Meiotic cell cycle | 51321 | 2.45 | < 0.001 |
| 9 | Chromosome segregation | 7059 | 2.41 | < 0.001 |
| 10 | Mitosis | 7067 | 2.39 | < 0.001 |
The top 10 enriched ontologies for positive and negative correlates of survival using DAVID and GSEA ranked according to p-value and NES, respectively.
Abbreviations: DAVID, Database for Annotation, Visualization and Integrated Discovery; GSEA, Gene Set Enrichment Analysis; NES, normalized enrichment score.
Immune-related genes associated positively correlated with survival in the Phillips were compared to those indentified in the HGA dataset. Fifteen “immune response” GOterm annotated genes were shared between the two datasets. These included myeloid-lineage expressed genes GPR183, IRF8, IL6R, KYNU, NCF4, STXBP2, TNFAIP8L2 and TNFSF13 (APRIL) based on BioGPS, suggesting a common myeloid-lineage enrichment in long-term survivors from the 2 datasets. Those immune-related genes that distinguished the Phillips from the HGA dataset included a number of Fc-receptors (CD16A and B, CD32, CD64) and MHC class-I HLA-A. MHC class-II and T-cell associated genes that had been identified in the HGA geneset were absent from the Phillips dataset. Negative regulators of immune function were studied to identify any association with shorter survival in the Phillips datasets. Interleukin-4 (IL-4) was found to be associated with shorter survival (p=0.0175) supporting a link between TH2 immune response polarization and a shorter survival. Conversely, FOXP3 and TGFB1 were significantly associated with long-term survival (p=0.0203 and p=0.0143 respectively). No significant survival association was observed for STAT3, IL-10, IL13 or TGFB2 gene expression.
In a histological validation cohort of HGA, immune cell infiltration and Karnofsky/Lansky performance score contribute to long term survival
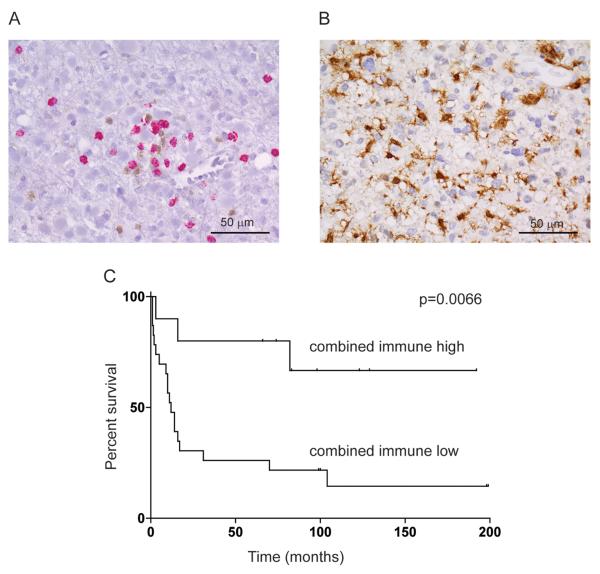
Based on the results of microarray analysis of long-term survivors of HGA, it was hypothesized that increased tumor infiltration of microglia and/or T-cells is associated with longer survival. To validate this hypothesis IHC was used to measure immune cell infiltration in a larger cohort of long-term survivors of HGA. The patient cohort for this study included 14 long-term survivors (median survival 8.5 years; range 5.5-17 years) and 19 patients whose survival was in the typical range (median survival 9 months; range 1-31 months). Immunostaining of FFPE diagnostic samples was performed for CD8 and CD4, specific markers for cytotoxic T-cells and helper T-cells respectively (Figure 1A). The association of a significant number of putative myeloid/monocyte function and lineage-specific genes with long term survival in HGA also implied the presence of infiltrating microglia/macrophages in these tumors. To confirm the presence of microglia/macrophages, the most abundant myeloid-linage cells in the central nervous system (CNS), immunostaining for AIF1was performed (Figure 1B). AIF1, also known as Iba1, is a specific marker for the microglia/macrophage population in the central nervous system (18). Frequency of infiltration of specific immune cells was scored as number of stained cells per 400x field of vision. Immunostaining representing high and low T-cell and microglia/macrophage infiltration are provided in Supplemental Figure 1. Additional known survival associated factors - age at diagnosis, Karnofsky/Lansky performance score, tumor grade, location, extent of surgery and therapy received - were included in the survival analyses to address any potential confounding factors.
Figure 1.
Representative histology of (A) greater than median tumor infiltration of cytotoxic T cells (CD8; red) and helper T-cells (CD4; brown) in long term survivor HGA11, and (B) greater than 75th percentile tumor infiltration of microglia/macrophages (AIF1; brown) in long term survivor HGA12. Immunohistochemistry was performed using FFPE tumor sections with hematoxylin counterstaining (400x magnification). (C) Kaplan-Meier survival analysis of combined immune cell infiltration. High combined immune cell infiltration was defined as greater than median cytotoxic T-cell or helper T-cell or greater than 75th percentile microglia/macrophage infiltration. Thirty three HGA samples were used.
Univariate Kaplan-Meier analysis was first used to identify factors that were significantly associated with survival. Factors addressed in this study consisted of non-continuous variables (tumor grade, adult/pediatric, radiation treatment, temozolomide treatment, supratentorial location, thalamic location and gross total resection) and continuous variables that were divided at the mean value into high / low (helper T-cell (CD4) infiltration, cytotoxic T-cell (CD8) infiltration, microglia/macrophage (AIF1) infiltration, age at diagnosis and Karnofsky/Lansky performance score). Factors significantly associated with survival were performance score greater than the mean of 80 (p=0.00387), cytotoxic T-cell infiltration greater than the mean (p=0.0154) and helper T-cell infiltration greater than mean (p=0.0272). Mean cytotoxic and helper T-cell infiltration was 1.26 (range 8.2-0) and 1.98 (range 37-0) cells per 400x field of view respectively. Association of survival with greater than mean microglia/macrophage infiltration showed a trend toward significance (p=0.089). To further investigate the influence of microglia/macrophage infiltration on survival the cutoff for high infiltration in this cell population was made more stringent by using > 75th percentile, which showed a significant association with survival (p=0.0146). The 75th percentile score for microglia/macrophage infiltration was 100 (range 220-0) cells per 400x field of view. None of the remaining factors showed a significant association with survival.
Significant variables were next subject to multivariate analysis. Each of the 3 immune cell population infiltrations showed a significant correlation with each other by either Pearson’s or Spearman’s approaches. Since these factors were not therefore independent it was considered appropriate to also combine them (microglia/macrophage > 75th percentile or cytotoxic T-cell > mean or helper T-cell > mean) into one measure as a single variable. By Kaplan-Meier analysis the combined immune cell variable was significantly associated with survival (p=0.0066) (Fig. 1C).
Combined immune cell and individual immune cell variables were modeled with Karnofsky/Lansky performance score and analyzed using Cox’s proportional hazard (Table IV). This analysis showed that when combined with performance score each of the immune variables apart from helper T-cell infiltration were independent variables for survival. The best model appeared to consist of performance and combined immune cell infiltration, with both factors contributing significantly to longer survival (p = 0.00268 and 0.00772, respectively) (Table IV).
Table IV.
Multivariate survival model of Karnofsky/Lansky performance score and immune cell infiltration using Cox’s proportional hazards regression analysis.
| Model | Variable | Hazard ratio |
95% Confidence interval |
p-value |
|---|---|---|---|---|
| Karnofsky/Lansky performance and microglia/macrophage infiltration (AIF1) | ||||
| Performance > mean | 0.216 | 0.086-0.538 | 0.00101 | |
| AIF1 > 75 percentile | 0.138 | 0.031-0.618 | 0.00963 | |
| Karnofsky/Lansky performance and helper T-cell infiltration (CD4) | ||||
| Performance > mean | 0.402 | 0.169-0.957 | 0.0394 | |
| CD4 > mean | 0.202 | 0.026-1.59 | 0.1282 | |
| Karnofsky/Lansky performance and cytotoxic T-cell (CD8) | ||||
| Performance > mean | 0.317 | 0.132-0.762 | 0.0102 | |
| CD8 > mean | 0.206 | 0.047-0.913 | 0.0375 | |
| Karnofsky/Lansky performance and combined immune (AIF1 > 75 percentile or CD4 > mean or CD8 > mean) | ||||
| Performance > mean | 0.253 | 0.103-0.620 | 0.00268 | |
| Combined immune | 0.183 | 0.0523-0.638 | 0.00772 | |
DISCUSSION
HGA are heterogeneous but almost uniformly fatal with only a small percentage of long-term survivors. An unbiased genome-wide microarray approach, with validation in an independent dataset, was employed to analyze this broad diagnostic class to determine whether increased enrichment of immune genes and cells was associated with better patient survival as had been seen in a similar study of another brain tumor, ependymoma (19). The results of this study showed that this was indeed the case, with immune function ontologies, immune cell lineage specific genes and infiltrating immune cell frequency being significantly associated with long term survival in HGA. These data provide additional support to the theory that host immunity can control tumor growth in a subset of HGA, and provides a potential explanation for instances of long-term survival in this otherwise uniformly and rapidly fatal disease.
A number of large-scale gene expression clustering studies have identified sets of genes reportedly predictive of prognosis in HGA. Rich et al. identified a migration/invasion molecular signature that was associated with a worse survival in GBM (6). Using an agglomerative approach, Phillips et al. defined three molecular subgroups of HGA (proliferative, proneural and mesenchymal) one of which conveyed a better prognosis (10). However, the gene sets indentified in these studies share few genes in common. To address this problem, Zhang et al. combined a number of independent data sets and identified a 23-gene classifier that out-performed previously reported classifiers from the independent cohorts (9). Of note, 2 of 23 genes were immune-related, the remaining 21 being related to cellular proliferation.
The majority of studies aimed at identifying prognostic factors in HGA have used standard survival patient cohorts that contain few long term survivors. The narrow survival range associated with these cohorts presents difficulty in assigning good and bad outcomes. The inclusion of a large proportion of long-term survivors in the present study, although not representative of natural survival distribution in HGA, provides a wider survival range and thus more clearly delineated good and bad outcomes. These more clearly defined survival cohorts allow for stronger inferences to be made from associated gene expression analyses. To explore this hypothesis one other study, in addition to the present, compared primary GBM from 7 long-term survivors (> 2 years) to 13 short-term survivors (< 9 months) (20). Using a genomic approach they identified a prognostic fingerprint of 43 genes. Consistent with the present study 6 survival-associated immune genes were identified (CD34, IGHG1, IL13RA1, IL17, IL22 and SERPING1) of which 2 were also associated with longer survival in the present analysis (IL17 and SERPING1).
Data from the present study which identified apparently reciprocal prognostic cell cycle and immune functions, supports the theory that an equilibrium may exist between immune system and tumor, known as immunoediting (21). A shift in this equilibrium can either result in complete elimination of tumor by the immune system, or alternatively escape from immune control leading to tumor progression. In long-term survivors of HGA, therapeutic intervention (surgery, radiation and/or chemotherapy) may have skewed this equilibrium in favor of the host-immune system, resulting in clinically significant tumor control. Opposing prognostic cell cycle and immune function gene sets have previously been observed in GBM and ependymoma (9, 19).
That genes related to immune functions are associated with long-term survivors of HGA imply that the host immune system may be involved with controlling tumor growth in these cases. Further details of the specific immunological mechanism through which this occurs is of significant interest, potentially providing a basis for the design of more effective immunotherapeutic strategies. A rational approach to improving immunotherapeutic approaches would be to design strategies based on data taken from direct clinical studies of human host anti-CNS tumor immune responses. This report potentially illustrates just such a response, underscoring the value and potential impact of these findings.
The data described in the present study can provide some mechanistic detail of a putative antitumor immune response in the human CNS. Biological inferences can be gained by examining the specific immune function genes and cell types that are associated with HGA long-term survivors and by comparison with the good-outcome associated immune genes found in other CNS tumor tumors. An earlier study by our group identified a similar association of immune gene and cell enrichment in EPN, a glial tumor of childhood (19). EPN commonly has a less aggressive phenotype than HGA, with a recurrence rate of approximately 50%. A number of T-cell specific genes are seen in the HGA survival associated genes that were not identified in EPN outcome associated immune genes. This includes multiple components of the T-cell immune synapse (CD2, CD3D, CD3E, CD3G, CD8B, TCRGC2, TRBC1, TARP and TRAT1), cytotoxic mediators (Granzyme B, H, K and M) and key signaling molecules restricted to activated T-cells (CD69, ZAP70, CARD11 and VAV1). The presence of CD8B and cytotoxic mediators, in addition to above listed T-cell specific genes, implies that cytotoxic T-cells (CTLs) are an important cellular correlate of survival in HGA. This theory is supported by multivariate analysis of a larger cohort of HGA which showed that frequency of tumor infiltrating cytotoxic T-cells (CD8) was significantly associated with longer survival.
Some of the immune genes up-regulated in long-term survival HGA are associated with specific T-cell functions. Prior studies have demonstrated that the majority of T cells within most human GBM are Th2-biased; however, these data were not generated from long-term survivors(22). Polarization of infiltrating T-cells in long-term survivors to the Th1 phenotype is implied by the presence of GIMAP4, HAVCR2 (TIM3) and STAT4 (23-25). Together, these data provide preliminary evidence that, beyond the simple presence of an immune infiltrate, the phenotype and function of that infiltrate may influence survival in HGA. This conclusion is consistent with the report by Galon et al. demonstrating that the type (specifically Th1), density, and location of immune cells within human colorectal tumors predict clinical outcome better than current staging criteria (26). A number of histological studies have been performed to assess the role of the host immune system in control of HGA growth and patient outcome. Three studies identified a positive correlation of lymphocyte infiltration with outcome (27-29). The results of three other studies however showed no correlation (30, 31) or a negative correlation(32). It should be noted that these studies relied on morphometry to identify immune infiltrates and unlike the present study were not able distinguish subtypes of T-cells nor identify microglia/macrophages.
Much attention in the study of the interaction of HGA with the host immune system has been devoted to immunosuppressive factors that have shown to contribute to a poor prognosis. A number of soluble factors released by HGA most notably TGFβ, IL-4, IL-10 and IL-13 have been shown to be immunosuppressive (33-35). The present study demonstrated that IL-4 expression correlated with shorter-survival but no other immunosuppressive factors were shown to be associated with a worse outcome. More recently, attention has been focused on a number of specific immune cells shown to contribute to an immunosuppressive milieu in the tumor. These include Tregs, myeloid-derived suppressive cells and most-recently, neutrophils (36-40). STAT3 induced signaling in gliomas has been implicated in induction of inflammation and mesenchymal transformation resulting in a poor prognosis (41, 42). No correlation of these immunosuppressive cells and signaling pathways with a poor prognosis was observed in either dataset in the present analysis.
The findings of the present study provide evidence that, despite the immunosuppressive nature of HGA, immunologically mediated control of tumor growth may arise in some instances, resulting in prolonged survival. Further characterization of the mechanism of immune control of HGA in long-term survivors is warranted and may aid in the design of more effective immunotherapies.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Patsy Ruegg at IHCtech for assistance with immunohistochemistry and Liza Litzenberger for photographic expertise.
1. Source of support: This work was supported by the Morgan Adams Foundation and National Institutes of Health grant 1 R01 CA140614-01A1.
2. Abbreviations
- HGA
high grade astrocytoma
- GBM
glioblastoma
- AA
anaplastic astrocytoma
- FFPE
formalin-fixed formalin-embedded
- DAVID
Database for Annotation, Visualization and Integrated Discovery
- GSEA
Gene Set Enrichment Analysis
- GO
Gene Ontology Project
- AIF1
allograft inhibitory factor-1
- IHC
immunohistochemistry
- CNS
central nervous system.
REFERENCES
- 1.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, Gorlia T, Allgeier A, Lacombe D, Cairncross JG, Eisenhauer E, Mirimanoff RO. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352:987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Prados MD, Scott C, Curran WJ, Jr., Nelson DF, Leibel S, Kramer S. Procarbazine, lomustine, and vincristine (PCV) chemotherapy for anaplastic astrocytoma: A retrospective review of radiation therapy oncology group protocols comparing survival with carmustine or PCV adjuvant chemotherapy. J Clin Oncol. 1999;17:3389–3395. doi: 10.1200/JCO.1999.17.11.3389. [DOI] [PubMed] [Google Scholar]
- 3.Curran WJ, Jr., Scott CB, Horton J, Nelson JS, Weinstein AS, Fischbach AJ, Chang CH, Rotman M, Asbell SO, Krisch RE, et al. Recursive partitioning analysis of prognostic factors in three Radiation Therapy Oncology Group malignant glioma trials. J Natl Cancer Inst. 1993;85:704–710. doi: 10.1093/jnci/85.9.704. [DOI] [PubMed] [Google Scholar]
- 4.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 5.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rich JN, Hans C, Jones B, Iversen ES, McLendon RE, Rasheed BK, Dobra A, Dressman HK, Bigner DD, Nevins JR, West M. Gene expression profiling and genetic markers in glioblastoma survival. Cancer Res. 2005;65:4051–4058. doi: 10.1158/0008-5472.CAN-04-3936. [DOI] [PubMed] [Google Scholar]
- 7.Liang Y, Diehn M, Watson N, Bollen AW, Aldape KD, Nicholas MK, Lamborn KR, Berger MS, Botstein D, Brown PO, Israel MA. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci U S A. 2005;102:5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serao NV, Delfino KR, Southey BR, Beever JE, Rodriguez-Zas SL. Cell cycle and aging, morphogenesis, and response to stimuli genes are individualized biomarkers of glioblastoma progression and survival. BMC Med Genomics. 2011;4:49. doi: 10.1186/1755-8794-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang J, Liu B, Jiang X, Zhao H, Fan M, Fan Z, Lee JJ, Jiang T, Song SW. A systems biology-based gene expression classifier of glioblastoma predicts survival with solid tumors. PLoS One. 2009;4:e6274. doi: 10.1371/journal.pone.0006274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phillips HS, Kharbanda S, Chen R, Forrest WF, Soriano RH, Wu TD, Misra A, Nigro JM, Colman H, Soroceanu L, Williams PM, Modrusan Z, Feuerstein BG, Aldape K. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9:157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 11.McLendon RE, W. O, Kros JM, Korshunov A, Ng H-K. In: Pathology and Genetics of Tumours of the Nervous System. Cavenee PKWK, editor. IARCPress; Lyon: 2007. [Google Scholar]
- 12.Wu Z, I. R, Gentleman R, Murillo F Martinez, Spencer F. A model-based background adjustment for oligonucleotide expression arrays. J Am Stat Assoc. 2004;99:909–917. [Google Scholar]
- 13.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dennis G, Jr., Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 15.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, Hodge CL, Haase J, Janes J, Huss JW, 3rd, Su AI. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biol. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada M, Ohsawa K, Imai Y, Kohsaka S, Kamitori S. X-ray structures of the microglia/macrophage-specific protein Iba1 from human and mouse demonstrate novel molecular conformation change induced by calcium binding. J Mol Biol. 2006;364:449–457. doi: 10.1016/j.jmb.2006.09.027. [DOI] [PubMed] [Google Scholar]
- 19.Donson AM, Birks DK, Barton VN, Wei Q, Kleinschmidt-Demasters BK, Handler MH, Waziri AE, Wang M, Foreman NK. Immune gene and cell enrichment is associated with a good prognosis in ependymoma. J Immunol. 2009;183:7428–7440. doi: 10.4049/jimmunol.0902811. [DOI] [PubMed] [Google Scholar]
- 20.Marko NF, Toms SA, Barnett GH, Weil R. Genomic expression patterns distinguish long-term from short-term glioblastoma survivors: a preliminary feasibility study. Genomics. 2008;91:395–406. doi: 10.1016/j.ygeno.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 21.Swann JB, Smyth MJ. Immune surveillance of tumors. J Clin Invest. 2007;117:1137–1146. doi: 10.1172/JCI31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Waziri A, Killory B, Ogden AT, 3rd, Canoll P, Anderson RC, Kent SC, Anderson DE, Bruce JN. Preferential in situ CD4+CD56+ T cell activation and expansion within human glioblastoma. J Immunol. 2008;180:7673–7680. doi: 10.4049/jimmunol.180.11.7673. [DOI] [PubMed] [Google Scholar]
- 23.Monney L, Sabatos CA, Gaglia JL, Ryu A, Waldner H, Chernova T, Manning S, Greenfield EA, Coyle AJ, Sobel RA, Freeman GJ, Kuchroo VK. Th1-specific cell surface protein Tim-3 regulates macrophage activation and severity of an autoimmune disease. Nature. 2002;415:536–541. doi: 10.1038/415536a. [DOI] [PubMed] [Google Scholar]
- 24.Filen JJ, Filen S, Moulder R, Tuomela S, Ahlfors H, West A, Kouvonen P, Kantola S, Bjorkman M, Katajamaa M, Rasool O, Nyman TA, Lahesmaa R. Quantitative proteomics reveals GIMAP family proteins 1 and 4 to be differentially regulated during human T helper cell differentiation. Mol Cell Proteomics. 2009;8:32–44. doi: 10.1074/mcp.M800139-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nishikomori R, Usui T, Wu CY, Morinobu A, O’Shea JJ, Strober W. Activated STAT4 has an essential role in Th1 differentiation and proliferation that is independent of its role in the maintenance of IL-12R beta 2 chain expression and signaling. J Immunol. 2002;169:4388–4398. doi: 10.4049/jimmunol.169.8.4388. [DOI] [PubMed] [Google Scholar]
- 26.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pages F. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 27.Brooks WH, Markesbery WR, Gupta GD, Roszman TL. Relationship of lymphocyte invasion and survival of brain tumor patients. Ann Neurol. 1978;4:219–224. doi: 10.1002/ana.410040305. [DOI] [PubMed] [Google Scholar]
- 28.Palma L, Di Lorenzo N, Guidetti B. Lymphocytic infiltrates in primary glioblastomas and recidivous gliomas. Incidence, fate, and relevance to prognosis in 228 operated cases. J Neurosurg. 1978;49:854–861. doi: 10.3171/jns.1978.49.6.0854. [DOI] [PubMed] [Google Scholar]
- 29.Boker DK, Kalff R, Gullotta F, Weekes-Seifert S, Mohrer U. Mononuclear infiltrates in human intracranial tumors as a prognostic factor. Influence of preoperative steroid treatment. I. Glioblastoma. Clin Neuropathol. 1984;3:143–147. [PubMed] [Google Scholar]
- 30.Schiffer D, Cavicchioli D, Giordana MT, Palmucci L, Piazza A. Analysis of some factors effecting survival in malignant gliomas. Tumori. 1979;65:119–125. doi: 10.1177/030089167906500114. [DOI] [PubMed] [Google Scholar]
- 31.Rossi ML, Jones NR, Candy E, Nicoll JA, Compton JS, Hughes JT, Esiri MM, Moss TH, Cruz-Sanchez FF, Coakham HB. The mononuclear cell infiltrate compared with survival in high-grade astrocytomas. Acta Neuropathol. 1989;78:189–193. doi: 10.1007/BF00688208. [DOI] [PubMed] [Google Scholar]
- 32.Safdari H, Hochberg FH, Richardson EP., Jr. Histological correlations with survival in malignant gliomas. Acta Neurochir Suppl (Wien) 1979;28:485–488. [PubMed] [Google Scholar]
- 33.Wrann M, Bodmer S, de Martin R, Siepl C, Hofer-Warbinek R, Frei K, Hofer E, Fontana A. T cell suppressor factor from human glioblastoma cells is a 12.5-kd protein closely related to transforming growth factor-beta. EMBO J. 1987;6:1633–1636. doi: 10.1002/j.1460-2075.1987.tb02411.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hishii M, Nitta T, Ishida H, Ebato M, Kurosu A, Yagita H, Sato K, Okumura K. Human glioma-derived interleukin-10 inhibits antitumor immune responses in vitro. Neurosurgery. 1995;37:1160–1166. doi: 10.1227/00006123-199512000-00016. discussion 1166-1167. [DOI] [PubMed] [Google Scholar]
- 35.Doherty TM, Kastelein R, Menon S, Andrade S, Coffman RL. Modulation of murine macrophage function by IL-13. Journal of Immunology. 1993;151:7151–7160. [PubMed] [Google Scholar]
- 36.Bettinger I, Thanos S, Paulus W. Microglia promote glioma migration. Acta Neuropathol. 2002;103:351–355. doi: 10.1007/s00401-001-0472-x. [DOI] [PubMed] [Google Scholar]
- 37.Hussain SF, Yang D, Suki D, Aldape K, Grimm E, Heimberger AB. The role of human glioma-infiltrating microglia/macrophages in mediating antitumor immune responses. Neuro Oncol. 2006;8:261–279. doi: 10.1215/15228517-2006-008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fecci PE, Mitchell DA, Whitesides JF, Xie W, Friedman AH, Archer GE, Herndon JE, 2nd, Bigner DD, Dranoff G, Sampson JH. Increased regulatory T-cell fraction amidst a diminished CD4 compartment explains cellular immune defects in patients with malignant glioma. Cancer Res. 2006;66:3294–3302. doi: 10.1158/0008-5472.CAN-05-3773. [DOI] [PubMed] [Google Scholar]
- 39.Heimberger AB, Abou-Ghazal M, Reina-Ortiz C, Yang DS, Sun W, Qiao W, Hiraoka N, Fuller GN. Incidence and prognostic impact of FoxP3+ regulatory T cells in human gliomas. Clin Cancer Res. 2008;14:5166–5172. doi: 10.1158/1078-0432.CCR-08-0320. [DOI] [PubMed] [Google Scholar]
- 40.Sippel TR, White JT, Nag K, Tsvankin V, Klaassen MK, Kleinschmidt-Demasters BK, Waziri A. Neutrophil Degranulation and Immunosuppression in Patients with Glioblastoma: Restoration of Cellular Immune Function by Targeting Arginase I. Clinical cancer research : an official journal of the American Association for Cancer Research. 2011 doi: 10.1158/1078-0432.CCR-11-1107. [DOI] [PubMed] [Google Scholar]
- 41.Justicia C, Gabriel C, Planas AM. Activation of the JAK/STAT pathway following transient focal cerebral ischemia: signaling through Jak1 and Stat3 in astrocytes. Glia. 2000;30:253–270. doi: 10.1002/(sici)1098-1136(200005)30:3<253::aid-glia5>3.0.co;2-o. [DOI] [PubMed] [Google Scholar]
- 42.Carro MS, Lim WK, Alvarez MJ, Bollo RJ, Zhao X, Snyder EY, Sulman EP, Anne SL, Doetsch F, Colman H, Lasorella A, Aldape K, Califano A, Iavarone A. The transcriptional network for mesenchymal transformation of brain tumours. Nature. 2010;463:318–325. doi: 10.1038/nature08712. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.