Abstract
Thymic-derived natural T regulatory cells (nTregs) are characterized by functional and phenotypic heterogeneity. Recently, a small fraction of peripheral Tregs have been shown to express Klrg1, but it remains unclear the extent Klrg1 defines a unique Treg subset. Here we show that Klrg1+ Tregs represent a terminally differentiated Treg subset derived from Klrg1− Tregs. This subset is a recent antigen-responsive and a highly activated short-lived Treg population that expresses enhanced levels of Treg suppressive molecules and that preferentially resides within mucosal tissues. The development of Klrg1+ Tregs also requires extensive IL-2R signaling. This activity represents a distinct function for IL-2, independent from its contribution to Treg homeostasis and competitive fitness. These and other properties are analogous to terminally differentiated short-lived CD8+ T effector cells. Our findings suggest that an important pathway driving antigen-activated conventional T lymphocytes also operates for Tregs.
Keywords: T regulatory cells, T cell subsets, Autoimmunity, IL-2R signaling, Klrg1
Introduction
Natural CD4+ Foxp3+ T regulatory (nTregs) cells are a dedicated thymic-derived population that critically function to maintain self-tolerance by actively suppressing autoreactive T cells that escape thymic negative selection (1). However, upon antigen stimulation, conventional peripheral CD4+ T cells sometimes express Foxp3 and adopt an induced Tregs (iTregs) fate (2). These iTregs are more numerous in mucosal tissues and are thought to limit immune responses to antigenic stimulation, especially to environmental antigens, such as food and allergens, and commensal microorganisms (3).
nTregs show extensive phenotypic heterogeneity with respect to surface markers of T cell activation (CD62L, CD69), adhesion molecules (CD103), and chemokine receptors (CCR7 and CCR6), analogous to subpopulations of naïve, effector and memory Tregs (4–10). More recently, Klrg1 has been found on a small proportion of Tregs in the periphery with enhanced suppressive function and these Klrg1+ Tregs exhibit a gene expression profile of “activated” effector Tregs (11–13). Tregs also exhibit functional heterogeneity in that they utilize the transcription factors, i.e. T-bet, IRF4, and Stat3, for distinctive suppressive programs to inhibit inflammatory Th1, Th2, and Th17 responses, respectively (14–16). However, the extent this phenotypic and functional heterogeneity defines unique vs. interrelated Treg subpopulations and the cell extrinsic factors favoring particular Treg subsets remain poorly defined.
IL-2 provides essential non-redundant signals at several levels for Tregs (17). First, IL-2 is essential for nTreg thymic development (18–20). Second, IL-2 induces proliferation of recent thymic emigrants in neonatal life to amplify nTreg numbers to establish an immune tolerant state (21). Third, IL-2 is the main cytokine for homeostasis and competitive fitness of peripheral nTregs in adult mice (22–24). Lastly, antigen-activated conventional T cells readily differentiate into iTregs when stimulated with IL-2 and TGFβ (25, 26). More recently, we showed that thymic Treg development and peripheral homeostasis were normal in mice that expressed mutations within signaling domains of IL-2Rβ that greatly diminished, but did not abrogate, Stat5 activation (27). Such mice were overly healthy, but with age exhibited inflammatory infiltrates in several tissues, particularly the salivary gland, lung, liver, and intestine. Consistent with this phenotype, gene expression profiling of nTregs with impaired IL-2R showed that several key IL-2-depenent genes of Tregs, e.g. Foxp3, and Il2ra, were normally expressed. However, many other genes remained IL-2-dependent, including Klrg1, Itgae (CD103), Il10, Gzmb, and Prdm1 (Blimp-1). (27). This finding suggests that IL-2 provides additional important functions in the periphery for Tregs besides homeostasis and raises the possibility that IL-2R is required for the development of Treg subsets marked by expression of Klrg1 and/or CD103.
Here we examined the properties associated with Klrg1+ Tregs and their relationship to the other Treg subsets. As extensive IL-2R signaling is necessary for optimal expression of Klrg1 (27), we also examined whether this reflected a distinct role for IL-2 in the persistence and development of a unique subset based on this surface marker. In comparison to other Treg subsets based on expression of CD62L and CD69, we find that Klrg1 defines a terminally differentiated Treg subpopulation with a distinctive molecular profile. Klrg1+ Tregs preferentially reside within mucosal sites and require strong IL-2R signaling for their development.
Material and Methods
Mice
C57BL6 (B6) mice, and TCRα−/− mice obtained from the JAX Laboratory. CD45.1-congenic B6 and IL-15−/− mice were obtained from Taconic. The reporter mice, Foxp3/GFP (28) (kindly provided by A. Rudensky), Foxp3/RFP (29) (kindly provided by R.A. Flavell), and Blimp-1/GFP (30) (kindly provided by S. L. Nutt) were previously described. IL-2Rβ transgenic mice, IL-2RβWT (Y0), IL-2RβY341 (Y1), and IL-2RβY341,395,498 (Y3) mice, on the IL2Rβ−/− genetic background, were previously described (27). Blimp-1/GFP mice and Y3 mice were each crossed to Foxp3/RFP mice in order to mark their Tregs with the RFP reporter. All mice were maintained in animal facility under VAF conditions at the University of Miami. Animal studies were approved by the Institutional Animal Care and Use Committee at the University of Miami.
Cell preparation and purification
Single cell suspensions from spleen, MLN and PP were prepared by mechanic disruption. LP cells were prepared as previously described (31) with minor modifications. In brief, the small intestine was obtained after cutting 0.5 cm below the stomach and 1 cm above the cecum and flushed with HBSS containing 5% FBS. After dissecting the PP, the intestines were cut into pieces (2–5 mm) and treated (shaking at 200 rpm for 20 min at 37°C) with Ca++-Mg++-free HBSS containing 5% FBS and 1.3 mM EDTA. Gut pieces were washed with RMPI 1640 and further treated (shaking at 200 rpm for 60 min at 37°C) with RPMI1640 containing 5% FBS, collagenase VIII (30 U/ml) and trypsin inhibitor (0.24 mg/ml). LP cells were collected after filtering using a 70 μm cell strainer and then purified on a 44%/67% Percoll gradient (800 g for 20 min at 20°C).
For adoptive transfer and microarray analyses, Tregs and Treg-depleted conventional CD4+ T cells were purified by sorting. In brief, splenic CD4+ T cells were enriched by positive selection using anti-CD4 magnetic-beads (Miltenyi Biotec) and sorted using a FACS Aria IIu cell sorter (Becton Dickenson). The purity of sorted populations was typically over 99% and never less than 98%.
Antibodies and Flow Cytometry
Conjugated antibodies were purchased from BD Pharmingen (San Jose, CA. USA), Biolegend (San Diego, CA. USA) and eBioscience (San Diego, CA. USA). Intracellular staining of Ki67, Bcl-2 and Foxp3 was performed together according to the manufacturer's instructions for Foxp3 (eBioscience). Flow-cytometric analyses were performed on an LSRII flow cytometer with FACS Diva software (Becton Dickenson). Typically at least 300,000 events were obtained for each sample.
Allergic Asthma Induction
Mice were sensitized on day 0 by i.p. injection of 66 μg ovalbumin (crystallized chicken egg albumin, grade V; Sigma-Aldrich, St. Louis, MO) adsorbed to 6.6 μg alum (aluminum potassium sulfate; Sigma-Aldrich) in 200 μl PBS with an i.p. boost on day 5. On day 12, mice were aerosol challenged with 0.5% w/v ovalbumin (Sigma-Aldrich) in PBS for 1 hour using a BANG nebulizer (CH Technologies, Westwood, NJ) into a Jaeger-NYU Nose-Only Directed-Flow Inhalation Exposure System (CH Technologies). Mice were sacrificed either on day 12 without aerosol or on day 15, i.e. 3 days after the aerosol challenge. Bronchoalveolar lavages obtained as well as lung lobes perfused and processed for single cell suspensions made from lung homogenate for flow cytometry analysis as described previously (32). Draining bronchial LN and spleens were also procured for subsequent flow cytometry analysis.
BrdU incorporation assay
Mice received 0.8mg/ml BrdU containing drinking water for 5 days. The incorporation of BrdU was assessed by FACS using anti-BrdU by a minor modification of the manufacturer's instructions (BD Pharmingen). Briefly, splenic cells were stained with surface markers for 15 min on ice. After washing, cells were fixed with Cytofix/Cytoperm Buffer for another 15 to 30 min and then incubated with Cytoperm Plus Buffer for 10 min on ice. Cells were washed again and re-fixed with Cytofix/Cytoperm Buffer for 5 min. After DNase (300μg/ml) treatment, cells were then stained with fluorescent anti-BrdU and other intracellular antibodies for 20 min at room temperature.
Treg suppression and survival assays in vitro
Suppression was assessed by the capacity of purified Treg subsets to inhibit anti-CD3 (0.25 μg/ml) induced proliferation by conventional CD4+ T cells as previously described (18). To assess Treg survival, magnetic bead enriched splenic CD4+ T cells from B6 mice were cultured in 24-well plates (1–2 × 106/well) in complete medium (33) for 3 days. At the indicated times, Tregs were enumerated by staining for Foxp3 and viability assessed by using the Live/Dead fixable stain kit (Invitrogen). Briefly, cells were harvested and incubated with the reconstituted fluorescent reactive dye for 30 min at RT. After washing by Hank's buffer, the cells were stained with surface markers and then fixed for intracellular Foxp3 staining. These data in conjunction with the number of recovered Tregs were used to calculate the survival of Tregs.
Microarry Analysis
Total RNA was isolated using TRizol and further purified with RNAeasy Minikit (QIAGEN). RNA quantity and quality was assessed by analysis with an Agilent 2100 BioAnalyzer. 20 to 50 ng RNA was used in a single round of linear RNA probe amplification and labeling using NuGEN Ovation Pico WTA system, WT-Ovation Exon module and Encore Biotin Module (NuGEN, San Carlos, CA, USA). Probe preparation and microarray analyses using Affymetrix Mouse Gene ST 1.0 arrays were performed at the Microarray and Gene Expression Core within the John P. Hussman Institute for Human Genomics at the University of Miami. Image analysis was performed using the Affymetrix Command Console Software (AGCC). Resulting data were normalized with the RMA method using software at GeneSifter (Seattle, WA). Multi-group comparisons of the transformed data of at least 2 independent biological replicates were performed using ANOVA applying the Benjamini Hochberg correction for false positives. Genes expressed ≥2.0-fold up or down (p<0.05) between groups were considered differentially expressed. GEA analyses for differentially expressed genes for GO processes or KEGG pathways were performed using GeneSifter. The expression data are available through the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/) accession number GSE36527.
Adoptive transfer
The indicated purified Treg population was adoptively transferred by i.v. injection through the tail vein into B6, Y3, CD45.1-B6, or TCRα−/− recipients, as indicated in each Figure. In most cases TCRα−/− recipients were co-injected with Treg-depleted conventional CD4+ T cells using Foxp3/GFP reporter mice to achieve ~1:10 ratio of Treg:T conventional cells. To study developmental progression, Tregs were labeled with CFSE using the Vibrant CFDA SE Cell Tracer Kit (Sigma-Aldrich) prior to transfer. The Tregs (5 × 105/ml) were incubated with 5 μM CFSE in RPMI1640 containing 5% FBS for 15 min at 37° C according to the manufacturer's instruction.
IL2-IC treatment
Mouse IL-2 and the JES6-1A12 mAb to mouse IL-2 were purchased from eBioscience. IL2-IC was prepared as previously described (34). In brief, IL-2 and JES6-1A12 were incubated at a molar ratio of 2 to 1 in PBS at room temperature for 30 min such that each mouse received 1 μg of IL-2 and 5 μg of JES6-1A12 in 200 μl PBS by daily i.p. injections for 3 consecutive days. As a control, mice received only 200 μl PBS.
Statistical analysis
Data were analyzed using Prism 5.0 software. All data are represented as the mean ± SD. Multi- and two-group statistical analyses were performed using in a one-way ANOVA with Tukey's multiple comparison test and unpaired t-test. P<0.05 was considered significant. Significant differences are designated as * P<0.05, ** P<0.01, *** P<0.001, **** P<0.0001. BrdU turnover data were subjected to linear regression analysis and assessed for significantly different slopes.
Results
Klrg1+ Tregs are a recent antigen-responsive highly activated subset
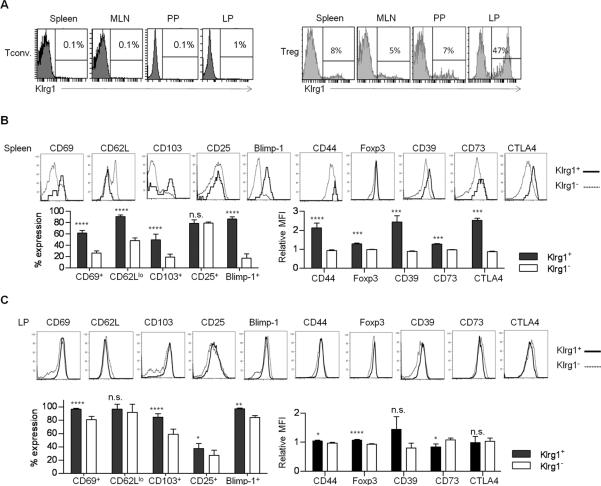
Klrg1 is minimally expressed on conventional CD4+ T cells but is found at a relatively high level on a small population of Tregs in peripheral immune tissues such as the spleen, mesenteric LN (MLN) and Peyer's patch (PP) and on nearly 50% of the Tregs from the lamina propria (LP) of the small intestine (Fig. 1A). Klrg1+ Tregs in the spleen (Fig. 1B) expressed a more activated phenotype (CD69+, CD62Llo, CD103+, and CD44hi) (Fig. 1B) when compared to Klrg1− Tregs. Blimp-1, which marks IL-10-producing activated Tregs (35), was also highly expressed by Klrg1+ Tregs (Fig. 1B).This comparison also revealed that Klrg1+ Tregs expressed higher levels of Foxp3 and several important Treg functional molecules (CTLA4, CD39, CD73) (Fig. 1B). In the LP, most Tregs expressed an activated phenotype, but expression of CD69, CD103, CD25, Blimp-1, and Foxp3 was slightly, but significantly, greater for Klrg1+ Tregs (Fig. 1C).
FIGURE 1. Properties of Klrg1+ Tregs from B6 mice.
(A) Expression of Klrg1 after gating CD4+ T conventional and CD4+ Foxp3+ Tregs from WT spleen, MLN, PP and the LP of the small intestine. (B, C) The expression of indicated markers on Klrg1+ and Klrg1− splenic (B) and LP (C) Tregs. The expression of Blimp-1 was analyzed in Tregs from WT Blimp/GFP Foxp3/RFP dual reporter mice. Data are representative of 3–8 mice and the % expression or mean fluorescent intensity (MFI) was determined. The MFI values in Klrg1+ or Klrg1− Tregs were normalized to total Tregs.
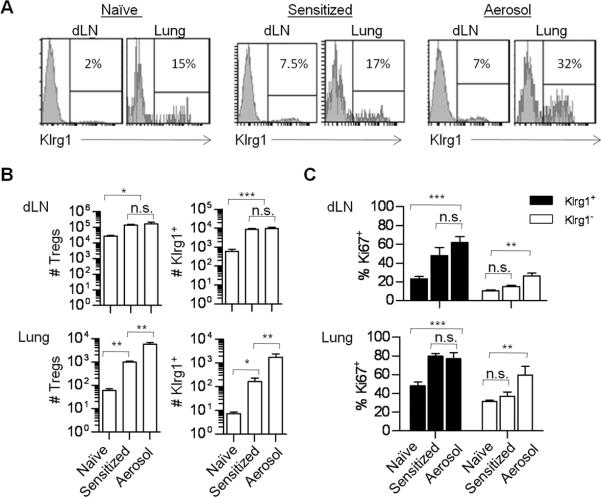
The high prevalence of Klrg1+ Treg cells within the LP at the steady state suggests that these Treg cells preferentially home and/or are activated within tissue sites. To test this notion, Klrg1 expression by Tregs in the lung and the bronchial draining LN (dLN) was examined after ice were either sensitized with OVA and alum or after aerosol challenge to induce allergic hypersensitivity (Fig. 2A). After sensitization, the number of total and Klrg1+ Tregs increased in the dLN and lung (Fig. 2B). Noticeably, three days after the aerosol challenge with OVA, an increased number of total and Klrg1+ Treg cells were detected in the lung but not the dLN in comparison to mice that were only sensitized (Fig.2B). Therefore, most of the responding Klrg1+ Treg cells are probably a result of a response in situ rather than migration into this mucosal tissue site. In addition, increased proliferation as assessed by Ki67 expression was noted by Klrg1+ Treg cells in the dLN and lung after OVA sensitization and aerosol challenge (Fig. 2C). Collectively, these data indicate that Klrg1+ Tregs are a highly activated Treg subpopulation with enhanced expression of several Treg functional molecules that readily respond to antigenic challenge.
FIGURE 2. Distribution and proliferative activity of Klrg1+ Tregs in allergic hypersensitive mice.
Allergy was induced in B6 mice as described in the Methods. Just before the aerosol challenge, the Treg composition was assessed for some of the sensitized mice, as indicated. (A) The expression of Klrg1 by Tregs from the indicated tissues before and after induction of acute asthma. (B) Enumeration of the number of total and Klrg1+ Tregs. (C) Proliferative activity of the indicated Klrg1+ and Klrg1− Tregs based on Ki67 expression. Data are representative of 3–8 mice/group.
Distinct gene expression by Klrg1+ Tregs
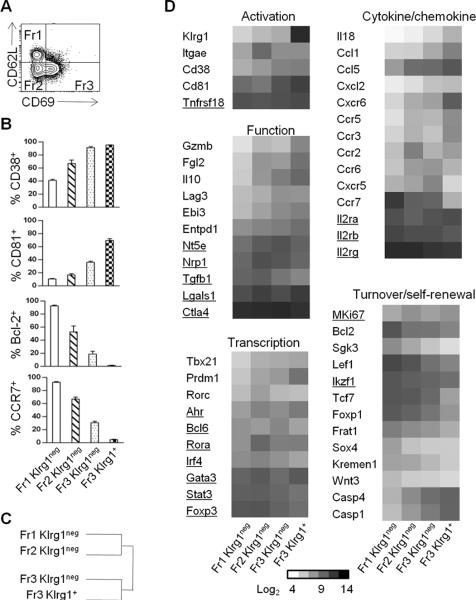
To further define the molecular properties of Klrg1+ Tregs and explore their relationship to other Tregs, gene expression profiling of ex vivo isolated splenic nTreg subsets was performed based on expression of CD62L, CD69 and Klrg1 (Fig. 3A). These markers discriminate conventional T cells with properties of resting naïve to activated effector and memory cells. Tregs from Foxp3/RFP reporter mice were FACS purified to obtain 4 subsets which are CD62Lhi CD69− (Fr1 Klrg1−), CD62Llo CD69− (Fr2 Klrg1−), and CD62Llo CD69+ (Fr3 Klrg1−) subsets. Klrg1+ Treg (Fr3 Klrg1+) cells were purified to co-express CD69 because most (60–70%) Klrg1+ Tregs are CD69+ (Fig. 1B).
FIGURE 3. Gene expression profile by Treg subsets from B6 mice.
Klrg1-depleted Fr1, Fr2, Fr3 and Fr3 Klrg1+ Tregs were purified from the spleen by FACS sorting. Total RNA was isolated and gene expression profiling was performed using Affymetrix Mouse Gene ST 1.0 arrays. Samples (n=2, except Fr3 Klrg1neg, n=4) represent independent biological replicates. (A) Representation of the 3 main Treg fractions (Fr) used for gene array analysis. (B) Protein expression of selected genes by FACS for Treg subsets. Data are mean ± SD for 4 mice per group. (C) Euclidean clustering of sample relatedness for genes that varied by ≥2-fold (over- or under-represented) between the indicated Treg subsets. (D) Genes differentially expressed by (over- or under-represented) only between the indicated Treg subsets. Shown are the expression levels (log2) of selected genes that varied by ≥2-fold between any two subsets except those underlined, which varied by a lower level. Data were analyzed by one-way ANOVA p<0.05, Benjamini and Hochberg correction.
When comparing genes differentially expressed by ≥2-fold only between the 4 Treg subsets, 745 unique Affymetrix targets were identified. To verify this list, we confirmed that several genes exhibited cell surface expression in accordance with the mRNA levels (Fig. 3B, 3D). In addition, our expression pattern for Klrg1+ Tregs resembled that of another study that compared Klrg1+ Tregs to total Tregs from distinct tissues (13.) Euclidian clustering of differentially expressed genes between Treg subsets was performed to globally evaluate the relationship between these subsets. This analysis revealed two clades, with Fr1 Klrg1− and Fr2 Klrg1− vs. Fr3 Klrg1− and Fr3 Klrg1+ Tregs more related to each other (Fig. 3C). Gene enrichment analyses (GEA) annotated 233 of 781 genes (29.8%) into 13 Gene Ontology (GO) functional classifications pathways with z scores >2.0. The 4 largest clusters of genes that varied between and distinguished these subsets are related to immune system processes, lymphocyte activation, proliferation, and cell death. Other notable characteristic that differed between subsets were tissue migration, cell localization and homing, cytoskeleton organization, and hemopoiesis.
When considering selected genes (Fig. 3D), Fr1 Klrg1− exhibits properties consistent with a resting Treg subset with preference for residing in lymphoid tissues. Besides lower levels of many Treg suppressive molecules [Gzmb, Fgl2, Il10, Lag3, Ebi3 (a component of IL-35), and Entpd1 (CD39)], Fr1 Klrg1− Tregs expressed lower levels of cytokines and chemokines (Il18, Ccl5, Ccl1), chemokine receptors targeting tissue migration (Ccr2, Ccr3, Ccr5, Ccr6, Cxcr6), activation markers (Cd38, Lag3, Tbx21, Rorc), and pro-apoptotic molecules (Casp1, Casp4). In contrast, Fr1 Klrg1− Tregs expressed increased levels of lymphoid homing receptor Ccr7 and anti-apoptotic Bcl2 and Sgk3. Fr1 Klrg1− also showed increased expression of genes related to hemopoiesis [Foxp1] and Wnt signaling (Sox4,Tcf7, Lef1, Wnt3, Kremen1, Frat1), suggesting that this Treg subset might be enriched in cells capable of self-renewal.
The genes mentioned above related to Treg function, chemokines, cytokines, tissues migration and cell death were often increased in Fr2 Klrg1−, Fr3 Klrg1− and Fr3 Klrg1+, with a gradient of increased expression from Fr1 Klrg1− (lowest) to Fr3 Klrg1+ (highest). In contrast, decreased expression for mRNAs that function in lymphoid tissue homing, survival, hemopoiesis, and Wnt signaling were found in these 3 subsets, again with a gradient of expression, but from Fr3 Klrg1+ (lowest) to Fr1 Klrg1− (highest). In addition, heightened levels of Tbx21 (T-bet) and especially Prdm1 (Blimp-1) were found in Klrg1+ Tregs. Thus, the extreme for high or low expression of many genes was often associated with Klrg1+ Tregs, illustrating the highly distinctive properties of this subset. Collectively, analogous to conventional T cells, these results indicate that CD62L, CD69, and Klrg1 represent useful phenotypic markers to distinguish functional Treg subsets and show that Klrg1+ Tregs exhibit distinctive properties, consistent with it being a short-lived end-stage tissue-seeking suppressor subset.
Varied suppressive function in Treg subsets
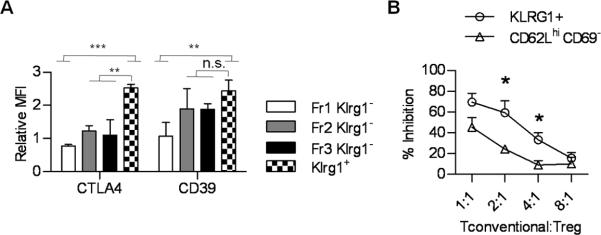
The potency of a T effector response is often enumerated based on the frequency and level of expression of molecules, e.g. cytokines, which are characteristic of T cell function. In this regard the level of CTLA4 was substantially increased in splenic Klrg1+ Tregs even when compared to activated Fr3 Klrg1− Tregs (Fig. 4A). As CTLA4 was not detected as a differentially expressed gene (Fig. 3D), this increase may represent a post-transcriptional mechanism or reflect an inability to detect a mRNA difference due to the high levels of CTLA4 mRNA in Tregs. In contrast, CD39 expression by Klrg1+ Tregs was significantly increased only when compared to “resting” Fr1 Klrg1− Tregs. Thus, all Treg suppressive molecules are not selectively over-represented in the Klrg1+ subset. In vitro suppression was also consistently greater for splenic Klrg1+ Tregs when compared to F1 Klrg1− Tregs (Fig. 4B), which expressed lower amounts of many Treg suppressive molecules (Fig. 3D). Collectively, these data in conjunction with our gene array analysis support the notion that Klrg1+ Tregs are armed to optimally deliver a number of molecules associated with Treg suppressive function.
FIGURE 4. Treg suppressive activity of Klrg1+ Tregs.
(A) Distribution of CTLA4 and CD39 by the indicated splenic Treg subsets. (B) In vitro suppression by the indicated splenic Treg subsets isolated from Foxp3/RFP reporter mice. The % inhibition was determined in comparison to [3H]thymidine incorporation by responder cells lacking Tregs. Results are from 3 experiments.
IL-2R signaling is essential for Klrg1+ Tregs
We previously showed that mutations of Y341 (Y1), Y395 and Y498 (Y2), or Y341, Y395, and Y498 (Y3) to phenylalanine in the cytoplasmic tail of IL-2Rβ resulted in a dose-dependent reduction of IL-2-dependent tyrosine Stat5 phosphorylation (pStat5) with lowest, but detectable, pStat5 associated with Y3 (27). Unlike IL-2Rβ-deficient mice, which develop rapid lethal systemic autoimmunity due to failed maturation of Tregs, mice expressing each mutant IL-2Rβ were overtly healthy and contained a normal ratio and number of Tregs and normal levels of Foxp3, but Y2 and Y3 mice eventually showed symptoms of autoimmune disease. Thus, nTreg development and homeostasis are readily supported by a low level of IL-2R signaling. Gene expression profiling, however, from Tregs with impaired IL-2R signaling revealed a substantial number of genes that were IL-2-dependent. Two such genes are Klrg1 and Itgae (CD103).
When Tregs were examined from mice bearing mutant IL-2Rβ, fewer Klrg1+ Tregs were found in the spleen (Fig. 5A). A similar decrease was detected in peripheral LN, MLN, and PP (not shown) and these effects were generally proportional to increased Y→F IL-2Rβ mutations. This decrease was especially striking in the LP where Klrg1 is normally expressed by 40–50% of the Tregs. The decrease in Klrg1+ Tregs occurred even though the number of total Tregs was not reduced in the LP of Y1 and Y3 mice (not shown). As a control, normal levels of Klrg1+ Tregs were found in the spleen and LP of mice expressing transgenic WT IL-2Rβ (Y0) on the IL-2Rβ−/− genetic background. Normal levels of Klrg1+ Tregs were also found in IL-15−/− mice. Thus, Klrg1+ Tregs depend on IL-2, but not IL-15 signaling, which also utilizes IL-2Rβ (Fig. 5A).
FIGURE 5. IL-2R signaling is required for the development of Klrg1+ Tregs.
The expression of Klrg1 (A) and other activation markers (B) by Tregs from the spleen and LP of the indicated B6 mice. Data are from 3–11 mice per group.
With respect to other T cell activation antigens expressed by Tregs from Y3 mice, a decrease in CD103+ Tregs was noted in the spleen and peripheral LN (not shown), but this effect was absent in the LP (Fig. 5B). Thus, the requirement for IL-2Rβ signaling differs for CD103+ and Klrg1+ Tregs. Noticeably, there was no reduction, but a relatively higher proportion, of CD69+ and CD62Llo activated Tregs (Fig. 5B) Thus, impaired IL-2R signaling, as supported by Y3, does not generally dampen Treg activation, but somewhat alters Treg subset composition.
Klrg1 marks a unique Treg subset
Fewer Tregs that express Klrg1 in Y3 mice might simply reflect decreased expression of these molecules by common related Tregs or might indicate the absence of a specific IL-2-dependent Treg subset. To address this issue, we examined whether WT Klrg1+ Tregs selectively repopulate Y3 mice after adoptive transfer of WT Tregs from Foxp3/GFP reporter mice. Since IL-2 contributes to peripheral Treg homeostasis, donor Tregs with WT IL-2R which delivers normal strong IL-2R signaling are expected to have a competitive advantage over the more numerous recipient-derived Y3 Tregs, which are maintained by suboptimal IL-2R signaling (27). Therefore, we reasoned that WT Klrg1+ Tregs should only be favored if this population was selectively diminished in Y3 mice.
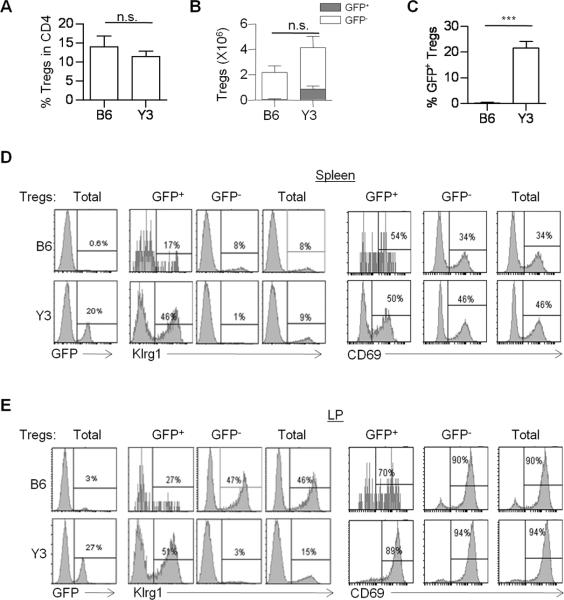
At 4 weeks post-transfer, the percentage (Fig. 6A) and number (Fig. 6B) of total splenic Tregs were similar in WT and Y3 recipient mice. Donor GFP+ Tregs, however, comprised a substantial fraction (20%) of the total Tregs in Y3 recipients while GFP+ Tregs represented <1% of the Tregs in WT recipients (Fig. 6B and 6C), demonstrating the competitive advantage of WT Tregs within the Y3 recipients. Importantly, the donor WT GFP+ Tregs in Y3 recipients were highly enriched in Klrg1+ Tregs such that the composition of the total Treg pool reflected that in normal WT mice. In contrast, the proportion of activated splenic Tregs expressing CD69, which is molecularly related to Klrg1+ Tregs (see Fig. 3), was largely unaltered (Fig. 6D). This relationship strikingly held for the LP in Y3 recipients where 27% of Tregs were GFP+ donor-derived and >50% of the GFP+ Tregs expressed Klrg1 (Fig. 6E) and nearly all Klrg1+ Tregs were of donor origin. Collectively, these data indicate that the Tregs expressing a WT IL-2R have a competitive advantage over Y3 Tregs and favor the development of Klrg1+ Tregs in Y3 recipients. This finding supports the notion that Klrg1 marks a distinct IL-2-dependent Treg subset and that this subset depends on more extensive IL-2R signaling.
FIGURE 6. Klrg1 marks a splenic Treg subset.
Purified Foxp3/GFP+ WT Tregs (4 × 105) were transferred into the B6 or Y3 adult recipient mice. 4 weeks post-transfer, (A) the percent of total Tregs in CD4+ T cells, (B,C) the number of host (GFP−) vs. donor (GFP+) Tregs, and the (C) percentage of donor GFP+ Tregs in total Tregs were determined. (D,E) Total CD4+ Foxp3+ Tregs were gated in spleen (D) and LP (E) and the expression of Klrg1 and CD69 in the GFP+, GFP− or total Tregs was determined. Data are representative of 3–4 mice per group.
IL-2 drives preferential development of Klrg1+ Tregs
The near lack of Klrg1+ Tregs in Y3 mice indicates that these cells depend on more extensive IL-2R signaling than other Tregs. To further test this notion, we evaluated the effect of extensive IL-2R signaling on Treg subsets by treating normal mice with agonist anti-IL-2/IL-2 complexes (IL2-IC) using the JES6-1A12 mAb that preferentially expands Tregs (34, 36). 3 days after the last injection of IL2-IC, a selective expansion of total and Klrg1+ splenic Tregs was noted in their relative percentage (Fig. 7A) and numbers (Fig. 7B). The increase was ~2.5-fold for total Tregs, but almost 4-fold for Klrg1+ Tregs. IL2-IC also markedly increased CD25 expression (Fig. 7A), which is known to be upregulated by IL-2 in conventional activated T cells (37). At the peak of the response, nearly all Tregs, including Klrg1+ cells, were recently in cell cycle as evident by the high percentage of Ki67+ cells, but Bcl-2 levels minimally varied (Fig. 7C), suggesting that this form of IL-2R signaling favors proliferation rather than enhanced cell survival. Unlike the transient increase in Klrg1+ Tregs (Fig. 7B), the proportion of splenic Tregs that expressed CD103, CD62L, and CD69 minimally varied (Fig. 7D). Thus, even though these latter Treg subsets proliferated in response to IL2-IC, they are stably maintained in their relative proportions. Thus, extensive IL-2R signaling through IL2-IC in lympho-replete mice is particularly efficient in driving development of Klrg1+ Tregs.
FIGURE 7. The effect of IL2-IC on Treg subsets.
(A–D) WT B6 mice received 3 daily injections of IL2-IC complex. 3, 5, or 7 days after the last injection, splenic Tregs were evaluated. Control represents mice that were injected with PBS and were analyzed only 3 days after the last injection. Representative FACS profiles of the indicated markers 3 days after the last injection of IL2-IC (A) and time course of Treg numbers (B) of the indicated populations. (C) Expression of Ki67 and Bcl-2 3 days after the last injection of IL2-IC. (D) Time course for the expression of the indicated markers. Data are from at least 3 mice per group per time point. (E) Splenic Klrg1+ or Klrg1− Tregs (1 × 105) from the Foxp3/RFP reporter mice were injected with splenic conventional CD4+ T cells (1 × 106) from the Foxp3/GFP reporter mice at the ratio of 1:10 into TCRα−/− mice. One day later the recipients received 3 daily injections of IL2-IC. 6 days after the cell transfer, the indicated cell populations from the spleen were analyzed for the expression of RFP and Klrg1. Data are 4–5 mice/group where each point on the graph represents an individual mouse
To test the extent that Klrg1+ Tregs directly expand in response to IL-2, Klrg1+ and Klrg1− RFP+ Tregs were purified, transferred into TCRα−/− mice with Treg-depleted conventional CD4+ T cells, and the recipients were treated with IL2-IC. Under these conditions only the Klrg1− Treg population expanded and ~20% developed into Klrg1+ Tregs (Fig. 7E). Notably, for recipients of Klrg1+ Tregs, the few donor cells detected still expressed Klrg1. Thus, the expansion of Klrg1+ Tregs by IL2-IC likely reflects proliferation by Klrg1− precursors that are driven by IL-2 to develop into Klrg1+ Tregs.
IL-2R signaling distinctively shapes the development of Klrg1+ Tregs in the LP
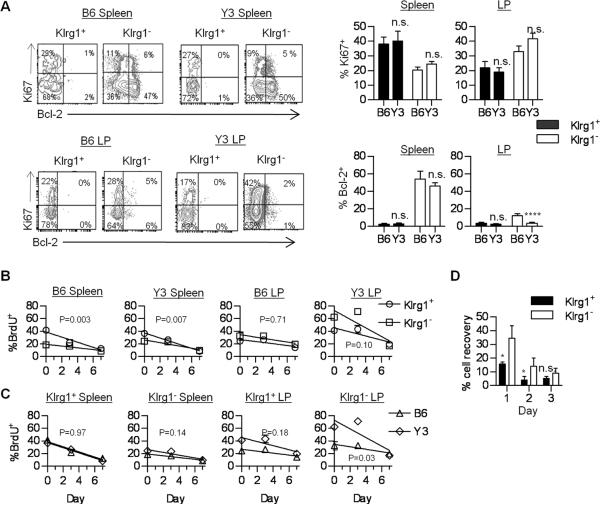
Given the marked reduction of Klrg1+ Tregs in the LP, we also examined the proliferation and turnover of Klrg1+ and Klrg1− Tregs within the spleen and LP of WT and Y3 mice. Proliferation as assessed by Ki67 expression of Klrg1+ and Klrg1− Tregs from the spleen and LP was similar between B6 and Y3 mice (Fig. 8A). This finding suggests that homeostatic proliferation of Tregs is maintained by low IL-2R signaling associated with Y3 IL-2Rβ. In both types of mice, higher Ki67 expression was associated with Klrg1+ Tregs in the spleen, but with Klrg1− Tregs in the LP. Thus, the environments associated within a peripheral immune tissue and the gut mucosa distinctively shape the proliferative behavior of Tregs. Bcl-2 expression was equivalently lower for Klrg1+ Tregs in B6 and Y3 spleen (Fig. 8A). For both Treg subsets in the LP, Bcl-2 levels were generally low, but the Klrg1− Tregs from Y3 mice contained an even lower fraction of Bcl-2+ cells.
FIGURE 8. Proliferative activity and turnover of Klrg1+ and Klrg1− Tregs.
(A) Bcl-2 and Ki67 expression by the indicated CD4+ Foxp3+ Tregs. Data are from 6–8 mice per group. (B,C) Mice received BrdU for 5 days and then were switched to normal water (Day 0). BrdU incorporation was determined for the indicated Treg populations. Linear regression analysis was performed for BrdU loss and compared between Klrg1+ or Klrg1− Treg subsets from the WT B6 and Y3 mice (B) or compared for an indicated individual subset between B6 and Y3 mice (C). The p values of whether the slopes are significantly different are shown within each graph. Data represent 3 mice/group. (D) Purified CD4+ T cells were cultured in medium for the indicated time. Cell viability and recovery were determined for the Klrg1+ and Klrg1− CD4+ Foxp3+ Tregs and compared to the number of Tregs at culture initiation. Data are from 3 experiments.
To study the turnover of proliferating Tregs, BrdU incorporation studies were performed where cohorts of WT and Y3 mice received BrdU in their drinking water for 5 days and then BrdU was chased by providing normal drinking water. Immediately after the 5 day labeling period (Day 0), BrdU incorporation was measured and this largely agreed with the expression of Ki67. Upon BrdU withdrawal, splenic B6 and Y3 Klrg1+ Tregs showed greater turnover as reflected by a more rapid loss of BrdU (Fig. 8B) and the rate of loss was identical when directly comparing each subset individually between B6 and Y3 mice (Fig. 8C). These data indicate that weak IL-2R signaling is largely effective in supporting homeostasis for splenic Tregs. The lower expression of Bcl-2 by Klrg1+ Tregs suggests that these cells may be more susceptible to apoptosis. Consistent with this view, this Treg subset from WT B6 mice was more susceptible to cell death than Klrg1− Tregs after in vitro culture in media (Fig. 8D). Overall, these data indicate that Klrg1 marks a Treg subset within a peripheral lymphoid tissue that has undergone substantial recent proliferation and is short-lived with very similar behavior in both B6 and Y3 mice. Thus, the relative lack of this subset within the spleen of Y3 mice most likely reflects a requirement for relatively high IL-2R signaling to promote their development.
For the LP, the turnover of between Klrg1+ and Klrg1− Tregs was similar when examined in B6 or Y3 mice (Fig. 8B). However, Klrg1− LP Tregs from Y3 mice showed significantly greater turnover (p=0.03) than Klrg1− LP Tregs from B6 mice (Fig. 8C). Thus, there is a requirement for stronger IL-2R signaling to properly support the homeostasis of Tregs in the LP. As Treg proliferation between B6 and Y3 subsets was similar (Fig. 8A), this likely reflects a role for IL-2 in contributing to Treg survival in the LP. Thus, the striking lack of Klrg1+ Treg cell within the LP (Fig. 5A) may reflect failed IL-2-dependent development that is due in part to poor survival of Klrg1− cells that serve as precursor cells.
Klrg1+ Tregs are terminally differentiated
The preceding experiments demonstrate that Klrg1+ Tregs exhibit phenotypic and molecular properties of a late and perhaps terminally differentiated Treg subset. A hallmark of terminally differentiated cells is a lack of plasticity to express traits of other cells related within its lineage. To determine whether Klrg1+ Tregs lack developmental heterogeneity, Klrg1+ and Klrg1− nTreg subsets and Treg cell-depleted CD4 T conventional cells were purified by sorting using Foxp3/RFP and Foxp3/GFP reporter mice, respectively. These cells were mixed at a ~1:10 ratio (Foxp3/RFP+:Foxp3/GFP−) and transferred into untreated TCRα−/− recipients (Fig. 9A). The addition of conventional T cells provides a potential source of growth factors, e.g. IL-2, required by the donor nTregs and permits assessment of iTreg development through detection of GFP+ cells.
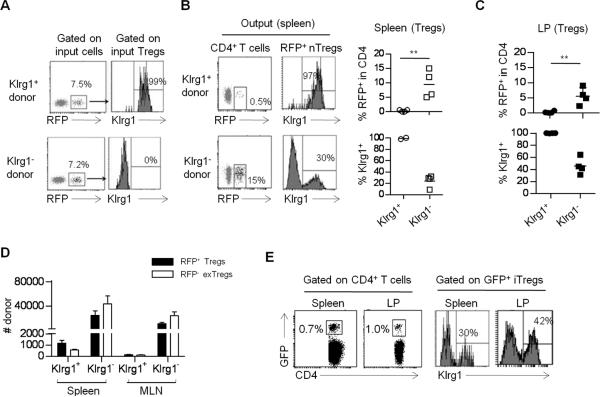
FIGURE 9. Klrg1+ Tregs are terminally differentiated and derived from distinct Klrg1−subsets.
(A–C) Klrg1+ or Klrg1− Treg subsets (1 × 105) were purified from Thy-1.1+ Foxp3/RFP reporter mice and adoptively transferred with (A-C, F) conventional CD4+ T cells (1 × 106) from Foxp3/GFP reporter mice at 1:10 ratio. The purity of injected cells was verified by FACS analysis (A). 4 weeks post-transfer, the spleen (B) and LP (C) of TCRα−/− recipients were examined for % donor RFP+ nTreg and their expression of Klrg1+. (D) RFP+ Thy-1.1+ Klrg1+ and Klrg1− Treg subsets were transferred alone into Thy-1.2+ TCRα−/− mice and Treg stability was assessed 2 weeks later by enumerating the indicated RFP+ Treg and RFP− exTreg Thy-1.1+donor cells from the spleen and MLN. (E) Foxp3/GFP-marked conventional CD4+ T cells (depleted of Tregs) were transferred with Klrg1− Foxp3/RFP Tregs into TCRα−/− recipients and iTreg development was assessed by enumerating CD4+ GFP+ T cells (left) and their development into Klrg1+ iTregs (right) from the spleen and LP 4 weeks post-transfer. All the data were derived from 3–4 mice per group.
At 4 weeks post-transfer, RFP+ nTregs were readily found in the spleen (Fig. 9B) and LP (Fig. 9C) that received Klrg1−, but not Klrg1+, nTregs. The few RFP+ nTregs in the LP from recipients that received Klrg1+ Tregs indicate that the overall low number of donor Klrg1+ cells does not simply reflect preferentially homing of this subset to tissue sites. In these TCRα−/− recipients where a few RFP+ Tregs were found, they remained as Klrg1+ Tregs (Fig. 9B, C, upper panel). However, a substantial proportion of the Klrg1− nTregs developed into Klrg1+ cells (Fig. 9B, C lower panel). Furthermore, in the absence of conventional T cells, Thy-1.1 RFP+ donor Tregs were transferred into TCRα−/− mice, an environment favoring de-differentiation of Tregs into Foxp3− cells (exTregs) (38). In this setting Klrg1−, but not Klrg1+, donor Tregs readily generated a large population of exTregs (Fig. 9D). Thus, low number of donor cells detected after the transfer of Klrg1+ nTregs is not because Klrg1+ Tregs express an unstable phenotype and then expand as exTregs. Collectively, these findings indicate that Klrg1+ Tregs are derived from Klrg1− cells and are terminally differentiated. The poor recovery of Klrg1+ Tregs in TCRα−/− recipients is consistent with them being mostly short-lived. However, their lack of expansion suggests that the high Ki67 expression and BrdU incorporation associated with Klrg1+ Tregs (Fig. 7) may actually represent proliferation by precursors to the Klrg1+ Treg subset.
When Foxp3/GFP-depleted conventional CD4+ T cells were transferred with Klrg1+ and Klrg1− nTregs into TCRα−/− mice, a readily measurable (~1%) population of GFP+ iTregs was also detected in the spleen and LP (Fig. 9E). A substantial fraction (30–60%) of these iTregs also expressed Klrg1. This distribution and the high purity of donor conventional CD4+ T cells (>99.7% GFP−) make it unlikely that these GFP+ cells post-transfer are solely accounted for by a few contaminating GFP+ nTregs in the conventional T cells inoculum. These data indicate that iTregs express Klrg1 and are consistent with the possibility that both nTregs and iTregs develop into the Klrg1+ subset.
Developmental progression of Klrg1+ Tregs
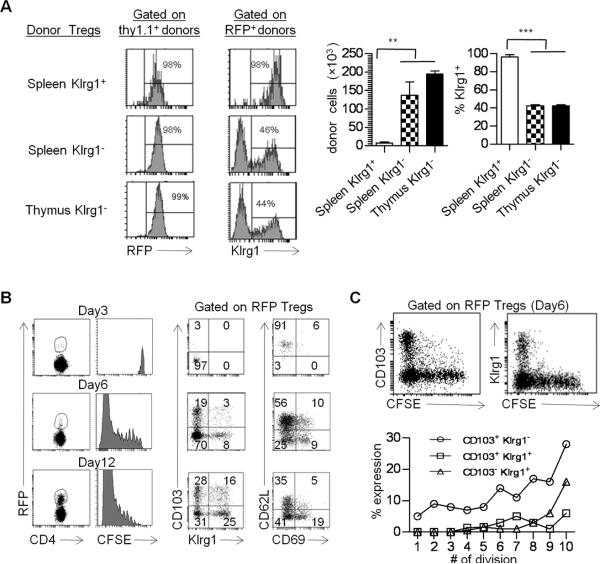
Klrg1+ Tregs are not found in the thymus (11, 12), raising the possibility that splenic Klrg1+ nTregs might actually reflect selective development by iTregs. To directly examine the development and stability of Klrg1+ Tregs by nTregs in a lympho-replete setting, Y3 mice were adoptively transferred with spleen- or thymus-derived Tregs. Y3 mice have normal numbers of conventional and regulatory T cells (27), but transferred WT Tregs have a competitive advantage over recipient Y3 Tregs due to normal IL-2R signaling and homeostasis, greatly facilitating their identification (Fig. 6). By transferring Foxp3/RFP+ Thy-1.1+ Tregs (>98% pure) into Thy-1.2+ Y3 recipients, we followed the fraction of Thy-1.1+ donor Tregs that lost expression of RFP+ and became exTregs.
When donor thymic or splenic Klrg1− Tregs were transfer into Y3 mice, both populations substantially engrafted (Fig. 10A, right) and readily (>40%) developed into Klrg1+ Tregs (Fig. 10A). The development of Klrg1+ Tregs from thymic donor cells directly shows that nTregs readily yield Klrg1+ cells. Thus, nTregs and iTregs (Fig. 9F) develop into the Klrg1+ subset. Analogous to TCRα−/− recipients (Fig. 9), very few splenic Klrg1+ donor Tregs were found in Y3 recipients, and nearly all of these remained as Klrg1+ Tregs. Furthermore, in all 3 type of transfers, virtually all (>98%) input Tregs were stable and remained RFP+ cells (Fig. 10A, left). This contrasts with TCRα−/− recipients where exTregs were readily detected and indicates that the lymphopenic environment preferentially supports exTreg production.
FIGURE 10. Developmental progression of Klrg1+ Tregs.
(A) The indicated Thy-1.1+ RFP+ Tregs (5 × 104) were transferred into Thy-1.2+ Y3 recipients. Two weeks post-transfer the donor cells were examined from the spleen of each recipient. Data are from 3 mice/group. (B) CFSE-labeled CD62Lhi CD69− CD103− Klrg1– Fr1 Tregs were transferred into Y3 mice and the phenotype of the donor cells was determined on the indicated day post transfer (B). The relationship of CD103 and Klrg1 expression as a function of cell division (CFSE-dilution) was determined 6 days post-transfer (C). Data are representative 3 mice/time point.
If Klrg1+ Tregs represent an end stage terminally differentiated cell, expression of Klrg1 should be acquired late during Treg their activation in vivo. To test this notion, the developmental progression of Klrg1+ Tregs was examined in vivo by following the cell surface phenotype and cell division status of CFSE-labeled CD62Lhi CD69− CD103− Klrg1− “resting” splenic nTregs after adoptively transfer into Y3 mice. The donor nTregs nearly fully diluted the CFSE 12 days post transfer (Fig. 10B). With increased time fewer CD62Lhi cells were detected while more active CD62Llo CD69+ and CD62Llo CD69− as well as CD103+ and Klrg1+ nTregs developed. Furthermore, Klrg1 was not detected until the donor cells nearly fully diluted CFSE whereas CD103+ cells were detected at all division steps (Fig. 10C). Thus, these data further support the notion that Klrg1 marks a Treg subset that has undergone substantial proliferation and is an end stage developmental product.
Discussion
Our understanding concerning effector T cell responses and the development of memory has been greatly facilitated by defining phenotypic and molecular properties that delineate important cell subsets and by establishing extrinsic and intrinsic factors favoring development of one subset over another. Several lines of recent work indicate that this paradigm also holds for Tregs. First, transcription factors important for Th development also function in Tregs in a manner to distinctively coordinate Treg suppressive programs toward the effector subtype that they inhibit (14–16). Second, the gene profiles of Treg subtypes, particularly as they relate to nTreg vs. iTreg, were distinguished from each other by their transcriptional profile (13). Lastly, TCR-transgenic Tregs give rise to memory-like Tregs after response to a model self antigen (39). By examining polyclonal Tregs, our current study extends this notion by establishing that ex vivo derived phenotypic distinct nTregs express distinctive functional and gene expression profiles and that IL-2R signaling represents a critical developmental signal for one such subset, i.e. Klrg1+ Tregs.
In a manner analogous to conventional T cells, expression of CD62L, CD69, and Klrg1 delineates nTreg subsets. Gene expression analyses give a picture of Treg subsets that uniquely function within the immune system largely defined based on varied activation, growth, death, self-renewal, and migratory properties. Similar trends in gene expression were also found for Klrg1+ Tregs in another study (13). Gene expression and immunological analysis indicate that these subsets generally exhibit a progressively greater trend (Fr1 Klrg1−→Fr2 Klrg1−→Fr3 Klrg1−→Fr3 Klrg1+) toward increased proliferation, expression of chemokine receptors targeting inflammatory sites and molecules associated with suppressive function, but lower expression of Bcl-2hi. Fr1 Klrg1− Tregs exhibited properties akin to “resting” lymphoid tissue residing long-lived Tregs. Our precursor-product studies indicate that at least some Fr1 Tregs readily support the development of these other more highly activated Treg subsets culminating in production of Klrg1+ Tregs.
Klrg1 has been shown to mark conventional T cells with a history of high proliferation (40). Expression of Klrg1 by CD8+ T cells delineates important steps during an immune response. Expression of a high level of Klrg1 by CD8+ T cells only occurs after extensive antigen-dependent proliferation (41, 42). The CD8+ Klrg1hi cells are associated with Blimp-1-dependent terminally differentiated CD62Llo tissue-seeking effector cells while the Klrg1low cells are enriched in precursors of memory cells (41, 43). For Klrg1+ Tregs, a large majority are Klrg1hi with approximately 30%–40% as Ki67hi, >95% Bcl-2lo and CD62Llo, indicative of a highly proliferative short-lived population. After adoptive transfer of “resting” CD62Lhi Tregs into Y3 recipients, development of Klrg1+ Tregs requires at least 8 cell divisions. Collectively, these observations are consistent with Klrg1 marking a Treg population with a high replicative history. Key genes associated with Treg function, particularly Il10, Fgl2, and Entpd1 (CD39), as well as chemokine receptors associated with cells within inflammatory sites were most highly expressed in the Klrg1+ Treg subset. Furthermore, Blimp-1 expression was highly enriched in Klrg1+ Tregs with essentially all as Blimp-1+. By evaluating naïve and antigen-challenged airway hypersensitive mice and the patterns of Ki67 expression and BrdU incorporation, most Klrg1+ Tregs behave as recent antigen-stimulated highly proliferative, but short-lived, cells. In marked contrast to Klrg1− Tregs, transferred purified Klrg1+ Tregs did not undergo homeostatic expansion in lymphopenic TCRα−/− or lympho-replete Y3 recipients and for the few cells found, the Klrg1+ phenotype was stable. Overall, these properties are similar to short-lived terminally differentiated CD8+ CTL. This characteristic and their high prevalence in the LP and the lung of allergic hypersensitive mice are consistent with the notion that most Klrg1+ Tregs are terminally differentiated tissue-residing suppressor cells. However, we cannot exclude that a small fraction of Klrg1+ Tregs are long-lived and might be analogous to memory-precursor cells.
IL-2 is well known to provide essential homeostatic signals for nTregs (21, 23). IL-2 also shapes the competitive fitness of Tregs through its ability to maintain Foxp3 levels (24). Our past work showed that these key activities of IL-2 readily occur under conditions that support weak suboptimal IL-2R signaling for Tregs within peripheral immune tissues (27). As shown here, weak IL-2R signaling by Tregs bearing Y3 mutant IL-2Rβ supports relatively normal homeostasis but failed to support Klrg1+ and CD103+ Tregs, the latter to a lesser extent. One explanation for this finding is that high IL-2R signaling is required for developmental activities distinct from IL-2-dependent growth/survival. Alternatively, more extensive proliferation that is linked to high IL-2R signaling is required for production of Klrg1+ Tregs and this effect is distinct from IL-2-mediated homeostasis. These two possibilities are not mutually exclusive. Furthermore, after transfer into TCRα−/− recipients, IL2-IC readily stimulated Klrg1− Tregs to proliferate and develop into Klrg1+ cells whereas IL2-IC did not stimulate proliferation of purified Klrg1+ Tregs. This finding suggests that IL-2 may drive the development of the Klrg1+ subset from Klrg1− Tregs and raises the possibility that the selective increased of Klrg1+ Tregs in B6 mice after IL2-IC may at least in part reflect development by Klrg1− Tregs.
An important new finding from this study is that we have defined another function for IL-2R signaling in the periphery, i.e. the development of terminally differentiated Klrg1+ Tregs. This IL-2-dependent activity on Tregs is also highly analogous to the contribution of IL-2 for CD8+ T effector cells (44–46). Thus, Tregs use a similar strategy as effector CD8+ T cells to promote their development into cells with heightened suppressive effector function. Additionally, this study has uncovered a distinctive tissue-specific requirement for IL-2R signaling for Treg homeostasis. Within a peripheral lymphoid tissue, weak IL-2R signaling readily supported Treg homeostasis as Ki67 expression as well as BrdU uptake and loss were very similar for WT B6 and Y3 Tregs. However, the turnover of Tregs, particularly Klrg1− cells, in the LP was accelerated in Y3 mice, indicating that Tregs in the LP require stronger IL-2R signaling for increased life-span. Other signals, therefore, must compensate for lower IL-2R activity to maintain these cells. This altered homeostasis coupled with failed development of Klrg1+ Tregs likely contributes to tissue specific inflammation that occurs in older Y3 mice (27). Some Teff activity is also attenuated in Y3 mice which might offset IL-2R-dependent defects associated with Y3 Tregs (27).
Based on our gene profiling and direct analysis of Klrg1+ and Klrg1− Tregs, we favor a model where distinct expression of CD62L, CD69, and Klrg1 represents distinct activation states. The heterogeneity in expression of these and other cells surface molecules has been appreciated for some time (4–12), but little is known concerning their potential inter-relationships. This study establishes one such relationship by showing that high IL-2R signaling supports the proliferation of Klrg1− Tregs and causes them to develop into short-lived Klrg1+ Treg population with heightened expression of a number of molecules associated with suppressive function. These Klrg1+ Tregs are also poorly proliferative, unresponsive to IL-2, and lack plasticity. These properties are highly analogous to the association of Klrg1 expression with CD8+ T cells and NK cells that exhibit replicative senescence (47–50) and behave as terminally differentiated cells (41, 42). As illustrated for Treg suppressive activity (14–16), our findings indicate that Tregs also co-opt strategies used by conventional antigen-activated T cells to drive development of Tregs of distinct activation states. The linkage of high IL-2R signaling for the production of Klrg1+ Tregs suggests that this subset is favored under conditions where a stronger or more persistent autoreactive T cell response occurs leading to greater levels of IL-2. High IL-2 levels by the autoreactive T cells may then be counterbalanced by the development of Klrg1+ Tregs that over-express Treg suppressive molecules. This may occur in peripheral lymphoid tissues but is likely important in inflamed tissues which contain pathogenic effector cells. The short-lived nature of most Klrg1+ Tregs leads to a diminished contribution by these Tregs, if the autoreactive response is restrained. However, chronic immune stimulation, as occurs in within the gut mucosa, appears to favor continued development of Klrg1+ Tregs. Thus, lower levels of Klrg1+ Tregs may represent a risk factor for autoimmunity. Indeed, the autoimmune symptoms associated with older Y3 mice may be due in part to the lower level of Klrg1+ Tregs in these mice. In any case this model provides a frame-work for future studies to more fully understand the inter-relationship and development of Treg subpopulations that encounter self or foreign antigens.
Acknowledgments
We thank I. Castro for assistance with the adoptive transfer experiments, and L. Nathanson and E. Echandia for the microarrays.
Abbreviations used in this article
- nTregs
natural T regulatory cells
- iTregs
induced T regulatory cells
- MLN
mesenteric lymph node
- PP
Peyer's patches
- LP
lamina propria
- MFI
mean fluorescent intensity
Footnotes
This work was supported by grants from the NIH (R01CA45957, R01AI055815 and R56 AI100289).
The authors have no competing financial interests
References
- 1.Wing K, Sakaguchi S. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat. Immunol. 2010;11:7–13. doi: 10.1038/ni.1818. [DOI] [PubMed] [Google Scholar]
- 2.Zhou L, Chong MM, Littman DR. Plasticity of CD4+ T cell lineage differentiation. Immunity. 2009;30:646–655. doi: 10.1016/j.immuni.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 3.Izcue A, Coombes JL, Powrie F. Regulatory lymphocytes and intestinal inflammation. Annu. Rev. Immunol. 2009;27:313–338. doi: 10.1146/annurev.immunol.021908.132657. [DOI] [PubMed] [Google Scholar]
- 4.Itoh M, Takahashi T, Sakaguchi N, Kunioyasu Y, Shimizu J, Otsuka F, Sakaguchi S. Thymus and autoimmunity: Production of CD25+ CD4+ naturally anergic and suppressive T cells as a key function of the thymus in maintaining immunological self-tolerance. J. Immunol. 1999;162:5317–5326. [PubMed] [Google Scholar]
- 5.Thornton AM, Shevach EM. Suppressor effector function of CD4+ CD25+ immunoregulatory T cells is antigen nonspecific. J. Immunol. 2000;164:183–190. doi: 10.4049/jimmunol.164.1.183. [DOI] [PubMed] [Google Scholar]
- 6.Banz A, Peixoto A, Pontoux C, Cordier C, Rocha B, Papiernik M. A unique subpopulation of CD4+ regulatory T cells controls wasting disease, IL-10 secretion and T cell homeostasis. Eur. J. Immunol. 2003;33:2419–2428. doi: 10.1002/eji.200324205. [DOI] [PubMed] [Google Scholar]
- 7.Huehn J, Siegmund K, Lehmann JC, Siewert C, Haubold U, Feuerer M, Debes GF, Lauber J, Frey O, Przybylski GK, Niesner U, de la Rosa M, Schmidt CA, Brauer R, Buer J, Scheffold A, Hamann A. Developmental stage, phenotype, and migration distinguish naive- and effector/memory-like CD4+ regulatory T cells. J. Exp. Med. 2004;199:303–313. doi: 10.1084/jem.20031562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kleinewietfeld M, Puentes F, Borsellino G, Battistini L, Rotzschke O, Falk K. CCR6 expression defines regulatory effector/memory-like cells within the CD25+ CD4+ T-cell subset. Blood. 2005;105:2877–2886. doi: 10.1182/blood-2004-07-2505. [DOI] [PubMed] [Google Scholar]
- 9.Suffia I, Reckling SK, Salay G, Belkaid Y. A role for CD103 in the retention of CD4+ CD25+ Treg and control of Leishmania major infection. J. Immunol. 2005;174:5444–5455. doi: 10.4049/jimmunol.174.9.5444. [DOI] [PubMed] [Google Scholar]
- 10.Zhang N, Schroppel B, Lal G, Jakubzick C, Mao X, Chen D, Yin N, Jessberger R, Ochando JC, Ding Y, Bromberg JS. Regulatory T cells sequentially migrate from inflamed tissues to draining lymph nodes to suppress the alloimmune response. Immunity. 2009;30:458–469. doi: 10.1016/j.immuni.2008.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beyersdorf N, Ding X, Tietze JK, Hanke T. Characterization of mouse CD4 T cell subsets defined by expression of KLRG1. Eur. J. Immunol. 2007;37:3445–3454. doi: 10.1002/eji.200737126. [DOI] [PubMed] [Google Scholar]
- 12.Stephens GL, Andersson J, Shevach EM. Distinct subsets of FoxP3+ regulatory T cells participate in the control of immune responses. J. Immunol. 2007;178:6901–6911. doi: 10.4049/jimmunol.178.11.6901. [DOI] [PubMed] [Google Scholar]
- 13.Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc. Natl. Acad. Sci. U. S. A. 2010;107:5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chaudhry A, Rudra D, Treuting P, Samstein RM, Liang Y, Kas A, Rudensky AY. CD4+ regulatory T cells control TH17 responses in a Stat3-dependent manner. Science. 2009;326:986–991. doi: 10.1126/science.1172702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koch MA, Tucker-Heard G, Perdue NR, Killebrew JR, Urdahl KB, Campbell DJ. The transcription factor T-bet controls regulatory T cell homeostasis and function during type 1 inflammation. Nat. Immunol. 2009;10:595–602. doi: 10.1038/ni.1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zheng Y, Chaudhry A, Kas A, deRoos P, Kim JM, Chu TT, Corcoran L, Treuting P, Klein U, Rudensky AY. Regulatory T-cell suppressor program co-opts transcription factor IRF4 to control Th2 responses. Nature. 2009;458:351–356. doi: 10.1038/nature07674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malek TR, Castro I. Interleukin-2 receptor signaling: at the interface between tolerance and immunity. Immunity. 2010;33:153–165. doi: 10.1016/j.immuni.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Malek TR, Yu A, Vincek V, Scibelli P, Kong L. CD4 regulatory T cells prevent lethal autoimmunity in IL-2Rβ-deficient mice. Implications for the nonredundant function of IL-2. Immunity. 2002;17:167–178. doi: 10.1016/s1074-7613(02)00367-9. [DOI] [PubMed] [Google Scholar]
- 19.Burchill MA, Yang J, Vang KB, Moon JJ, Chu HH, Lio CW, Vegoe AL, Hsieh CS, Jenkins MK, Farrar MA. Linked T cell receptor and cytokine signaling govern the development of the regulatory T cell repertoire. Immunity. 2008;28:112–121. doi: 10.1016/j.immuni.2007.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lio CW, Hsieh CS. A two-step process for thymic regulatory T cell development. Immunity. 2008;28:100–111. doi: 10.1016/j.immuni.2007.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bayer AL, Yu A, Adeegbe D, Malek TR. Essential role for interleukin-2 for CD4+ CD25+ T regulatory cell development during the neonatal period. J. Exp. Med. 2005;201:769–777. doi: 10.1084/jem.20041179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat. Immunol. 2005;6:1142–1151. doi: 10.1038/ni1263. [DOI] [PubMed] [Google Scholar]
- 23.Setoguchi R, Hori S, Takahashi T, Sakaguchi S. Homeostatic maintenance of natural Foxp3+ CD25+ CD4+ regulatory T cells by interleukin (IL)-2 and induction of autoimmune disease by IL-2 neutralization. J. Exp. Med. 2005;201:723–735. doi: 10.1084/jem.20041982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gavin MA, Rasmussen JP, Fontenot JD, Vasta V, Manganiello VC, Beavo JA, Rudensky AY. Foxp3-dependent programme of regulatory T-cell differentiation. Nature. 2007;445:771–775. doi: 10.1038/nature05543. [DOI] [PubMed] [Google Scholar]
- 25.Davidson TS, DiPaolo RJ, Andersson J, Shevach EM. Cutting Edge: IL-2 is essential for TGF-β-mediated induction of Foxp3+ T regulatory cells. J. Immunol. 2007;178:4022–4026. doi: 10.4049/jimmunol.178.7.4022. [DOI] [PubMed] [Google Scholar]
- 26.Zheng SG, Wang J, Wang P, Gray JD, Horwitz DA. IL-2 is essential for TGF-β to convert naive CD4+ CD25− cells to CD25+Foxp3+ regulatory T cells and for expansion of these cells. J. Immunol. 2007;178:2018–2027. doi: 10.4049/jimmunol.178.4.2018. [DOI] [PubMed] [Google Scholar]
- 27.Yu A, Zhu L, Altman NH, Malek TR. A low interleukin-2 receptor signaling threshold supports the development and homeostasis of T regulatory cells. Immunity. 2009;30:204–217. doi: 10.1016/j.immuni.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fontenot JD, Rasmussen JP, Williams LM, Dooley JL, Farr AG, Rudensky AY. Regulatory T cell lineage specification by the forkhead transcription factor foxp3. Immunity. 2005;22:329–341. doi: 10.1016/j.immuni.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 29.Wan YY, Flavell RA. Regulatory T-cell functions are subverted and converted owing to attenuated Foxp3 expression. Nature. 2007;445:766–770. doi: 10.1038/nature05479. [DOI] [PubMed] [Google Scholar]
- 30.Kallies A, Hawkins ED, Belz GT, Metcalf D, Hommel M, Corcoran LM, Hodgkin PD, Nutt SL. Transcriptional repressor Blimp-1 is essential for T cell homeostasis and self-tolerance. Nat. Immunol. 2006;7:466–474. doi: 10.1038/ni1321. [DOI] [PubMed] [Google Scholar]
- 31.Laky K, Lefrancois L, Puddington L. Age-dependent intestinal lymphoproliferative disorder due to stem cell factor receptor deficiency: parameters in small and large intestine. J. Immunol. 1997;158:1417–1427. [PubMed] [Google Scholar]
- 32.Fang L, Adkins B, Deyev V, Podack ER. Essential role of TNF receptor superfamily 25 (TNFRSF25) in the development of allergic lung inflammation. J. Exp. Med. 2008;205:1037–1048. doi: 10.1084/jem.20072528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malek TR, Porter BO, Codias EK, Scibelli P, Yu A. Normal lymphoid homeostasis and lack of lethal autoimmunity in mice containing mature T cells with severely impaired IL-2 receptors. J. Immunol. 2000;164:2905–2914. doi: 10.4049/jimmunol.164.6.2905. [DOI] [PubMed] [Google Scholar]
- 34.Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311:1924–1927. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- 35.Cretney E, Xin A, Shi W, Minnich M, Masson F, Miasari M, Belz GT, Smyth GK, Busslinger M, Nutt SL, Kallies A. The transcription factors Blimp-1 and IRF4 jointly control the differentiation and function of effector regulatory T cells. Nat. Immunol. 2011;12:304–311. doi: 10.1038/ni.2006. [DOI] [PubMed] [Google Scholar]
- 36.Letourneau S, van Leeuwen EM, Krieg C, Martin C, Pantaleo G, Sprent J, Surh CD, Boyman O. IL-2/anti-IL-2 antibody complexes show strong biological activity by avoiding interaction with IL-2 receptor α subunit CD25. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:2171–2176. doi: 10.1073/pnas.0909384107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Malek TR, Ashwell JD. Interleukin 2 upregulates expression of its receptor on a T cell clone. The Journal of experimental medicine. 1985;161:1575–1580. doi: 10.1084/jem.161.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Komatsu N, Mariotti-Ferrandiz ME, Wang Y, Malissen B, Waldmann H, Hori S. Heterogeneity of natural Foxp3+ T cells: a committed regulatory T-cell lineage and an uncommitted minor population retaining plasticity. Proc. Natl. Acad. Sci. U. S. A. 2009;106:1903–1908. doi: 10.1073/pnas.0811556106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rosenblum MD, Gratz IK, Paw JS, Lee K, Marshak-Rothstein A, Abbas AK. Response to self antigen imprints regulatory memory in tissues. Nature. 2011;480:538–542. doi: 10.1038/nature10664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Voehringer D, Blaser C, Brawand P, Raulet DH, Hanke T, Pircher H. Viral infections induce abundant numbers of senescent CD8 T cells. J. Immunol. 2001;167:4838–4843. doi: 10.4049/jimmunol.167.9.4838. [DOI] [PubMed] [Google Scholar]
- 41.Joshi NS, Cui W, Chandele A, Lee HK, Urso DR, Hagman J, Gapin L, Kaech SM. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity. 2007;27:281–295. doi: 10.1016/j.immuni.2007.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sarkar S, Kalia V, Haining WN, Konieczny BT, Subramaniam S, Ahmed R. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 2008;205:625–640. doi: 10.1084/jem.20071641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kaech SM, Wherry EJ. Heterogeneity and cell-fate decisions in effector and memory CD8+ T cell differentiation during viral infection. Immunity. 2007;27:393–405. doi: 10.1016/j.immuni.2007.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kalia V, Sarkar S, Subramaniam S, Haining WN, Smith KA, Ahmed R. Prolonged interleukin-2Rα expression on virus-specific CD8+ T cells favors terminal-effector differentiation in vivo. Immunity. 2010;32:91–103. doi: 10.1016/j.immuni.2009.11.010. [DOI] [PubMed] [Google Scholar]
- 45.Obar JJ, Molloy MJ, Jellison ER, Stoklasek TA, Zhang W, Usherwood EJ, Lefrancois L. CD4+ T cell regulation of CD25 expression controls development of short-lived effector CD8+ T cells in primary and secondary responses. Proc. Natl. Acad. Sci. U. S. A. 2010;107:193–198. doi: 10.1073/pnas.0909945107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mitchell DM, Ravkov EV, Williams MA. Distinct Roles for IL-2 and IL-15 in the Differentiation and Survival of CD8+ Effector and Memory T Cells. J. Immunol. 2010;184:6719–6730. doi: 10.4049/jimmunol.0904089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huntington ND, Tabarias H, Fairfax K, Brady J, Hayakawa Y, Degli-Esposti MA, Smyth MJ, Tarlinton DM, Nutt SL. NK cell maturation and peripheral homeostasis is associated with KLRG1 up-regulation. J. Immunol. 2007;178:4764–4770. doi: 10.4049/jimmunol.178.8.4764. [DOI] [PubMed] [Google Scholar]
- 48.Ibegbu CC, Xu YX, Harris W, Maggio D, Miller JD, Kourtis AP. Expression of killer cell lectin-like receptor G1 on antigen-specific human CD8+ T lymphocytes during active, latent, and resolved infection and its relation with CD57. J. Immunol. 2005;174:6088–6094. doi: 10.4049/jimmunol.174.10.6088. [DOI] [PubMed] [Google Scholar]
- 49.Thimme R, Appay V, Koschella M, Panther E, Roth E, Hislop AD, Rickinson AB, Rowland-Jones SL, Blum HE, Pircher H. Increased expression of the NK cell receptor KLRG1 by virus-specific CD8 T cells during persistent antigen stimulation. J. Virol. 2005;79:12112–12116. doi: 10.1128/JVI.79.18.12112-12116.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Voehringer D, Koschella M, Pircher H. Lack of proliferative capacity of human effector and memory T cells expressing killer cell lectinlike receptor G1 (KLRG1) Blood. 2002;100:3698–3702. doi: 10.1182/blood-2002-02-0657. [DOI] [PubMed] [Google Scholar]